Abstract
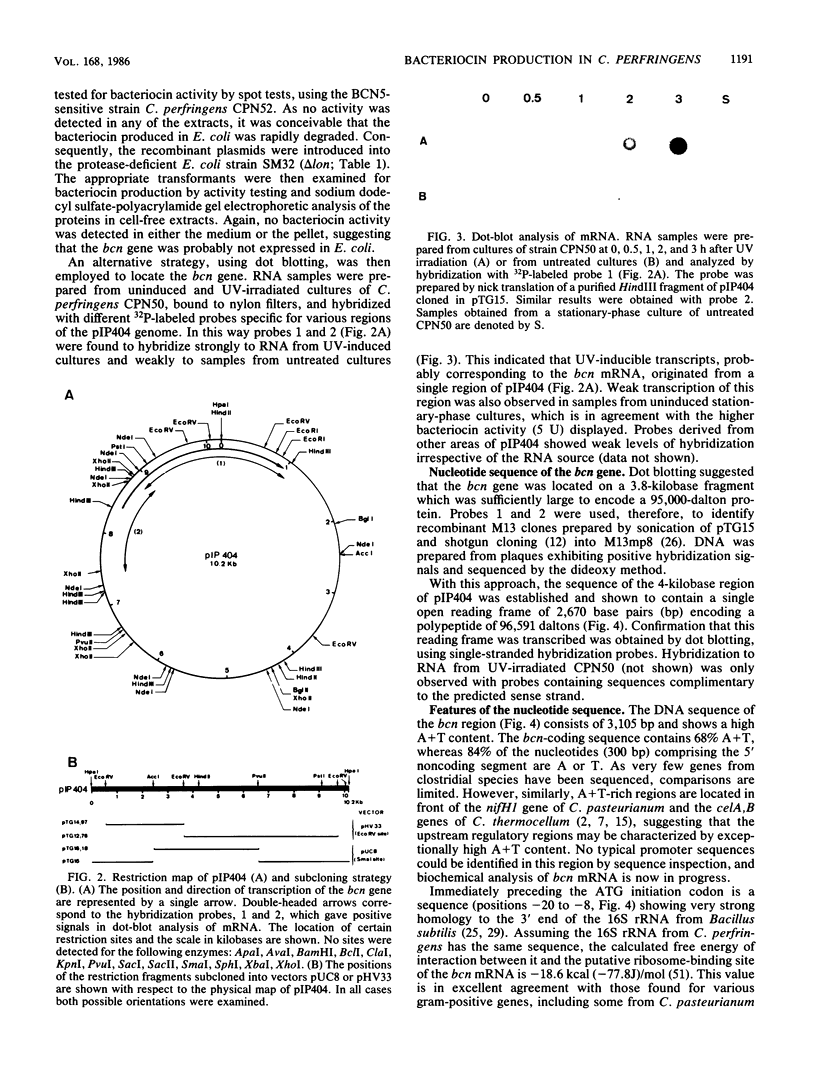
The bacteriocinogenic plasmid pIP404 from Clostridium perfringens was isolated and cloned in Escherichia coli, and its physical map was deduced. Expression of the bcn gene, encoding bacteriocin BCN5, is inducible by UV irradiation of C. perfringens and thus resembles the SOS-regulated bacteriocin genes of enteric bacteria. The location of bcn on pIP404 was established by a dot-blot procedure, using specific hybridization probes to analyze mRNA samples from induced and uninduced cultures. From the nucleotide sequence of its gene, the molecular weight of BCN5 was deduced to be 96,591, and a protein of this size was secreted by bacteriocin-producing cultures of C. perfringens. The primary structure of the protein suggests that it may function as an ionophore, since a hydrophobic domain, resembling those of the ionophoric colicins, is present at the COOH terminus. No bacteriocin activity could be detected in E. coli harboring plasmids bearing the bcn gene, even when the transcriptional and translational signals were replaced by those of lacZ. A possible explanation may be found in the unusual codon usage of the adenine-thymine-rich bcn gene, as this shows a preference for codons with a high adenine-plus-thymine content, especially in the wobble position. Many of the frequently used codons correspond to those recognized by minor tRNA species in E. coli. Consequently, bcn expression might be limited by tRNA availability in this bacterium.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham L. J., Wales A. J., Rood J. I. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985 Jul;14(1):37–46. doi: 10.1016/0147-619x(85)90030-7. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschek H. P., Solberg M. Isolation of a plasmid responsible for caseinase activity in Clostridium perfringens ATCC 3626B. J Bacteriol. 1981 Jul;147(1):262–266. doi: 10.1128/jb.147.1.262-266.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort G., Magot M., Ionesco H., Sebald M. Characterization and transferability of Clostridium perfringens plasmids. Plasmid. 1977 Nov;1(1):52–66. doi: 10.1016/0147-619x(77)90008-7. [DOI] [PubMed] [Google Scholar]
- Béguin P., Cornet P., Aubert J. P. Sequence of a cellulase gene of the thermophilic bacterium Clostridium thermocellum. J Bacteriol. 1985 Apr;162(1):102–105. doi: 10.1128/jb.162.1.102-105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi R. A., Vanderkooi G. The low polarity of many membrane proteins. Proc Natl Acad Sci U S A. 1972 Apr;69(4):930–932. doi: 10.1073/pnas.69.4.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. C., Chen J. S., Johnson J. L. Structural features of multiple nifH-like sequences and very biased codon usage in nitrogenase genes of Clostridium pasteurianum. J Bacteriol. 1986 Apr;166(1):162–172. doi: 10.1128/jb.166.1.162-172.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S. T. Characterisation of the promoter for the LexA regulated sulA gene of Escherichia coli. Mol Gen Genet. 1983;189(3):400–404. doi: 10.1007/BF00325901. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Grundström T., Jaurin B., Robinson J. J., Weiner J. H. Location and nucleotide sequence of frdB, the gene coding for the iron-sulphur protein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Aug;126(1):211–216. doi: 10.1111/j.1432-1033.1982.tb06768.x. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Guest J. R. Production of a soluble form of fumarate reductase by multiple gene duplication in Escherichia coli K12. Eur J Biochem. 1979 Dec;102(1):65–71. doi: 10.1111/j.1432-1033.1979.tb06263.x. [DOI] [PubMed] [Google Scholar]
- Deininger P. L. Approaches to rapid DNA sequence analysis. Anal Biochem. 1983 Dec;135(2):247–263. doi: 10.1016/0003-2697(83)90680-2. [DOI] [PubMed] [Google Scholar]
- Dougan G., Sherratt D. The transposon Tn1 as a probe for studying ColE1 structure and function. Mol Gen Genet. 1977 Mar 7;151(2):151–160. doi: 10.1007/BF00338689. [DOI] [PubMed] [Google Scholar]
- Edlund T., Grundström T., Normark S. Isolation and characterization of DNA repetitions carrying the chromosomal beta-lactamase gene of Escherichia coli K-12. Mol Gen Genet. 1979 Jun 7;173(2):115–125. doi: 10.1007/BF00330301. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Grépinet O., Béguin P. Sequence of the cellulase gene of Clostridium thermocellum coding for endoglucanase B. Nucleic Acids Res. 1986 Feb 25;14(4):1791–1799. doi: 10.1093/nar/14.4.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Ionesco H., Bieth G., Dauguet C., Bouanchaud D. Isolement et identification de deux plasmides d'une souche bactériocinogène de Clostridium perfringens. Ann Microbiol (Paris) 1976 Oct;127B(3):283–294. [PubMed] [Google Scholar]
- Ionesco H., Wolff A., Sebald Production induite de bactériocine et d'un bactériophage par la souche BP6K-N-5 de Clostridium perfringens. Ann Microbiol (Paris) 1974 Oct-Nov;125B(3):335–346. [PubMed] [Google Scholar]
- Karube I., Urano N., Yamada T., Hirochika H., Sakaguchi K. Cloning and expression of the hydrogenase gene from Clostridium butyricum in Escherichia coli. FEBS Lett. 1983 Jul 11;158(1):119–122. doi: 10.1016/0014-5793(83)80689-9. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mizusawa S., Ward D. F. A bacteriophage lambda vector for cloning with BamHI and Sau3A. Gene. 1982 Dec;20(3):317–322. doi: 10.1016/0378-1119(82)90200-1. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Morlon J., Lloubès R., Varenne S., Chartier M., Lazdunski C. Complete nucleotide sequence of the structural gene for colicin A, a gene translated at non-uniform rate. J Mol Biol. 1983 Oct 25;170(2):271–285. doi: 10.1016/s0022-2836(83)80148-x. [DOI] [PubMed] [Google Scholar]
- Ogasawara N. Markedly unbiased codon usage in Bacillus subtilis. Gene. 1985;40(1):145–150. doi: 10.1016/0378-1119(85)90035-6. [DOI] [PubMed] [Google Scholar]
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J. 1984 Dec 1;3(12):2895–2898. doi: 10.1002/j.1460-2075.1984.tb02227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primrose S. B., Ehrlich S. D. Isolation of plasmid deletion Mutants and study of their instability. Plasmid. 1981 Sep;6(2):193–201. doi: 10.1016/0147-619x(81)90066-4. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P. The ins and outs of colicins. Part I: Production, and translocation across membranes. Microbiol Sci. 1984 Oct;1(7):168–175. [PubMed] [Google Scholar]
- Robinson M., Lilley R., Little S., Emtage J. S., Yarranton G., Stephens P., Millican A., Eaton M., Humphreys G. Codon usage can affect efficiency of translation of genes in Escherichia coli. Nucleic Acids Res. 1984 Sep 11;12(17):6663–6671. doi: 10.1093/nar/12.17.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rood J. I., Scott V. N., Duncan C. L. Identification of a transferable tetracycline resistance plasmid (pCW3) from Clostridium perfringens. Plasmid. 1978 Sep;1(4):563–570. doi: 10.1016/0147-619x(78)90013-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rüther U., Koenen M., Otto K., Müller-Hill B. pUR222, a vector for cloning and rapid chemical sequencing of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4087–4098. doi: 10.1093/nar/9.16.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires C. H., Heefner D. L., Evans R. J., Kopp B. J., Yarus M. J. Shuttle plasmids for Escherichia coli and Clostridium perfringens. J Bacteriol. 1984 Aug;159(2):465–471. doi: 10.1128/jb.159.2.465-471.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Truffaut N., Sebald M. Plasmid detection and isolation in strains of Clostridium acetobutylicum and related species. Mol Gen Genet. 1983;189(1):178–180. doi: 10.1007/BF00326075. [DOI] [PubMed] [Google Scholar]
- Varley J. M., Boulnois G. J. Analysis of a cloned colicin Ib gene: complete nucleotide sequence and implications for regulation of expression. Nucleic Acids Res. 1984 Sep 11;12(17):6727–6739. doi: 10.1093/nar/12.17.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain-Hobson S., Sonigo P., Danos O., Cole S., Alizon M. Nucleotide sequence of the AIDS virus, LAV. Cell. 1985 Jan;40(1):9–17. doi: 10.1016/0092-8674(85)90303-4. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff A., Ionesco H. Purification et caractérisation de la bactériocine N5 de Clostridium perfringens BP6K-N5 TYPE A. Ann Microbiol (Paris) 1975 Oct-Nov;126(3):343–356. [PubMed] [Google Scholar]
- Yamada M., Ebina Y., Miyata T., Nakazawa T., Nakazawa A. Nucleotide sequence of the structural gene for colicin E1 and predicted structure of the protein. Proc Natl Acad Sci U S A. 1982 May;79(9):2827–2831. doi: 10.1073/pnas.79.9.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]