Abstract
The chemokine stromal cell-derived factor 1, SDF-1, is an important regulator of leukocyte and hematopoietic precursor migration and pre-B cell proliferation. The receptor for SDF-1, CXCR4, also functions as a coreceptor for T-tropic HIV-1 entry. We find that mice deficient for CXCR4 die perinatally and display profound defects in the hematopoietic and nervous systems. CXCR4-deficient mice have severely reduced B-lymphopoiesis, reduced myelopoiesis in fetal liver, and a virtual absence of myelopoiesis in bone marrow. However, T-lymphopoiesis is unaffected. Furthermore, the cerebellum develops abnormally with an irregular external granule cell layer, ectopically located Purkinje cells, and numerous chromophilic cell clumps of abnormally migrated granule cells within the cerebellar anlage. Identical defects are observed in mice lacking SDF-1, suggesting a monogamous relationship between CXCR4 and SDF-1. This receptor-ligand selectivity is unusual among chemokines and their receptors, as is the function in migration of nonhematopoietic cells.
Chemokines comprise a large number of structurally related proteins that regulate migration and activation of leukocytes through G protein-coupled cell-surface receptors (1, 2). Chemokines are important in directing emigration of specific leukocyte subsets into inflammatory sites, as well as in determining trafficking patterns of recirculating lymphocyte subsets. The chemokine stromal cell-derived factor 1 (SDF-1) has complex effects on the migration, proliferation, and differentiation of leukocytes. SDF-1 is a highly efficacious chemoattractant for T lymphocytes and monocytes in vitro and in vivo and also a chemoattractant for CD34+ hematopoietic progenitors (3, 4). Furthermore, SDF-1 regulates B lymphocyte maturation; its first known biological activity was stimulation of pre-B cell proliferation (5). Consistent with this proliferative effect, mice deficient in SDF-1 have severely reduced B-lymphopoiesis but additionally show an absence of bone marrow myelopoiesis (6). SDF-1-deficient mice die perinatally and have ventricular septal defects. Thus, SDF-1 may be involved in directing progenitor cells into appropriate microenviroments to receive expansion and differentiation signals, as well as in delivering such signals itself.
SDF-1 is the biological ligand for CXCR4, a G protein-coupled seven-transmembrane receptor (7, 8). CXCR4 also functions as a coreceptor for the entry of T-tropic strains of HIV-1 into CD4+ cells (9). SDF-1 specifically blocks viral entry and infection of CD4+ T cells through CXCR4 (7, 8). The occurrence of T-tropic primary HIV-1 isolates in infected individuals is associated with the decline of CD4+ cells and clinical progression to AIDS.
SDF-1 and CXCR4 have several unusual features for a chemokine and receptor. First, SDF-1 is extraordinarily conserved in evolution, with only one amino acid substitution between the human and mouse proteins (10). Based on the presence of an intervening amino acid between the two N-terminal cysteines, SDF-1 has been grouped with the CXC chemokine subfamily; however, the protein sequence of SDF-1 appears to be equally related evolutionarily to both the CXC and CC chemokines. Therefore, it has been proposed that SDF-1 is a primordial chemokine (3). Most chemokine receptors bind more than one chemokine, and most chemokines bind to more than one receptor (1, 2). To date, SDF-1 is the only known ligand for CXCR4, and CXCR4 is the only known receptor for SDF-1. However, because SDF-1 has such widely varying biological activities, multiple receptors or ligands could be readily envisioned. Unlike the induced expression of many chemokines and their receptors, SDF-1 and CXCR4 are expressed constitutively in a wide range of tissues including brain, heart, kidney, liver, lung, and spleen (11–13). CXCR4 is not only expressed on lymphocytes and monocytes in peripheral blood (11, 14, 15) but also in the brain on a variety of cell types, including microglia, astrocytes, and neurons (16, 17). During early development, CXCR4 is expressed in both hematopoietic organs and proliferative areas of brain (18).
The widespread distribution and constitutive expression pattern of CXCR4 and SDF-1 suggest roles in diverse cellular aspects during development. To examine the biological function of CXCR4 in developing leukocytes as well as nonhematopoietic cell lineages, and as a genetic approach to test the receptor-ligand relationship between CXCR4 and SDF-1, we have generated CXCR4-deficient mice and compared their phenotype with those of SDF-1-deficient mice.
MATERIALS AND METHODS
Generation of CXCR4-Deficient Mice.
A CXCR4 genomic clone was isolated from a 129/Sv mouse genomic λ phage library (Stratagene). A targeting construct was made by ligating 5′-SalI-KpnI and 3′-BamHI-KpnI genomic fragments to a pgk-neomycin-resistance gene cassette (Fig. 1A). The linearized construct was introduced into RW4 embryonic stem (ES) cells (Genome Systems, St. Louis) by electroporation (19). Homologous recombinant ES clones were obtained after G418 selection and PCR identification by using primers PN1 5′-AAGAACGAGATCAGCAGCCT-3′, PS5 5′-AGGCAGGTCAGTCTGAGAAT-3′, and PS2 5′-CAAGGAAACTGCTGGCTGAA-3′. ES cells heterozygous for the CXCR4 gene were injected into C57BL/6J blastocysts (19) and gave rise to chimeric mice that transmitted the CXCR4 mutation in the germline.
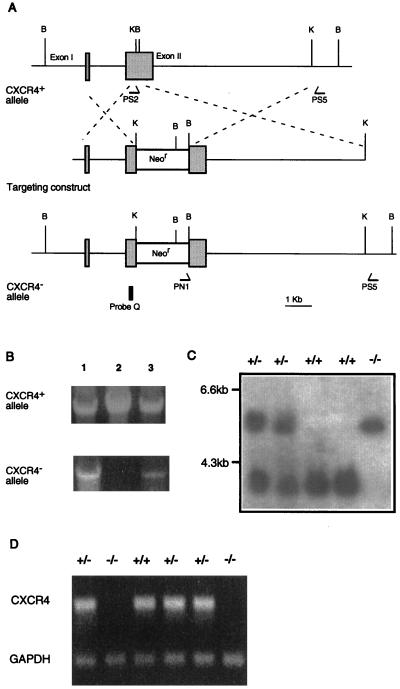
Figure 1.
Generation of CXCR4−/− mice. (A) Schematic diagram of the CXCR4 genomic locus, the targeting construct, and the targeted CXCR4 allele. B, BamHI; K, KpnI. Shadowed boxes represent the two coding exons of the CXCR4 gene. The 80-bp KpnI-BamHI fragment of exon II is replaced by the pgk-neomycin-resistance gene cassette (Neor, open box) resulting in disruption of the fourth transmembrane domain of CXCR4-coding sequences. (B) Three representative ES cell clones screened by PCR using PS5, a 3′ common primer, in combination with PS2 for detection of the CXCR4+ allele or PN1 for detection of CXCR4− allele. (C) Southern blots with probe Q on BamHI-digest DNA to genotype representative mutant and littermate mice. (D) Reverse transcription–PCR analysis of brain demonstrates the absence of CXCR4 transcript in CXCR4−/− mice. Glyceraldehyde-3-phosphate dehydrogenase is the control transcript.
SDF-1-deficient mice were described previously (6).
Fetal Liver B Cell Culture.
For B cell culture (20), E15.5 fetal liver cells (1 × 106 cells per 2 ml in 12-well plates) were suspended in RPMI 1640 medium supplemented with 5% fetal calf serum, 5 × 10−5 M 2-mercaptoethanol, 50 units/ml penicillin, 50 μg/ml streptomycin, and 10 ng/ml recombinant murine IL-7 (R & D Systems) and cultured for 5 days. The nonadherent cells were collected for flow cytometry.
Fetal Thymus Organ Culture.
Fetal thymic lobes from E15.5 embryos were placed on 12-mm diameter culture plate inserts (Millicell-HA 0.45 μm from Millipore) that had been preincubated for 12 hr in 24-well tissue culture plates filled with 300 μl of DMEM supplemented with 10% fetal calf serum (21). After 7 days of culture, single cell suspensions were prepared from the thymic lobes for flow cytometry.
Flow Cytometry.
Single cell suspensions prepared from mouse lymphoid organs were stained with antibodies as recommended by the manufacturer (PharMingen). mAbs were conjugated with fluorescein isothiocyanate (FITC), R-phycoerythrin-RM4–5, or Cy-Chrome: anti-B220-Cy-Chrome, anti-CD43-phycoerythrin, anti-IgM-FITC, anti-CD4-phycoerythrin, anti-CD8-Cy-Chrome, anti-CD3-FITC, anti-CD11b-phycoerythrin, anti-CD18-FITC, and anti-Gr-1-FITC. Cells were subjected to FACScan (Becton Dickinson). Propidium iodide staining was used to exclude dead cells.
Histology and Immunohistochemistry.
Histologic evaluation was performed on formalin-fixed paraffin-embedded sections. Cytochemical stains of lymphoid organs were performed on paraffin sections with naphthol AS-D chloroacetate (Sigma) as the substrate for neutrophil-specific esterase and on cryostat sections with diaminobenzidine tetrahydrochloride (Vector Laboratories) as the chromogenic substrate for neutrophil myeloperoxidase. Immunohistochemical staining of lymphoid organs was performed on cryostat sections by using mAbs (PharMingen): anti-B220, anti-CD34, anti-Ter-119, anti-CD62P, and anti-CD11b. Detection was with an appropriate biotinylated secondary antibody and the Elite Vectastain avidin-biotin-horseradish peroxidase reagent with diaminobenzidine tetrahydrochloride. Fetal brain was fixed in 10% formalin and cryoprotected in 15% and then 30% sucrose in PBS, and 10–12 μm sections were cut by cryostat. Immunostaining was performed by overnight incubations with anti-calbindin (Sigma) or anti-BrdUrd (Boehringer Mannheim) in PBS with 5% goat serum and 0.1% Nonidet P-40 followed by incubation with a Cy3-linked goat anti-mouse antibody (Jackson Laboratories).
BrdUrd Labeling of Proliferating Cells.
Pregnant females were injected i.p. with BrdUrd (Sigma) (50 μg/g body weight) and killed 2 hr later. Sections were postfixed in 70% ethanol, permealized in 0.4% Triton X-100, and treated with 2 M HCl followed by 0.1 M NaB2O7 before immunostaining.
RESULTS
Inactivation of the CXCR4 gene.
We replaced the fourth transmembrane domain and a portion of the third extracellular domain of the CXCR4 gene with a neomycin resistance gene. The structure of the mutant locus was confirmed by PCR and Southern blots of the ES cell and germline-transmitted mouse DNA (Fig. 1A–C). CXCR4 transcripts were absent in homozygous mutant mice (Fig. 1D).
Mice heterozygous for the CXCR4 mutation are viable, fertile, and appear normal. CXCR4-deficient mice die perinatally, with ≈one-third dead at E18.5. Viable CXCR4-deficient embryos were slightly smaller than control embryos. Histological analysis revealed severe abnormalities in bone marrow and cerebellum, which will be described in detail below. The remainder of the organs appeared grossly and microscopically normal, except that the lungs were collapsed, and the kidneys had vascular congestion and prominent interstitial hemorrhage. Four-chamber cardiac development was seen in six mice examined, with no ventricular septal defects noted (data not shown) in contrast to SDF-1-deficient mice (6).
B-Lymphopoiesis in CXCR4-Deficient Liver.
Mouse hematopoiesis begins in the fetal liver after day 11 of embryonic life and the fetal liver remains the major hematopoietic organ until the first postnatal week (22). To investigate the role of CXCR4 in early hematopoiesis, fetal liver cell suspensions from E18.5 embryos were analyzed by flow cytometry for the development of B lymphocytes and myeloid progenitors. In comparison with wild-type and heterozygous littermates, the B220+/CD43+ pro-B population was severely reduced in CXCR4-deficient fetal liver (7.0% vs. 0.2% respectively) (Fig. 2A). E15.5 fetal liver cells were cultured in vitro with IL-7 to induce the proliferation of the pro-B population (20, 23). Wild-type fetal liver yielded a large proportion of pro-B cells upon IL-7 stimulation (58.8%), whereas CXCR4-deficient fetal liver yielded few pro-B cells (1.6%) (Fig. 2B).
Figure 2.
Defects in B-lymphopoiesis and myelopoiesis and normal T lymphopoiesis in CXCR4−/− mice determined by flow cytometry. (A and B) Pro-B cells (B220+CD43+) within lymphocyte scatter gates were enumerated in E18.5 fetal liver (A) and 5 day Whitlock–Witte culture with IL-7 of E15.5 fetal liver cells (B). (C) Staining of Gr-1+CD11b+ (Upper) and CD18+CD11b+ (Lower) myeloid cells within scatter gates for myelomonocytic cells in E18.5 fetal liver. E18.5 fetal thymocytes (D) or cells from thymic organ culture of E15.5 thymus (E) were stained with CD4 and CD8 mAb. The percentages of gated cells within the boxes are indicated.
T Lymphopoiesis in CXCR4-Deficient Thymus.
The thymus of E18.5 CXCR4-deficient embryos appeared histologically normal (data not shown). Flow cytometry confirmed that CD4+CD8+ T cell progenitors were not affected (Fig. 2D). Although CXCR4 expression was shown to be modulated during T cell differentiation in a maturation-dependent pattern in thymus (24), CXCR4-deficient thymocytes were able to differentiate from immature CD4+/CD8+ cells to mature single positive cells in in vitro fetal thymic organ culture (Fig. 2E) (21).
Hematopoiesis in CXCR4-Deficient Bone Marrow.
During mouse fetal development, bone marrow becomes the hematopoietic site after E15 and remains the major definitive hematopoietic organ throughout life (22, 25). Sections of E15.5 wild-type bone marrow revealed nearly 100% cellularity with a predominance of early myeloid and erythroid elements. In contrast, the marrow from E15.5 CXCR4-deficient embryo is hypocellular with marked reduction in hematopoiesis. The entire marrow is composed predominantly of stromal cells and osteoblasts with a severe reduction in all hematopoietic lineages (data not shown). Sections from E18.5 CXCR4-deficient bone marrow also showed reduced hematopoiesis but with cellularity comparable with wild-type control and normal numbers of maturing erythrocytes and megakaryocytes (Fig. 3B).
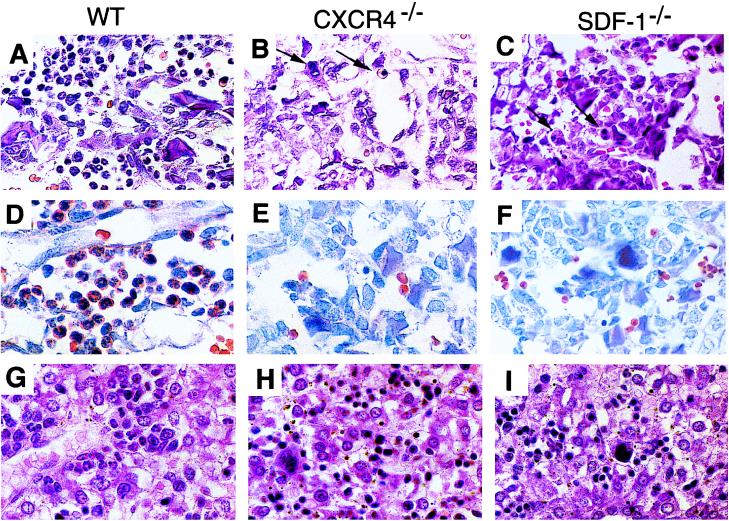
Figure 3.
Defects in B-lymphopoiesis and myelopoiesis in CXCR4−/− and SDF-1−/− mice. (A–C) Sections of femur bone marrow stained with H&E. Arrows indicate erythroid and megakaryocytic cells. (D–F) Sections of bone marrow stained for chloroacetate esterase activity. (G–I) Sections of liver stained with H&E.
Myelopoiesis Defects in CXCR4- and SDF-1-Deficient Mice.
The development of myeloid cells in CXCR4-deficient fetal liver was monitored with two sets of surface markers, CD11b/CD18 and CD11b/Gr-1. Myeloid cells in E18.5 CXCR4-deficient fetal liver were reduced to 25% of the numbers seen in wild-type control (Fig. 2C). This result was confirmed by histologic evaluation of E18.5 liver, which revealed noticeable reduction of myelopoiesis with marked erythroid predominance and many fewer mature myeloid elements as compared with wild-type (Fig. 3 G and H). SDF-1-deficient mice showed an identical reduction in liver myelopoiesis by histologic (Fig. 3I), cytochemical and flow cytometry analyses (data not shown). Markedly reduced myeloid elements also were noted in the spleen of both CXCR4- and SDF-1-deficient E18.5 mice (data not shown).
The most dramatic effect of CXCR4 deficiency on myelopoiesis was seen in the bone marrow. Sections of E18.5 wild-type bone marrow were hypercellular with a predominance of myeloid elements (Fig. 3 A and D). In contrast, the CXCR4-deficient marrow showed a virtual absence of myeloid forms as determined by hematoxylin/eosin (H&E) staining (Fig. 3B), cytochemical stains for chloroacetate esterase (Fig. 3E) and myeloperoxidase (data not shown). An identical phenotype was observed in SDF-1-deficient bone marrow (Fig. 3 C and F) as previously reported (6).
Abnormal Neuron Migration in CXCR4- and SDF-1-Deficient Cerebellum.
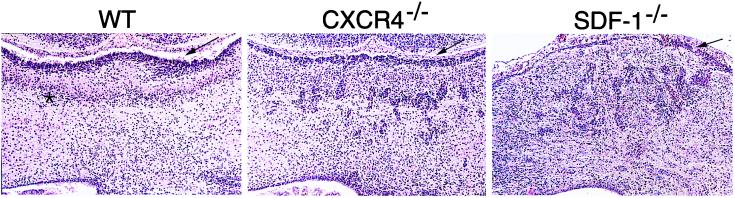
During embryonic development, CXCR4 is expressed in the proliferative neuroepithelium by E14 (18). Initial histological exam revealed a disorganized cerebellum in both CXCR4- and SDF-1-deficient mice, in contrast to normally formed cerebrum, basal ganglia, midbrain, and spinal cord (data not shown). The E18.5 cerebellum has a distinct cytoarchitecture with two principal neuronal populations. The most peripheral external granule cell layer (EGL), comprises proliferative granule cell precursors, with an underlying diffuse Purkinje cell layer (26). As compared with the well-developed wild-type cerebellum with distinct layers, H&E staining of E18.5 CXCR4- and SDF-1-deficient cerebellum revealed a markedly attenuated EGL and the presence of chromophilic cell clumps within the cerebellar anlage (Fig. 4).
Figure 4.
Cerebellum defects in CXCR4−/− and SDF-1−/− mice. Coronal sections of E18.5 cerebellum stained with H&E. Arrows indicate EGL and asterisk indicates Purkinje cell layer.
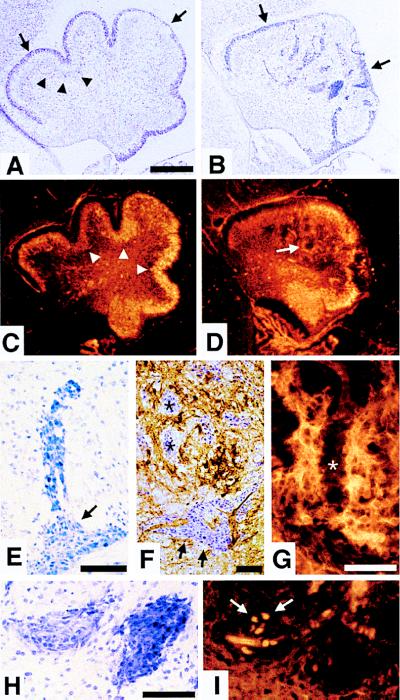
Detailed analysis of CXCR4-deficient cerebellum revealed a highly distorted architecture (Fig. 5 A and B). The EGL is irregular, and the overall shape of the cerebellum is altered, with an absence of foliation (Fig. 5 A and B). The Purkinje cells, rather than immediately underlying the EGL, are located ectopically (Fig. 5 C and D). The vast majority of cells within the cerebellar chromophilic aggregates appear viable, although individual pyknotic nuclei could be seen in some of the clumps (data not shown). The cellular morphology, the fusiform cells migrating from the EGL (Fig. 5E), and the absence of calbindin staining of the cells in clumps (Fig. 5F) are consistent with the chromophilic clusters being comprised primarily of early granule cell progenitors. Most of the cell clumps (asterisks in Fig. 5F) are surrounded by Purkinje cells with few exceptions (arrow in Fig. 5F). It appears that the Purkinje cell neurites do not extend into the clumps (Fig. 5G). The cell clumps displayed heterotypic Nissl stain (Fig. 5H) and variable BrdUrd labeling (arrows in Fig. 5I)
Figure 5.
Abnormal neuronal cell migration in CXCR4−/− cerebellum. Parasagittal sections of E18.5 cerebellum with anterior to the left and dorsal to the top. (A and B) Nissl staining. The EGL (arrows) of wild-type cerebellum (A) lies between the meninges and the diffuse Purkinje cell layer (partially marked by arrowheads). The abnormal cerebellum in CXCR4−/− mice (B) has an irregular EGL (arrows) and large chromophilic cell clumps within the cerebellar anlage. (C and D) Calbindin immunohistochemistry. In comparison with the Purkinje cell layer (arrowheads) in wild-type cerebellum (C), Purkinje cells are located ectopically (arrow) in CXCR4−/− cerebellum (D). (E–I) Chromophilic cell clumps in CXCR4−/− cerebellar anlage. Streaming cells from the EGL (E). Combined Nissl and calbindin staining (F). Purkinje cell neurites do not extend into the clumps (asterisks) (G). Adjacent sections stained by Nissl technique (H) or BrdUrd incorporation (I). Scale bar is 200 μm (A–D) and 50 μm (E–H).
DISCUSSION
We have demonstrated an important function for the chemokine receptor CXCR4 in development of widely different cell lineages. Furthermore, our studies extend previous results reported for SDF-1-deficient mice (6), by demonstrating a defect in cerebellar development and a delay in hematopoietic development, that parallel those seen in CXCR4-deficient mice. Both CXCR4 and SDF-1 play important roles in hematopoiesis. The severe reduction of B-lymphopoiesis and lack of bone marrow myelopoiesis in CXCR4-deficient mice parallels that in SDF-1-deficient mice (6). Few B220+CD43+ pro-B cells are present in both CXCR4-and SDF-1-deficient mice, showing a block at an early stage in B cell development. Furthermore, we find a quantitative decrease of myeloid lineage cells in fetal liver and spleen of both CXCR4- and SDF-1-deficient mice. The data are compatible with several possible functions for CXCR4 and SDF-1 in hematopoiesis.
To date, chemokines and their receptors have been largely studied for their roles in directing leukocyte migration and activation (1, 2). CXCR4 and SDF-1 have been shown to mediate the chemotaxis of hematopoietic progenitor cells, lymphocytes, and monocytes in vitro (3, 4) and lymphocyte and monocyte accumulation in vivo (3). Recently, it has been shown that CXCR4 can mediate the migration of early stage B cell precursors toward SDF-1 (27). The severe reduction of B-lymphopoiesis and the absence of bone marrow myelopoiesis in both CXCR4- and SDF-1-deficient mice may reflect a defect in directing the migration of several hematopoietic cell types to the correct microenvironments, which in turn provides necessary growth and maturation factors.
Alternatively, the SDF-1 and CXCR4 pathway may directly deliver essential proliferation and differentiation signals to hematopoietic cells. This idea is supported by the finding that SDF-1 can stimulate pre-B cell proliferation in vitro (5). The downstream effects of CXCR4 activation are largely unknown. It has been shown recently that the binding of SDF-1 to CXCR4 can induce tyrosine phosphorylation of the Pyk2 tyrosine kinase (28). Whether the signals transduced through CXCR4 can lead to activation of transcription factors such as E2A, Ikaros, and Pax5, which have been shown to be essential for pro-B cell development, remains to be explored (29).
Mice lacking either CXCR4 or SDF-1 display abnormal migration of granule cells, a type of neuronal cell in the cerebellum. Perhaps secondary to this, the cerebellum is grossly malformed, with an absence of foliation. Normally, granule cell progenitors are generated between E13 and E15 in the rhombic lip and then migrate rostromedially along the surface of the cerebellar anlage, forming the EGL. Postnatally, the post-mitotic granule cell precursors migrate into the cerebellum past Purkinje cells forming the internal granule cell layer (26). Two lines of reasoning suggest that the ectopic granule cell clusters in both CXCR4- and SDF-1-deficient mice result from the abnormal inward migration of EGL cells. First, at E15.5 cells can be seen streaming from the EGL with fewer clusters within the anlage than at E18.5 (data not shown), suggesting cluster formation is an ongoing process between E15 and E18. Second, there is an identifiable, although irregular, EGL present in CXCR4-deficient mice at both E15.5 and E18. In particular, some rostral portions of the EGL that require the longest distance of migration appear normally formed, which would seem unlikely if cells from the rhombic lip failed to migrate subpially. An intriguing hypothesis is that inactivation of CXCR4 or SDF-1 may lead EGL cells to migrate prematurely into the cerebellum, before a supportive environment or appropriate cues for the localization in the internal granule cell layer within the cerebellum are in place.
Although Purkinje cell organization within the cerbellum is irregular, many cells seem to migrate appropriately. This result is comparable with the phenotype of Math1-deficient mice, in which complete dysgenesis of the EGL does not preclude the migration of Purkinje cells (30). Thus, the ectopic location of the Purkinje cells in CXCR4-deficient mice is likely to be secondary to the misplaced EGL, which may cause Purkinje cell migration to stall subcortically.
As with the defect in hematopoiesis, we do not know whether normal granule cell migration in the cerebellum requires a gradient of SDF-1 to guide cell localization or whether SDF-1 and CXCR4-signaling pathways regulate development or differentiation of neural cells required for coordination of cell migration. The mechanism of neural patterning in developing cerebellum is complicated and not well understood. Multiple environmental cues have been proposed to be involved in the migratory pathway of developing neuronal cells in cerebellum (31, 32). Our results demonstrate that CXCR4 and SDF-1 specifically regulate neuronal migration in cerebellum because other regions of the central nervous system such as the cerebral cortex appear to develop normally. Consistent with this difference, distinctive neural migration routes are used for establishment of the subcortical cell layers in the cerebrum and cerebellum. A selective cerebellar defect also is seen in the rostral cerebellar malformation (rcm) mutant in which the migration of EGL is abnormal postnatally (33).
There is evidence for other developmental defects in CXCR4- and SDF-1-deficient mice. Lack of closure of the ventricular septum has been noted in SDF-1-deficient mice, and SDF-1 was expressed in endocardium (6). The earliest we have been able to observe newborn CXCR4-deficient mice is <2 hr after birth, at which time they are dead. Neither the myocardial, cerebellar, or hematopoietic defects explain lack of viability. As well as brain and heart, CXCR4 and SDF-1 are well expressed in lung and kidney (12, 13), and we have found that the lungs are collapsed and the kidneys have vascular congestion in CXCR4-deficient embryos. These organs require further examination.
Previously, it has been unclear whether SDF-1 is the only ligand for CXCR4 or CXCR4 is the only receptor for SDF-1. The nearly identical phenotype of CXCR4- and SDF-1-deficient mice provides genetic evidence that suggests a monogamous relationship between CXCR4 and SDF-1. In particular, the findings strongly argue that SDF-1 acts through a single receptor, CXCR4, in the development of mouse hematopoietic and neuronal cells. Although septal defects were observed in SDF-1-deficient mice (6) and not in the CXCR4-deficient mice studied here, septal defects are common in a variety of knockout mice strains and may reflect differences in genetic background rather than in specific function of the altered locus.
Our results demonstrate that CXCR4 and SDF-1 are important in development. Although chemokine receptors and the chemokine family are largely known for their role in leukocyte migration, activation, and homing, recent studies demonstrate expression of several chemokine receptors in the central nervous system (34). SDF-1 and CXCR4 provide the first example of a chemokine-receptor interaction directly involved in regulating neuronal cell migration. CXCR4 is expressed in the embryonic nervous system (18) as well as in adult brain (17). It has recently been suggested that CXCR4 may directly mediate HIV-1 infection of the central nervous system and lead to viral-associated neuronal apoptosis (35–38). Further study of the function of CXCR4 in the central nervous system will advance understanding of the pathogenesis of AIDS dementia (39).
Acknowledgments
We thank Drs. Frederick Alt, Craig Gerard, Joe Sodroski, and Li-Huei Tsai for reviewing this manuscript. Q.M. and D.J. are supported by the Irvington Institute, Cancer Research Institute and the National Institutes of Health. This work was supported by National Institutes of Health Grant HL-48675.
ABBREVIATIONS
- SDF-1
stromal cell-derived factor 1
- EGL
external granule cell layer
- FITC
fluorescein isothiocyanate
- ES
embryonic stem
- H&E
hematoxylin/eosin
Note Added in Proof
Subsequent to the submission of this manuscript, the results of two independently derived lines of CXCR4-deficient mice were published (40, 41), which show similar defects in B-lymphopoiesis. One group reported an essentially identical cerebellar defect to that described herein (41). A second group detailed a defect in vascularization of the gastrointestinal tract (40). Although we have not done detailed studies on vascularization of different organs, we can confirm the presence of attenuated, thin-walled mesenteric blood vessels and occasional intestinal hemorrhagic foci in our CXCR4-deficient mice. Nevertheless, the esophagus, stomach, small bowel, and colon appeared normally formed with only occasional foci of hemorrhage noted in some CXCR4-deficient mice. The vascularization defect may be related to the prominent vascular congestion and interstitial hemorrhage in the kidneys and the collapsed lungs we have noted in CXCR4-deficient mice. Both of the other groups reported lack of closure of the ventricular septum in their CXCR4-deficient mice (40, 44), similar to that seen in SDF-1 deficient mice (6). It is possible that the penetrance of ventricular septal defects is dependent on the genetic background of the mice.
References
- 1.Luster A D. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 2.Baggiolini M. Nature (London) 1998;392:565–568. doi: 10.1038/33340. [DOI] [PubMed] [Google Scholar]
- 3.Bleul C C, Fuhlbrigge R C, Casasnovas J M, Aiuti A, Springer T A. J Exp Med. 1996;184:1101–1110. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiuti A, Webb I J, Bleul C, Springer T A, Gutierrez-Ramos J C. J Exp Med. 1997;185:111–120. doi: 10.1084/jem.185.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagasawa T, Kikutani H, Kishimoto T. Proc Natl Acad Sci USA. 1994;91:2305–2309. doi: 10.1073/pnas.91.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S-I, Kitamura Y, Yoshida N, Kikutani H, Kishimoto T. Nature (London) 1996;382:635–638. doi: 10.1038/382635a0. [DOI] [PubMed] [Google Scholar]
- 7.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. Nature (London) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 8.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J-L, Arenzana-Seisdedos A, Schwartz O, Heard J-M, Clark-Lewis I, Legler D F, et al. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 10.Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, Shinohara T, Honjo T. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 11.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 13.Federsppiel B, Melhado I G, Duncan A M, Delaney A, Schappert K, Clark-Lewis I, Jirik F R. Genomics. 1993;16:707–712. doi: 10.1006/geno.1993.1251. [DOI] [PubMed] [Google Scholar]
- 14.Hori T, Sakaida H, Sato A, Nakajima T, Shida H, Yoshie O, Uchiyama T. J Immunol. 1998;160:180–188. [PubMed] [Google Scholar]
- 15.Forster R, Kremmer E, Schubel A, Breitfeld D, Kleinschmidt A, Nerl C, Bernhardt G, Lipp M. J Immunol. 1998;160:1522–1531. [PubMed] [Google Scholar]
- 16.Tanabe S, Heesen M, Yoshizawa I, Berman M A, Luo Y, Bleul C C, Springer T A, Okuda K, Gerard N, Dorf M E. J Immunol. 1997;159:905–911. [PubMed] [Google Scholar]
- 17.Lavi E, Strizki J M, Ulrich A M, Zhang W, Fu L, Wang Q, O’Connor M, Hoxie J A, Gonzalez-Scarano F. Am J Pathol. 1997;151:1035–1042. [PMC free article] [PubMed] [Google Scholar]
- 18.Jazin E E, Soderstrom S, Ebendal T, Larhammar D. J Neuroimmunol. 1997;79:148–154. doi: 10.1016/s0165-5728(97)00117-3. [DOI] [PubMed] [Google Scholar]
- 19.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 20.Whitlock C A, Robertson D, Witte O N. J Immunol Methods. 1984;67:353–369. doi: 10.1016/0022-1759(84)90475-7. [DOI] [PubMed] [Google Scholar]
- 21.Ceredig R. J Immunol. 1988;141:355–362. [PubMed] [Google Scholar]
- 22.Ikuta K, Uchida N, Friedman J, Weissman I L. Annu Rev Immunol. 1992;10:759–783. doi: 10.1146/annurev.iy.10.040192.003551. [DOI] [PubMed] [Google Scholar]
- 23.Lee G, Namen A E, Gillis S, Ellingsworth L R, Kincade P W. J Immunol. 1989;142:3875–3883. [PubMed] [Google Scholar]
- 24.Kitchen S G, Zack J A. J Virol. 1997;71:6928–6934. doi: 10.1128/jvi.71.9.6928-6934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kee B L, Paige C J. Int Rev Cytol. 1995;157:129. doi: 10.1016/s0074-7696(08)62158-0. [DOI] [PubMed] [Google Scholar]
- 26.Altman J, Bayer S A. Development of the Cerebellar System in Relation to its Evolution, Structure, and Functions. Boca Raton, FL: CRC; 1997. [Google Scholar]
- 27.D’Apuzzo M, Rolink A, Loetscher M, Hoxie J A, Clark-Lewis I, Melchers F, Baggiolini M, Moser B. Eur J Immunol. 1997;27:1788–1793. doi: 10.1002/eji.1830270729. [DOI] [PubMed] [Google Scholar]
- 28.Davis C B, Dikic I, Unutmaz D, Hill C M, Arthos J, Siani M A, Thompson D A, Schlessinger J, Littman D R. J Exp Med. 1997;186:1793–1798. doi: 10.1084/jem.186.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorshkind K. Cell. 1994;79:751–753. doi: 10.1016/0092-8674(94)90065-5. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Arie N, Bellen H J, Armstrong D L, McCall A E, Gordadze P R, Guo Q, Matzuk M M, Zoghbi H Y. Nature (London) 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 31.Hatten M E. Curr Opin Neurobiol. 1993;3:38–44. doi: 10.1016/0959-4388(93)90033-u. [DOI] [PubMed] [Google Scholar]
- 32.Hatten M E, Heintz N. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 33.Ackerman S L, Kozak L P, Przyborski S A, Rund L A, Boyer B B, Knowles B B. Nature (London) 1997;386:838–842. doi: 10.1038/386838a0. [DOI] [PubMed] [Google Scholar]
- 34.Horuk R, Martin A W, Wang Z, Schweitzer L, Gerassimides A, Guo H, Lu Z, Hessselgesser J, Perez H D, Kim J, et al. J Immunol. 1997;158:2882–2890. [PubMed] [Google Scholar]
- 35.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature (London) 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 36.Vallat A-V, De Girolami U, He J, Mhashikar A, Marasco W, Shi B, Gray F, Bell J, Keohane C, Smith T W, et al. Am J Pathol. 1998;152:167–178. [PMC free article] [PubMed] [Google Scholar]
- 37.Hesselgesser J, Halks-Miller M, DelVecchio V, Peiper S C, Hoxie J, Kolson D L, Taub D, Horuk R. Curr Biol. 1997;7:112–121. doi: 10.1016/s0960-9822(06)00055-8. [DOI] [PubMed] [Google Scholar]
- 38.Hesselgesser J, Taub D, Baskar P, Greenberg M, Hoxie J, Kolson D L, Horuk R. Curr Biol. 1998;8:595–598. doi: 10.1016/s0960-9822(98)70230-1. [DOI] [PubMed] [Google Scholar]
- 39.Price R W, Brew B, Sidtis J, Rosenblum M, Scheck A C, Cleary P. Science. 1988;239:586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- 40.Tachibana K, Hirota S, Lizasa H, Yoshida H, Kawabata K, Kataoka Y, Kitamura Y, Matsushima K, Yoshida N, Nishikawa S-I, Kishimoto T, Nagasawa T. Nature (London) 1998;393:591–594. doi: 10.1038/31261. [DOI] [PubMed] [Google Scholar]
- 41.Zou Y-R, Kittmann A H, Kuroda M, Taniuchi I, Littman D R. Nature (London) 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]