Abstract
The majority of T cells develop in the thymus and exhibit well characterized phenotypic changes associated with their maturation. Previous analysis of intestinal intraepithelial lymphocytes (IEL) from nude mice and a variety of experimentally manipulated models led to the view that at least a portion of these cells represent a distinct T cell population that matures extrathymically. The IEL that are postulated to mature within the intestine include both T cell receptor (TCR) αβ- and γδ-bearing subpopulations. They can be distinguished from conventional thymically derived T cells in that they express an unusual coreceptor, a CD8α homodimer. In addition, they can utilize the Fc receptor γ-chain in place of the CD3-associated ζ-chain for TCR signaling and their maturation depends on the interleukin 2 receptor β-chain. Moreover, TCRαβ+CD8αα+ IEL are not subject to conventional thymic selection processes. To determine whether CD3−CD8αα+ IEL represent precursors of T cells developing extrathymically, we examined IEL from knockout mice lacking the recombination activating gene-1 (rag-1), CD3ɛ, or both Lck and Fyn, in which thymic T cell development is arrested. CD3−CD8αα+CD16+ IEL from all three mutant strains, as well as from nude mice, included cells that express pre-TCRα transcripts, a marker of T cell commitment. These IEL from lck−/−fyn−/− animals exhibited TCR β-gene rearrangement. However, CD3−CD8αα+CD16+ IEL from ɛ-deficient mice had not undergone Dβ-Jβ joining, despite normal rearrangement at the TCRβ locus in thymocytes from these animals. These results revealed another distinction between thymocytes and IEL, and suggested an unexpectedly early role for CD3ɛ in IEL maturation.
Most T cells mature in the thymus and use several polypeptide chains to transmit signals from their T cell receptors (TCRs) to the cell interior, including the CD3 γ, δ, and ɛ chains and the associated ζ homodimer. Intestinal intraepithelial lymphocytes (IEL) of the small intestine contain a complex mixture of T cells that includes both TCRαβ- and TCRγδ-bearing populations. A subset of these cells is unique in that the cells within that subset express an unusual form of the CD8 coreceptor, a CD8α homodimer (1, 2). This property distinguishes these IEL from conventional CD8αβ-expressing T cells that mature within the thymus. In addition, CD3+CD8αα+ IEL, which can utilize the Fc receptor γ-chain in place of the CD3-associated ζ-chain for TCR signaling, develop in mice deficient for ζ, whereas no mature CD4+ or CD8αβ+ T cells expressing high TCR levels emerge from the thymuses of these animals (3–6). Moreover, TCRαβ+CD8αα+ IEL are not subject to conventional thymic positive and negative selection processes (1, 7–12) as are their thymically derived counterparts. These differences, as well as their dependence on the interleukin 2 receptor β-chain for development (13), have led to the view that at least a portion of IEL represents a distinct T cell population that matures extrathymically within the gut epithelium (14, 15). The detection of recombination activating gene (RAG-1) and pre-TCRα (pre-Tα) transcripts in IEL from severe combined immunodeficient (SCID) and nude mice, respectively (16, 17), supports this scenario and suggests that immature IEL rearrange their TCR genes and express pre-TCRs in situ.
Phenotypic changes associated with thymopoiesis have been documented extensively. It is proposed that a signal emanating from the pre-TCR/CD3 complex results in maturation of CD4−CD8− thymocytes to the CD4+CD8+ stage and the subsequent proliferation of these cells (18–21). Genetically manipulated mice deficient for pre-TCRα or TCRβ only inefficiently generate CD4+CD8+ thymocytes and exhibit a 10- to 100-fold reduction in thymocyte number (18, 20). During thymocyte development, the pre-TCR signal is thought to be mediated by the nonreceptor protein tyrosine kinase p56lck (Lck) or, in its absence, by p59fyn (Fyn). In mice lacking both these src-family protein tyrosine kinases, maturation of TCRαβ+ thymocytes is blocked at the CD44loCD25+ CD4−CD8− stage (22, 23). Finally, positive selection of CD4+CD8+ thymocytes into mature CD4+ or CD8+ cells generally requires a TCRαβ signal resulting from recognition of a complex of self-peptide and major histocompatibility complex product expressed in the thymic cortex (24).
Although signals that regulate thymopoiesis thus are well characterized, little is known regarding the development of TCRαβ+ intestinal IEL maturing in situ. CD3−CD8αα+ IEL are abundant in RAG-deficient and SCID mice (2, 15) and are enriched in young, wild-type mice (25, 26), suggesting that these cells are precursors of CD3+ IEL. Kinetic studies suggest that these IEL might arise from CD3−CD8α−CD44+ IEL precursors and yield TCRγδ+CD8αα+ IEL (25). These immature IEL may derive from lymphoid progenitors, expressing CD117 (c-kit) and interleukin 7R, resident in cryptopatches within the lamina propria (27). Such cells recently have been shown to repopulate the IEL and mesenteric lymph nodes in irradiated SCID mice (28).
Our recent study of IEL in mice deficient for both Lck and Fyn demonstrated that inefficient maturation of TCRγδ+CD8αα+ IEL could proceed, whereas appearance of mature TCRαβ+CD8αα+ IEL was completely abrogated (29). IEL from these mice were slightly enriched for CD3−CD8α−CD44+ and CD3−CD8αα+CD16+B220+ cells, which may be precursors of CD3+ IEL. To determine whether CD3−CD8αα+ IEL represent committed T cells, we evaluated the expression of pre-Tα transcripts and TCR β-gene rearrangement in these IEL sorted from nude mice and mutant mice in which T cell development is arrested. The data presented indicate that the CD3−CD8αα+CD16+ IEL population includes T cell progenitors. Moreover, analyses of IEL from ζ-deficient mice revealed a unique maturation intermediate for TCRαβ+CD8αα+ IEL that expresses CD8α homodimers and cytoplasmic TCR β-chains, but lacks detectable surface TCR expression. Overall, these data are consistent with the view that extrathymic T cell maturation in the small intestine proceeds with TCR gene rearrangement within the gut environment.
MATERIALS AND METHODS
Mice.
Mice deficient for CD3ɛ or ζ (3, 21) or both Lck and Fyn (22) have been reported previously. RAG-1-deficient (30) and C57BL/6J mice were purchased from The Jackson Laboratory. All mice were maintained under specific pathogen-free conditions.
Cell Preparation and Staining.
IEL were isolated and purified as described (29, 31) and were surface-stained as reported (29). For cytoplasmic staining, surface-stained cells were washed and fixed by incubation in 100 μl of 4% paraformaldehyde in PBS for 20 min in the dark. After three washes, the cells were permeablized in FACS buffer (balanced salt solution/2% fetal calf serum/0.8% sodium azide) plus 0.1% saponin and blocked for 5 min on ice with anti-FcγRII/III in saponin. Intracellular staining for TCRβ was performed by incubation on ice with anti-TCRβ-fluorescein isothiocyanate (FITC), followed by three washes in 0.1% saponin in FACS buffer.
Flow Cytometric Analysis and Sorting.
Multiparameter cytometric analysis was performed on a FACScan flow cytometer with lysis ii software (Becton Dickinson), and flow cytometric analyses were plotted by using reproman software (Truefacts Software, Seattle, WA).
Cells for analysis of pre-Tα expression and TCRβ gene rearrangement were stained by using anti-CD3-FITC, anti-CD16-PE, and anti-CD8α-TC. They were sorted on a FACStar Plus (Becton Dickinson) into two populations, by first gating on the CD3-negative lymphocytes and then collecting individually the CD3−CD8α+CD16+ cells and the remaining CD3− cells.
DNA–PCR Assay for Detection of TCR Gene Rearrangement.
High-molecular-weight DNA was prepared from frozen cell pellets by using a QIAmp Blood Kit (Qiagen, Chatsworth, CA). For unsorted cells, quantitation of DNA was performed by spectrographic analysis. For sorted cells, starting cell number was used to equilibrate the input template between samples. Parallel analyses using Dβ2 and Jβ2 PCR primers, which amplify even the germ-line, unrearranged DNA, were used to approximate the relative amount of template DNA in each sample. Analysis of TCR β-chain gene rearrangement was performed by using PCR primers, oligonucleotide probes, and the conditions described previously (32, 33). Detection of TCR γ- and δ-gene rearrangement using Vγ5 and Jγ1, Vδ4 and Jδ1, and Vδ5 and Jδ1 primer pairs was as reported previously (34).
Reverse Transcription–PCR (RT-PCR) for Detection of Pre-Tα Transcripts.
Total cellular RNA was isolated from sorted and unsorted cell populations by using RNAzol B (Tel-Test, Friendswood, TX). cDNA was generated by using random hexamers and reverse transcriptase, AMV-RT (GIBCO/BRL), at 37°C for 1 hr. Primers used for PCR amplification were: 5′ primer, 5′-TAGCGGATCCCTGCAACTGGGATCATGCTTC-3′, which recognizes the extracellular end of pTα, and 3′ primer, 5′-CAGAGGATCCTCAGAGGGGAGGGTAAGATC-3′, which recognizes the cytoplasmic portion of pTα (35). Thirty-five cycles of PCR amplification were performed (2 min at 94°C, 2 min at 58°C, 3 min at 72°C) in 50-μl reactions. PCR products were hybridized with a 32P-end-labeled oligonucleotide probe, 5′-CAGAGGATCCCTACTTGCAGGTCAGGAGCACATCGA-3′ (17), which spans the transmembrane region of pTα.
Amplification of cDNA for hypoxanthine-guanine phosphoribosyltransferase was performed in parallel reactions as described previously (36).
RESULTS
Small Numbers of CD3−CD8αα+ IEL Expressing Cytoplasmic TCR β-Chains Are Present in ζ−/− Mice.
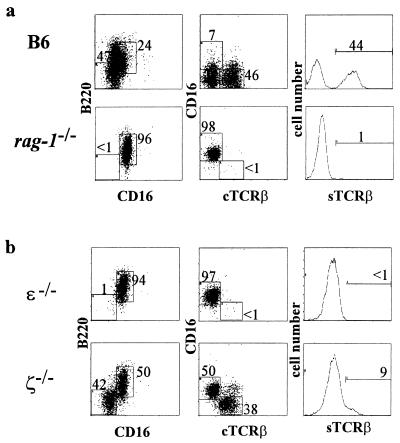
Development of thymocytes of the TCRαβ lineage is blocked at the CD4−CD8− stage in mice unable to express the components of the pre-TCR or transmit its downstream signals because of gene ablation (18, 20–23). Our previous analyses of the cell surface phenotype of IEL from mice lacking RAG-1, or both the src-family protein tyrosine kinases Lck and Fyn, suggested that the maturation of TCRαβ IEL is arrested in these animals (29). IEL lacking surface TCRαβ but expressing CD8α, B220, and CD16 were readily detected in rag-1−/− mice (Fig. 1a and ref. 29) and in lck−/−fyn−/− animals (29). IEL with a similar surface phenotype also were seen in mice lacking the CD3 component ɛ (Fig. 1b). It has been suggested that such CD3−CD8αα+ IEL represent precursors of CD3+CD8αα+ IEL (14, 15, 25).
Figure 1.
Some CD3−CD8αα+ IEL from ζ−/− mice express cytoplasmic TCR β-chains. IEL were isolated from the indicated mice and stained for three-color flow cytometry by using anti-CD8α-TC, anti-B220-PE, or anti-TCRβ-PE, and anti-CD16-FITC. (a) IEL from 4- to 8-week-old mice were analyzed. Data are shown for lymphocytes pregated on CD8α expression and are presented on a logarithmic scale. cTCRβ and sTCRβ represent cytoplasmic and surface TCRβ expression, respectively. The percentage of CD8α+ cells falling within each rectilinear gate is shown. (b) The data shown are for 6-week-old ɛ−/− and ζ−/− mice. Similar results were seen for 4- to 16-week-old ζ-deficient mice analyzed in at least four independent experiments.
Previous studies of ζ−/− mutant mice demonstrated that CD3-associated ζ-chains are needed for normal levels of TCR surface expression and for normal development or survival of CD4+CD8+ thymocytes (3–6). Although no CD4+ or CD8+ thymocytes are detectable in these animals, a few CD3−CD4+ or CD8+ T cells are present in their peripheral lymph nodes. In contrast, TCRαβ+CD8αα+ IEL development is diminished but not eliminated in mice deficient for the CD3-associated ζ-chains, with a few mature TCRαβ+ cells present (3–5). We reasoned that ζ−/− mice, which substitute the perhaps less efficient TCR signaling via FcR γ-chains, would provide a source of IEL precursors uncontaminated with conventional T cells and might allow us to identify additional intermediates of IEL maturation.
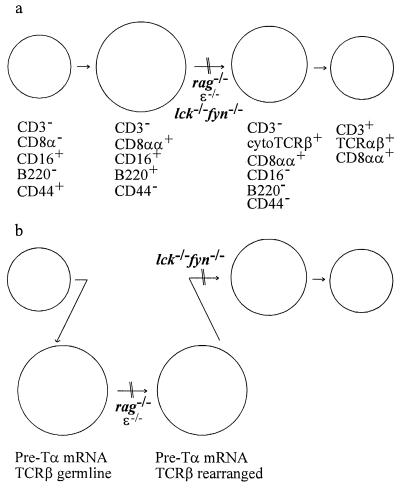
As previously reported, comparison of IEL from ζ−/− mice with those from their heterozygous littermates revealed a lack of TCRαβ+CD8αβ+ and TCRαβ+CD4+ cells, as well as a paucity of TCRαβ+CD8αα+ and TCRγδ+CD8αα+ IEL (data not shown and refs. 3–6). The small number of TCRαβ+CD8αα+ IEL in ζ−/− mice expressed only low levels of surface TCRαβ (Fig. 1b and ref. 3). CD8α+ IEL in the ζ−/− animals fell largely into two populations based on CD16 and B220 expression: one expressing both, and the other lacking expression of these markers (Fig. 1b). Furthermore, the majority of the CD8αα+CD16−B220− IEL express cytoplasmic TCR β-chains but lack detectable surface TCRβ expression (Fig. 1b), indicating that these cells may represent intermediates along the TCRαβ+CD8αα+ IEL maturation pathway. Such cells were not detected in mice lacking ɛ (Fig. 1b), consistent with the existence of an early developmental block in these animals. One possible scheme for the developmental pathway of TCRαβ+CD8+ IEL is shown in Fig. 2a.
Figure 2.
Models of lineage relationships for developing TCRαβ+CD8αα+ IEL. (a) This model is based on published studies (2, 15, 25, 29) and the data for surface and cytoplasmic staining shown in Fig. 1. Arrows indicate postulated developmental transitions. Cell-size relationships are depicted relative to wild-type IEL. (b) This model, where the surface phenotypes are as depicted in a, also incorporates the data for pre-Tα expression and TCRβ gene rearrangement presented in Figs. 4 and 5.
CD3−CD8αα+CD16+ IEL Express Pre-Tα Transcripts.
There has been much speculation that CD3−CD8αα+ IEL represent precursors of mature IEL (14, 15); however, because these cells lack surface TCR but express CD8αα and CD16, the FcR α-chain, it is formally possible that these cells are natural killer or dendritic cells, or their progenitors. To determine whether the CD3−CD8αα+ IEL population includes T cell precursors, we investigated whether these cells express transcripts for pre-Tα. This surrogate TCR α-chain is expressed by immature CD4−CD8− thymocytes, but not by TCRγδ thymocytes, splenic NK cells, or other nonlymphoid tissues (17). Recently, Bruno and coworkers demonstrated expression of pre-Tα transcripts among bulk IEL prepared from nude mice (17), but these studies did not focus on the CD3−CD8αα+ possible precursor population.
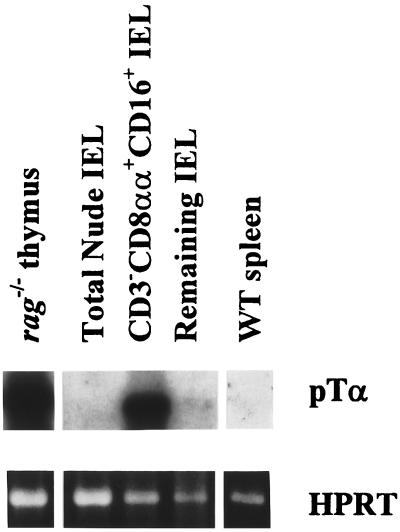
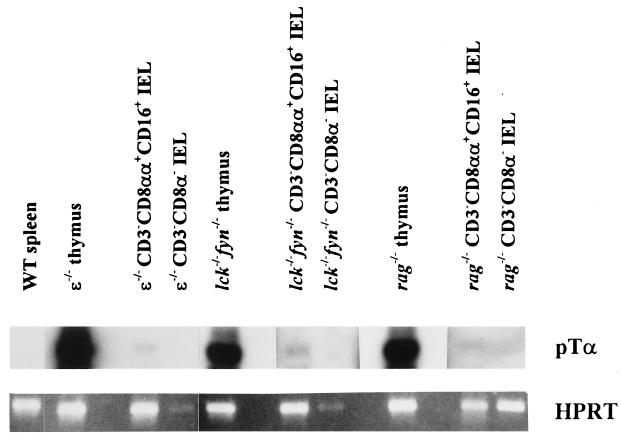
To detect pre-Tα mRNA in sorted IEL subpopulations from nude mice, we used a sensitive RT-PCR technique. Pre-Tα transcripts were detected in CD3−CD8αα+CD16+ IEL (Fig. 3), indicating that this subpopulation includes T cell precursors. Consistent with earlier reports (17), pre-Tα transcripts were abundant in the immature thymocytes from the RAG-1-deficient mice but were not detected in the mature T cells among the C57BL/6 splenocytes (Fig. 3). Subsequent analyses of sorted IEL from several mutant mouse strains that exhibit a block in T cell maturation demonstrated pre-Tα transcripts in CD3−CD8αα+CD16+ IEL from ɛ−/−, lck−/−fyn−/−, and rag-1−/− mice (Fig. 4). This indicated that the CD3−CD8αα+CD16+ IEL include committed T cells, although the pre-Tα message levels were markedly lower than in thymocytes from these mice.
Figure 3.
Pre-Tα transcripts are expressed in CD3−CD8αα+CD16+ IEL from nude mice. RNA was extracted from sorted CD3−CD8αα+CD16+ and CD3−CD8α− IEL isolated from nude mice, and from thymocytes from rag-1−/− mice and splenocytes from wild-type C57BL/6 mice. RT-PCR was performed by using pre-Tα-specific or hypoxanthine phosphoribosyltransferase (HPRT)-specific primers. Pre-Tα PCR products were detected by Southern blot analysis with an oligonucleotide probe specific for the pre-Tα transmembrane region. HPRT reaction products were visualized by ethidium bromide staining.
Figure 4.
Pre-Tα transcripts are expressed in CD3−CD8αα+CD16+ IEL from mice exhibiting a block in T cell development. RNA was extracted from sorted CD3−CD8αα+CD16+ and CD3−CD8α− IEL isolated from pools of six 5- to 10-week-old mice of the indicated genotypes. Thymocytes from the same animals and splenocytes from wild-type C57BL/6 mice served as positive and negative controls, respectively. RT-PCR and detection of the amplification products were performed as for Fig. 3.
TCR Gene Rearrangement in CD3−CD8αα+CD16+ IEL.
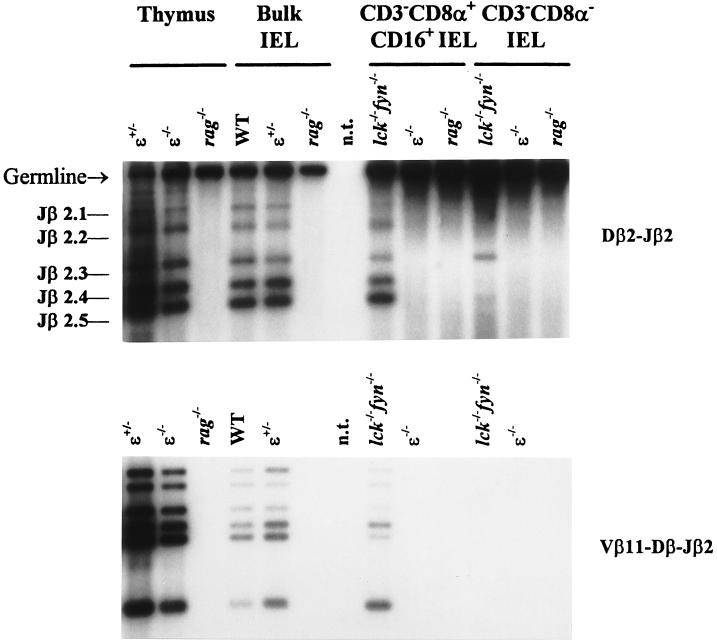
TCR gene rearrangements are detected among thymocytes as early as fetal day 11 and are a prerequisite for the expression of TCR chains (37). Variable gene assembly is a lymphocyte-specific process, in part because expression of products encoded by the recombination activation genes (rag-1 and rag-2) is restricted to lymphoid precursor cells (38). To test further the hypothesis that CD3−CD8αα+CD16+ IEL include lymphoid precursors committed to the T cell lineage, we used a DNA–PCR strategy to examine the status of TCR gene rearrangements among these cells. Vγ-Jγ and Vδ-DδJδ rearrangements were detected in sorted CD3−CD8αα+CD16+ IEL from ɛ-deficient mice (data not shown). It should be noted that a few mature TCRγδ+ IEL were present in lck−/−fyn−/− mice (29).
Although both Dβ-Jβ and Vβ-DβJβ joining at the TCRβ locus occur predominantly in T cell precursors (39), detection of Dβ-Jβ but not Vβ-DβJβ rearrangements among B cells suggests that Vβ segment recombination to a DβJβ complex is a regulated, T lineage-specific step in TCRβ gene rearrangement (40, 41). Thymocytes from both lck−/−fyn−/− and ɛ−/− mice are blocked at the CD4−CD8− stage, and undergo both Dβ-Jβ and Vβ-DβJβ gene rearrangements (Fig. 5; data not shown and refs. 21 and 42). However, sorted CD3−CD8αα+CD16+ IEL from lck−/−fyn−/− and ɛ−/− mice exhibited distinct gene rearrangement patterns at the TCRβ locus. Both Dβ-Jβ and Vβ-DβJβ gene rearrangements were readily detected among CD3−CD8αα+CD16+ IEL from lck−/−fyn−/− mice (Fig. 5). In contrast, as with sorted IEL from RAG-deficient mice that are unable to rearrange their TCR genes, Dβ-Jβ gene rearrangement was not detected among purified CD3−CD8αα+CD16+ IEL from ɛ−/− mice (Fig. 5). These data suggest that TCRβ gene rearrangement is controlled differentially in the thymus and intestine. Moreover, they imply that CD3ɛ may have an unexpectedly early role in IEL maturation.
Figure 5.
TCRβ gene rearrangement is detected in CD3− IEL from lck−/−fyn−/− but not ɛ−/− mice. DNA was purified from the indicated sorted cell type and amplified in parallel reactions by using a 5′ Dβ2- or Vβ11-specific primer paired with a 3′ primer immediately downstream of Jβ2.6. PCR products were detected by Southern blot analysis using a Jβ2-specific oligonucleotide probe.
DISCUSSION
The data reported here confirm that CD3−CD8αα+CD16+ IEL include cells that exhibit characteristics of T cells that distinguish them from natural killer and dendritic cells. We demonstrated that pre-Tα transcripts are expressed in CD3−CD8αα+CD16+ IEL (Figs. 3 and 4), extending the earlier finding that pre-Tα-expressing cells are present in bulk IEL from nude mice (17). Both Dβ-Jβ and Vβ11-DβJβ gene rearrangements were readily detected among CD3−CD8αα+CD16+ IEL from lck−/−fyn−/− mice (Fig. 5), confirming that this IEL population includes T cell precursors. However, we have not excluded the possibility that this entire population, as defined by its surface phenotype, may also include natural killer cells, dendritic cells, or their precursors. Indeed, one interpretation of our data, indicating a low level of pre-Tα transcripts in CD3−CD8αα+CD16+ IEL relative to that seen for thymocytes from the same mice (Fig. 4), would be that only a small subset of these IEL is expressing pre-Tα.
Surprisingly, although TCRβ gene rearrangement was readily observed among sorted CD3−CD8αα+CD16+ IEL from lck−/−fyn−/− mice, it was not detected in similar cells from ɛ−/− animals (Fig. 5). In contrast, the block in thymocyte development in these mice appears to be equivalent, because CD4−CD8− thymocytes from lck−/−fyn−/− and ɛ−/− mice are CD44loCD25+ and have undergone TCRβ gene rearrangement (Fig. 5; refs. 21–23 and 42 and data not shown). We cannot rule out the possibility that a small proportion of the cells in these purified cell populations from ɛ−/− animals has undergone TCRβ gene rearrangement. However, the strong germ-line band in the ɛ−/− lanes (Fig. 5), which demonstrates ample DNA template for PCR amplification in this experiment, and the fact that we could easily detect Dβ-Jβ DNA rearrangements among bulk ɛ−/− IEL that had been spiked with 2% ɛ−/− thymocytes (data not shown), suggest that this is an unlikely explanation for these results. Thus, at the level of TCRβ gene rearrangement, CD3−CD8αα+CD16+ IEL from lck−/−fyn−/− and ɛ−/− mice are not equivalent populations.
It is possible that CD3−CD8αα+CD16+ IEL may include at least two distinct intermediates along the T lineage pathway, the more mature of which has undergone TCRβ gene rearrangement. The developmental block manifest in IEL from ɛ-deficient mice may be earlier than that in lck−/−fyn−/− mice, before the onset of Dβ-Jβ rearrangement, but after expression of pre-Tα (Fig. 2b) and TCRγ and δ gene rearrangement (ref. 29 and data not shown). It was surprising that expression of a component of the TCR-signaling complex, CD3ɛ, should influence DNA rearrangement at the TCRβ locus. However, this is not without precedent, because analyses of knockout mice deficient for Igβ, one of the signaling molecules associated with Ig heavy chains, have indicated that B cell development is blocked at the immature CD43+B220+ stage, after DH-JH recombination but before VH-DHJH joining (43).
Analyses of thymocytes from mice deficient for both CD3ɛ and ζ carrying a constitutively active Lck transgene (ɛ−/−ζ−/−LckF505) indicate that expression of the transgene results in the maturation of CD4−CD8− thymocytes to the CD4+CD8+ stage (K. Forbush, M.M., and R.M.P., unpublished data). However, our studies of IEL subsets in these ɛ−/−ζ−/−LckF505 mice indicate no evidence for the LckF505 transgene driving IEL maturation (data not shown), whereas in similar mice expressing CD3ɛ (ɛ+/−ζ−/−LckF505), cells mature to the CD3−CD8αα+ stage expressing cytoplasmic TCRβ chains (S.T.P., L.Y.B., J.A.H., M.M., R.M.P., and A.M.P., unpublished data). Taken together these studies further support our contention that CD3ɛ expression influences early IEL maturation.
The nature of the signal that drives the appearance of CD3−CD8αα+CD16+ IEL that have undergone Vβ-DβJβ gene rearrangement in lck−/−fyn−/− mice, but that is absent in their ɛ−/− counterparts, is not immediately apparent. These more mature IEL that have completed TCRβ gene rearrangement are unlikely to arise from CD4−CD8− thymocytes that have undergone TCRβ gene rearrangement, because thymopoiesis appears to advance to an identical point in both strains of mice and such an explanation would predict, therefore, that such cells should also be found in the intestine of CD3ɛ-deficient mice. Because TCRγδ+ IEL are present in lck−/−fyn−/− but not ɛ−/− animals (ref. 29 and data not shown), it is conceivable that these IEL may facilitate this maturation step, perhaps via an unidentified mechanism similar to that exhibited when TCRγδ+ T cells drive the CD4−CD8− to CD4+CD8+ transition of host thymocytes upon injection into SCID hosts (44).
The above data demonstrated the T cell commitment of at least a subset of the CD3−CD8αα+CD16+ IEL, and our examination of IEL from ζ−/− mice revealed more mature IEL committed to the TCRαβ lineage. Evidence suggests that ζ-deficient IEL substitute the FcR γ-chain for TCR signaling (3–6), and perhaps because γ includes only one immune receptor tyrosine-based activation motif (ITAM) whereas ζ has three, these ζ−/− IEL mature only inefficiently (45). We report here that ζ−/− animals include significant numbers of CD3−CD8αα+cytoTCRβ+ IEL lacking CD16 and B220 expression (Fig. 1b), which may represent developmental intermediates of the TCRαβ lineage maturing within the intestinal epithelium. Terhorst and coworkers (46) have reported recently the maturation of IEL accompanied by the loss of CD16 and B220 in response to anti-CD3ɛ treatment or transfer of CD16+ IEL into rag-2−/− mice. IEL from wild-type mice include negligible numbers of these CD3−CD8αα+cytoTCRβ+ IEL lacking CD16 and B220 expression, which might rapidly rearrange their TCRα genes and mature to express surface TCRs. Cells with a similar phenotype are found neither in the thymus nor peripheral lymph nodes (data not shown). Moreover, these IEL do not appear to be derived from CD4+CD8+ thymocytes that have extinguished CD4 and CD8β expression because they are not present in ZAP70−/− mice (S.T.P., Q. Kong, A. C. Chan, and A.M.P., unpublished observations), which include abundant populations of CD4+CD8+ thymocytes. This view is supported further by analysis of the methylation state of the CD8β gene among CD8αα+ IEL, which suggests these cells do not arise from CD8β-expressing precursors (47).
Overall, these data support the existence of discrete developmental steps, identifiable by TCR gene rearrangement status and cell surface phenotype, in the extrathymic maturation sequence that underlies the emergence of TCRαβ+CD8αα+ IEL from CD3−CD8αα+ cells within the small intestine. Although a body of evidence indicates distinct T cell differentiation pathways operate in the intestine and thymus, our data highlight an additional difference between IEL maturation and thymocyte development and suggest an unexpectedly early role for CD3ɛ in IEL maturation.
Acknowledgments
We thank Deborah Wilson, Katherine Forbush, Ethan Ojala, and Xiao Cun Pan for maintaining our animal colony at the University of Washington and Edward Chung for technical assistance. We are grateful to Drs. Arthur Weiss and Nicolai van Oers (Howard Hughes Medical Institute, University of California at San Francisco) for their helpful discussions and for providing access to lck−/−fyn−/− mice, to Dr. Brian Iritani for PCR primers and the probe to detect pre-Tα transcripts, to Kathi Prewitt and Christina Nef for assistance in preparing the manuscript, and to Dr. Andrew Herman for his critical review. S.T.P. was supported by the Poncin Scholarship Fund and Grants GM-07226 and CA-09537 from the National Institutes of Health. M.M. is supported by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, and Association pour la Recherche sur le Cancer. R.M.P. and A.M.P. were Investigators of the Howard Hughes Medical Institute. This work was supported in part by Grant 051518 from The Wellcome Trust.
ABBREVIATIONS
- IEL
intraepithelial lymphocytes
- TCR
T cell receptor
- pTα
pre-TCR α-chain
- SCID
severe combined immunodeficient
- RT-PCR
reverse transcription–PCR
References
- 1.Lefrancois L. J Immunol. 1991;147:1746–1751. [PubMed] [Google Scholar]
- 2.Guy-Grand D, Cerf-Bensussan N, Malissen B, Malassis-Seris M, Broittet C, Vassalli P. J Exp Med. 1991;173:471–481. doi: 10.1084/jem.173.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malissen M, Gillet A, Rocha B, Trucy J, Vivier E, Boyer C, Kontgen F, Brun N, Mazza G, Spanopoulou E, Guy-Grand D, Malissen B. EMBO J. 1993;12:4347–4355. doi: 10.1002/j.1460-2075.1993.tb06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu C, Ueda R, She J, Sancho J, Wang B, Weddell G, Loring J, Kurahara C, Dudley E, Hayday A, Terhorst C, Huang M. EMBO J. 1993;12:4863–4875. doi: 10.1002/j.1460-2075.1993.tb06176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno H, Aoe T, Taki S, Kitamura D, Ishida Y, Rajewsky K, Saito T. EMBO J. 1993;12:4357–4366. doi: 10.1002/j.1460-2075.1993.tb06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Love P E, Shores E W, Johnson M D, Tremblay M L, Lee E J, Grinberg A, Huang S P, Singer A, Westphal H. Science. 1993;261:918–921. doi: 10.1126/science.7688481. [DOI] [PubMed] [Google Scholar]
- 7.Rocha B, Vassalli P, Guy-Grand D. J Exp Med. 1991;173:483–486. doi: 10.1084/jem.173.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poussier P, Edouard P, Lee C, Binnie M, Julius M. J Exp Med. 1992;176:187–199. doi: 10.1084/jem.176.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murosaki S, Yoshikai Y, Ishida A, Nakamura T, Matsuzaki G, Takimoto H, Yuuki H, Nomoto K. Intern Immunol. 1991;3:1005–1013. doi: 10.1093/intimm/3.10.1005. [DOI] [PubMed] [Google Scholar]
- 10.Poussier P, Teh H S, Julius M. J Exp Med. 1993;178:1947–1957. doi: 10.1084/jem.178.6.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rocha B, von Boehmer H, Guy-Grand D. Proc Natl Acad Sci USA. 1992;89:5336–5340. doi: 10.1073/pnas.89.12.5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croitoru K, Bienenstock J, Ernst P. Intern Immunol. 1994;6:1467–1473. doi: 10.1093/intimm/6.10.1467. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki H, Duncan G S, Takimoto H, Mak T W. J Exp Med. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poussier P, Julius M. Annu Rev Immunol. 1994;12:521–553. doi: 10.1146/annurev.iy.12.040194.002513. [DOI] [PubMed] [Google Scholar]
- 15.Rocha B, Guy-Grand D, Vassalli P. Curr Opin Immunol. 1995;7:235–242. doi: 10.1016/0952-7915(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 16.Guy-Grand D, Vanden Broecke C, Briottet C, Malassis-Seris M, Selz F, Vassalli P. Eur J Immunol. 1992;22:505–510. doi: 10.1002/eji.1830220232. [DOI] [PubMed] [Google Scholar]
- 17.Bruno L, Rocha B, Rolink A, von Boehmer H, Rodewald H. Eur J Immunol. 1995;25:1877–1882. doi: 10.1002/eji.1830250713. [DOI] [PubMed] [Google Scholar]
- 18.Mombaerts P, Clarke A, Rudnicki M, Iacomini J, Itohara S, Lafaille J, Wang L, Ichikawa Y, Jaenisch R, Hooper M, Tonegawa S. Nature (London) 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 19.Levelt C, Mombaerts P, Iglesias A, Tonegawa S, Eichmann K. Proc Natl Acad Sci USA. 1993;90:11401–11405. doi: 10.1073/pnas.90.23.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fehling H J, Krotkova A, Saint-Ruf C, von Boehmer H. Nature (London) 1995;375:795–798. doi: 10.1038/375795a0. [DOI] [PubMed] [Google Scholar]
- 21.Malissen M, Gillet A, Ardouin L, Bouvier G, Trucy J, Ferrier P, Vivier E, Malissen B. EMBO J. 1995;14:4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Oers N, Lowin-Kropf B, Finlay D, Connolly K, Weiss A. Immunity. 1996;5:429–436. doi: 10.1016/s1074-7613(00)80499-9. [DOI] [PubMed] [Google Scholar]
- 23.Groves T, Smiley P, Cooke M P, Forbush K A, Perlmutter R M, Guidos C J. Immunity. 1996;5:417–428. doi: 10.1016/s1074-7613(00)80498-7. [DOI] [PubMed] [Google Scholar]
- 24.Robey E, Fowlkes B J. Annu Rev Immunol. 1994;12:675–705. doi: 10.1146/annurev.iy.12.040194.003331. [DOI] [PubMed] [Google Scholar]
- 25.Lin T, Matsuzaki G, Yoshida H, Kobayashi N, Kenai H, Omoto K, Nomoto K. Eur J Immunol. 1994;24:1080–1087. doi: 10.1002/eji.1830240511. [DOI] [PubMed] [Google Scholar]
- 26.Poussier P, Julius M. Semin Immunol. 1995;7:321–334. doi: 10.1016/1044-5323(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 27.Kanamori Y, Ishimaru K, Nanno M, Maki K, Ikuta K, Nariuchi H, Ishikawa H. J Exp Med. 1996;184:1449–1459. doi: 10.1084/jem.184.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito H, Kanamori Y, Takemori T, Nariuchi H, Kubota E, Takahashi-Iwanaga H, Iwanaga T, Ishikawa H. Science. 1998;280:275–278. doi: 10.1126/science.280.5361.275. [DOI] [PubMed] [Google Scholar]
- 29.Page S T, van Oers N S C, Perlmutter R M, Weiss A, Pullen A M. Eur J Immunol. 1997;27:554–562. doi: 10.1002/eji.1830270229. [DOI] [PubMed] [Google Scholar]
- 30.Mombaerts P, Iacomini J, Johnson R, Herrup K, Tonegawa S, Papaioannou V. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 31.Morrissey P, Charrier K, Horovitz D, Fletcher F, Watson J. J Immunol. 1995;154:2687–2686. [PubMed] [Google Scholar]
- 32.Levin S D, Anderson S J, Forbush K, Perlmutter R M. EMBO J. 1993;12:1671–1680. doi: 10.1002/j.1460-2075.1993.tb05812.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson S J, Abraham K M, Nakayama T, Singer A, Perlmutter R M. EMBO J. 1992;11:4877–4886. doi: 10.1002/j.1460-2075.1992.tb05594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke A R, Hooper M, Farr A, Tonegawa S. Cell. 1993;72:337–348. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- 35.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling H J, von Boehmer H. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 36.Reiner S, Zheng S, Corry D, Locksley R. J Immunol Methods. 1993;165:37–46. doi: 10.1016/0022-1759(93)90104-f. [DOI] [PubMed] [Google Scholar]
- 37.Ohki-Hamazaki H, Makino Y, Kanno M, Koseki H, Akasaka T, Taniguchi M. Int Immunol. 1995;7:493–499. doi: 10.1093/intimm/7.3.493. [DOI] [PubMed] [Google Scholar]
- 38.Lieber M R, Hesse J E, Mizuuchi K, Gellert M. Genes Dev. 1987;1:751–761. doi: 10.1101/gad.1.8.751. [DOI] [PubMed] [Google Scholar]
- 39.Okazaki K, Davis D D, Sakano H. Cell. 1987;49:477–485. doi: 10.1016/0092-8674(87)90450-8. [DOI] [PubMed] [Google Scholar]
- 40.Ferrier P, Covey L R, Suh H, Winoto A, Hood L, Alt F W. Int Immunol. 1989;1:66–74. doi: 10.1093/intimm/1.1.66. [DOI] [PubMed] [Google Scholar]
- 41.Ferrier P, Krippl B, Blackwell T K, Furley A J W, Suh H, Winoto A, Cook W D, Hood L, Costantini F, Alt F W. EMBO J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ardouin L, Ismaili J, Malissen B, Malissen M. J Exp Med. 1998;187:105–116. doi: 10.1084/jem.187.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong S, Nussenzweig M C. Science. 1996;272:411–414. doi: 10.1126/science.272.5260.411. [DOI] [PubMed] [Google Scholar]
- 44.Lynch F, Shevach E. Intern Immunol. 1993;5:991–995. doi: 10.1093/intimm/5.8.991. [DOI] [PubMed] [Google Scholar]
- 45.Simpson S J, Hollander G, She J, Levelt C, Huang M, Terhorst C. Intern Immunol. 1995;7:287–293. doi: 10.1093/intimm/7.2.287. [DOI] [PubMed] [Google Scholar]
- 46.She J, Simpson S J, Gupta A, Hollander G, Levelt C, Liu C, Allen D, van Houten N, Wang B, Terhorst C. J Immunol. 1997;158:4678–4687. [PubMed] [Google Scholar]
- 47.Hamerman J A, Page S T, Pullen A M. J Immunol. 1997;159:1240–1246. [PubMed] [Google Scholar]