Abstract
Although much progress has been made in identifying the signaling pathways that mediate the initial responses to interferons (IFNs), much less is known about how IFN-stimulated genes (ISGs) are kept quiescent in untreated cells, how the response is sustained after the initial induction, and how ISG expression is down-regulated, even in the continued presence of IFN. We have used the cell sorter to isolate mutant cells with constitutively high ISG expression. A recessive mutant, P2.1, has higher constitutive ISG levels than the parental U4C cells, which do not respond to any IFN. Unexpectedly, P2.1 cells also are deficient in the expression of ISGs in response to double-stranded RNA (dsRNA). Electrophoretic mobility-shift assays revealed that the defect is upstream of the activation of the transcription factors NFκB and IFN regulatory factor 1. Analysis of the pivotal dsRNA-dependent serine/threonine kinase PKR revealed that the wild-type kinase is present and is activated normally in response to dsRNA in P2.1 cells. Together, these data suggest that the defect in P2.1 cells is either downstream of PKR or in a component of a distinct pathway that is involved both in activating multiple transcription factors in response to dsRNA and in regulating the basal expression of ISGs.
Interferons (IFNs) are pleiotropic mediators of antiviral, antiproliferative, and immunomodulatory responses in target cells (1). Binding to cognate receptors leads to dimerization of heteromeric receptor subunits (2) and to activation of receptor-associated cytoplasmic Janus tyrosine kinases (JAKs). The activated JAKs phosphorylate IFN receptor subunits on specific tyrosine residues, thereby providing docking sites for downstream signaling molecules, including signal transducers and activators of transcription (STATs). STATs associated with the activated receptor complex become phosphorylated on tyrosine themselves, dimerize, and migrate to the nucleus, alone or in conjunction with other DNA binding proteins, to promote expression of specific IFN-stimulated genes (ISGs) (reviewed in refs. 3–6).
Although much progress has been made in identifying the major factors involved in the initial induction of ISGs after IFN stimulation (the JAKs and STATs), other aspects of ISG regulation are understood less well. For example, ISG transcription continues for many hours after factors involving STATs have disappeared (7, 8), suggesting that additional factors sustain the response. Although IFN regulatory factor-1 (IRF-1) is involved in this process (9, 10), other factors must also be able to carry out this function because ablation of IRF-1 in mice does not affect the kinetics of IFN-induced ISG expression (11). The down-regulation of ISGs also is not understood well. ISG expression is transient, even in the continued presence of IFN, declining to basal levels within 24–48 hr (12). Repressors are likely to be involved in terminating ISG transcription because treating cells with IFN in the presence of protein synthesis inhibitors prolongs ISG transcription (12). Although IFN-2, which binds to IFN-stimulated response elements (ISREs), has been proposed to repress ISG expression after IFN induction (13), IRF-2-null mouse embryo fibroblasts (MEFs) are not defective in down-regulating ISGs (11), again suggesting redundant mechanisms. Other ISRE-specific transcriptional repressors have been identified, including the lymphoid-specific factors ICSBP (14) and PIP (ICSAT) (15). However, these factors cannot account for ISG silencing in nonlymphoid cells. In addition to regulation at the transcriptional level, cytoplasmic or nuclear phosphatases are likely to influence the duration of IFN responses by dephosphorylating the activated IFN receptor subunits, the JAKs, and the STATs, leading to down-regulation of the signaling pathways (16–18).
Double-stranded RNA (dsRNA) accumulates in cells infected by many different viruses (19) and is instrumental in activating expression of the interferon genes (20). dsRNA also activates incompletely defined pathways that induce expression of some ISGs directly (21). This activation is independent of IFN because induction is retained in cells that lack IFN genes or the ability to respond to IFN (22). One well studied component of this signaling cascade is the dsRNA-dependent serine/threonine kinase PKR (23), a dimer that binds to dsRNA directly, leading to its activation and autophosphorylation (ref. 24, reviewed in ref. 23). Active PKR then can phosphorylate cellular substrates, including eukaryotic protein synthesis initiation factor 2, resulting in a global inhibition of protein synthesis (25). PKR also is involved in phosphorylating the NFκB inhibitor IκB (26), leading to the release of active NFκB, thus contributing to activation of the IFNβ gene (27, 28) and some ISGs (29, 30). dsRNA-dependent induction of these ISGs involves ISRE-binding factors, and both IRF-1 (22, 31) and the recently identified dsRNA-activated factor (DRAF) (32) have been implicated in this process. Unlike the IFN-dependent induction of ISGs, the direct induction of ISGs by dsRNA does not require JAKs, STAT2, or p48, although STAT1 is required (22). However, because additional components of the dsRNA-dependent signaling cascade have remained elusive, a complete picture of the mechanism of PKR-dependent regulation of ISGs has yet to emerge.
We have described mutant human cell lines unresponsive to type I and/or type II IFNs (33–36) and mutants that secrete IFN constitutively (37). To isolate constitutive mutants that are independent of IFN secretion, we mutagenized the JAK1-minus cell line, U4C, which is unresponsive to all IFNs (35, 38). By using this approach, we now have isolated mutants with defects in ISG regulatory components. One of these, P2.1, has higher constitutive levels of ISG transcripts, indicating that the regulation of basal ISG expression is altered. Of interest, P2.1 is also deficient in dsRNA-dependent responses, including the activation of NFκB and IRF-1 and the transcriptional induction of ISGs. Together, these data reveal the existence of a pivotal factor that is involved in regulating multiple aspects of ISG expression.
MATERIALS AND METHODS
Cell Lines, Mutagenesis, Use of the Fluorescent Activated Cell Sorter (FACS), dsRNA Treatment, and Transfections.
The parental 2C4 and IFN-unresponsive mutant human cell lines used in these studies have been described elsewhere (34, 35, 38). All cells were cultured in DMEM supplemented with 10% fetal calf serum. For mutagenesis, U4C cells were treated with the intercalating mutagen ICR191 at 10–15 μg/ml, which provided 50–70% lethality (33, 39). In these cells, the IFN-responsive 9–27 gene promoter is 5′ of a cDNA encoding the human cell surface protein CD2. This promoter can be induced by both type I and II IFNs, and cells harboring the 9–27/CD2 construct have been used to isolate mutants unresponsive to either type of IFN (34). Four pools of 1 × 107 cells each were mutagenized twice and then were sorted for cells with high CD2 expression. The cells were stained with a phycoerythrin-conjugated anti-CD2 mAb (Dako). FACScan analysis was performed with a Becton-Dickinson instrument by using the lysys ii software package. For dsRNA-dependent induction of ISGs, cells in serum-free medium for 12–15 hr were treated with poly (IC)·poly (IC) (Pharmacia) at a final concentration of 100 μg/ml. Stable transfection of U4C and P2.1 cells with pRKmJAK1 (35) was carried out by the calcium phosphate procedure (40), followed by selection for puromycin resistance, from the co-transfected pSV2puro, by using 1 μg/ml of puromycin.
Electrophoretic Mobility-Shift Assays (EMSA), Western Transfers, RNA Analyses, and in Vitro Analysis of PKR Activity.
EMSA experiments (41) were performed by using whole-cell extracts. Complementary oligonucleotides representing the PRDII binding site for NFκB (28) and the ISRE binding site for ISG15 (42) were annealed and end-labeled by using standard procedures (40). For Western analyses, proteins were resolved in an 8% polyacrylamide gel, were transferred to polyvinylidenedifluoride membranes, and were incubated with antibodies. Bands were visualized by enhanced chemiluminescence by using Renaissance reagents (DuPont). Antisera to IRF-1 and IRF-2 were from Santa Cruz Biotechnology, and a polyclonal antiserum to PKR was kindly provided by Michael Katze (University of Washington). For in vitro analysis of activity, PKR was immunoprecipitated from 100 μg of whole cell extract and was analyzed in vitro for kinase activity (43). Duplicate samples were analyzed for PKR protein to assure equal loading.
RNA was isolated by the Trizol method according to the manufacturer’s specifications (GIBCO/BRL). Total RNA (10 μg) was used for RNase protection analysis of endogenous ISG mRNA (44). Probes for γ-actin and the IFN-responsive genes ISG54, 561, 6–16, 9–27, IRF-1, and guanylate-binding protein (GBP) have been described (36). Relative ISG transcript levels were calculated by normalizing to actin controls determined by PhosphorImager analysis (Molecular Dynamics). Reverse transcriptase–PCR analysis of IFNβ gene expression was performed according to standard procedures (40).
RESULTS
FACS-Based Separation of Mutant Cells with Constitutive ISG Expression.
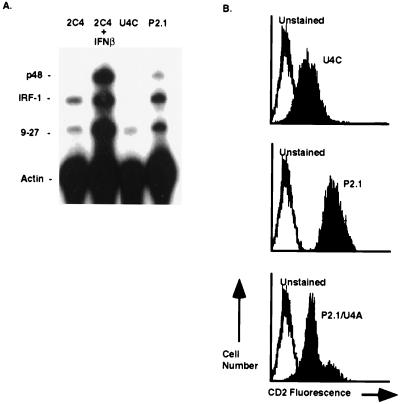
Previous attempts to isolate mutant cells with constitutively high ISG expression yielded three distinct complementation groups, each constitutively expressing IFNα or IFNβ (37). To avoid mutants with defects in regulating IFN gene expression, we used U4C cells that lack JAK1 and thus are unresponsive to any IFN (38). Because U4C were heavily mutagenized previously, only two additional rounds of mutagenesis were performed, followed by four rounds of sorting for high CD2 expression. Individual clones were isolated from the resulting population, and the basal expression of endogenous ISGs was evaluated by RNase protection analysis. As shown in Fig. 1A, clone P2.1 had 3- to 5-fold higher constitutive expression of several ISG when compared with parental U4C cells or to wild-type 2C4 cells (34). This constitutive expression was, however, much lower than the IFNα-induced ISG expression observed in 2C4 cells (20- to 100-fold increase; Fig. 1A). To test for dominance, puromycin-resistant P2.1 cells were fused to hygromycin-resistant U4A cells (also lacking JAK1; see ref. 14). After selection for hybrid cells by using both drugs, the levels of ISG transcripts were examined by RNase protection analysis. ISG mRNA levels in the fused population were comparable to U4A basal levels (data not shown), revealing that the new mutation in P2.1 cells is recessive. Furthermore, CD2 expression in the fused population was also at a low, wild-type level (Fig. 1B), confirming that CD2 expression correlates with the expression of the endogenous ISGs.
Figure 1.
RNase protection analysis of constitutive ISG expression in P2.1 cells. (A) Analysis of RNA from untreated 2C4 cells (lane 1), IFNα-treated 2C4 cells (lane 2), untreated U4C cells (lane 3), and untreated P2.1 cells (lane 4). Total cellular RNA (10 μg) was analyzed with probes for p48, IRF-1, 9–27, and γ-actin. (B) Analysis of the dominance of the constitutive phenotype. Puromycin-resistant P2.1 cells were fused with hygromycin-resistant U4A cells, and heterokaryons were selected with both drugs. FACS analysis of CD2 expression in U4C cells (Top), P2.1 cells (Middle), and P2.1/U4A heterokaryons (Bottom).
P2.1 Cells Are Deficient in dsRNA-Dependent Responses.
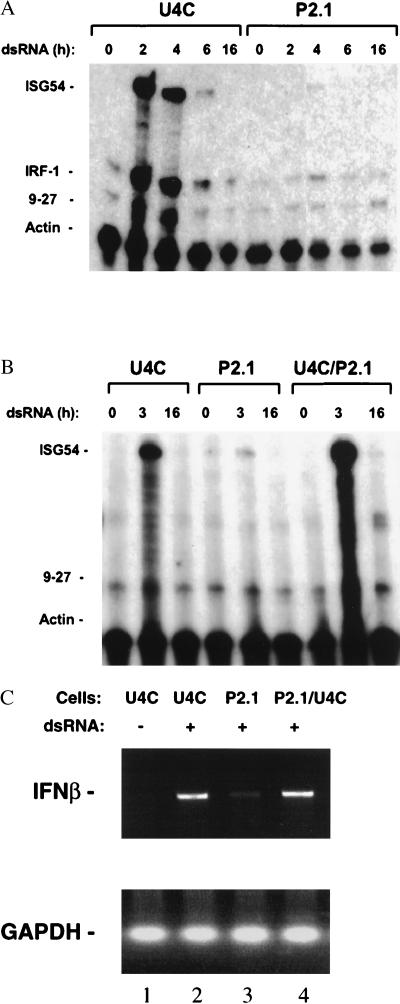
A recessive phenotype resulting in elevated basal ISG expression could arise through the constitutive activation of transcriptional inducers, such as STATs, or through the loss or down-regulation of ISG-specific transcriptional repressors. Because we found no detectable increase in the basal activation of STATs (data not shown), NFκB, or IRF-1 (see below), the possibility of a defect in ISG repression was examined in more detail. We reasoned that the defect in ISG regulation might be more pronounced in P2.1 than in U4C cells after ISGs were induced because the loss or misregulation of a transcriptional repressor might alter the kinetics of ISG down-regulation, resulting in a sustained or augmented response. dsRNA was used because neither U4C nor P2.1 cells respond to IFN. Treatment of U4C cells with dsRNA induced ISG54 expression by 100- to 150-fold within 2 hr (Fig. 2A). Unexpectedly, dsRNA hardly induced ISG54 expression in P2.1 cells at all (2- to 3-fold in 2 hr; see Fig. 2A). Similar observations were made when the expression of IRF-1 (Fig. 2A), 561, and guanylate-binding protein (GBP) was evaluated (see later). To test for dominance, puromycin-resistant P2.1 and hygromycin-resistant U4C cells were fused. As shown in Fig. 2B, the responsiveness to dsRNA was restored fully in the hybrid population indicating that, like the constitutive phenotype, the impairment of the dsRNA response is recessive. IFNβ gene induction by dsRNA was also defective in P2.1 cells but was restored on fusion of P2.1 with parental U4C cells (Fig. 2C).
Figure 2.
dsRNA-dependent induction of ISGs and IFNβ in U4C and P2.1 cells. (A) Serum-deprived U4C and P2.1 cells were untreated or treated with 100 μg/ml of dsRNA for the times indicated. The levels of ISG54, IRF-1, 9–27, and γ-actin mRNAs were determined by RNase protection. (B) Dominance of the dsRNA-resistant phenotype in P2.1 cells. Puromycin-resistant P2.1 cells were fused with hygromycin-resistant U4C cells, and the inducibility of ISG54 mRNA was analyzed in the cell population resistant to both drugs by RNase protection. The cells were untreated (lanes 1, 4, and 7) or treated with 100 μg/ml of dsRNA for 3 or 16 hr. (C) IFNβ gene induction by dsRNA in P2.1 and U4C cells. The cells were left untreated (lane 1) or treated with 100 μg/ml of dsRNA for 7 h (lanes 2–4). IFNβ mRNA levels were analyzed by reverse transcription–PCR. Glyceraldehyde-3-phosphate dehydrogenase mRNA levels were analyzed as an internal control.
Transcription Factor Activation by dsRNA Is Defective in P2.1 Cells.
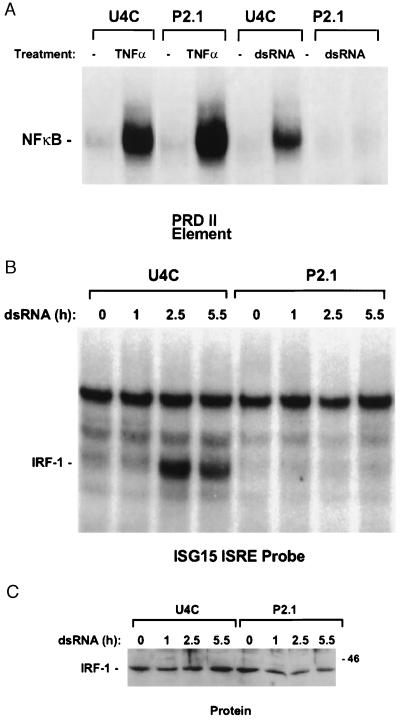
Several dsRNA-induced transcriptional activators have been identified, including NFκB (28) and the ISRE-binding factors IRF-1 (22, 31) and dsRNA-activated factor (32). EMSA of NFκB activation, using the PRDII promoter element of the IFNβ promoter, revealed that treatment of U4C cells with dsRNA and tumor necrosis factor α (TNFα) strongly induced NFκB (Fig. 3A). In P2.1 cells, NFκB was induced by TNFα but not by dsRNA (Fig. 3A), indicating that the defect in dsRNA-dependent signaling in P2.1 was likely to be upstream of NFκB and in a component not shared with the TNFα signaling cascade.
Figure 3.
Deficient activation of NFκB and IRF-1 by dsRNA in P2.1 cells. (A) Activation of NFκB for binding to a PRDII element. EMSAs were performed with whole cell extracts prepared from U4C and P2.1 cells, untreated or treated for 3 hr with 40 ng/ml of TNFα or 100 μg/ml of dsRNA. (B) Activation of IRF-1 for binding to an ISG15 ISRE. EMSAs were performed with whole cell extracts from U4C and P2.1 cells, untreated or treated with 100 μg/ml of dsRNA for the times shown above each lane. (C) Western analysis for IRF-1 protein levels in the extracts used for the EMSA experiments. The sizes of molecular weight standards are indicated in kilodaltons.
Recent data have suggested that IRF-1 activity can be regulated through activation of the latent protein independently of protein synthesis (31, 45, 46). Analysis by EMSA revealed that dsRNA did activate IRF-1 for binding to an ISG15 ISRE probe in U4C but not in P2.1 cells (Fig. 3B). The activation was rapid (within 2 hr) and did not involve an increase in IRF-1 protein levels (Fig. 3C). Western analysis of extracts from P2.1 cells indicated that the amounts of IRF-1 in P2.1 and U4C cells were similar and that it migrated identically (Fig. 3C). Taken together with the above results, these data reveal that the defect in P2.1 is caused by loss of a dsRNA-regulated factor required for activation of both IRF-1 and NFκB. The defect is recessive, as shown by cell fusion studies similar to those described above (data not shown). We were not able to detect dsRNA-induced dsRNA-activated factor (DRAF) (32) in any of the cell lines tested, including parental U4C or wild-type 2C4 (data not shown), and thus could not determine whether activation of this factor was defective in P2.1 cells.
PKR Activation by dsRNA is Normal in P2.1 Cells.
Although dsRNA signaling is understood incompletely, PKR is certainly a major component (23). Previous studies, performed with MEFs from PKR-null mice, demonstrated that PKR is required for most responses to dsRNA, including the activation of transcription factors such as NFκB and IRF-1 and the induction of IFNβ and ISG transcription (31). The status of PKR in P2.1 was evaluated by Western and in vitro kinase analyses. The protein is present and is the appropriate size in P2.1 cells (see Fig. 4B), and it can be activated by dsRNA in vitro (data not shown). In addition, the sequence of PKR mRNA from P2.1 cells, analyzed after reverse-transcriptase PCR, is wild-type (data not shown). Therefore, the failure of P2.1 to support dsRNA-dependent transcription is not caused by a mutation in the PKR gene or by loss of its expression.
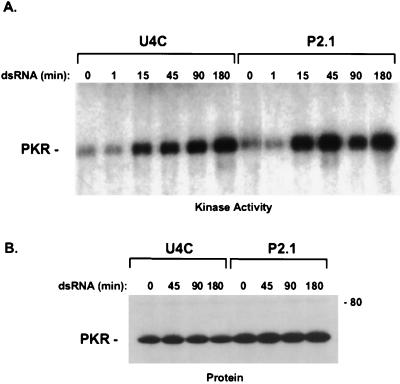
Figure 4.
Activation of endogenous PKR by dsRNA. (A) U4C and P2.1 cells were untreated or treated with 100 μg/ml of dsRNA for the times shown above each lane. The cells were washed with PBS to remove bound dsRNA and were lysed in assay buffer. PKR was immunoprecipitated from 100 μg of total cell lysate and was incubated with 10 μCi of γ-32P-labeled ATP for 10 min at 30°C. (B) Western analysis of PKR protein by using a mAb. The sizes of molecular weight standards are indicated in kilodaltons.
We next determined whether PKR is activated in dsRNA-treated P2.1 cells. U4C and P2.1 cells were treated with dsRNA, and whole-cell lysates were prepared. PKR was immunoprecipitated, and an in vitro kinase assay was used to evaluate its activation. In this assay, activation of PKR would have to occur in vivo because no activator was added to the in vitro kinase reaction, and the cells were washed rigorously before lysis to remove any adherent dsRNA. To control for the possibility that tightly bound cell-associated dsRNA might carry through into the in vitro assay, a “one-minute” time point was included in which dsRNA was added to the cells just before isolation. PKR was activated equally by dsRNA in U4C and P2.1 cells (Fig. 4A). The activation was evident within 15 min and increased further over time. There was no activity at the one-minute time point, arguing against carry over of dsRNA into the in vitro kinase reaction. Little change in PKR protein levels was observed (Fig. 4B), indicating that the increase in PKR phosphorylation reflects changes in its activation status. Taken together, these data reveal that the dsRNA signaling defect in P2.1 is downstream of the activation of PKR.
IFN Responses in JAK1-Complemented P2.1 Cells.
The IFNγ response was identical in wild-type 2C4 and in JAK1-complemented P2.1 cells (Fig. 5A). This finding is distinct from observations made with MEFs from PKR-null mice, where the IFNγ-dependent induction of the guanylate-binding protein (GBP) gene is defective (31). The IFNα-dependent induction of 6–16 (Fig. 5B) and 561 (data not shown) mRNA was examined in pools of JAK1 reconstituted P2.1 and U4C cells. Although small differences in the kinetics of the IFNα response were observed, the overall induction of these genes in the two populations was very similar, suggesting that the defective component in P2.1 is not required for IFN-dependent responses. Importantly, the restoration of JAK1 had no effect on the dsRNA signaling defect in P2.1 (Fig. 5A).
Figure 5.
ISG induction in response to IFN in JAK1-complemented U4C and P2.1 cells. (A) IFNγ-induced mRNA synthesis in P2.1/JAK1 and U4C/JAK1 cells was examined by RNase protection. The cells were untreated or treated for 3 hr with 100 μg/ml of dsRNA or 500 units/ml of IFNγ. (B) Time course of 6–16 induction by IFNα. JAK1-complemented U4C or P2.1 cells were untreated or treated with 2,500 units/ml of IFNα for the times indicated. The levels of 6–16 and actin mRNAs were analyzed by RNase protection.
DISCUSSION
Isolation of a Mutant Cell Line Defective in Regulating Basal ISG Expression.
P2.1 cells were isolated by using a FACS-based strategy designed to enrich pools of mutagenized U4C cells for clones with high constitutive expression of 9–27, monitored with a 9–27-regulated CD2 cell surface marker. The finding that P2.1 cells were also deficient in dsRNA-induced responses was unexpected. Knowledge of whether the constitutive and dsRNA-unresponsive phenotypes are caused by the same or by different mutations will have to await complementation of the defects. Once complemented, P2.1 cells should provide important new information about signaling molecules involved in maintaining the appropriate regulation of ISG expression.
The strategy described here extends previous studies aimed at isolating constitutive mutants (37). The initial work used a drug-based selection strategy in which mutagenized wild-type 2fTGH cells were selected for constitutive expression of the 6–16 ISG promoter. Because the clones identified by this approach had mutations that led to the constitutive production of type I IFNs, we used a different strategy. First, we reasoned that redundant mutations leading to subtle changes in basal ISG expression might be detected more efficiently through a FACS-based approach than by selection strategies, which might require higher levels of basal expression. Second, to avoid responses to IFN, JAK1-null cells were used. Of the mutant cell lines now available, only those lacking STAT1 or JAK1 are completely unresponsive to all IFNs (3). Because we did not want to rule out the possibility of detecting mutants in which STAT1 was constitutively activated (see below), we used the U4C cells.
Constitutive signaling mutants could arise, for example, through the loss of regulatory factors required to suppress ISG expression (repressors) or through the loss of phosphatases required to inactivate IFN-dependent signaling by dephosphorylating critical components. Because the basal expression of ISGs is very low, it seems likely that repressors associate constitutively with the ISG promoters. Whether the same repressors are involved both in terminating responses and in maintaining transcriptional quiescence is not known. Although the loss of a transcriptional repressor could account for the constitutive phenotype of P2.1 cells, if the constitutive phenotype is linked to the signaling defect in response to dsRNA, then loss of a repressor cannot easily account for both phenotypes. It is more reasonable to propose that regulation of the activity of a repressor, perhaps through a secondary modification such as phosphorylation, may be defective in P2.1 cells. Indeed, the loss of a cellular kinase that regulates both ISG-specific repressor activity and dsRNA-inducible transcription factor activity is the most likely explanation for the P2.1 phenotype, as described in more detail below.
The Defect in Response to dsRNA.
Many dsRNA-dependent actions are mediated by the IFNs that are induced in response to virus infection. In addition, dsRNA can activate the transcription of some ISGs independently of IFN, a process that is understood poorly but that may be mediated, in part, by NFκB and IRF-1 (22). Binding to dsRNA activates PKR, enabling it to phosphorylate downstream substrates such as eukaryotic protein synthesis initiation factor-2 (23, 25). The identities of other components that contribute to dsRNA-dependent signaling events have remained elusive.
With respect to dsRNA-dependent effects, the phenotypes of P2.1 cells and PKR-null cells are very similar. PKR-null cells resist the effects of dsRNA, both in terms of ISG induction and transcription factor activation (31, 47). Nonetheless, several lines of evidence suggest that the P2.1 phenotype is not caused by inactivation of PKR. PKR protein is wild-type in P2.1 cells, as demonstrated by Western and in vitro kinase assays and by the sequence of the PKR message from P2.1 cells. Significantly, the kinetics of PKR activation by dsRNA are similar in P2.1 and U4C parental cells. These data argue against a defect in the dsRNA-dependent activation of PKR and also against a defect in the uptake of dsRNA. Differences in the ability of JAK1-complemented P2.1 cells and PKR-null MEFs to respond to IFN also serve to distinguish the two phenotypes. We believe that P2.1 cells are defective in a novel component of the dsRNA response pathway.
Models To Explain the P2.1 Phenotype.
The defect in P2.1 cells affects the dsRNA-dependent induction of both NFκB-regulated genes (e.g., IRF-1 and IFNβ; Fig. 2 A and C and Fig. 5A) and of genes regulated by ISRE-binding factors (e.g., 561 and ISG54, Fig. 2 and Fig. 3D), implicating the loss of a pivotal factor in the dsRNA signaling cascade. Although few dsRNA-dependent signaling factors have been identified, data from PKR-null mice have revealed that PKR is required for the dsRNA-dependent activation of NFκB and IRF-1 and for the induction of certain ISGs (31). Therefore, one model that can explain the P2.1 phenotype places the defect downstream of PKR but upstream of the activation of IRF-1 and NFκB. With respect to NFκB, it has been shown that PKR can phosphorylate IκB directly in an in vitro kinase reaction (26) although it may not do so in vivo. Indeed, for this model to be correct, the PKR-dependent activation of transcription factors cannot be direct but must require a phosphorylation cascade. The missing component in P2.1 cells, possibly a cellular kinase, would be required to relay the PKR-dependent signal to NFκB and perhaps to other transcriptional regulators such as IRF-1.
A second model would implicate multiple signaling pathways in the activation of transcriptional regulators that act downstream of dsRNA. In this case, the defect in P2.1 would not be linked necessarily to the PKR-dependent pathway but would be in a separate pathway that is coordinately required to activate effector molecules such as NFκB and IRF-1. Multiple mechanisms of NFκB activation have been identified, and a cellular kinase involved in IkB phosphorylation, IkappaB Kinase or IKK, has been isolated recently (48–50). This kinase is involved in the activation of NFκB after stimulation of cells with TNFα or IL-1, but a role in dsRNA-dependent signaling has not been demonstrated (48–50). Of interest, human cells defective in the activation of NFκB also have been isolated. These cells, 1.3E2, differ from P2.1 cells in that they are deficient not only in dsRNA-dependent induction of NFκB but also in its activation by other stimuli, including TNFα (51). The defect in 1.3E2 cells is clearly upstream of IkB phosphorylation, although it is unknown whether the 1.3E2 defect is in IKK. It is unlikely that P2.1 cells are defective in IKK or in a component identical to the defect in 1.3E2 cells because the TNFα response in P2.1 cells is intact. Nonetheless, it is possible that the defective protein in P2.1 cells is an analogous kinase that is involved specifically in dsRNA signaling and perhaps also in regulating basal ISG expression.
A growing body of evidence suggests that IRF-1 must be activated, probably by phosphorylation on serine or threonine residues, to enable it to bind DNA and stimulate transcription (31, 45, 46). The pertinent site(s) of phosphorylation and the identity of the kinase(s) involved is currently unknown. PKR is involved in some aspect of this activation because PKR-null MEFs are impaired severely in their ability to support the dsRNA- or IFN-dependent activation of IRF-1 (31). In P2.1 cells, we observed that the dsRNA-dependent but not the IFN-dependent activation of IRF-1 was defective (data not shown). The lack of a defect in the IFN-dependent induction of target genes in the P2.1 cells with JAK1 restored further distinguishes mutant P2.1 from PKR-null mouse cells, in which the responsiveness to both types of IFN is partially impaired (31), and lends credence to the idea that P2.1 cells are defective in a component of a PKR-independent pathway. Alternatively, the role of PKR in mediating IFN responses may differ between murine and human cells. We observed minor differences in the kinetics of the IFNα response in P2.1 cells with JAK1 restored compared with U4C cells, but these differences may well be caused by different levels of JAK1. Identification of the defective component in P2.1 awaits complementation with DNA encoding the wild-type protein, which is currently under way.
Acknowledgments
We thank Bryan Williams, Sandy Der, Sudip Bandyopadhyay, George Leonard, Aseem Kumar, and Suzanne Kadereit for helpful comments and suggestions and Michael Katze for the polyclonal PKR antiserum. This work was supported by Grant P01-CA62220 from the National Institutes of Health and Training Grant F32-AI 08956 (to D.W.L.). We gratefully acknowledge the support of the W. M. Keck Foundation for our fluorescence-activated cell sorting facility.
ABBREVIATIONS
- IFNs
interferons
- ISGs
IFN-stimulated genes
- dsRNA
double-stranded RNA
- JAKs
Janus tyrosine kinases
- STATs
signal transducers and activators of transcription
- ISRE
IFN-stimulated response element
- IRF-1
IFN regulatory factor 1
- MEFs
mouse embryo fibroblasts
- FACS
fluorescent activated cell sorter
- EMSA
electrophoretic mobility-shift assays
- TNFα
tumor necrosis factor α
References
- 1.De Maeyer E, De Maeyer-Guignard J. Interferons and Other Regulatory Cytokines. New York: Wiley; 1988. [Google Scholar]
- 2.Uzé G, Lutfalla G, Mogensen K E. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- 3.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 4.Ihle J N, Kerr I M. Trends Genet. 1995;11:69–74. doi: 10.1016/s0168-9525(00)89000-9. [DOI] [PubMed] [Google Scholar]
- 5.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 6.Leaman D W, Leung S, Li X, Stark G R. FASEB J. 1996;10:1578–1588. [PubMed] [Google Scholar]
- 7.Imam A M A, Ackrill A M, Dale T C, Kerr I M, Stark G R. Nucleic Acids Res. 1990;18:6573–6580. doi: 10.1093/nar/18.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy D E, Kessler D S, Pine R, Darnell J E., Jr Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 9.Briken V, Ruffner H, Schultz U, Schwarz A, Reis L F L, Strehlow I, Decker T, Staeheli P. Mol Cell Biol. 1995;15:975–982. doi: 10.1128/mcb.15.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reis L F L, Harada H, Wolchok J D, Taniguchi T, Vilcek J. EMBO J. 1992;11:185–193. doi: 10.1002/j.1460-2075.1992.tb05041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig T M, Amakawa R, Kishihara K, Wakeham A, et al. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 12.Friedman R L, Manly S P, McMahon M, Kerr I M, Stark G R. Cell. 1984;38:745–755. doi: 10.1016/0092-8674(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 13.Harada H, Fujita T, Miyamoto M, Kimurak Y, Murayama M, Furia A, Miyata T, Taniguchi T. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 14.Nelson N, Marks M S, Driggers P H, Ozato K. Mol Cell Biol. 1993;13:588–599. doi: 10.1128/mcb.13.1.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamagata T, Nishida J, Tanaka T, Sakai R, Mitani K, Yoshida M, Taniguchi T, Yazaki Y, Hirai H. Mol Cell Biol. 1996;16:1283–1294. doi: 10.1128/MCB.16.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haque S J, Flati V, Deb A, Williams B R G. J Biol Chem. 1995;270:25709–25714. doi: 10.1074/jbc.270.43.25709. [DOI] [PubMed] [Google Scholar]
- 17.Igarashi K-I, David M, Larner A C, Finbloom D S. Mol Cell Biol. 1993;13:3984–3989. doi: 10.1128/mcb.13.7.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David M, Chen H E, Goelz S, Larner A C, Neel B G. Mol Cell Biol. 1995;15:7050–7058. doi: 10.1128/mcb.15.12.7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welsh R M, Sen G C. In: Nonspecific Host Responses to Viral Infection. Nathanson N, editor. Philadelphia: Lippincott; 1997. 109–141. [Google Scholar]
- 20.Thanos D, Maniatis T. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 21.Tiwari R K, Kusari J, Kumar R, Sen G C. Mol Cell Biol. 1988;8:4289–4294. doi: 10.1128/mcb.8.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bandyopadhyay S K, Leonard G T, Jr, Bandyopadhyay T, Stark G R, Sen G S. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 23.Williams B R G. Semin Virol. 1995;6:191–202. [Google Scholar]
- 24.Patel R C, Stanton P, McMillan N M, Williams B R, Sen G C. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samuel C E. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 26.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R G. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenardo M J, Fan C M, Maniatis T, Baltimore D. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 28.Visvanathan K V, Goodbourne S. EMBO J. 1989;8:1129–1138. doi: 10.1002/j.1460-2075.1989.tb03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drew P D, Franzoso G, Becker K G, Bours V, Carlson L M, Siebenlist U, Ozato K. J Interferon Cytokine Res. 1995;15:1037–1045. doi: 10.1089/jir.1995.15.1037. [DOI] [PubMed] [Google Scholar]
- 30.Ten R M, Blank V, Bail O L, Kourilsky P, Israel A. Immunology. 1993;316:496–501. [PubMed] [Google Scholar]
- 31.Kumar A, Yang Y-L, Flati V, Der S, Kadereit S, Deb A, Haque J, Reis L, Weissmann C, Williams B R. EMBO J. 1997;16:406–416. doi: 10.1093/emboj/16.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daly C, Reich N C. Mol Cell Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellegrini S, John J, Shearer M, Kerr I M, Stark G R. Mol Cell Biol. 1989;9:4605–4612. doi: 10.1128/mcb.9.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn B A, Quelle F W, Rogers N C, Schindler C, Stark G R, Ihle J N, Kerr I M. Nature (London) 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 35.Müller M, Briscoe J, Laxton C, Guschin D, Ziemiecki D, Silvennoinen O, Harpur A G, Barbieri G, Witthuhn B A, Schindler C, et al. Nature (London) 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 36.Müller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKendry R, Pellegrini S, Kerr I M, Stark G R. J Virol. 1994;68:4057–4062. doi: 10.1128/jvi.68.6.4057-4062.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kohlhuber F, Rogers N C, Watling D, Feng J, Guschin D, Briscoe J, Witthuhn B A, Kotenko S V, Pestka S, Stark G R, et al. Mol Cell Biol. 1997;17:695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKendry R, John J, Flavell D, Muller M, Kerr I M, Stark G R. Proc Natl Acad Sci USA. 1991;88:11455–11459. doi: 10.1073/pnas.88.24.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 41.Sadowski H B, Gilman M Z. Nature (London) 1993;362:79–83. doi: 10.1038/362079a0. [DOI] [PubMed] [Google Scholar]
- 42.Reich N, Evans B, Levy D, Knight E, Jr, Darnell J E., Jr Proc Natl Acad Sci USA. 1987;84:6394–6398. doi: 10.1073/pnas.84.18.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel R C, Stanton P, Sen G C. J Biol Chem. 1994;269:18593–18598. [PubMed] [Google Scholar]
- 44.Melton D A, Krieg P A, Rebagliati M R, Maniatis T, Zinn K, Green M R. Nucleic Acids Res. 1984;12:7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirchhoff S, Koromilas A E, Schaper F, Grashoff M, Sonenberg N, Hauser H. Oncogene. 1995;11:439–445. [PubMed] [Google Scholar]
- 46.Watanabe N, Sakakibara J, Hovanessian A G, Taniguchi T, Fujita T. Nucleic Acids Res. 1991;19:4421–4428. doi: 10.1093/nar/19.16.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Y-L, Reis L F, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R, Aguet M, Weissmann C. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J, Young D B, Barbosa M, Mann M, Manning A, Rao A. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 49.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 50.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 51.Courtois G, Whiteside S T, Sibley C H, Israel A. Mol Cell Biol. 1997;17:1441–1449. doi: 10.1128/mcb.17.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]