Abstract
Both DNA methylation and hypoacetylation of core histones are frequently associated with repression of gene expression. Possible connections between these processes were investigated by taking advantage of genes controlled by methylation in Neurospora crassa. Trichostatin A (TSA), a potent inhibitor of histone deacetylase, derepressed a copy of hph that was repressed by DNA methylation which resulted from repeat-induced point mutation (RIP) acting on sequences flanking hph. Derepression by TSA was comparable to derepression by the inhibitor of DNA methylation, 5-azacytidine. TSA treatment also repressed an allele of am whose expression depends on methylation of an adjacent transposon, Tad. DNA methylation in the hph and Tad/am regions was greatly reduced by TSA treatment. TSA also caused hypomethylation of other methylated alleles of am generated by RIP. In contrast, TSA did not affect methylation of several other methylated genomic sequences examined, including the nucleolar rDNA and the inactivated transposon PuntRIP1. Several possible models are discussed for the observed selective demethylation induced by TSA. The implication that acetylation of chromatin proteins can directly or indirectly control DNA methylation raises the possibility that connections between protein acetylation and DNA methylation result in self-reinforcing epigenetic states.
Keywords: histone acetylation/silencing/epigenetics/5-azacytidine/deacetylase
DNA is modified by methylation of cytosines in many higher organisms, including mammals, plants, and some fungi. DNA methylation can silence genes (see refs. 1 and 2) and may serve in genome defense systems (3, 4) and in the regulation of certain endogenous genes, such as genes subjected to genomic imprinting or dosage compensation in mammals (5). Although DNA methylation does not appear to interfere directly with transcription, it can indirectly prevent transcription initiation (6) or elongation (7, 8). How methylation exerts its repressive effect remains largely undefined, but proteins that bind specifically to methylated DNA have been identified (9), and methylation appears to cause assembly of an inactive form of chromatin (6, 10). Histones H3 and H4 are hypoacetylated on the heavily methylated inactive X chromosome (11–14) and hyperacetylated in the unmethylated “CpG islands” in animal genomes (15). In a study on sequences introduced into animal cells as episomes, it was found that 5-azacytidine (5-AC) and sodium butyrate, which cause hypomethylation of DNA and hyperacetylation of histones, respectively, could both relieve repression (16). Butyrate has pleiotropic effects at the high concentrations at which it must be used (see ref. 17), but this observation raised the possibility that methylation operates through an effect on histone acetylation, or vice versa. Recently, a potent direct inhibitor of histone deacetylases, (R)-trichostatin A (TSA) (18), was found to substitute for 5-AC to derepress silent, methylated rDNA genes in interspecific plant hybrids (19). Changes in methylation and/or acetylation in the rDNA, or at an undefined regulatory locus, may have caused the derepression. Effects on DNA methylation were not assessed. Two key questions are (i) Can DNA methylation affect histone acetylation? and (ii) Can acetylation affect DNA methylation? Either possibility could account for the observed correlations. If both occur, this should produce a self-reinforcing cycle that could account for stable epigenetic states.
The fungus Neurospora crassa offers an attractive system to investigate these possibilities. Most of the Neurospora genome is unmethylated and DNA methylation is nonessential (20, 21), but it is clear that DNA methylation can control some genes in this organism (8, 22, 23). Most specifically, we know that methylation of alleles of the am (glutamate dehydrogenase) and mtr (methyltryptophan-resistant) genes prevent transcription elongation (8). Inhibition of DNA methylation by the drug 5-AC or by the dim-2 mutation, which prevents all methylation in Neurospora, activates methylated am and mtr alleles. Similarly, inhibition of methylation by using 5-AC or other means reactivates a methylated copy of the bacterial hph (hygromycin B phosphotransferase) gene (23) and causes repression of an allele of am whose expression depends on methylation of a transposon inserted in its upstream region (22). As a step to investigate the mechanism of methylation-dependent effects on gene expression, I investigated the effect of the histone deacetylase inhibitor TSA on expression of these genes. TSA was found to reverse the effects attributable to methylation. Analyses of DNA methylation demonstrated that TSA can cause selective loss of demethylation in Neurospora, implying that acetylation of histones or other proteins can somehow control DNA methylation.
MATERIALS AND METHODS
Strains.
The following strains from our laboratory collection were used in this study: N220 (am∷Tad3-2 ure-2 mat a) (22), N644 (am132 [(am/hph/am)ec42 pJI2]RIP77 inl mat A) (23), N669 (amRIP4 amRIPec4 lys-1 mat a), N672 (amRIP5 amRIPec5 lys-1 mat A), N617 (amRIP8MM mat a), N676 (amRIP7 amec7 lys-1 mat A) (24). The am/hph/am region of N644 was from pJI2, and consists of a direct repeat of the am gene separated by a 1.7-kb segment including the bacterial hph gene from pDH25 driven by the Aspergillus nidulans trpC promoter (25, 26). The am132 allele contains a deletion that removes all sequences homologous to the am probe and to am sequences introduced by transformation.
Cultures.
Liquid cultures of Neurospora, inoculated with 5–10 × 104 fresh conidia per ml, were grown at 32°C with shaking in sucrose (1.5–2.0%) Vogel’s medium (27) supplemented with alanine and inositol to support growth of am, inl strains. TSA (Wako) was added to the medium immediately prior to inoculation. Plate tests were performed with solidified Vogel’s medium containing sorbose (2%), fructose (0.05%), and glucose (0.05%) in place of sucrose to cause colonial growth. Glycine (20 mM) was included in some plates to tighten the selection for am+ strains. Conidia were routinely plated in 5 ml of 0.7% agar on plates with 25 ml of 1.5% agar medium. TSA (1 μl of 10 mg/ml in dimethyl sulfoxide) was administered from 4-mm-diameter Whatman no. 1 paper discs placed in the middle of plates shortly after plating. For some experiments, an additional 5 ml of top agar with or without hygromycin B (hyg; 600 μg/ml; Calbiochem) was added after 18 hr at 32°C. Plates were typically photographed 40 hr later.
Southern Hybridizations.
DNA was isolated from 1- to 3-day liquid cultures as previously described. DNA samples (1 μg) were digested for at least 4 hr with 10 units of restriction enzyme (NEB), fractionated on 1% agarose gels, transferred to nylon membranes, and probed sequentially as previously described (23). Probes were prepared by priming with random hexamers using the 2.6-kb BamHI am fragment, the 650-bp XbaI–BamHI upstream am fragment, the 1.0-kb ClaI Ψ63 fragment, the 9.2-kb KpnI rDNA fragment, the 1.0-kb ClaI-BamHI hph fragment, or a 2.5-kb his-3 fragment (to verify that digests were complete; not shown).
RESULTS
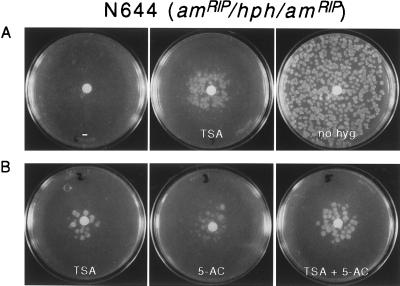
The possibility that TSA would activate a gene repressed by DNA methylation was first tested with the bacterial gene hph, which is present as a single chromosomal copy in N. crassa strain N644 (23). This strain was derived from a transformant in which hph was unmethylated and conferred resistance to hyg (26). The approximately 1-kb gene, driven by the A. nidulans trpC promoter (see Materials and Methods), lies between two copies of the Neurospora am (glutamate dehydrogenase) gene. This allowed us to render hph, and the trpC promoter, methylated by induction of RIP in the flanking repeated am sequences (23). RIP is a sexual-phase-specific genome defense system that results in multiple G⋅C to A⋅T transition mutations in duplicated sequences (4, 28–31). Remaining cytosines are frequently methylated after the action of RIP and the methylation can extend into adjacent unmutated sequences, such as those of the hph gene flanked by mutated copies of am. We had identified progeny of the original transformant in which RIP had indeed caused heavy methylation of the two copies of am plus the intervening sequences, including hph. The hph gene was silenced in most strains (e.g., N644) but expression could be restored if methylation was inhibited by treatment with 5-AC or by limiting production of the methyl-group donor, S-adenosylmethionine (AdoMet), using a conditional mutation in the AdoMet synthetase gene (23). The effect of TSA on expression of the methylated hph gene was tested by plating conidia (asexual spores) of strain N644 in the presence or absence of TSA (10 μg = 33 nmol) applied to paper discs in the middle of the plates. Because hyg quickly kills strains not expressing hph (23), this drug was added 18 hr after plating the conidia. One day later, hyg-resistant colonies appeared near the source of TSA; no colonies were seen on the control plate without TSA (Fig. 1A). The density of colonies near the disc appeared equivalent to the density on a similar plate lacking hyg and TSA, suggesting that TSA caused derepression in virtually every colony. Other plate tests revealed that 20 nmol (5 μg) of 5-AC induced hph roughly as well as 33 nmol of TSA (Fig. 1B), although 5-AC inhibited growth considerably more than did TSA (data not shown). Interestingly, TSA and 5-AC together seemed to result in greater induction than either drug alone (Fig. 1B). To test the stability of the apparent derepression by TSA, colonies from plates with TSA and hyg, and colonies from control plates lacking the drugs, were picked and tested on plates containing hyg but no TSA. Colonies from the TSA plus hyg plate, but not the control plate, grew vigorously on hyg medium, but showed loss of resistance after conidiation (data not shown). Thus TSA caused a long-term, but not permanent, derepression of the hph gene.
Figure 1.
Reactivation of silenced hph gene by TSA and 5-AC. N. crassa strain N644 (am132, inl, amRIP/hph/amRIP, mat A) harbors a single copy of the Escherichia coli hph gene that was inactivated by methylation because of the action of RIP on flanking direct repeats of the am gene (23, 26). Sets A and B were from separate experiments using independent solutions and cultures. In each experiment shown, ≈1,000 conidia were plated on each of the plates, 1 μl of TSA (33 mM in dimethyl sulfoxide), 5-AC (20 mM), both, or neither (plates marked − and no hyg) was applied to the paper discs, and, except for the right-most plate in A, hyg was added in 0.7% agar medium after 17–18 hr at 32°C. Control plates with TSA or 5-AC but lacking hyg revealed slight inhibition of growth by TSA and somewhat greater inhibition by 5-AC (not shown).
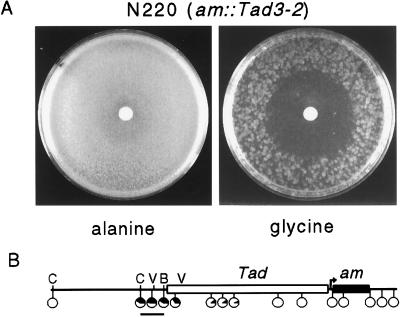
A case in which methylation affects gene expression in the opposite direction from that normally observed provided an attractive opportunity for a second genetic test of the effect of TSA on methylated DNA. Expression of the am gene in strain N220 relies somehow on methylation spanning the 5′ end of a LINE-like transposon, Tad, inserted upstream of the am basal promoter (22) (Fig. 2B). am expression is required for growth on minimal medium supplemented with glycine. Normally, ≈2% of N220 conidia plated on restrictive (glycine) medium form colonies (ref. 22 and data not shown). When methylation is prevented, however, by using either 5-AC or a strain harboring the dim-2 mutation, which prevents all methylation in vegetative tissue of Neurospora (20), the am gene is fully silenced. Loss of methylation somehow allows Tad to silence am. Although the cause of this methylation is not known, it is apparently not a result of RIP (22).
Figure 2.
TSA accentuates silencing of the am gene caused by the adjacent transposon Tad. (A) Approximately 4 × 105 conidia of N. crassa strain N220 (am∷Tad3–2, ure-2, mat a) were plated on permissive (alanine) or restrictive (glycine) sorbose plates. One microliter of TSA (33 mM in dimethyl sulfoxide) was applied to the paper discs and the plates were incubated 3 days at 32°C. The difference in colony density in the portions of the plates not affected by TSA reflects the ratio of Am+ and Am− colonies characteristic of this strain. (B) Map of am∷Tad3–2 region of strain N220. Tad (open rectangle) is inserted 70 bp upstream of the transcription start sites (arrow) of am (black rectangle). The approximate methylation status of 15 sites in the region, as determined in a previous study (22), is depicted in black in pie charts placed close to the sites examined. Those sites for BamHI (B), ClaI (C), and EcoRV (V) that are relevant to Fig. 5 are indicated. The bar beneath the map represents a 650-bp XbaI–BamHI fragment used as a probe for the blot shown in Fig. 5.
The effect of TSA on am expression in strain N220 was tested by plating ≈400,000 conidia on permissive (alanine) or restrictive (glycine) media and then adding TSA (10 μg) to the center of the dishes, as before. A slight inhibition of growth was detected near the source of the TSA on alanine medium, but the strain produced essentially confluent growth, as expected (Fig. 2A). TSA did not appreciably inhibit a wild-type strain on either alanine or glycine medium (not shown). In striking contrast, growth of N220 on glycine medium was limited to the region of the plate at least 2.5 cm from the source of the TSA. Thus the deacetylase inhibitor accentuated the inhibitory effect of Tad on am expression, as occurs when DNA methylation is prevented (22).
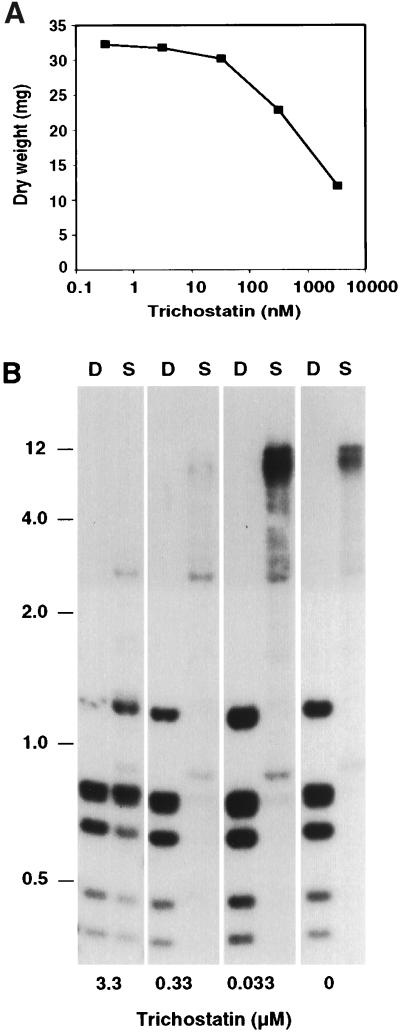
The effects of TSA on expression of hph and am suggested that histone hypoacetylation and DNA methylation may operate in a common silencing pathway. One possibility was that the silencing resulted from hypoacetylation directed by the DNA methylation. It was also possible that hypoacetylation triggered DNA methylation, which then more directly affected gene expression. I therefore investigated whether TSA affected DNA methylation. The methylation state of the amRIP/hph/amRIP region was examined in N644 grown in nonselective medium containing up to 1 μg/ml (3.3 μM) TSA, a concentration that retarded growth somewhat under the conditions of the experiment (Fig. 3A). Methylation was assessed by comparing Sau3AI and DpnII digests by Southern hybridization. Sau3AI fails to cut DNA when the C in its recognition site (GATC) is methylated, whereas its isoschizomer DpnII is not inhibited by cytosine methylation. A substantial reduction in methylation in the amRIP/hph/amRIP region was caused by growth in 3.3 μM TSA, as shown by the fact that Sau3AI gave nearly complete digestion of am and hph sequences (Fig. 3B and data not shown).
Figure 3.
Effect of TSA on growth and DNA methylation. (A) Liquid cultures inoculated with N. crassa strain N644 (7 × 104 conidia per ml) were supplemented with up to 1 μg/ml (3.3 μM) TSA and grown 27 hr. The untreated culture yielded 32.8 mg of dry tissue. (B) Southern hybridization of selected samples from the cultures. Samples of DNA were prepared and digested with DpnII (D) or Sau3AI (S) and probed for am sequences. Stronger signals in the 0.33 μM lanes is partially due to 2× heavier loadings of DNA in these lanes. The positions of selected size standards (kb) are indicated.
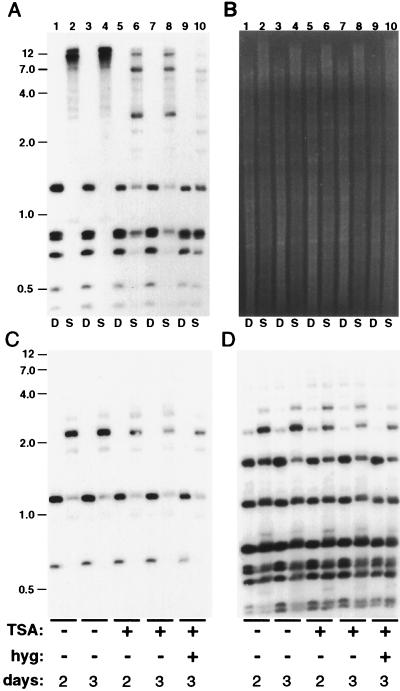
Although this finding suggested that TSA may have inhibited methylation, it seemed possible that the reduced methylation was simply the result of inhibited growth, because young cultures of Neurospora show some reduction in overall methylation (32). Therefore DNA from N644 cultures grown for longer periods of time with and without TSA were examined. Derepression of hph was also tested in the same set of cultures by challenging them to grow in the liquid medium after addition of hyg. Cultures with 0.1 μg/ml (0.33 μM) TSA, or less, failed to grow appreciably in the presence of hyg (data not shown). In contrast, cultures supplemented with 3.3 μM TSA grew well in hyg medium. Although all cultures with this level of TSA showed somewhat retarded growth initially, as in the previous experiment, by 46 hr they appeared to have “caught up” to the cultures without TSA. This observation was confirmed by measuring tissue weights (Fig. 4 legend and data not shown). Most interestingly, DNA prepared from mature (2- and 3-day) cultures grown nonselectively in the presence of TSA showed marked reduction in methylation in the amRIP/hph/amRIP region (Fig. 4A), as observed with the younger culture (Fig. 3). No difference in methylation was evident between the 2- and 3-day cultures, reinforcing the conclusion that the reduction in methylation observed here and in the previous experiment was not somehow due to differences in growth. DNA from a culture grown in TSA and hyg showed the greatest reduction in methylation (Fig. 4A), presumably because selection for hyg favored cells with reduced methylation of hph (23).
Figure 4.
TSA causes selective hypomethylation of DNA. Strain N644 was grown for 2 or 3 days, as indicated, with or without TSA (1 μg/ml; lanes 5–10) and hyg (added to a concentration of 0.1 mg/ml 17 hr after inoculation; lanes 9 and 10). The dry weights of the cultures illustrated were, from left to right, 31.1 mg, 38.5 mg, 37.5 mg, 37.3 mg, and 32.8 mg. DNA was isolated and analyzed by digestion with DpnII (D) or Sau3AI (S) and by probing for am (A), Ψ63 (C), rDNA (D), or hph (not shown). (B) Total genomic DNA visualized by staining with ethidium bromide. The positions of selected size standards (kb) are indicated.
The marked reduction in methylation caused by TSA in the amRIP/hph/amRIP region was particularly surprising because no change in overall methylation was apparent from inspection of the ethidium bromide-stained genomic DNA samples (Fig. 4B). We had previously noted that comparisons of Sau3AI and DpnII (or MboI) digests of N. crassa DNA provide a simple indicator of overall methylation (33). Thus it seemed possible that TSA caused a highly selective loss of methylation in the genome, in contrast to the rather uniform reduction in methylation achieved by 5-AC treatment, by use of any of several mutants defective in methylation (refs. 20, 32, and 34; H. Foss, C. Roberts, and E.S., unpublished results; and A. Hagemann, M. Freitag, and E.S., unpublished results) or by limiting methylation by using strains harboring mutations in genes required for S-adenosylmethionine biosynthesis (20, 34). To test directly whether TSA caused differential hypomethylation, the blot shown was reprobed for two other regions known to be methylated in the N. crassa genome. These were Ψ63, a 5S rRNA pseudogene interrupted by a transposon that has been inactivated by RIP (20, 35), and the tandemly repeated rRNA genes (36). Hybridization results revealed that neither Ψ63 (Fig. 4C) nor rDNA (Fig. 4D) sequences were hypomethylated. Thus the histone deacetylase inhibitor caused striking hypomethylation in the amRIP/hph/amRIP region but caused no apparent change in methylation of the other regions.
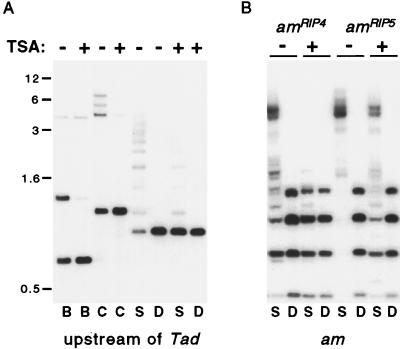
The effect of TSA on methylation was also examined in the upstream am/Tad boundary in strain N220 and in several additional alleles of am that were methylated as a result of RIP. Digestion of N220 DNA with BamHI, Bsp106I, or Sau3AI revealed TSA-induced hypomethylation at the upstream am/Tad boundary (Fig. 5A), as found in the amRIP/hph/amRIP region of N644. The amRIP/hph/amRIP construct of N644 was built using only sequences downstream of the BamHI site (Fig. 2B); thus the methylated upstream am/Tad sequences include less than 180 bp found in strain N644.
Figure 5.
TSA-induced hypomethylation of am∷Tad3–2 and amRIP alleles. Strains containing am∷Tad3-2 (N220), amRIP4 and amRIPec4 [N669 (24)] or amRIP5 and amRIPec5 [N672 (24)] were grown from conidia for 2 days in the presence or absence of 1 μg/ml TSA, as indicated. (A) DNA samples of N220 were digested with EcoRV (which is not inhibited by cytosine methylation) plus BamHI (B), Bsp106I (an isoschizomer of ClaI; C), Sau3AI (S), or DpnII (D) and probed for sequences upstream of Tad (see Fig. 2B). The positions of selected size standards (kb) are indicated. (B) DNA of N669 or N672 was digested with Sau3AI or DpnII and probed sequentially for am, Ψ63 (not shown), and rDNA (not shown).
Strains bearing am alleles inactivated by RIP showed similar results. Several strains examined each contain two amRIP alleles, one at the native am locus and another at the unlinked site where the 2.6-kb BamHI am fragment that triggered RIP had integrated (24, 29, 37). Strain N669 was chosen because the methylation of both am sequences (amRIP4 and amRIPec4) in this strain appears to be dependent on maintenance methylation. That is, if the methylation of these alleles is removed by treatment with 5-AC, or if one of these alleles is isolated and reintroduced into Neurospora in an unmethylated state, the methylation is not reestablished (24). In contrast, in strain N672, the amRIP5 and amRIPec5 alleles become methylated de novo, as is most common for sequences bearing moderate or heavy damage by RIP (4). Interestingly, both of the amRIP alleles in both strains N669 and N672 showed striking reduction of methylation in response to TSA treatment (Fig. 5B). The alleles that do not trigger de novo methylation and therefore must depend on maintenance methylation (amRIP4 and amRIPec4) were most affected. Curiously, in strain N672, the allele at the native am locus appeared more sensitive to TSA than the ectopic allele, as indicated by the relatively weak signal of the Sau3AI fragment diagnostic of the ectopic copy [1.4-kb fragment matching the largest DpnII fragment (24)]. A strain (N617) containing only a single am allele (amRIP8), at its native locus, also showed dramatic reduction in methylation (data not shown). Reprobings of the blot with Ψ63 and rDNA (data not shown) revealed no apparent loss of methylation from these regions, as with strain N644 (Fig. 4).
DISCUSSION
The exposed N-terminal tails of each of the four core histones are subject to a variety of posttranslational modifications that may affect chromatin function. Early evidence that acetylation of lysines in the tails is associated with gene expression has been consistently supported and extended (see refs. 38–40). The general picture that has emerged is that hyperacetylation is a prerequisite for transcription, whereas hypoacetylation can result in transcriptional repression (see refs. 41–43). Several transcription factors and transcriptional coactivators have been demonstrated to possess histone acetyltransferase (HAT) activity (44, 45), and a number of transcriptional repressors have been shown to recruit histone deacetylases (HDACs) (46–51). Studies with antibodies specific to histones acetylated at particular sites (52) or with probes for sequences in hyper- or hypoacetylated chromatin (53) revealed gross variation in histone acetylation in different chromosomal regions, presumably reflecting differential availability of HATs and HDACs. DNA sequences associated with epigenetic silencing, such as on the inactive X chromosome of mammals (11–14), in heterochromatin of insects (54), in the silent mating type genes in yeasts (55, 56), and in the centromere regions of fission yeast (57), are typically associated with hypoacetylated histones. Methylated sequences in animals have also been found associated with hypoacetylated histones (15).
Direct evidence that histone acetylation can affect gene expression came from studies in which genes for HDACs were mutated (58) or inhibited with drugs (16, 17, 19, 49, 57, 59, 60). Curiously, two cases were found in which genes could be activated by using either an inhibitor of DNA methylation (5-AC) or an inhibitor of HDACs (butyrate or TSA). In the first, Hsieh (16) showed that butyrate could enhance expression of a methylated episome transfected into human cells. No effect on methylation was detected. In the second study, both butyrate and a specific inhibitor of HDACs, TSA, were found to substitute for 5-AC to derepress silent, methylated rDNA genes in interspecific plant hybrids (19). Effects on DNA methylation were not assessed. Thus, changes in methylation and/or acetylation in the rDNA, or at an undefined regulatory locus, may have caused the derepression. Nevertheless, these findings raised two possibilities: the repressive effect typical of DNA methylation may be mediated by deacetylation of histones, or deacetylation may lead to DNA methylation. These possibilities were investigated by using genes in Neurospora whose expression is known to be negatively or positively controlled by DNA methylation: a bacterial transgene (hph) repressed by methylation (23), an allele of the Neurospora am gene that is controlled indirectly by methylation of a transposon (Tad) inserted upstream of the am basal promoter (22), and several methylated alleles of am generated by RIP (24). TSA treatment induced expression of hph and silenced expression of the am allele downstream of Tad. Surprisingly, DNA methylation was dramatically reduced in the hph and Tad/am regions, as well as in all amRIP alleles examined. Thus, the changes in gene expression may have resulted directly from histone hyperacetylation, as has been observed in other systems, or may have resulted indirectly from an effect of hyperacetylation on methylation. Although it is conceivable that TSA affected a process other than histone acetylation, TSA is a direct, noncompetitive inhibitor of HDACs and no other effects of the drug have yet been found (18). As far as I know, this is the first indication that DNA methylation may depend, directly or indirectly, on the acetylation state of histones. Interestingly, TSA did not affect methylation of other genomic sequences examined, including the nucleolar rDNA and a transposon inactivated by RIP.
What is the most straightforward interpretation of these results? In every system examined, TSA has been found to cause hyperacetylation of the core histones, apparently because their state of acetylation reflects the balance of HATs and HDACs working in opposition (18, 19, 57, 59, 61). Variation in the distribution of HATs and HDACs in different chromosomal regions should result in regional differences in the degree of hyperacetylation in response to TSA, but this possibility has not been carefully examined. In principle, the observed loss of methylation caused by TSA in our system could have resulted either from hyperacetylation of the methylated regions or from hyperacetylation elsewhere in the genome. For example, if a hypothetical negative regulator of a DNA methyltransferase were activated by increased acetylation in the vicinity of its gene, this might lead to decreased DNA methylation. This is not the most economical model, however, especially considering that TSA strongly affected methylation in some chromosomal regions but left other areas unaffected. There are several possible explanations for the differential effect of TSA on DNA methylation. TSA may not have inhibited the action of HDACs in all regions. Alternatively, deacetylation may have been inhibited globally but only some regions had access to HATs and were therefore reacetylated. Consistent with this line of reasoning, it has recently been shown that TSA activates the WAF1/Cip1 promoter in a human cell line through Sp1 sites (59). This is particularly interesting in light of evidence that deletion or mutations in Sp1 sites of the mouse and hamster aprt genes lead to methylation of their CpG islands (62–64). Thus, it does not seem unreasonable to suppose that in our system the am enhancer region upstream of Tad3-2 would recruit a HAT and thereby increase acetylation in the region after TSA treatment, whereas the Ψ63 pseudogene, with its heavily mutated transposon (35), would lack sites to which HATs are recruited and thus remain hypoacetylated. It is worth noting in this context that the upstream region of the Neurospora am gene includes a CCAAT site that binds a protein (AAB) equivalent to HAP5 of yeast, which is a member of an activation complex dependent on the HAT encoded by GCN5 (65, 66). The distribution of regulatory sites and DNA methylation in rDNA is not yet well defined in Neurospora, but it is interesting that butyrate was not found to influence the gross acetylation level of histones in rDNA of human chromosomes (53).
The observed loss of methylation could be a direct or indirect result of TSA-induced hyperacetylation. It is possible that hyperacetylation released transcription in some chromosomal regions and the transcription, rather than hyperacetylation, per se, inhibited DNA methylation. We know that absence of transcription is not sufficient to trigger methylation and that methylation of am prevents transcription elongation (8), but it is conceivable that activation by hyperacetylation could overcome this effect. A second possibility consistent with the observations is that acetylation directly controls DNA methylation. Hypoacetylation could trigger DNA methylation or hyperacetylation could inhibit methylation (or both). Either DNA methylation or hypoacetylation could be responsible for repression of transcription in this model. The observation of interchangeable effects of 5-AC and TSA implies, however, that hypoacetylation cannot be solely responsible for repression unless the state of methylation feeds back on the state of acetylation. Recent suggestions that methyl-DNA binding proteins may recruit HDACs are consistent with this possibility (67, 68). If methylated sequences recruit HDACs, which then cause deacetylation, and deacetylation promotes methylation, this should produce a rather stable repressed state. This self-reinforcing epigenetic state could account for observed maintenance of methylation, including maintenance of heterogeneous methylation at nonsymmetrical sites (69). To distinguish between these models it will be necessary to determine whether transcription is required for TSA-induced changes in methylation and whether DNA methylation can directly affect histone acetylation. A variety of studies will be required to discover all the connections between modifications of DNA and chromatin and between these modifications and gene expression.
Acknowledgments
I thank K. Sprague, A. Bird, and all members of my laboratory for discussing this work, M. Freitag, S. Hays, B. Margolin, G. Kothe, E. Kuzminova, and J. Selker for comments on the manuscript, and members of my laboratory for sharing materials. I thank A. Klar for suggesting the use of the paper discs. This study was supported by a grant from the National Institutes of Health (GM 35690).
ABBREVIATIONS
- 5-AC
5-azacytidine
- TSA
trichostatin A
- HAT
histone acetyltransferase
- HDAC
histone deacetylase
- RIP
repeat-induced point mutation
- hyg
hygromycin
References
- 1.Bird A. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 2.Kass S U, Pruss D, Wolffe A P. Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 3.Yoder J A, Walsh C P, Bestor T H. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 4.Selker E U. Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- 5.Jaenisch R. Trends Genet. 1997;13:323–329. doi: 10.1016/s0168-9525(97)01180-3. [DOI] [PubMed] [Google Scholar]
- 6.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barry C, Faugeron G, Rossignol J-L. Proc Natl Acad Sci USA. 1993;90:4557–4561. doi: 10.1073/pnas.90.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rountree M R, Selker E U. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tate P H, Bird A P. Curr Biol. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 10.Keshet I, Lieman-Hurwitz J, Cedar H. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 11.Jeppesen P, Turner B M. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 12.O’Neill L P, Turner B M. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keohane A M, O’Neill L P, Belyaev N D, Lavender J S, Turner B M. Dev Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 14.Boggs B A, Connors B, Sobel R E, Chinault A C, Allis C D. Chromosoma. 1996;105:303–309. doi: 10.1007/BF02524648. [DOI] [PubMed] [Google Scholar]
- 15.Tazi J, Bird A. Cell. 1990;60:909–920. doi: 10.1016/0092-8674(90)90339-g. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh C L. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruh J. Mol Cell Biochem. 1982;42:65–82. doi: 10.1007/BF00222695. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 19.Chen Z J, Pikaard C S. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foss H M, Roberts C J, Claeys K M, Selker E U. Science. 1993;262:1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- 21.Foss H M, Roberts C J, Claeys K M, Selker E U. Science. 1995;267:316. doi: 10.1126/science.7824923. [DOI] [PubMed] [Google Scholar]
- 22.Cambareri E B, Foss H M, Rountree M R, Selker E U, Kinsey J A. Genetics. 1996;143:137–146. doi: 10.1093/genetics/143.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irelan J T, Selker E U. Genetics. 1997;146:509–523. doi: 10.1093/genetics/146.2.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer M J, Marcotte B A, Selker E U. Mol Cell Biol. 1995;15:5586–5597. doi: 10.1128/mcb.15.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen D, Leong S A, Wilson L J, Henner D J. Gene. 1987;57:21–26. doi: 10.1016/0378-1119(87)90172-7. [DOI] [PubMed] [Google Scholar]
- 26.Irelan J T, Hagemann A T, Selker E U. Genetics. 1994;138:1093–1103. doi: 10.1093/genetics/138.4.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis R H, De Serres F J. Methods Enzymol. 1970;17A:47–143. [Google Scholar]
- 28.Selker E U, Cambareri E B, Jensen B C, Haack K R. Cell. 1987;51:741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- 29.Selker E U, Garrett P W. Proc Natl Acad Sci USA. 1988;85:6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cambareri E B, Jensen B C, Schabtach E, Selker E U. Science. 1989;244:1571–1575. doi: 10.1126/science.2544994. [DOI] [PubMed] [Google Scholar]
- 31.Selker E U. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- 32.Foss, H. M., Roberts, C. J. & Selker, E. U. (1998) Mol. Gen. Genet., in press. [DOI] [PubMed]
- 33.Selker E U, Richardson G A, Garrett-Engele P W, Singer M J, Miao V. Cold Spring Harbor Symp Quant Biol. 1993;58:323–329. doi: 10.1101/sqb.1993.058.01.038. [DOI] [PubMed] [Google Scholar]
- 34.Roberts C J, Selker E U. Nucleic Acids Res. 1995;23:4818–4826. doi: 10.1093/nar/23.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margolin, B. S., Garrett-Engele, P. W., Stevens, J. N., Yen-Fritz, D., Garrett-Engele, C., Metzenberg, R. L. & Selker, E. U. (1998) Genetics, in press. [DOI] [PMC free article] [PubMed]
- 36.Perkins D D, Metzenberg R L, Raju N B, Selker E U, Barry E G. Genetics. 1986;114:791–817. doi: 10.1093/genetics/114.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selker E U, Fritz D Y, Singer M J. Science. 1993;262:1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- 38.Csordas A. Biochem J. 1990;265:23–38. doi: 10.1042/bj2650023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wade P A, Pruss D, Wolffe A P. Trends Biochem Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 40.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 41.Hampsey M. Trends Genet. 1997;13:427–429. doi: 10.1016/s0168-9525(97)01292-4. [DOI] [PubMed] [Google Scholar]
- 42.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 43.Hebbes T R, Thorne A W, Clayton A L, Crane-Robinson C. Nucleic Acids Res. 1992;20:1017–1022. doi: 10.1093/nar/20.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brownell J E, Zhou J, Ranalli T, Kobayashi R, Edmondson D G, Roth S Y, Allis C D. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- 45.Mizzen C A, Yang X J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, Nakatani Y, Allis C D. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 46.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, et al. Nature (London) 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 47.Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber Agus N, DePinho R A. Nature (London) 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 48.Hassig C A, Fleischer T C, Billin A N, Schreiber S L, Ayer D E. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 49.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 50.Kadosh D, Struhl K. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 51.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 52.Turner B M, Birley A J, Lavender J. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- 53.Breneman J W, Yau P M, Swiger R R, Teplitz R, Smith H A, Tucker J D, Bradbury E M. Chromosoma. 1996;105:41–49. doi: 10.1007/BF02510037. [DOI] [PubMed] [Google Scholar]
- 54.Bone J R, Lavender J, Richman R, Palmer M J, Turner B M, Kuroda M I. Genes Dev. 1994;8:96–104. doi: 10.1101/gad.8.1.96. [DOI] [PubMed] [Google Scholar]
- 55.Braunstein M, Rose A B, Holmes S G, Allis C D, Broach J R. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 56.Braunstein M, Sobel R E, Allis C D, Turner B M, Broach J R. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ekwall K, Olsson T, Turner B M, Cranston G, Allshire R C. Cell. 1997;91:1021–1032. doi: 10.1016/s0092-8674(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 58.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sowa Y, Orita T, Minamikawa S, Nakano K, Mizuno T, Nomura H, Sakai T. Biochem Biophys Res Commun. 1997;241:142–150. doi: 10.1006/bbrc.1997.7786. [DOI] [PubMed] [Google Scholar]
- 60.Chen W Y, Bailey E C, McCune S L, Dong J Y, Townes T M. Proc Natl Acad Sci USA. 1997;94:5798–5803. doi: 10.1073/pnas.94.11.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sheridan P L, Mayall T P, Verdin E, Jones K A. Genes Dev. 1997;11:3327–3340. doi: 10.1101/gad.11.24.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Nature (London) 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 63.Macleod D, Charlton J, Mullins J, Bird A P. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 64.Mummaneni P, Walker K A, Bishop P L, Turker M S. J Biol Chem. 1995;270:788–792. doi: 10.1074/jbc.270.2.788. [DOI] [PubMed] [Google Scholar]
- 65.Chen H, Crabb J W, Kinsey J A. Genetics. 1998;148:123–130. doi: 10.1093/genetics/148.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Georgakopoulos T, Thireos G. EMBO J. 1992;11:4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 68.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 69.Hagemann A T, Selker E U. In: Control and Function of DNA Methylation in Neurospora crassa. Russo V E A, Martienssen R A, Riggs A D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 335–344. [Google Scholar]