Abstract
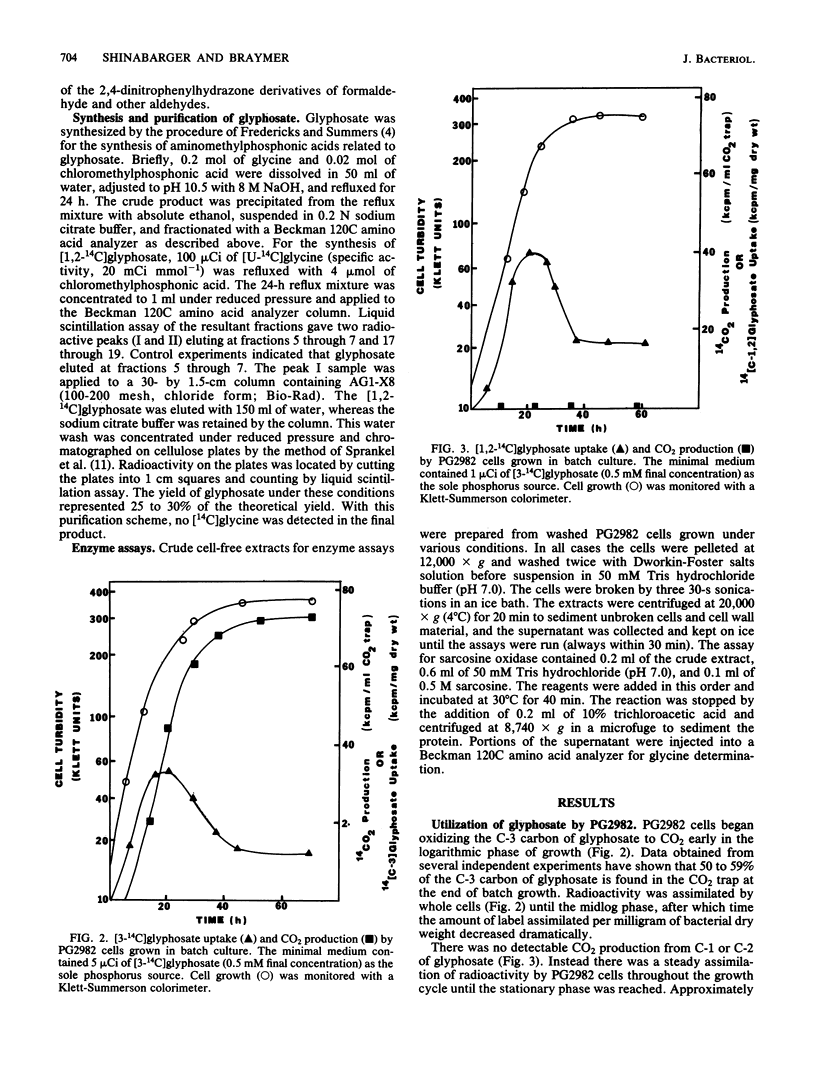
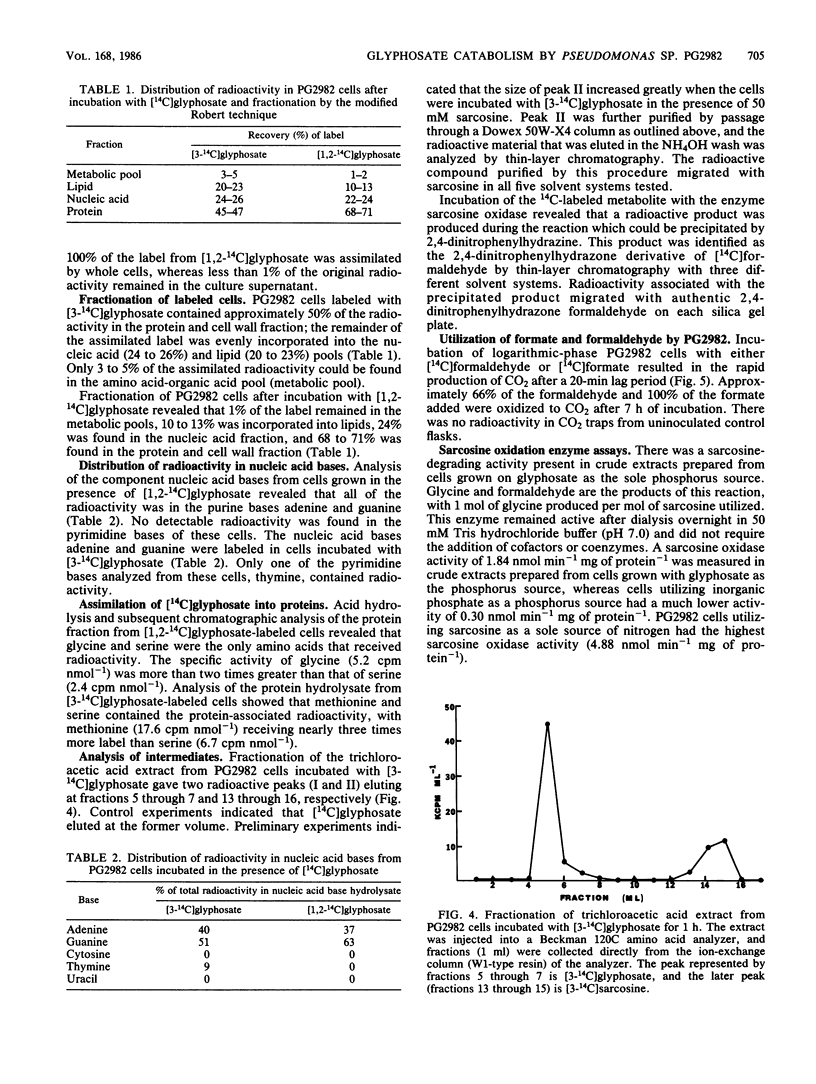
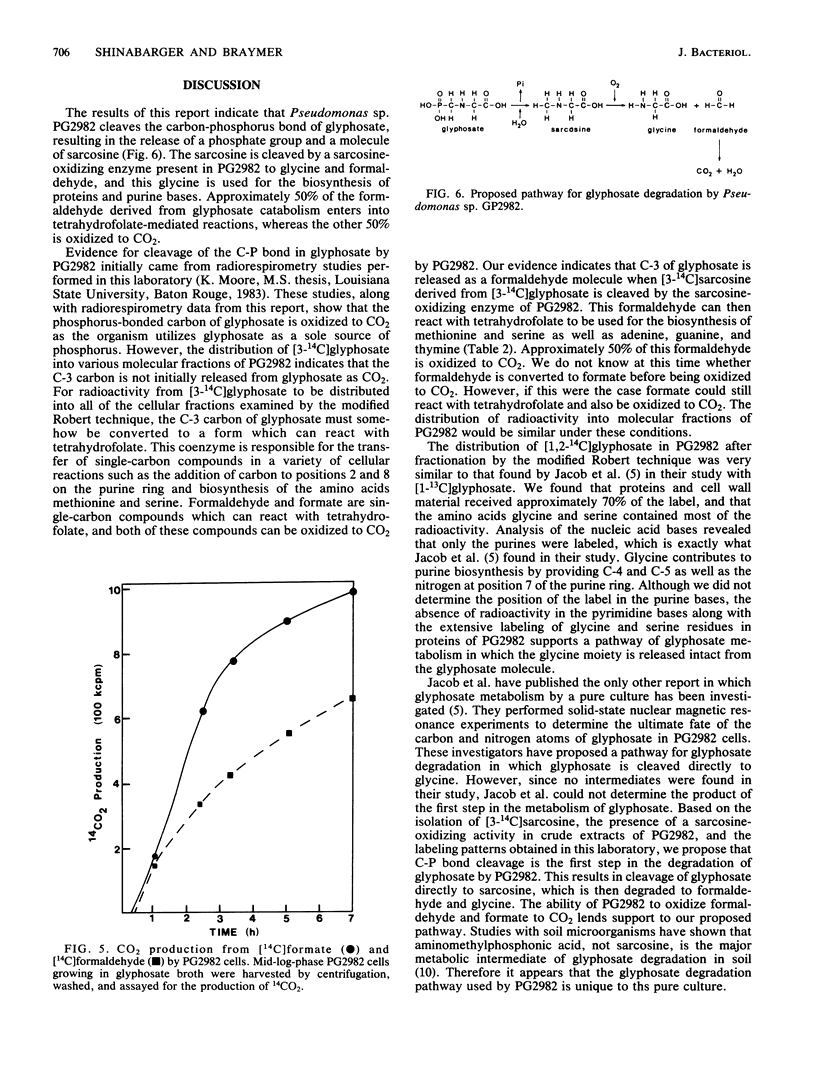
The pathway for the degradation of glyphosate (N-phosphonomethylglycine) by Pseudomonas sp. PG2982 has been determined by using metabolic radiolabeling experiments. Radiorespirometry experiments utilizing [3-14C]glyphosate revealed that approximately 50 to 59% of the C-3 carbon was oxidized to CO2. Fractionation of stationary-phase cells labeled with [3-14C]glyphosate revealed that from 45 to 47% of the assimilated label is distributed to proteins and that the amino acids methionine and serine are highly labeled. Adenine and guanine received 90% of the C-3 label found in the nucleic acid fraction, and the only pyrimidine base labeled was thymine. These results indicated that C-3 of glyphosate was at some point metabolized to a C-1 compound whose ultimate fate could be both oxidation to CO2 and distribution to amino acids and nucleic acid bases that receive a C-1 group from the C-1-donating coenzyme tetrahydrofolate. Pulse-labeling of PG2982 cells with [3-14C]glyphosate resulted in the isolation of [3-14C]sarcosine as an intermediate in glyphosate degradation. Examination of crude extracts prepared from PG2982 cells revealed the presence of a sarcosine-oxidizing enzyme that oxidizes sarcosine to glycine and formaldehyde. These results indicate that the first step in glyphosate degradation by PG2982 is cleavage of the carbon-phosphorus bond, resulting in the release of sarcosine and a phosphate group. The phosphate group is utilized as a source of phosphorus, and the sarcosine is degraded to glycine and formaldehyde. This pathway is supported by the results of [1,2-14C]glyphosate metabolism studies, which show that radioactivity in the proteins of labeled cells is found only in the glycine and serine residues.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Comai L., Sen L. C., Stalker D. M. An Altered aroA Gene Product Confers Resistance to the Herbicide Glyphosate. Science. 1983 Jul 22;221(4608):370–371. doi: 10.1126/science.221.4608.370. [DOI] [PubMed] [Google Scholar]
- Jacob G. S., Schaefer J., Stejskal E. O., McKay R. A. Solid-state NMR determination of glyphosate metabolism in a Pseudomonas sp. J Biol Chem. 1985 May 25;260(10):5899–5905. [PubMed] [Google Scholar]
- Moore J. K., Braymer H. D., Larson A. D. Isolation of a Pseudomonas sp. Which Utilizes the Phosphonate Herbicide Glyphosate. Appl Environ Microbiol. 1983 Aug;46(2):316–320. doi: 10.1128/aem.46.2.316-320.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers S. G., Brand L. A., Holder S. B., Sharps E. S., Brackin M. J. Amplification of the aroA gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl Environ Microbiol. 1983 Jul;46(1):37–43. doi: 10.1128/aem.46.1.37-43.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueppel M. L., Brightwell B. B., Schaefer J., Marvel J. T. Metabolism and degradation of glyphosphate in soil and water. J Agric Food Chem. 1977 May-Jun;25(3):517–528. doi: 10.1021/jf60211a018. [DOI] [PubMed] [Google Scholar]
- Steinrücken H. C., Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem Biophys Res Commun. 1980 Jun 30;94(4):1207–1212. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]



