Abstract
Stimulation of human blood cultures with bacterial lipopolysaccharide (LPS) shows large inter-individual variation in interleukin 10 (IL-10) secretion, which has been shown to have a genetic component of over 70%. Alleles at two microsatellite loci in the 4 kb immediately upstream of the human IL-10 transcription initiation site in 132 individuals from 56 Dutch families were defined and assigned as haplotypes. LPS-induced IL-10 secretion was measured by ELISA and related to the IL-10 promoter haplotypes present in 78 unrelated individuals obtained from these families. Analysis showed that LPS-induced IL-10 secretion from unrelated individuals varied with IL-10 promoter haplotypes (P = 0.024; Kruskal-Wallis test). Two observations were made in relation to secreted IL-10 levels and promoter haplotypes; first, those haplotypes containing the allele IL10.R3 were associated with lower IL-10 secretion than haplotypes containing any other IL10.R allele. Second, the haplotype IL10.R2/IL10.G14 was associated with highest IL-10 secretion overall, whereas the haplotype IL10.R3/IL10.G7 was associated with lowest IL-10 secretion. These data demonstrate that the ability to secrete IL-10 can vary in man according to the genetic composition of the IL-10 locus.
Keywords: human monocytes/macrophages/cytokines/lipopolysaccharide/gene regulation
Interleukin 10 (IL-10) is an important immunoregulatory cytokine in man (1). It is involved in the regulation of inflammatory responses through direct influence over tumor necrosis factor production (2, 3). IL-10 is also involved in the pathology of human autoimmune disease (4–6), particularly in the dysregulation of B-cell function in systemic lupus erythematosus leading to autoantibody production (7, 8). In addition, its ability to induce T-cell anergy (9) and inhibit major histocompatibility complex class-I expression (10) may be important in its apparent contribution to tumor-related immunosuppression (11–13).
IL-10 also plays an important role in the development of infectious disease. Recently, it has been shown to affect macrophage responses during mycobacterial infections (14). Furthermore, the severity to which meningitis progresses is associated with serum IL-10 levels, such that high serum IL-10 was observed in patients with a poor or fatal outcome (15–17), whereas patients who had mild disease and a good prognosis had lower serum IL-10 levels. A recent study demonstrated striking differences between individuals in their ability to produce IL-10 following lipopolysaccharide (LPS) stimulation of whole blood cultures in vitro from first-degree family members (18), suggesting that differences in IL-10 production contained a considerable hereditary component. When it was considered whether inter-individual differences in secreted IL-10 might be related to corresponding differences in tumor necrosis factor (TNF) secretion, this was not the case (18); thus, differences in IL-10 secretion are not simply reflecting differences in the individual’s ability to produce TNF (3). Analysis of differences in IL-10 production between monozygotic or dizygotic twins and nonrelated individuals allowed the heritability of differences in IL-10 secretion to be estimated as 74% (18). Furthermore, preliminary experiments have shown that differences in LPS-induced IL-10 secretion are related to corresponding differences in mRNA production (rather than mRNA stability), implicating differential transcription as the principle mechanism of this difference in IL-10 production (T.H., unpublished observations). This therefore suggests that the nature of the 5′-flanking region of the IL-10 gene may contribute to the observed differences.
Having previously demonstrated the heritability of high or low IL-10 secretion (18), we wished to determine the extent to which the structure of the IL-10 promoter contributed to this phenomenon. Given the fact that the differences in IL-10 production are most likely transcriptionally regulated, it could either be cis- or trans-regulated. One straightforward way to demonstrate cis-regulation would be to determine whether an association existed between IL-10 production and polymorphic elements within the IL-10 promoter. In a recent study, Turner et al. (19) demonstrated a difference in IL-10 secretion, in association with the presence or absence of an “A” at position −1082 of the human IL-10 promoter, following Con A stimulation of peripheral blood mononuclear cells. The marked, heritable, variation in IL-10 secretion following LPS stimulation in our own studies (18) and the association of variable IL-10 levels with a range of disease states led us to search for evidence of additional genetic markers correlating with secretion in the LPS system. The 5′-flanking region of the human IL-10 gene contains two dinucleotide repeats (microsatellites), which we have shown to be highly polymorphic (20). These are located 1.1 and 4.0 kb 5′ of the transcription initiation site (21) and can therefore be considered to be very close to the gene, if not actually within it. We have also demonstrated a specific association between particular alleles at the IL-10 locus and the presence of systemic lupus erythematosus (22), directly implicating this locus in disease pathogenesis. In the present study, we have used these microsatellites to define the haplotypic nature of the human IL-10 gene in individuals for whom the IL-10 secretion in response to LPS in vitro had been defined. In this way, we anticipated a greater chance of “capturing” a putative functional mutation within a given haplotype than by finding it in association with individual alleles alone. In addition to such a cis-effect, variation in IL-10 secretion might result from variation in the levels of some external factor that influenced IL-10 production (a trans-effect). TNF is a major inducer of IL-10 following LPS stimulation (3), but it has already been shown that differences in TNF production do not account for the differences in IL-10 secretion seen in our test population (18). Therefore, we investigated the relationship between variation in IL-10 secretion and different IL-10 promoter structures, using our highly polymorphic IL-10 microsatellite markers, in an LPS stimulation system.
MATERIALS AND METHODS
LPS-Stimulated IL-10 Secretion.
Although many studies have demonstrated that IL-10 can be secreted by T-cells, B-cells, and monocytes (1, 2, 7), we felt it was important to consider that infections take place in the context of all peripheral blood cells (including erythrocytes) and accordingly have elected to use whole blood assays in this study and others (18, 23); this approach has recently found favor elsewhere (24). The most important aspect of a functional assay used to define genetic variation is minimal intra-individual, day-to-day variation. In comparisons between whole blood assays and those performed on isolated peripheral blood mononuclear cells, whole blood assays were shown to exhibit less day-to-day variation (25). With specific regard to the stimulation of IL-10 secretion, van der Linden and colleagues showed that the coefficient of variation in a series of 10 individuals was about 10% on a day-to-day basis, suggesting that intra-individual variation was stable (M.W. van der Linden, T.W.J.H., D. Sloeken, A. Sturk, and R.G.J.W., submitted for publication). Thus, this assay system combines low variation with the advantages of minimal cell manipulation and a more representative modeling of in vivo infection to present the whole blood assay as a satisfactory method of examining cytokine secretion. In addition, the low variation of IL-10 secretion from any one individual (10%, above; intra-individual variation) contrasts with the wide (but stable) range of IL-10 secretion levels seen across the whole group tested (inter-individual variation), demonstrating that this test forms a good basis for the study of genetically defined variation.
Blood samples were obtained in endotoxin-free heparin tubes (Chromogenix AM, Molndal, Sweden) from 132 individuals in 56 families, between 09.00 a.m. and 11.00 a.m. Samples were immediately diluted with an equal volume of endotoxin-free RPMI-1640 culture medium (Flow Laboratories). One-milliliter aliquots of diluted blood were incubated for 24 h with 1000 ng/ml LPS in 24-well cell culture plates at 37°C in 5% CO2. The supernatants were collected by centrifugation (1,500 × g, 10 min) and stored at −70°C until assay. The concentration of IL-10 was determined by ELISA using a polyclonal capture anti-human IL-10 Ab (JES3–9D7, PharMingen) and a biotinylated monoclonal anti-human IL-10 for detection (JES3–12G8, PharMingen), as previously described (18).
Genotyping at the IL-10 Microsatellites.
Genotyping at the IL10.G and IL10.R microsatellites (located approximately 1.2 and 4.0 kb upstream of the IL10 gene, respectively) was performed as previously described in detail (20–22). In addition, full details of primers and reaction conditions are present in the Genome Data Base entry for the human IL-10 gene. Briefly, genotyping at the IL10.G microsatellite was performed as previously described (20) using the following primer sequences: IL-10.1 (upstream), 5′ GTCCTTCCCCAGGTAGAGCAACACTCC 3′; IL-10.2 (downstream), 5′ CTCCCAAAGAAGCCTTAGTAGTGTTG 3′. The final reaction volume was 25 μl. After an initial melting time of 5 min at 95°C, samples were subjected to 29 rounds of 94°C, 15 s; 65°C, 1 min; 72°C, 1 min, with a final extension time of 5 min at 72°C.
Genotyping at the IL-10.R microsatellite was carried out as initially described (21) using the following primer sequences: IL-10.3 (upstream), 5′ CCCTCCAAAATCTATTTGCATAAG 3′; IL-10.4 (downstream), 5′ CTCCGCCCAGTAAGTTTCATCAC 3′. The final reaction volume was 20 μl. After an initial melting time of 5 min, samples were subjected to 30 rounds of 94°C, 15 s; 61°C, 15 s; 72°C, 15 s.
Biometra “Uno” thermoblocks were used in each case. For both the IL10.G and IL10.R microsatellites, in addition to the test DNA, each reaction contained 1 μM each primer; 200 μM each dATP, dGTP, and dTTP and 20 μM dCTP (Pharmacia); 0.5 units Primezyme (Biometra, Gottingen, Germany), in 1× reaction buffer with 1.5 mM MgCl2 (Biometra); [α-32P]dCTP (Amersham) was added to label the reaction product. Amplified products were mixed with formamide loading buffer, heated to 80°C for 10 min, and then cooled immediately on ice prior to separation on a sequencing gel containing 6% acrylamide (19:1, Life Sciences, Paisley, Scotland) and 7 M urea (Appligene, Strasbourg, France) at 75 W on a Stratagene Baseace apparatus (Stratagene), usually for 3 h. After drying, gels were allowed to expose x-ray film, and the alleles were assigned by direct sizing against an end-labeled 10-bp ladder (Life Sciences), run in two or three lanes of the gel.
Statistical Analysis.
IL-10 secretion in different haplotype groups was not distributed normally, and so nonparametric testing has been used throughout. IL-10 secretion is quoted as the median (± semi-interquartile range) for individual haplotypes. To determine whether IL-10 secretion varied with haplotype, all haplotypes with four or more observations of IL-10 secretion levels were compared simultaneously by the Kruskal-Wallis test of variance analysis. In some cases, strictly where indicated by the Kruskal-Wallis testing, individual groups were compared by the Mann-Whitney u test. In all cases, P values of <0.05 were taken as indicating significance. Minitab software and Apple Macintosh computers were used throughout.
RESULTS
Haplotype Frequency at the Human IL-10 Locus in Unrelated Individuals.
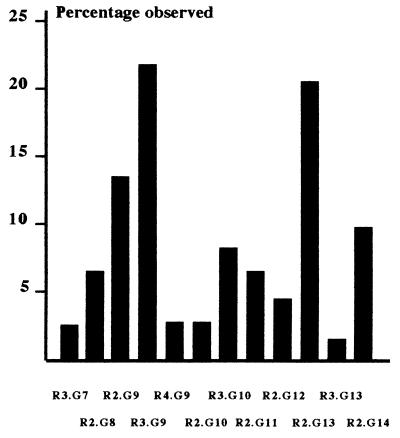
Given their close proximity (2, 800 bp apart), it would be misleading to consider the two IL-10 promoter microsatellites as independent. In addition, it was likely that putative functional mutations might be more clearly inferred in the context of unambiguously defined stretches of DNA. Families were therefore utilized to determine IL10.R/IL10.G haplotypes. Haplotypes were determined by considering the inheritance of IL10.R and IL10.G alleles from parents to children. In all cases, haplotypes were assigned unambiguously from complete family structures except in one case, where it was still possible to assign haplotypes unambiguously even though no DNA sample was available from the father. Seventy-eight unrelated individuals were identified from the total test population of 132. The haplotypes observed in these unrelated individuals and their frequencies are shown in Fig. 1. The most common were IL10.R3/IL10.G9 and IL10.R2/IL10.G13 with frequencies of 21.8 and 20.5%, respectively. This was in agreement with our previous observation that alleles IL10.R2 and IL10.G13 were strongly associated, as were IL10.R3 and IL10.G9, in a population of unrelated individuals from the West of Scotland (21).
Figure 1.
Frequency of IL-10 haplotypes. The number of times each haplotype was observed was expressed as a percentage of the total number of haplotypes (n = 156).
LPS-Induced IL-10 Secretion Varies with IL10.R/G Haplotype.
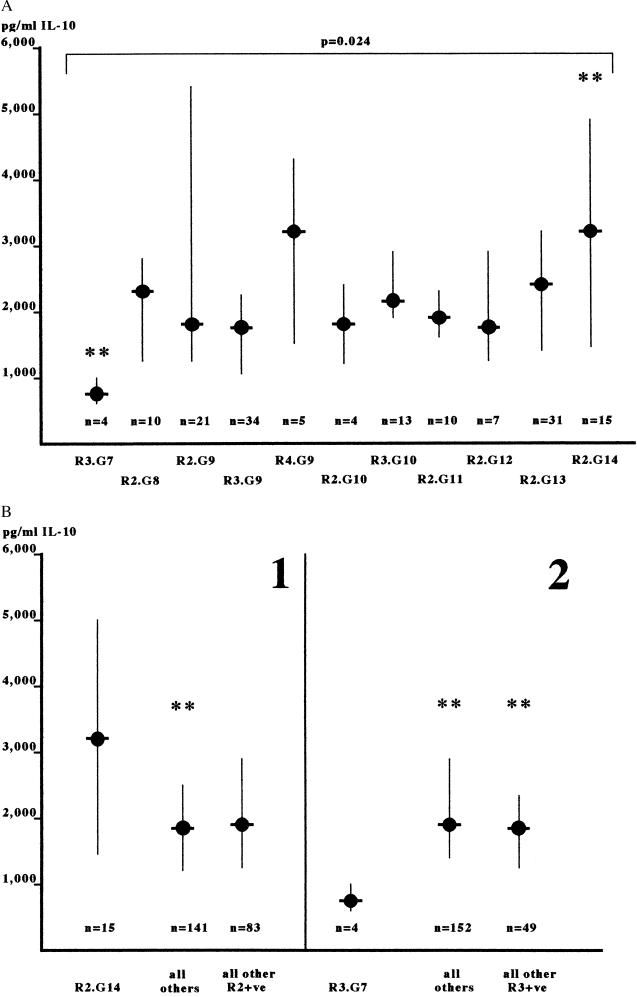
We first investigated whether any association was present between the IL10.R/G haplotypes observed in the pool of 78 unrelated individuals and the levels of IL-10 secreted following LPS stimulation in vitro. The level of IL-10 secreted following in vitro LPS stimulation did indeed vary according to the IL-10 promoter haplotype present. As shown in Fig. 2, IL-10 secretion was differentially distributed between IL-10 promoter haplotypes (P = 0.024, Kruskal-Wallis test). The haplotype IL10.R3/G7 had the lowest median IL-10 secretion [762 pg/ml (578–1000); z value = −2.77, Kruskal-Wallis test], whereas the IL10.R2/G14 haplotype had the greatest enhancement in IL-10 secretion (3108 pg/ml (1391–4990); z = +2.18, Kruskal-Wallis test). When compared against the IL-10 secretion associated with all other haplotypes combined, these differences were significant [for IL10.R3/G7 versus all others, 762(578–1000) versus 1972(1238–3018), P = 0.006, Mann-Whitney u test; for IL10.R2/G14 versus all others, 3108(1391–4990) versus 1876(1228–2770), P = 0.024, Mann-Whitney u test]. In addition, when the IL10.R3/G7 haplotype was considered against all other haplotypes containing allele R3, it was associated with lower IL-10 secretion in the unrelated individuals [IL10R3/G7, 762(578–1000) versus IL10.R3/G7−, 1972(1238–3018), P = 0.02, Mann-Whitney u test]. When all available haplotypes were considered, only in the case of IL10.R2.G14 was IL-10 secretion higher than all other haplotypes [3233(1813–5215) versus 2262(1365–3223), P = 0.011, Mann-Whitney u test] and all those containing the IL10.R2 allele [3233(1813–5215) versus 2383(1238–3393) P = 0.027, Mann-Whitney u test]. Therefore, the variation in IL-10 secretion observed between unrelated individuals following LPS stimulation in vitro was accounted for, in part, by differences in the IL-10 promoter haplotype related to two particular alleles, IL10.G7 (lower IL-10 secretion) and IL10.G14 (higher IL-10 secretion).
Figure 2.
(A) IL-10 secretion in relation to IL-10 locus haplotype. IL-10 secretion was determined following LPS stimulation of whole blood cultures as described in the text. IL-10 haplotypes were also so determined and considered against the levels of secreted IL-10. The medians and semi-interquartile ranges of IL-10 secretion are shown for each haplotype, which was observed four or more times. Levels of IL-10 secretion in these groups were examined simultaneously by the Kruskal-Wallis test of variance analysis; the given P value (P = 0.024) shows that the IL-10 secretion varied significantly between the haplotypes. The indicated haplotypes (∗∗) had the largest positive (R2.G14) or negative (R3.G7) variation from the overall median, as indicated by their z value. (B) Haplotypes associated with high or low IL-10 secretion. The haplotypes indicated from the Kruskal-Wallis analysis were compared with other haplotypes in respect to associated IL-10 secretion. In panel 1, the R2.G14 haplotype is shown to be associated with significantly higher IL-10 secretion than all other haplotypes (P = 0.027) and is strongly suggested to be associated with higher IL-10 secretion than all other IL10.R2-containing haplotypes (P = 0.065). In panel 2, the R3.G7 haplotype is shown to be associated with significantly lower IL-10 secretion than all other haplotypes (P = 0.006) and all other R3-containing haplotypes (P = 0.021). Data are the medians and semi-interquartile ranges. Asterisks indicate statistical significance.
The IL10.R3 Allele Is Associated with Lower IL-10 Secretion.
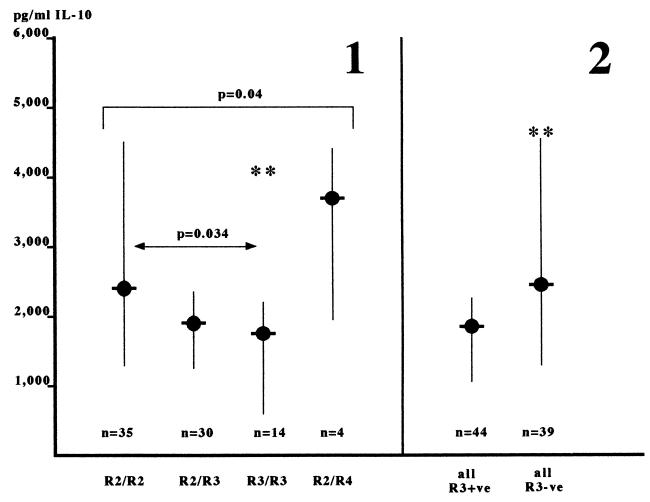
Given that the haplotype analysis had shown differences in IL-10 secretion between haplotypes and that in some cases these seemed to have independence from the IL10.R allele, it was of interest to investigate whether the IL10.R locus alone had an influence on IL-10 secretion. When the IL-10 secretion associated with the four observed genotypes at IL10.R was examined, a trend to their being different was observed (P = 0.04, Kruskal-Wallis test). This seemed to be due to the presence of allele IL10.R3 because the R3 homozygous genotypes had a strong negative variance from the median (z = −1.96), which was also observed to a lesser extent in the R2/R3 heterozygotes (z = −1.16). This was supported by the fact that the IL-10 secretion in the R3 homozygotes was significantly lower than that observed in the R2 homozygotes [R3/R3, 1735 (636–2192) versus R2/R2, 2369 (1238–4457), P = 0.034, Mann-Whitney u test] in the unrelated individuals. Furthermore, comparison of those genotypes containing the R3 allele with those not containing R3 also demonstrated lower IL-10 secretion in the R3+ genotypes [R3+, 1850 (1055–2265) versus R3-, 2456 (1296–4559), P = 0.011, Mann-Whitney u test]. Taken together, these data demonstrate that the IL10.R3 allele is associated with lower IL-10 secretion than other IL10.R alleles and is able, at least in part, to diminish IL-10 secretion whenever present. Finally, when all available haplotypes were compared, this observation was supported in that the IL-10 secretion associated with haplotypes containing the individual IL10.R alleles was significantly different (P = 0.007, Kruskal-Wallis test), again due to lower IL-10 secretion in association with the R3 allele [R3+ve, 2028(1316–2827) versus R3−ve, 2514(1391–3747); P = 0.0079)].
DISCUSSION
IL-10 is an important regulatory cytokine that is involved in many aspects of the immune response, and it seems dysregulated in human autoimmune (8), malignant (9–13), and infectious (15–17) disease (e.g., refs. 8,13,15). In addition, it has recently been shown that heritably higher levels of IL-10 secretion are an important component of the genetic background to systemic lupus erythematosus (26) and to the outcome of infectious disease (18). We therefore felt that it was important to determine whether these differences might have a genetic basis, particularly from within the IL-10 gene itself.
It has previously been demonstrated that the IL-10 secretion known to result from in vitro stimulation of human peripheral blood leukocytes with LPS varies markedly between individuals. This variation is a strong candidate for having a genetic basis. A recent report has demonstrated that the genetic component to the innate variation in IL-10 production was greater than 70% (18). In addition, preliminary experiments showed that variation in IL-10 secretion between identical twins was <10%, that the level of IL-10 secreted was directly proportional to the level of IL-10 mRNA synthesized, and that IL-10 mRNA half-life from low IL-10 secretors was equal to that from high IL-10 secretors. Thus, differential IL-10 secretion was likely to have its origins in differing rates of IL-10 mRNA synthesis, which in turn might reasonably be ascribed to differences in the structure of the IL-10 promoter.
We made the following observations: (a) IL-10 secretion in unrelated individuals did vary according to IL-10 promoter haplotypes; (b) the IL10R3/G7 haplotype was particularly associated with lower IL-10 secretion, and the IL10.R2/G14 haplotype was particularly associated with higher IL-10 secretion; the data suggested that these associations might be carried with the G7 and G14 alleles independently of the R allele present; (c) independently of events at the IL10.G microsatellite, the IL10.R3 allele was associated with lower IL-10 secretion than either the IL10.R2 or IL10.R4 alleles.
These results show clearly that IL-10 gene haplotypes are associated with differential production of IL-10. Thus, the heritable differences in IL-10 production that have previously been described are strongly linked to the gene’s structure.
Our data reveal the presence of three possible genotypes associated with differences in IL-10 production, as marked by the IL10.R3, IL10.G7, and IL10.G14 microsatellite alleles. First, the presence of the allele IL10R.3 marks an apparent predisposition to lower IL-10 secretion. This was true for the whole population of IL10.R3+ve haplotypes versus all other IL10.R3−ve haplotypes, irrespective of the IL10.G allele present and was typified by the lower IL-10 secretion from R3 homozygotes compared with R2 homozygotes (Fig. 3). Furthermore, the possibility that one particular haplotype might be biasing this conclusion was excluded by the observation that this was a general trend for individual IL10.G alleles. We have recently (27) reported an analysis of potential transcription factor binding sites within the area immediately 5′ of the IL-10 transcription site and extending beyond the IL10.R microsatellite; those sites potentially able to transmit cytokine-induced signals to the human IL-10 gene are clustered in the 1,500 bp immediately downstream of the IL10.R microsatellite. The IL-10 gene is known to be responsive to a number of cytokines, including tumor necrosis factor and interferon-gamma (28, 29); response elements for these and other cytokines were present in this area. Thus, we speculate that the region of the IL-10 gene immediately downstream of the IL10.R microsatellite may confer responsiveness to the transcription of human IL-10 and that differences in this property, resulting in lower IL-10 secretion, are marked by the IL10.R3 allele.
Figure 3.
IL10.R3-containing genotypes are associated with lower IL-10 secretion. Genotypes containing various IL10.R alleles were compared in respect of their association with IL-10 secretion, independently of the IL10.G allele present. In panel 1, all available genotypes at IL10.R were compared simultaneously by the Kruskal-Wallis test of variance analysis. This test showed that the IL-10 secretion varied between the groups, with a strong trend to significance (P = 0.04) and that the IL10.R3-homozygous genotypes (∗∗) varied most from the overall median (z = −1.96). In panel 2, IL10.R3-containing genotypes are shown to be associated with significantly lower IL-10 secretion than all other IL10.R-containing genotypes (∗∗; P = 0.011). Data are the medians and semi-interquartile ranges. This analysis included five additional unrelated individuals for whom IL10.R genotypes were known, but unambiguous haplotypes were unavailable.
We also observed significantly lower IL-10 secretion with the IL10.R3/G7 haplotype and higher IL-10 secretion in association with the IL10.R2/G14 haplotype (Fig. 2). Furthermore, comparison of these haplotypes with others containing the same R-allele but different G-alleles suggested that these differences were due to events associated to the G7 and G14 alleles themselves. Such a high-resolution examination would not have been possible without the knowledge of haplotypes.
Monocytes are the main source of IL-10, although many cell types can be stimulated to IL-10 secretion (1). LPS stimulation affects monocytes predominantly, and therefore our results are particularly relevant to genetic differences in monocyte IL-10 secretion. In this regard, it will be of interest to determine how our data fit together with those described at the −1082 G/A mutation (19), which, being defined by Con A stimulation, may be of more relevance to T-cells and thereby raising the possibility that genetic elements exist that govern differences in IL-10 secretion between cell types. It should be noted that the overall variation in IL-10 secretion could not be accounted for wholly by the differences related to IL-10 promoter haplotypes, suggesting that additional factors remain to be defined.
After antigen recognition has taken place, much of the magnitude and direction of immune responses is controlled by cytokines. If the degree to which cytokines were secreted varied between individuals, this might alter immune responses; in particular, once a potentially pathogenic event had occurred, differential cytokine secretion might alter the degree of pathogenesis. The question of whether cytokine genes might form a new class of quantitative trait loci has recently been formally posed (30, 31). Heterogeneity in genetic elements has been identified, associated with differences in secretion, in a number of cytokines, including tumor necrosis factor (32, 33) and interleukin-4 (34); in the case of the IL-1 locus, this may be quite complex (35). Such cytokine polymorphisms are now becoming associated with specific immunological diseases. For example, differences in the IL-1 gene cluster promote severity in peridontal disease (36), whereas the genes for both IL-4 (37) and IL-9 (38) are associated with various elements of atopy and asthma. Indeed, the relationship between IL-10 and IL-4 is important in determining the balance between IgG4 and IgE production (39). In addition, differences in serum IL-10 concentrations and in vitro secretion from patients and their first-degree family members in infectious (15–18), autoimmune (4, 5, 8, 9, 26), and malignant disease (9, 13, 40) have combined with differences in allele distribution (22) to implicate the IL-10 gene as an important quantitative trait loci for human disease. Furthermore, recently a new class of regulatory T-cells, the TR1 cell, has been identified that depends on high levels of IL-10 for growth (41, 42). These T-cells down-regulate the immune response in an antigen-specific way. Because these TR1 cells are dependent on high levels of IL-10 for growth, individuals with a genetic predisposition for higher IL-10 production are more likely to develop a functioning population of Tr1 cells (and thereby become antigen-tolerant) earlier than those whose characteristic is for lower IL-10 production. Thus, understanding genetically defined differences in IL-10 production allow understanding of differences in mechanisms responsible for immune tolerance between individuals.
Here, we present evidence that differences in the secretion of interleukin-10 is strongly associated with different haplotypes at the human IL-10 locus, thereby linking these two sets of observations. Taken together, these studies provide evidence that genetic heterogeneity not only controls whether antigen responsiveness occurs (via the major histocompatibility complex) but also, through cytokines and their receptors, the extent and direction in which that response may develop once antigen recognition has occurred.
Acknowledgments
J.E. is supported in part by the Breast Cancer Campaign.
ABBREVIATIONS
- LPS
lipopolysaccharide
- IL-10
interleukin 10
- TNF
tumor necrosis factor
References
- 1.Mosmann T R. Adv Immunol. 1994;56:1–26. [PubMed] [Google Scholar]
- 2.Malefyt R D, Abrams J, Bennet B, Figdor C G, de Vries J E. J Exp Med. 1991;174:1209–1220. doi: 10.1084/jem.174.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wanidworanun C, Strober W. J Immunol. 1993;151:6853–6861. [PubMed] [Google Scholar]
- 4.Llorente L, Richaud-Patin Y, Fior R, Alcocer-Varela J, Wijdnes J, Morel-Fourrier B, Galanaud P, Emilie P. Arthritis Rheum. 1994;37:1647–1655. doi: 10.1002/art.1780371114. [DOI] [PubMed] [Google Scholar]
- 5.Cash J J, Splawski J B, Thomas R, McFarlin J F, Schulze-Koops H, Davis L S, Fujita K, Lipsky P E. Arthritis Rheum. 1995;38:96–104. doi: 10.1002/art.1780380115. [DOI] [PubMed] [Google Scholar]
- 6.Perez L, Orte J, Brieva J A. Arthritis Rheum. 1995;38:1771–1776. doi: 10.1002/art.1780381210. [DOI] [PubMed] [Google Scholar]
- 7.Itoh K, Hirohata S. J Immunol. 1995;154:4341–4350. [PubMed] [Google Scholar]
- 8.Llorente L, Zou W, Levy Y, Richaud-Patin Y, Wijdenes Y, Alcocer-Varela J, Morel-Fourrier B, Brouet J-C, Alarcon-Segovia D, Galanaud P, et al. J Exp Med. 1995;181:839–844. doi: 10.1084/jem.181.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luscher U, Filgueira L, Juretic A, Zuber M, Luscher L J, Heberer M, Spagnoli G C. Int J Cancer. 1994;57:612–619. doi: 10.1002/ijc.2910570428. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda M, Salazar F, Petersson M, Masucci G, Hansson J, Pisa, Zhang Q C, Masucci M G, Kiessling R. J Exp Med. 1994;180:2371–2376. doi: 10.1084/jem.180.6.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Modlin R L, Moy R L, Dubinett S M, McHugh T, Nickloff B J, Uyemura K. J Immunol. 1995;155:2240–2247. [PubMed] [Google Scholar]
- 12.Suzuki T, Tahara H, Narula S, Moore K W, Robbins P D, Lotze M T. J Exp Med. 1995;182:447–486. doi: 10.1084/jem.182.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fortis C, Foppoli M, Gianotti L, Galli L, Citterio G, Consogno G, Gentilini O, Braga M. Cancer Lett. 1996;104:1–5. doi: 10.1016/0304-3835(96)04213-9. [DOI] [PubMed] [Google Scholar]
- 14.Murray P J, Wang L, Onufry R C, Tepper R I, Young R A. J Immunol. 1997;158:315–232. [PubMed] [Google Scholar]
- 15.Derkx B, Marchant A, Goldman M, Bijlmer R, van de Venter S. J Infect Dis. 1995;171:229–232. doi: 10.1093/infdis/171.1.229. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann A K, Halstenen A, Sornes S, Rokke O, Waage A. Infect Immun. 1995;63:2109–2112. doi: 10.1128/iai.63.6.2109-2112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.VanFurth A M, Seijmonsbergen E M, Langermans J A M, Groeneveld P H P, Debel C E, VanFurth R. Clin Infect Dis. 1995;21:220–222. doi: 10.1093/clinids/21.1.220. [DOI] [PubMed] [Google Scholar]
- 18.Westendorp R G J, Langermans J A M, Huizinga T W G, Elouali A H, Verweij C L, Vandenbroucke J P. Lancet. 1997;349:170–173. doi: 10.1016/s0140-6736(96)06413-6. [DOI] [PubMed] [Google Scholar]
- 19.Turner D M, Williams D M, Sankaran D, Lazarus M, Sinnott P J, Hutchinson I V. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 20.Eskdale J, Gallagher G. Immunogenetics. 1995;42:444–445. doi: 10.1007/BF00179416. [DOI] [PubMed] [Google Scholar]
- 21.Eskdale J, Kube D, Gallagher G. Immunogenetics. 1996;45:82–83. doi: 10.1007/s002510050174. [DOI] [PubMed] [Google Scholar]
- 22.Eskdale J, Wordsworth P, Bowman S, Field M, Gallagher G. Tissue Antigens. 1997;49:635–639. doi: 10.1111/j.1399-0039.1997.tb02812.x. [DOI] [PubMed] [Google Scholar]
- 23.Derkx H H F, Bruin K F, Jongeneel C V, de Waal L P, Brinkman B M N, Verweij C L, Houwing-Duistermaat J J, Rosendaal F R, van deVenter S J H. J Endotoxin Res. 1995;2:19–25. [Google Scholar]
- 24.Wing M G, Waldmann H, Isaacs J, Compston D A, Hale G. Ther Immunol. 1995;2:183–190. [PubMed] [Google Scholar]
- 25.de Groote D, Zangerle P F, Gevaert Y, Fassotte M F, Beguin Y, Noizat-Pirenne F, Pirenne J, Gathy R, Lopez M, Dehart I, et al. Cytokine. 1992;4:239–248. doi: 10.1016/1043-4666(92)90062-v. [DOI] [PubMed] [Google Scholar]
- 26.Llorente L, Richaud-Patin Y, Couderc J, Alarcon-Segovia D, Ruiz-Soto R, Alcocer-Castillejos N, Alcocer-Varela J, Granados J, Bahena S, Galanaud P, et al. Arthritis Rheum. 1997;38:1429–1435. doi: 10.1002/art.1780400810. [DOI] [PubMed] [Google Scholar]
- 27.Eskdale J, Kube D, Tesch H, Gallagher G. Immunogenetics. 1997;46:120–128. doi: 10.1007/s002510050250. [DOI] [PubMed] [Google Scholar]
- 28.Darnell J E, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 29.Platzer C, Meisel C H, Vogt K, Platzer K, Volk H D. Int Immunol. 1995;7:517–523. doi: 10.1093/intimm/7.4.517. [DOI] [PubMed] [Google Scholar]
- 30.Dasser A, Mitchison H, Mitchison A, Müller B. Cytokine. 1996;8:593–597. doi: 10.1006/cyto.1996.0079. [DOI] [PubMed] [Google Scholar]
- 31.Mitchison A. Immunogenetics. 1997;46:46–52. doi: 10.1007/s002510050241. [DOI] [PubMed] [Google Scholar]
- 32.Pociot F, Briant L, Jongeneel C V, Molvig J, Worsaae H, Abbal M, Thomsen M, Nerup J, Cambon-Thomsen A. Eur J Immunol. 1993;23:224–231. doi: 10.1002/eji.1830230135. [DOI] [PubMed] [Google Scholar]
- 33.Wilson A G, Symons J A, McDowell T L, McDevitt H O, Duff G W. Proc Natl Acad Sci USA. 1997;94:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Z M, Casolaro V, Chen V R, Georas S N, Monos D, Ono S J. J Immunol. 1996;156:424–429. [PubMed] [Google Scholar]
- 35.Guasch J F, Bertina R M, Reitsma P H. Cytokine. 1996;8:598–602. doi: 10.1006/cyto.1996.0080. [DOI] [PubMed] [Google Scholar]
- 36.Kornman K S, Crane A, Wang H Y, di Giovine F S, Newman M G, Pirk F W, Wilson T G, Jr, Higginbottom F L, Duff G W. J Clin Peridontol. 1997;24:72–77. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 37.Walley A J, Cookson W O. J Med Genet. 1996;33:689–692. doi: 10.1136/jmg.33.8.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholaides N C, Holroyd K J, Ewart S J, Eleff S M, Kiser M B, Dragwa C R, Sullivan C D, Grasso L, Zhang L-Y, Messier C J, Zhou T, Kleberger S R, Buetow K H, Levitt R C. Proc Natl Acad Sci USA. 1997;94:13175–13180. doi: 10.1073/pnas.94.24.13175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeannin P, Lecoanet S, Delneste Y, Gauchat J F, Bonnefoy J Y. J Immunol. 1998;160:3555–3561. [PubMed] [Google Scholar]
- 40.Dummer W, Becker J C, Schwaaf A, Leverkus M, Moll T, Brocker E B. Melanoma Res. 1995;5:67–68. doi: 10.1097/00008390-199502000-00008. [DOI] [PubMed] [Google Scholar]
- 41.Groux H, O’Garra A, Bigler M, Rouleau M, Antonenko S, de Vries J E, Roncarolo M G. Nature (London) 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 42.Asseman C, Powrie F. Gut. 1998;42:157–158. doi: 10.1136/gut.42.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]