Abstract
We report the identification and characterization of ERS-24 (Endoplasmic Reticulum SNARE of 24 kD), a new mammalian v-SNARE implicated in vesicular transport between the ER and the Golgi. ERS24 is incorporated into 20S docking and fusion particles and disassembles from this complex in an ATP-dependent manner. ERS-24 has significant sequence homology to Sec22p, a v-SNARE in Saccharomyces cerevisiae required for transport between the ER and the Golgi. ERS-24 is localized to the ER and to the Golgi, and it is enriched in transport vesicles associated with these organelles.
Newly formed transport vesicles have to be selectively targeted to their correct destinations, implying the existence of a set of compartment-specific proteins acting as unique receptor–ligand pairs. Such proteins have now been identified (Söllner et al., 1993a ; Rothman, 1994): one partner efficiently packaged into vesicles, termed a v-SNARE,1 and the other mainly localized to the target compartment, a t-SNARE. Cognate pairs of v- and t-SNAREs, capable of binding each other specifically, have been identified for the ER–Golgi transport step (Lian and Ferro-Novick, 1993; Søgaard et al., 1994), the Golgi–plasma membrane transport step (Aalto et al., 1993; Protopopov et al., 1993; Brennwald et al., 1994) in Saccharomyces cerevisiae, and regulated exocytosis in neuronal synapses (Söllner et al., 1993a ; for reviews see Scheller, 1995; Südhof, 1995). Additional components, like p115, rab proteins, and sec1 proteins, appear to regulate vesicle docking by controlling the assembly of SNARE complexes (Søgaard et al., 1994; Lian et al., 1994; Sapperstein et al., 1996; Hata et al., 1993; Pevsner et al., 1994).
In contrast with vesicle docking, which requires compartment-specific components, the fusion of the two lipid bilayers uses a more general machinery derived, at least in part, from the cytosol (Rothman, 1994), which includes an ATPase, the N-ethylmaleimide–sensitive fusion protein (NSF) (Block et al., 1988; Malhotra et al., 1988), and soluble NSF attachment proteins (SNAPs) (Clary et al., 1990; Clary and Rothman, 1990; Whiteheart et al., 1993). Only the assembled v–t-SNARE complex provides high affinity sites for the consecutive binding of three SNAPs (Söllner et al., 1993b ; Hayashi et al., 1995) and NSF. When NSF is inactivated in vivo, v–t-SNARE complexes accumulate, confirming that NSF is needed for fusion after stable docking (Søgaard et al., 1994).
The complex of SNAREs, SNAPs, and NSF can be isolated from detergent extracts of cellular membranes in the presence of ATPγS, or in the presence of ATP but in the absence of Mg2+, and sediments at ∼20 Svedberg (20S particle) (Wilson et al., 1992). In the presence of MgATP, the ATPase of NSF disassembles the v–t-SNARE complex and also releases SNAPs. It seems likely that this step somehow initiates fusion.
To better understand vesicle flow patterns within cells, it is clearly of interest to identify new SNARE proteins. Presently, the most complete inventory is in yeast, but immunolocalization is difficult in yeast compared with animal cells, and many steps in protein transport have been reconstituted in animal extracts (Rothman, 1992) that have not yet been developed in yeast. Therefore, it is important to create an inventory of SNARE proteins in animal cells. The most unambiguous and direct method for isolating new SNAREs is to exploit their ability to assemble together with SNAPs and NSF into 20S particles and to disassemble into subunits when NSF hydrolyzes ATP. Similar approaches have already been successfully used to isolate new SNAREs implicated in ER to Golgi (Søgaard et al., 1994) and intra-Golgi transport (Nagahama et al., 1996), in addition to the original discovery of SNAREs in the context of neurotransmission (Söllner et al., 1993a ).
Using this method, we now report the isolation and detailed characterization of ERS-24 (Endoplasmic Reticulum SNARE of 24 kD), a new mammalian v-SNARE that is localized to the ER and Golgi. ERS-24 is found in transport vesicles associated with the transitional areas of the ER and with the rims of Golgi cisternae, suggesting a role for ERS-24 in vesicular transport between these two compartments.
Materials and Methods
Preparation of Membranes, Recombinant NSF, and GST–α-SNAP
Triton X-100–solubilized total membrane extracts were prepared from CHO and RBL–2H3 cells as previously described (Söllner et al., 1993a ). Salt-washed dog pancreas microsomes were prepared as described (Walter and Blobel, 1983) and were a kind gift of Dr. Martin Wiedmann (Memorial Sloan-Kettering Cancer Center, New York). Recombinant (His)6-NSF-myc and (His)6-α-SNAP were prepared as previously described (Söllner et al., 1993a ). Recombinant glutathione-S-transferase (GST)–α-SNAP was constructed by inserting α-SNAP cDNA (Whiteheart et al., 1993) into a pGEX-2T vector (Pharmacia Fine Chemicals, Piscataway, NJ). The GST–α-SNAP fusion protein was expressed in Escherichia coli (XL1-BLUE) (Stratagene, La Jolla, CA) and purified from the bacterial extract on glutathione agarose beads (Sigma Chemical Co., St. Louis, MO).
Isolation of SNAREs from 20S Particles and Peptide Sequencing
Procedures used to purify SNAREs were essentially as described (Söllner et al., 1993a ), except that Triton X-100–solubilized total membrane extracts were prepared from RBL-2H3 cells and the reaction was scaled up by ∼10-fold. The detergent-solubilized total membrane extract was incubated with recombinant (His)6-α-SNAP and (His)6-NSF-myc in the reaction buffer as previously described (Söllner et al., 1993a ), except that recombinant (His)6-γ-SNAP was omitted. Subsequent binding of 20S complexes to mouse anti-myc mAb (9E10) (Evan et al., 1985) and specific elution of putative SNAREs from 20S particle in the presence of MgATP were performed as previously described (Söllner et al., 1993a ).
One of the released proteins migrated with an apparent molecular mass of 22 kD in 18% SDS-High-Tris-Urea-PAGE (Schlenstedt et al., 1990), and this protein was excised from a nitrocellulose filter after electroblotting and staining with Ponceau S (Sigma Chemical Co.) This protein band was further processed for internal amino acid sequence analysis essentially as described (Tempst et al., 1990; Erdjument-Bromage et al., 1994). Briefly, nitrocellulose-bound protein was in situ digested with trypsin, and the resulting peptides were separated by microbore reverse phase–HPLC (Lui et al., 1996; Elicone et al., 1994). Selected peptides were then subjected to chemical microsequencing as described (Tempst et al., 1994).
Isolation of cDNA Clones Encoding ERS-24
Peptide sequences from two tryptic fragments, T3 and T7, were used to generate degenerate PCR primers. These peptide sequences are underlined in Fig. 1. Degenerate oligonucleotide representing the sense primer (5′) ATGCA(A,G)GA(A,G)GA(C,T)GA(A,G)CA (3′) encoded the peptide sequence MQEDEQ in tryptic fragment T3. The antisense primer (3′) AA(A,G)TA(A,G,T)CT(C,T)AA(A,G)CT(C,T)TG (5′) was designed on the basis of the peptide sequence FIEFET in tryptic fragment T7. 0.5 μM each of sense and antisense PCR primers was used for PCR amplification. 5 μg of total cellular RNA prepared from RBL-2H3 cells was reverse transcribed as described (Sambrook et al., 1989), and the product was used as a template for PCR amplification. A major 300-bp amplification product was subcloned into pBluescript KS+ vector (Stratagene), subsequently randomly labeled, and used as a probe to screen a CHO cDNA phage library constructed with the Uni-ZAP XR vector (Stratagene).
Figure 1.
Peptide sequences of a putative SNARE, with an apparent molecular mass of 22 kD. Using detergent-solubilized total membrane fractions prepared from RBL-2H3 cells, putative SNAREs were isolated from 20S particles after MgATP hydrolysis by NSF. Peptide sequences obtained from a SNARE, with an apparent molecular mass of 22 kD (see Materials and Methods) are shown. An “x” means that no residue could be identified at this position. The peptide sequences used to design PCR primers are underlined.
Hybridization was done in duplicate filters at 60°C for 16–20 h in 5 × SSPE (1 × = 0.18 M NaCl, 1 mM EDTA, 10 mM NaH2PO4, pH 7.4), 5 × Denhardt's solution, and 0.5% SDS, in the presence of 20 mg/ml sonicated herring sperm DNA. The filters were washed twice in 2 × SSC (1 × = 0.15 M NaCl, 15 mM sodium citrate, pH 7.0) for 30 min at room temperature, followed by two washes at 60°C in 2 × SSC containing 0.1% SDS for 30 min, and finally washed twice at 60°C in 1 × SSC containing 0.1% SDS for 15 min each. About 50 positive clones were identified when 500,000 plaques were screened. Positive clones were excised from the Uni-ZAP XR vector as inserts in pBluescript SK− by coinfection with the helper phage, ExAssist, as described by the manufacturer (Stratagene), and then sequenced for both sense and antisense strands via the dideoxynucleotide method (Sanger et al., 1977).
Preparation of Antibodies and Western Blot Analysis
A fusion protein of ERS-24, ERS-24–myc(His)6, with a carboxy-terminal extension containing a myc epitope (EQKLISEEDL) (Evan et al., 1985) and six histidine residues, was constructed by inserting PCR-amplified ERS-24 cDNA into a modified pET3a vector (Novagen, Madison, WI; Søgaard et al., 1994). After overexpression in E. coli BL 21 (Novagen), recombinant ERS-24–myc(His)6 protein was purified by affinity chromatography using Ni-NTA-agarose as described by the manufacturer (Qiagen, Chatsworth, CA). ERS-24–myc(His)6 was further purified using a MonoQ column (Pharmacia Fine Chemicals) in a loading buffer containing 10 mM Bis-Tris-Propane, pH 7.0, and retrieved from the flow-through. The purified recombinant ERS-24–myc(His)6 protein was injected into rabbits for immunization. Positive antisera were purified by affinity chromatography with recombinant ERS-24–myc(His)6 conjugated to CNBr-activated Sepharose 4B (Pharmacia Fine Chemicals) (Harlow and Lane, 1988). The eluated antibodies were dialyzed against 10 mM MOPS buffer, pH 7.4, containing 50 mM KCl and 5% glycerol. The polyclonal anti–syntaxin 5 antibody was raised against the cytoplasmic domain of the hamster syntaxin 5 (residues 1–284) and affinity purified using the covalently coupled antigen as described above.
For Western blot analysis, detergent-solubilized total membrane extracts were fractionated on 18% High-Tris-Urea-SDS-PAGE (Schlenstedt et al., 1990; Laemmli, 1970) and blotted onto nitrocellulose filters. Filters were blocked with 5% milk powder in TBS and incubated with primary antibodies as indicated (affinity-purified polyclonal anti–ERS-24 antibody diluted 1:100; affinity-purified polyclonal anti–GOS-28 antibody [Nagahama et al., 1996] diluted 1:500; mAb (M3A5) recognizing β-COP [Duden et al., 1991] diluted 1:1,000; polyclonal anti–SSRα antibody [Wada et al., 1991] diluted 1:500; affinity-purified polyclonal anti–syntaxin 5 antibody diluted 1:100; polyclonal anti-p58 antibody [Saraste et al., 1987; Lahtinen et al., 1992] diluted 1:1,000; monoclonal anti-BiP (grp 78) antibody (StressGen Biotechnologies Corp., Victoria, British Columbia, Canada) diluted 1:200; and a polyclonal antibody against an amino-terminal peptide of BiP diluted 1:1,000). Immunoreactive bands were visualized with HRP-conjugated secondary antibodies (BioRad Laboratories, Hercules, CA) and enhanced chemiluminescence (ECL kit; Amersham Corp., Arlington Heights, IL).
Expression and Immunolocalization of the ERS-24–myc Fusion Protein in CHO Cells
A stable CHO cell line overexpressing epitope-tagged ERS-24 proteins (ERS-24–myc) was generated for immunolocalization studies. A myc epitope (EQKLISEEDL; Evan et al., 1985) encoding oligonucleotide was added by PCR to the 3′ end of the coding region with a flexible linker encoding three repetitive units of a Ser-Gly-Gly peptide (Weissman and Kim, 1992). The resulting constructs were subcloned into the mammalian cell expression vector, pcDNA-3 (Invitrogen, San Diego, CA). CHO cells were grown in MEM-α medium containing 10% FCS and were transfected with the epitope-tagged ERS-24 (ERS-24–myc in pcDNA-3) using the Lipofectamine reagent as described by the manufacturer (GIBCO BRL, Gaithersburg, MD). Cells were incubated in selection medium containing 1.0 mg/ml G418 sulfate (GIBCO BRL) 48 h after transfection, and subsequently subcloned using cloning cylinders 10–14 d after the initial transfection. These stably transfected cell lines were maintained in the presence of 0.5 mg/ml of G418 sulfate and examined for the expression of ERS-24–myc by Western blot analysis using both the affinity-purified polyclonal anti–ERS-24 antibody (1:100) and mAb 9E10 (1:500) (Evan et al., 1985).
Immunolocalization of ERS-24 was performed using a stable CHO cell line overexpressing the fusion ERS-24–myc protein. Indirect immunofluorescence localization was performed as described (Louvard et al., 1982; Warren et al., 1984). Cells were fixed with 3% paraformaldehyde (Polysciences, Inc., Warrington, PA) and stained with antibodies as indicated: affinity-purified polyclonal anti–ERS-24 antibody diluted 1:20, mAb antiBiP (grp-78) (StressGen) diluted 1:100, and mAb 9E10 diluted 1:100. The secondary antibodies, FITC-conjugated goat anti–rabbit IgG (Vector Labs) and Texas red–conjugated goat anti–mouse IgG (Molecular Probes, Eugene, OR), were diluted 1:100. The stained cells were observed with an Axiophot microscope using a ×100 objective oil immersion lens and a ×10 eye piece (Carl Zeiss, Inc., Thornwood, NY).
For EM immunolocalization studies, cells were fixed with 2% glutaraldehyde (Polysciences, Inc.) in phosphate buffer (pH 7.4), infiltrated with 2.3 M sucrose, and processed for cryoultramicrotomy according to the method of Tokuyasu (1986). Affinity-purified anti–ERS-24 antibodies (diluted 1:20) were localized with the protein A–gold method (Roth et al., 1978) or by goat anti–rabbit IgG-coupled gold. Double immunolabeling of ERS-24 and BiP was carried out by incubating thin cryosections with rabbit anti–ERS-24 (dilution 1:20) and mouse anti–BiP (dilution 1:50) antibodies, followed by gold-tagged goat anti–rabbit IgG (size of gold particles = 15 nm) and goat anti–mouse IgG (size of gold particles = 10 nm). After immunolabeling, sections were absorption stained with uranyl acetate (Tokuyasu, 1986) and examined in the electron microscope.
Cell Fractionation by Equilibrium Sucrose Density Gradient Centrifugation
Wild-type CHO cells and a stable CHO cell line overexpressing ERS-24– myc were used for the cell fractionation experiment. Postnuclear supernatants were prepared essentially as described (Dunphy and Rothman, 1983), layered on top of continuous sucrose density gradients (20–50% sucrose [wt/wt]), and centrifuged in an SW 28 rotor (Beckman Instruments, Inc., Palo Alto, CA) at 25,000 rpm (85,000 g) for 16 h. Gradient fractions were collected, and membranes were pelleted at 100,000 g for 1 h and resuspended in 2 × Laemmli buffer (Laemmli, 1970). Proteins were separated by 18% SDS-High-Tris-Urea-PAGE and processed for Western blot analysis.
Results
Isolation and Cloning of ERS-24
SNAREs were isolated from Triton X-100–solubilized whole cell membrane extracts prepared from RBL-2H3 cells, relying on their ability to assemble with SNAPs and NSF into 20S particles in the presence of ATPγS (Wilson et al., 1992; Söllner et al., 1993a ). Subsequent addition of MgATP resulted in the disassembly of the 20S complex and release of individual SNAREs. One of the released SNAREs migrated with an apparent molecular mass of 22 kD in 18% SDS-High-Tris-Urea-PAGE (Schlenstedt et al., 1990). After electroblotting, this protein band was excised from the nitrocellulose filter and processed for internal amino acid sequence analysis essentially as described (Tempst et al., 1990; Erdjument-Bromage et al., 1994; Söllner et al., 1993a ). Seven major tryptic fragments were analyzed by microsequencing (Fig. 1).
To isolate a cDNA clone encoding this protein, degenerate PCR primers were designed based on the peptide sequence MQEDEQ in tryptic fragment T3 and FIEFET in tryptic fragment T7 (Fig. 1 and Materials and Methods). These degenerate primers were used for PCR amplification from RBL-2H3 cDNA. One major PCR amplification product of 300 bp in length was obtained and used as a probe to screen a phage cDNA library from the CHO cell line (see Materials and Methods) to obtain full-length cDNA clones.
Four clones were analyzed by DNA sequencing. All four clones contain an identical open reading frame that encodes a polypeptide of 215 amino acids (Fig. 2). The predicted molecular mass of this protein is 24 kD and is in close agreement with the apparent size of 22 kD in an 18% SDS-High-Tris-Urea-PAGE. All seven tryptic peptide sequences previously identified by microsequencing were encoded within the open reading frame, clearly demonstrating that the cDNA clones encode the protein originally isolated from the 20S particles. A search of databases revealed no identical matches; hence, this protein is a novel mammalian protein. We now refer to this protein as ERS-24.
Figure 2.

Nucleotide and amino acid sequence of ERS-24. The nucleotide and amino acid sequence of a full-length cDNA clone encoding ERS-24 is shown. The amino acid sequence is shown in single-letter code above the nucleotide sequence; both are numbered on the right. The initiator methione is underlined; the asterisk after the amino acid sequence denotes a stop codon. The carboxy-terminal transmembrane region is boxed. The peptide sequences that were obtained from the SNARE isolated from the 20S particles (see Fig. 1) are underlined.
ERS-24 Is Homologous to Sec22p and rsec22
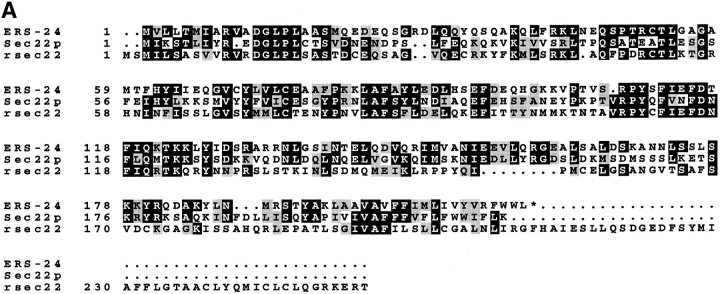
Comparison of the deduced polypeptide sequence of ERS-24 with the protein sequence data banks using the BLAST search program revealed that ERS-24 shares significant sequence homology to a known yeast v-SNARE, Sec22p (Newman et al., 1990, 1992; Dascher et al., 1991) (Fig. 3 A). These two proteins are overall 35% identical.
Figure 3.

ERS-24 and Sec22p share significant sequence homology and similar hydropathy profile. (A) A colinear alignment of polypeptide sequences of ERS-24, Sec22p, and rsec22. Identical amino acids are boxed in black; conservative changes are shaded in gray. ERS-24 and Sec22p are 35% identical; ERS-24 (from hamster) and rsec22 (from rat) are 35% identical; Sec22p and rsec22 are 32% identical. Of note, the long carboxy-terminal extension beyond the predicted transmembrane domain of rsec22 (Hay et al., 1996) is not present in ERS-24 and Sec22p. These sequence data are available from EMBL/DDBJ/GenBank under accession numbers P22214 (Sec22p), U42209 (rsec22), and U91742 (ERS-24). (B) Hydrophilicity/hydrophobicity profiles of ERS-24, Sec22p, and rsec22.
Furthermore, Kyte-Doolittle plots reveal that the carboxy-terminal two-thirds of these two proteins show almost identical hydrophobicity/hydrophilicity profiles (Fig. 3 B). Both proteins contain a carboxy-terminal hydrophobic segment that presumably acts as a membrane anchor. ERS-24 has a short stretch of hydrophobic amino acids at its amino-terminal end not found in Sec22p (Fig. 3, A and B). Both ERS-24 and Sec22p have heptad repeats of aliphatic residues in the carboxy-terminal region (residues 130–190) with a predicted propensity to form coiled-coils. ERS-24 has an additional region (amino acids 16–48) that has the potential to form coiled-coil structures. These regions are presumably important for protein–protein interactions (Lupas et al., 1991).
Recently, a rat homologue of Sec22p, rsec22 (Hay et al., 1996), that shares 32% sequence identity with yeast Sec22p (Fig. 3 A) was identified. Sequence alignment of ERS-24 (from hamster) and rsec22 (from rat) reveals that they are 35% identical, clearly demonstrating that ERS-24 and rsec22 are different proteins. rsec22 has at its carboxy terminus, beyond the putative transmembrane domain, a 46– amino acid–long extension that is absent in both ERS-24 and Sec22p (Fig. 3 A). The striking similarity in the hydrophobicity/hydrophilicity profiles of ERS-24 and Sec22p is not observed with rsec22 (Fig. 3 B).
ERS-24 is also homologous to a number of other putative v-SNAREs (18–28% sequence identity and 40–50% similarity). Homologous v-SNAREs include the SAR1 gene product of Arabidopsis thaliana (Schena and Davis, 1992) (no relation to the yeast GTPase also termed SAR1 [Barlowe et al., 1993]); p26/Ykt6, a prenylated SNARE in yeast (Søgaard et al., 1994); the yeast proteins Snc1p and Snc2p (Protopopov et al., 1993); the mammalian cellubrevin (McMahon et al., 1993); and the neuronal synaptobrevins/VAMPs 1 and 2 from many different sources (Trimble et al., 1988; Elferink et al., 1989; Baumert et al., 1989; Archer et al., 1990; Südhof et al., 1989). Interestingly, most of the homology resides within the carboxy-terminal onethird of these proteins (amino acid residues 130–190), while the amino-terminal halves of these proteins are widely divergent. In contrast, the identities between ERS-24, Sec22p, and rsec22 are evenly distributed over the entire proteins (Fig. 3 A).
ERS-24 Is a Type II Integral Membrane Protein
According to the amino acid sequence (Fig. 2), ERS-24 should behave as an integral membrane protein with its carboxy terminus anchored in the lipid bilayer. To test this, a polyclonal antibody was generated using a bacterially overexpressed recombinant ERS-24 protein. The affinitypurified antibody recognized only one band with an apparent molecular mass of 22 kD both in CHO and RBL-2H3 cells (Fig. 4 A), and one major band of 22 kD in dog pancreas microsomes (Fig. 4 B and C). When translocationcompetent dog pancreas microsomes were subjected to either high salt (1 M NaCl) or high pH (100 mM Na2CO3, pH 11.3) extraction (Fujiki et al., 1982), ERS-24 was found exclusively in the pellet fraction of a high speed centrifugation (100,000 g) (Fig. 4 B), indicating that it is an integral membrane protein. In contrast, BiP, a protein resident in the lumen of the ER (Haas and Wabl, 1983), was (as expected) found in the supernatant fraction under the same experimental conditions.
Figure 4.
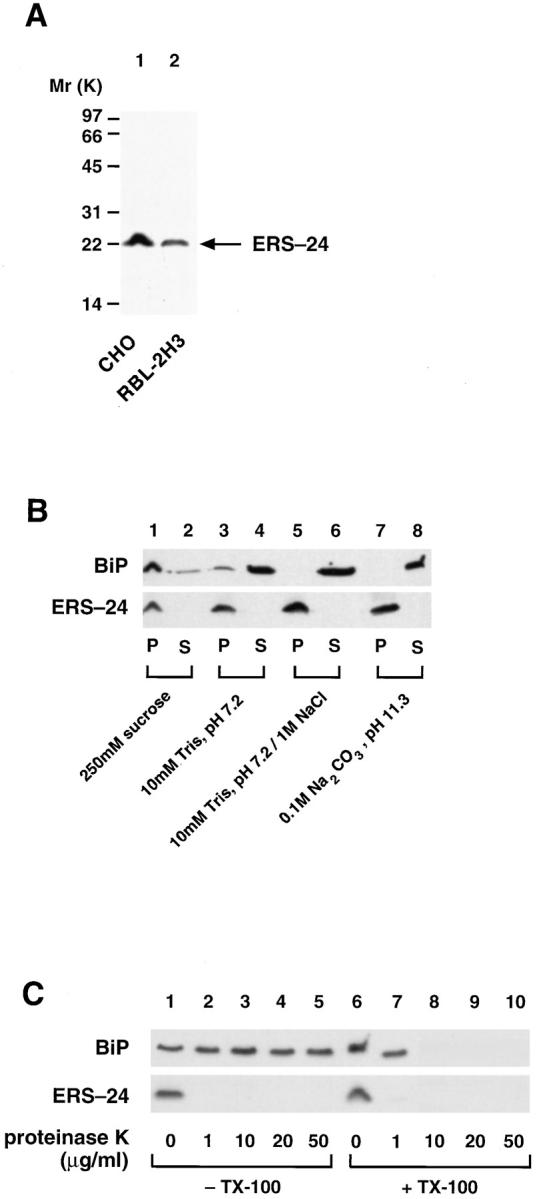
ERS-24 is a type II integral membrane protein with the bulk of its mass exposed to the cytoplasm. (A) Affinity-purified anti–ERS-24 antibodies recognize one distinct band in CHO and RBL-2H3 cells. Triton X-100–solubilized total membrane extracts (100 μg protein) from CHO and RBL-2H3 cells were fractionated on 18% SDS-High-Tris-Urea-PAGE, transferred onto nitrocellulose, and immunodecorated with affinity-purified anti– ERS-24 antibody. (B) ERS-24 is an integral membrane protein. Translocation-competent dog pancreas microsomes were incubated for 30 min on ice in the presence of the following buffers: 250 mM sucrose, 50 mM KOAc, 10 mM Hepes/KOH, pH 7.2 (lanes 1 and 2); 10 mM Tris-HCl, pH 7.2 (lanes 3 and 4); 1 M NaCl, 10 mM Tris-HCl, pH 7.2 (lanes 5 and 6); and 100 mM Na2CO3, pH 11.3 (lanes 7 and 8). Membranes were isolated by centrifugation at 100,000 g and supernatants were precipitated with TCA. Membranes (P) and supernatants (S) were analyzed by 18% SDS-High-Tris-Urea-PAGE, transferred onto nitrocellulose, and immunodecorated with either monoclonal anti-BiP (grp 78) antibodies (StressGen) (top) or affinity-purified polyclonal anti–ERS-24 antibodies ( bottom). (C) The bulk of ERS-24 is exposed to the cytoplasm. Translocation-competent, salt-washed dog pancreas microsomes (25 μg) were incubated with the indicated amounts (μg/ml) of proteinase K either in the presence (lanes 1–5) or absence (lanes 6–10) of Triton X-100. The proteinase K digest was carried out at room temperature for 10 min and terminated by addition of PMSF (1 mM). The reaction mixtures were TCA precipitated, fractionated on 18% SDS-High-Tris-Urea-PAGE, transferred onto nitrocellulose, and immunodecorated either with monoclonal anti-BiP (grp 78) antibody (StressGen) (top) or affinity-purified polyclonal anti–ERS-24 antibody (bottom).
Also, as expected for a SNARE (see Kutay et al., 1993), the NH2 terminus of ERS-24 is exposed to the cytoplasm. ERS-24 was sensitive to proteinase K (1 μg/ml) and was degraded either in the absence or presence of Triton X-100 (Fig. 4 C). As a control for the integrity of the microsomes used, BiP was found to be entirely protected from proteinase K digestion in the absence but not the presence of Triton X-100. These results demonstrate that the bulk of ERS-24 is cytoplasmically oriented, and that ERS-24 is membrane anchored by its hydrophobic COOH terminus as a type II membrane protein, a feature commonly noted in many SNAREs (Kutay et al., 1993).
ERS-24 Is Localized to the ER and the Golgi
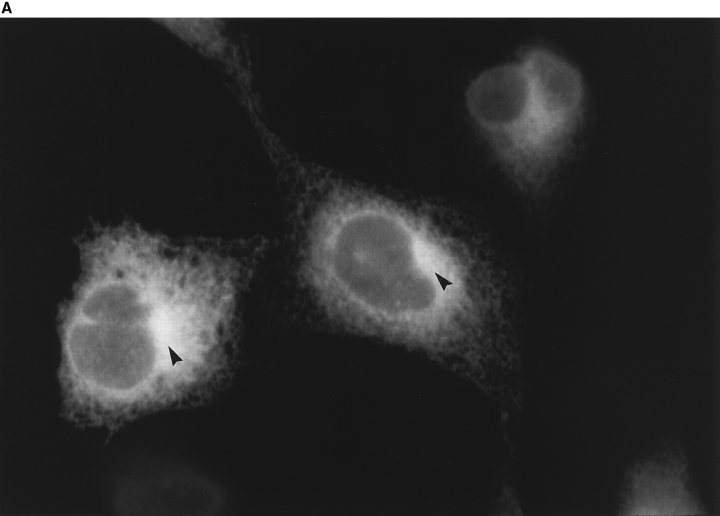
The sequence homology between ERS-24 and yeast Sec22p suggests that ERS-24, like Sec22p, plays a role in vesicle traffic between the ER and the Golgi, and thus should be associated, at least temporarily, with both compartments. Sec22p (along with Bos1p) has been found in the transport vesicles derived from the ER (Lian and Ferro-Novick, 1993; Barlowe et al., 1994), and the transport vesicles containing Bos1p and Sec22p have been shown to be required for fusion with a Golgi compartment (Lian and FerroNovick, 1993; Lian et al., 1994). Sec22p therefore has been previously assigned to the ER in yeast. Immunolocalization studies are difficult in yeast because of the absence of a morphologically well-defined Golgi. The isolation of ERS-24 in mammalian cells now provides an opportunity to perform more detailed localization studies. For these studies, it was necessary to overexpress ERS-24 since our polyclonal anti–ERS-24 antibody was not sensitive enough to detect ERS-24 in normal cells. We therefore used a stable CHO cell line that overexpresses an ERS-24 fusion protein, ERS-24–myc, containing a carboxy-terminal myc epitope (EQKLISEEDL; Evan et al., 1985). Immunofluorescence microscopy using the affinity-purified polyclonal antibody against ERS-24 showed a reticular staining pattern characteristic of the ER (Fig. 5 A). In addition, however, a clear juxtanuclear labeling pattern reminiscent of the Golgi (Fig. 5 A, arrowheads) was noted, suggesting that ERS-24–myc may reside in both of these compartments. Similar results were obtained with the monoclonal anti-myc (9E10) antibody (Evan et al., 1985; data not shown). In double labeling, a control monoclonal anti-BiP (grp 78) antibody (StressGen) directed against the peptide KSEKDEL found at the carboxy terminus of BiP and other luminal ER proteins (Munro and Pelham, 1987) highlighted the reticular network of the ER, but not the juxtanuclear region containing the Golgi (Fig. 5 B).
Figure 5.
Immunofluorescence localization of ERS-24 in CHO cells. Indirect immunofluorescence localization of ERS-24–myc and BiP in the stably transfected CHO cell line. Cells were double labeled with affinity-purified polyclonal anti–ERS-24 antibody and monoclonal anti-BiP (grp 78) antibody (StressGen). (A) ERS-24–myc–containing intracellular structures were visualized by incubating with FITC-conjugated, goat anti–rabbit IgG. (B) BiP-containing ER was visualized using Texas red–conjugated goat anti–mouse IgG. The distinct juxtanuclear regions, characteristic of the Golgi (arrowheads), are exclusively labeled by anti–ERS-24 antibody.
The subcellular localization of ERS-24–myc to the ER and the Golgi was firmly established by the immunoEM analysis. Fig. 6 A shows the colocalization of ERS-24–myc and BiP to the ER (left side). In comparison, only ERS24–myc is seen in the Golgi complex (Fig. 6 A, upper right). While ERS-24–myc is qualitatively present in multiple Golgi cisternae (Fig. 6 B) it seems to be concentrated at the fenestrated pole of the cis-most Golgi cisternae (Fig. 6 C, arrows). In addition, ERS-24–myc is found in vesicles associated with the terminal rim of the Golgi cisternae (Fig. 6 C, arrowhead). Quantitative analysis of the immuno-EM localization studies showed (see Table I) that the ERS-24–myc signal was enriched in these vesicles positioned lateral to the Golgi stack and in transfer vesicles found in the transition area of the ER. This labeling pattern is highly suggestive of an efficient incorporation of ERS-24–myc into transport vesicles, and is consistent with its predicted function as a v-SNARE involved in vesicle transport between the ER and the Golgi.
Figure 6.

Immuno-EM localization of ERS-24 to the ER and the Golgi region. (A) ImmunoEM visualization of the ER– Golgi area in a CHO cell line stably transfected with ERS-24– myc. Ultrathin cryosections of the glutaraldehyde-fixed cells were double labeled for BiP (10 nm gold) and ERS-24 (15 nm gold). The ER compartment to the middle left shows labeling for both BiP and ERS-24. By comparison, in the Golgi complex (G) to the upper right, only the ERS-24 label is present. B and C are single labeled for ERS-24. (B) Gold particles are present at various levels of a Golgi stack (G). (C) ERS-24 labeling is seen on both the fenestrated plate on the cis-Golgi pole and on vesicles associated with the terminal rim of the cisternae (right, arrowhead). Tubular profiles of the cis-Golgi plate in cross-section are indicated by arrows. Bars, 200 nm.
Table I.
Enrichment of ERS-24 in Transport Vesicles
| Cell compartment* | Density of ERS-24–myc | |
|---|---|---|
| number of gold particles per μm2 | ||
| Rough ER‡ | 36 ± 10 | |
| Transfer vesicles in the | ||
| transition area of ER | 63 ± 11 | |
| Vesicles positioned | ||
| lateral to the Golgi stack | 55 ± 10 | |
| Golgi stack | 20 ± 4 | |
| Nucleus§ | 5 ± 1 |
A CHO cell line stably transfected with ERS-24–myc was used for immuno-EM localization studies. Cells were fixed with 2% glutaraldehyde, infiltrated with 2.3 M sucrose, and processed for cryoultramicrotomy. Ultrathin cryosections were incubated with affinity-purified anti–ERS-24 antibody (diluted 1:20), followed by incubation with goat anti–rabbit IgG–gold (gold size 15 nm). Shown is the mean value of the number of gold particles per μm2 followed by the SEM.
Only the cells showing ERS-24 immunolabeling on the Golgi complex were included in this study. It was not infrequent to see cells with no labeling of the Golgi complex.
Including the nuclear envelope.
Except the nuclear envelope.
The localization of ERS-24 to the ER and the Golgi was independently confirmed by subcellular fractionation studies. In these studies, wild-type CHO cells and the stable CHO cell line overexpressing ERS-24–myc (the same cells used for immuno-EM) were used to control for potential changes in localization of ERS-24, resulting from overexpression or epitope tagging. Postnuclear fractions were fractionated by equilibrium density centrifugation using continuous sucrose gradients. Fig. 7, A and B, shows that resident proteins of the ER, such as BiP (Munro and Pelham, 1986) and SSR-α (Wada et al., 1991), and markers of the Golgi, such as syntaxin 5 (Banfield et al., 1994) and GOS-28 (Nagahama et al., 1996; Subramaniam et al., 1996), can be clearly separated on sucrose density gradients after equilibrium centrifugation. The endogenous ERS-24 in wild-type CHO cells has a broad distribution in this sucrose gradient, but nevertheless can be grouped into two populations (Fig. 7 A). One population of ERS-24 in the heavier sucrose density fractions shows overlapping, but not identical, distribution with proteins that had been previously localized to the ER, such as SSR-α (Wada et al., 1991) and BiP (Munro and Pelham, 1986). The second and most abundant population of ERS-24 is in the lighter sucrose density fractions and shows an overlapping distribution with several proteins previously localized to the Golgi region, including syntaxin 5 (Bennett et al., 1993; Banfield et al., 1994) and GOS-28 (Nagahama et al., 1996; same as GS-28, Subramaniam et al., 1996).
Figure 7.
Cofractionation of ERS-24 with markers of the ER and the Golgi. A Western blot analysis of the sucrose density gradient fractions of cell lysates prepared from the wild-type CHO cells (A) and a stable CHO cell line overexpressing ERS-24–myc fusion protein (B). Nitrocellulose filters were immunodecorated with polyclonal antibody against the amino-terminal portion of BiP, polyclonal anti-p58 antibody, polyclonal anti-SSRα antibody, affinity-purified polyclonal anti–syntaxin 5 antibody, affinity-purified polyclonal anti–GOS-28 antibody, and affinity-purified polyclonal anti–ERS-24 antibody.
The fusion protein, ERS-24–myc, and the endogenous ERS-24 share a similar bimodal subcellular distribution in the CHO cell line overexpressing the ERS-24–myc fusion protein (Fig. 7 B). Therefore, it is unlikely, although it cannot be excluded, that epitope tagging at the carboxy terminus per se alters the localization of ERS-24. However, it appears that the overexpression of total ERS-24 (endogenous plus recombinant ERS-24–myc) increases the relative amount of both the endogenous ERS-24 and the ERS24–myc fusion protein in the dense ER-containing sucrose fractions (Fig. 7 B). The reason for this is not clear. Nevertheless, endogenous ERS-24 (expressed at native levels) is largely Golgi localized, suggesting that the majority of native ERS-24 may be present in the Golgi at any one moment.
In sum, the morphological data and subcellular fractionation studies confirm that ERS-24 is localized to both the ER and the Golgi, suggesting a role for ERS-24 in the vesicle traffic between and possibly within these two compartments.
ERS-24 Interacts with Syntaxin 5 in 20S Particles and the ATPase Activity of NSF Destabilizes This Interaction
SNAREs are characterized by their ability to associate with SNAPs, particularly when they are assembled in v–t-SNARE complexes (Söllner et al., 1993b ; Hayashi et al., 1995). The resulting complex subsequently binds NSF, whose ATPase activity then releases SNAPs and the individual SNAREs (Söllner et al., 1993b ). Indeed, ERS-24 from Triton X-100 extracts of CHO cells likely assembled in a v–t-SNARE complex binds to α-SNAP (Fig. 8 A, lanes 3) and remains bound when NSF is added in the presence of ATPγS/ EDTA (Fig. 8 A, lane 4). In the presence of MgATP, NSF hydrolyzes ATP and ERS-24 is released from the immobilized α-SNAP (Fig. 8 A, lane 6). This confirms that ERS24 is an α-SNAP receptor and, when bound to SNAP (and possibly other proteins), is a substrate for NSF-dependent complex disruption.
Figure 8.
ERS-24 interacts with syntaxin 5 in 20S particles, and the ATPase activity of NSF disrupts this interaction. (A) The binding of ERS-24 from Triton X-100 extract of CHO cells to the recombinant GST–α-SNAP fusion protein and subsequent disassembly of ERS-24 from 20S particles in the presence of NSF and MgATP. Triton X-100–solubilized total CHO membranes (200 μg protein) and GST–α-SNAP fusion protein (1 μg) immobilized onto glutathione beads were incubated, either in the presence of ATPγS (lanes 1–4) or MgATP (lanes 5 and 6), in 1 ml buffer containing 20 mM Hepes-KOH, pH 7.0, 100 mM KCl, 1% glycerol, 1 mM DTT, 0.5% Triton X-100. To samples 2, 4, and 6, recombinant (His)6-NSF-myc (5 μg protein) was added. (Lanes 1 and 2) Controls for nonspecific binding to GST–glutathione beads in buffer containing ATPγS, either in the absence (lane 1) or presence (lane 2) of recombinant (His)6-NSF-myc. The samples were washed four times with 1 ml of the incubation buffer containing either ATPγS or MgATP, and then analyzed by Western blotting and immunodecoration with affinity-purified anti–ERS-24 antibody. Of note, the polyclonal anti–ERS-24 antibody, which was raised against ERS-24–myc (His)6, recognizes also the (His)6 tag or/ and myc tag of recombinant (His)6-NSF-myc. (B) The assembly of ERS-24 with syntaxin 5 in 20S particles in the presence of ATP/ EDTA and disruption of this interaction in the presence of the ATPase activity of NSF. Triton X-100–solubilized total CHO membranes (500 μg protein) were incubated with recombinant (His)6-α-SNAP and (His)6-NSF-myc (5 μg of each protein), either in the presence of ATP/EDTA (lane 1) or MgATP (lane 2), in the above-mentioned incubation buffer for 1 h at 4°C. Affinity-purified anti– ERS-24 antibody, covalently coupled to protein A–agarose (as described in Söllner et al., 1993), was added and the incubation was continued overnight at 4°C. The samples were washed four times with 1 ml of incubation buffer containing either ATP/EDTA (lane 1) or MgATP (lane 2), and the immunoprecipitate was analyzed by Western blotting. The blot was cut into two sections. The upper section, containing the higher M r proteins, was probed with affinity-purified anti–syntaxin 5 antibody, and the lower section containing the lower M r proteins was probed with affinity-purified anti–ERS-24 antibody. The anti–syntaxin 5 antibody recognized two protein bands. The band at 35 kD (syntaxin 5) comigrates with its bovine counterpart whose identity has been confirmed by peptide sequencing (data not shown). The nature of the 42 kD band (asterisk) is currently unknown.
In Fig. 8 B (lane 1), we show that ERS-24 interacts with syntaxin 5, the mammalian counterpart of Sed5p (Bennett et al., 1993; Banfield, 1994) in 20S docking and fusion particles that can be immunoprecipitated with antibody to ERS-24. Syntaxin 5 is released when NSF hydrolyzes ATP (Fig. 8 B, lane 2). This is in agreement with the previous observation that Sed5p forms a multimeric v- and t-SNARE complex in yeast containing Sec22p under nonpermissive temperature in NSF mutant sec18 strains (Søgaard, et al., 1994). The nature of the larger band (Fig. 8 B, asterisk), migrating at 42 kD is currently unknown, but it may represent a syntaxin homologue or a modified form of syntaxin 5. This protein also assembles into 20S particles with ERS-24 and undergoes ATP-dependent dissociation.
Discussion
In this paper we report the isolation and characterization of a new mammalian v-SNARE, ERS-24, that meets all of the known criteria for a SNARE including its ability to bind to α-SNAP and to assemble into a 20S particle. Its interaction with α-SNAP might be direct, or indirect with ERS-24 being assembled in a v–t-SNARE complex with other SNAREs. The ATPase activity of NSF disassembles the 20S particle and releases ERS-24. ERS-24 is primarily localized to the ER and the Golgi but may be mainly in the Golgi at any one moment when expressed at native levels. Overexpression for unknown reasons changes the distribution in favor of the ER.
ERS-24 is homologous to a large number of v-SNAREs but is most homologous to both yeast Sec22p (Newman et al., 1990; Dascher et al., 1991) and rsec22, another mammalian homologue of Sec22p (Hay et al., 1996) that is predominantly localized to the ER. These three proteins share sequence identities of ∼32–35%.
Sec22p is a v-SNARE required for the vesicle traffic between the ER and the Golgi (Lian and Ferro-Novick, 1993; Barlowe et al., 1994). The function of rsec22 has not been determined. Given this level of identity, it is not possible to say whether or not both ERS-24 and rsec22 are functionally equivalent (in mammalian cells) to Sec22p (in yeast), or whether they serve a different (but similar) role to the true homologue of Sec22p in mammals.
While there is no formal proof that ERS-24 is a v-SNARE needed for the vesicle traffic between the ER and the Golgi, the significant homology to Sec22p, its interaction with syntaxin 5, and its localization to these organelles suggests this possibility. Such proof awaits the demonstration that ERS-24 is packaged into the COPI- and/or COPIIcoated vesicles that depart the ER, as has been shown for Sec22p (Barlowe et al., 1994; Bednarek et al., 1995), and is needed for attachment to Golgi membranes, as has been shown with the transport vesicles containing Bos1p and Sec22p in yeast (Lian and Ferro-Novick, 1993). Nevertheless, we have shown that ERS-24 is enriched in vesicles in the transition area of the ER and in vesicles associated with the terminal rim of the Golgi cisternae in vivo, and it is incorporated into Golgi-derived COPI-coated transport vesicles formed in vitro (data not shown), suggesting that ERS-24 cycles between the ER and the Golgi, as expected for a v-SNARE shuttling cargo molecules between these two compartments.
EM shows that ERS-24 is not strictly restricted to the cis-pole of the Golgi, as one might simply expect for a v-SNARE exclusively involved uni- or bidirectionally in transport between ER and Golgi. Whether ERS-24 plays a functional role in intercisternal transport is unknown, even if it is localized in post–cis-Golgi vesicles to some extent. The presence of ERS-24 in later Golgi cisternae could easily be explained by a retrieval system that is <100% efficient, as for escaped ER resident proteins. Depending on the probability with which ERS-24 enters retrograde transport vesicles, a more or less steep concentration gradient of ERS-24 across the Golgi stack would be established in distillation (Rothman, 1981; Rothman and Wieland, 1996). Since ERS-24 is enriched in vesicles and buds at the rims of the Golgi, an exact assignment to individual cisternae is very difficult, thereby precluding the kind of quantitation that would be necessary to establish the definitive extent of an ERS-24 concentration gradient across the Golgi stack.
Regarding the question of whether ERS-24 could play an active role in targeting transport vesicles derived from the Golgi to their destination, or is rather just a passenger, i.e., a component of transport machinery that needs to be recycled, it is noteworthy, although not conclusive, that neither anti–ERS-24 antibodies nor the cytoplasmic domain of ERS-24 inhibited the cell-free Golgi transport reaction or the assembly of ERS-24 into 20S particles (data not shown). This in no way reduces the likelihood of the possibility that ERS-24 could serve as a v-SNARE targeting vesicles that budded from any level of the Golgi stack to the ER, among other possibilities. To address such issues, it will be necessary to identify additional components that are temporally associated with and/or regulate the activities of ERS-24. For example, in yeast Sed5p interacts with Sec22p and several other (predicted) v-SNAREs including Bos1p, Bet1p, and p26/Ykt6p, and is implicated in the vesicle traffic between the ER and the Golgi (Søgaard et al., 1994). ERS-24, a potential mammalian homologue of Sec22p, similarly interacts with syntaxin 5, a likely counterpart of Sed5p, in 20S docking and fusion particles. Whether syntaxin 5 is the sole partner t-SNARE interacting with ERS-24 remains to be determined. The identification of other potential interacting partners of ERS-24 shall provide insight into the function of ERS-24 in the vesicle flow between the ER and the Golgi and possibly within successive layers of the Golgi, increasing our understanding about how the activity of ERS-24 is regulated.
Acknowledgments
We are grateful to Dr. Masami Nagahama for anti–GOS-28 and anti–syntaxin 5 antibodies, as well as generous sharing of other reagents. We thank Drs. Mark Stamnes, James McNew, Thomas Weber, Michael Gmachl, and other members of the laboratory for helpful discussion and critical reading of the manuscript. We also thank Dr. Martin Wiedmann for dog pancreas microsomes and the anti-BiP antibody recognizing the amino-terminal portion of BiP; Dr. Michael Shia for the anti-SSRα (TRAPα) antibody; Dr. Jaakko Saraste for the anti-p58 antibody; Dr. Henry Metzger for RBL-2H3 cells; and Mary Lui for expert assistance in protein sequencing.
This work was supported by a National Institutes of Health (NIH) grant (to J.E. Rothman), the Mathers Charitable Foundation (to J.E. Rothman), the International Human Frontier Science Program (to J.E. Rothman and L. Orci), the Swiss National Science Foundation grant 3136665 (to L. Orci), a National Science Foundation grant BIR-9420123 (to P. Tempst), a National Cancer Institute core grant 5 P30 CA08748-29 (to P. Tempst), the Howard Hughes Medical Institute fellowship for physicians (to I. Paek), and the NIH clinical and molecular cancer research grant K12-CA-0172-01 (to Memorial Sloan-Kettering Cancer Center and to I. Paek).
Footnotes
1. Abbreviations used in this paper: GST, glutathione-S-transferase; NSF, N-ethylmaleimide–sensitive factor; SNAP, soluble NSF attachment protein; SNARE, SNAP receptor.
Please address all correspondence to James E. Rothman, Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, 1275 York Avenue, Box 251, New York 10021. Tel.: (212) 6398598. Fax: (212) 717-3604.
References
- Aalto MK, Ronne H, Keränen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO (Eur Mol Biol Organ) J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer, B.T., III, T. Ozcelik, R. Jahn, U. Francke, and T.C. Sudhof. 1990. Structures and chromosomal localizations of two human genes encoding synaptobrevins 1 and 2. J. Biol. Chem. 265:17267–17273. [PubMed]
- Banfield D, Lewis M, Rabouille C, Warren G, Pelham H. Localization of Sed5, a putative vesicle targeting molecule, to the cis-Golgi network involves both its transmembrane and cytoplasmic domains. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, d'Enfert C, Schekman R. Purification and characterization of SAR1p, a small GTP-binding protein required for transport vesicle formation. J Biol Chem. 1993;268:873–879. [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the ER. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Baumert M, Maycox PR, Navone F, DeCamilli P, Jahn R. Synaptobrevin: an integral membrane protein of 18,000 daltons present in small synaptic vesicles of rat brain. EMBO (Eur Mol Biol Organ) J. 1989;8:379–384. doi: 10.1002/j.1460-2075.1989.tb03388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek SY, Ravazzola M, Hosobuchi M, Amherdt M, Perrelet A, Schekman R, Orci L. COPI and COPII-coated vesicles bud directly from the ER in yeast. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Bennett MK, Garcia-Arrarás JE, Elferink LA, Peterson K, Fleming AM, Hazuka CD, Scheller RH. The syntaxin family of vesicular transport receptors. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci USA. 1988;85:7852–7856. doi: 10.1073/pnas.85.21.7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Sec9 is a SNAP-25-like component of a yeast SNARE complex that may be the effector of Sec4 function in exocytosis. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- Clary DO, Rothman JE. Purification of three related peripheral membrane proteins needed for vesicular transport. J Biol Chem. 1990;265:10109–10117. [PubMed] [Google Scholar]
- Clary DO, Griff IC, Rothman JE. SNAPs, a family of NSF attachment proteins involved in intracellular membrane fusion in animals and yeast. Cell. 1990;61:709–721. doi: 10.1016/0092-8674(90)90482-t. [DOI] [PubMed] [Google Scholar]
- Dascher C, Ossig R, Gallwitz D, Schmitt HD. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Griffiths G, Frank R, Argos P, Kreis TE. β-COP, a 110 kD protein associated with non-clathrin-coated vesicles and the Golgi complex, shows homology to β-adaptin. Cell. 1991;64:649–665. doi: 10.1016/0092-8674(91)90248-w. [DOI] [PubMed] [Google Scholar]
- Dunphy WG, Rothman JE. Compartmentation of asparaginelinked oligosaccharide processing in the Golgi apparatus. J Cell Biol. 1983;97:270–275. doi: 10.1083/jcb.97.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink LA, Trimble WS, Scheller RH. Two vesicle-associated membrane protein genes are differentially expressed in the rat central nervous system. J Biol Chem. 1989;264:11061–11064. [PubMed] [Google Scholar]
- Elicone C, Lui M, Geromanos S, Erdjument-Bromage H, Tempst P. Microbore reversed-phase high performance liquid chromatographic purification of peptides for combined chemical sequencing/laser-desorption mass spectrometric analysis. J Chromatogr. 1994;676:121–137. doi: 10.1016/0021-9673(94)00089-1. [DOI] [PubMed] [Google Scholar]
- Erdjument-Bromage H, Lui M, Sabatini DM, Snyder SH, Tempst P. High-sensitivity sequencing of large proteins: partial structure of the rapamycin-FKBP12 target. Protein Sci. 1994;3:2435–2446. doi: 10.1002/pro.5560031227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;4:2843–2850. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to ER. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas IG, Wabl M. Immunoglobulin heavy chain binding protein. Nature (Lond) 1983;306:387–389. doi: 10.1038/306387a0. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and O. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Hata Y, Slaughter CA, Südhof TC. Synaptic vesicle fusion complex conatins unc-18 homologue bound to syntaxin. Nature (Lond) 1993;366:347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- Hay JC, Hirling H, Scheller RH. Mammalian vesicle trafficking proteins of the ER and Golgi apparatus. J Biol Chem. 1996;271:5671–5679. doi: 10.1074/jbc.271.10.5671. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Yamasaki S, Nauenburg S, Binz T, Niemann H. Disassembly of the reconstituted synaptic vesicle membrane fusion complex in vitro. . EMBO (Eur Mol Biol Organ) J. 1995;14:2317–2325. doi: 10.1002/j.1460-2075.1995.tb07226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Rapoport TA. A class of membrane proteins with a C-terminal anchor. Trends Cell Biol. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lahtinen U, Dahllof B, Saraste J. Characterization of a 58 kDa cisGolgi protein in pancreatic exocrine cells. J Cell Sci. 1992;103:321–333. doi: 10.1242/jcs.103.2.321. [DOI] [PubMed] [Google Scholar]
- Lian JP, Ferro-Novick S. Bos1p, an integral membrane protein of the ER to Golgi transport vesicles, is required for their fusion competence. Cell. 1993;73:735–745. doi: 10.1016/0092-8674(93)90253-m. [DOI] [PubMed] [Google Scholar]
- Lian JP, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Ypt 1p implicated in v-SNARE activation. Nature (Lond) 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- Louvard D, Reggio H, Warren G. Antibodies to the Golgi complex and the ER. J Cell Biol. 1982;92:92–107. doi: 10.1083/jcb.92.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui M, Tempst P, Erdjument-Bromage H. Methodical analysis of protein-nitrocellulose interactions to design a refined digestion protocol. Anal Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled-coils from protein sequences. Science (Wash DC) 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Malhotra V, Orci L, Glick BS, Block MR, Rothman JE. Role of an N-ethylmaleimide-sensitive transport component in promoting fusion of transport vesicles with cisternae of the Golgi stack. Cell. 1988;54:221–227. doi: 10.1016/0092-8674(88)90554-5. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Ushkaryov YA, Edelmann L, Link E, Binz T, Niemann H, Jahn R, Südhof TC. Cellubrevin is a ubiquitous tetanus-toxin substrate homologous to a putative synaptic vesicle fusion protein. Nature (Lond) 1993;364:346–349. doi: 10.1038/364346a0. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HRB. An Hsp70-like protein in the ER: identity with the 78 kD glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 1986;46:291–300. doi: 10.1016/0092-8674(86)90746-4. [DOI] [PubMed] [Google Scholar]
- Munro S, Pelham HRB. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- Nagahama M, Orci L, Ravazzola M, Amherdt M, Lacomis L, Tempst P, Rothman JE, Söllner TH. A v-SNARE implicated in intraGolgi transport. J Cell Biol. 1996;133:507–516. doi: 10.1083/jcb.133.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the ER to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Graf J, Mancini P, Rossi G, Lian JP, Ferro-Novick S. SEC22 and SLY2are identical. Mol Cell Biol. 1992;12:3663–3664. doi: 10.1128/mcb.12.8.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J, Hsu SC, Braun JE, Calakas N, Ting AE, Bennett MK, Scheller RH. Specificity and regulation of a synaptic vesicle docking complex. Neuron. 1994;13:353–361. doi: 10.1016/0896-6273(94)90352-2. [DOI] [PubMed] [Google Scholar]
- Protopopov V, Govindan B, Novick P, Gerst JE. Homologs of the synaptobrevin/VAMP family of synaptic vesicle proteins function on the late secretory pathway in S. cerevisiae. . Cell. 1993;74:855–861. doi: 10.1016/0092-8674(93)90465-3. [DOI] [PubMed] [Google Scholar]
- Roth J, Bendayan M, Orci L. Ultrastructural localization of intracellular antigens by the use of protein A-gold complex. J Histochem Cytochem. 1978;26:1074–1081. doi: 10.1177/26.12.366014. [DOI] [PubMed] [Google Scholar]
- Rothman JE. The Golgi apparatus: two organelles in tandem. Science (Wash DC) 1981;213:1212–1219. doi: 10.1126/science.7268428. [DOI] [PubMed] [Google Scholar]
- Rothman, J.E., editor. 1992. Reconstitution of intracellular transport. In Methods in Enzymology. Academic Press, Inc., San Diego. 219.
- Rothman JE. Mechanisms of intracellular protein transport. Nature (Lond) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science (Wash DC) 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 545 pp.
- Sanger F, Nicklen S, Coulson A. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso 1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste J, Palade GE, Farquhar MG. Antibodies to rat pancreas Golgi subfractions: identification of a 58-kD cis-Golgi protein. J Cell Biol. 1987;105:2021–2029. doi: 10.1083/jcb.105.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Davis RW. HD-Zip proteins: members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci USA. 1992;89:3894–3898. doi: 10.1073/pnas.89.9.3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Gudmundsson GH, Boman HG, Zimmermann R. A large presecretory protein translocates both cotranslationally, using signal recognition particle and ribosome, and post-translationally, without these ribonucleoparticles, when synthesized in the presence of mammalian microsomes. J Biol Chem. 1990;265:13960–13968. [PubMed] [Google Scholar]
- Scheller RH. Membrane trafficking in the presynaptic nerve terminal. Neuron. 1995;14:893–897. doi: 10.1016/0896-6273(95)90328-3. [DOI] [PubMed] [Google Scholar]
- Søgaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Söllner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature (Lond) 1993a;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Söllner T, Bennett M, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993b;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Subramaniam VN, Peter F, Philp R, Wong SH, Hong W. GS28, a 28-kilodalton Golgi SNARE that participates in ER-Golgi transport. Science (Wash DC) 1996;272:1161–1163. doi: 10.1126/science.272.5265.1161. [DOI] [PubMed] [Google Scholar]
- Südhof TC. The synaptic vesicle cycle: a cascade of protein-protein interactions. Nature (Lond) 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- Südhof TC, Baumert M, Perin MS, Jahn R. A synaptic vesicle membrane protein is conserved from mammals to Drosophila. . Neuron. 1989;2:1475–1481. doi: 10.1016/0896-6273(89)90193-1. [DOI] [PubMed] [Google Scholar]
- Tempst P, Link AJ, Riviere LR, Fleming M, Elicone C. Internal sequence analysis of proteins separated on polyacrylamide gels at the submicrogram level: improved methods, applications, and gene cloning strategies. Electrophoresis. 1990;11:537–553. doi: 10.1002/elps.1150110704. [DOI] [PubMed] [Google Scholar]
- Tempst P, Geromanos S, Elicone C, Erdjument-Bromage H. Improvements in microsequence for low picomole sequence analysis. Methods. 1994;6:248–261. [Google Scholar]
- Tokuyasu KT. Application of cryo-ultramicrotomy to immunocytochemistry. J Microsc. 1986;143:143–149. doi: 10.1111/j.1365-2818.1986.tb02772.x. [DOI] [PubMed] [Google Scholar]
- Trimble WS, Cowan DM, Scheller RH. VAMP-1: a synaptic vesicle-associated integral membrane protein. Proc Natl Acad Sci USA. 1988;85:4538–4542. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada I, Rindress D, Cameron PH, Ou WJ, Doherty J, Louvard D, Bell AW, Dignard D, Thomas DY, Bergeron JJ. SSRa and associated calnexin are major calcium binding proteins of the ER membrane. J Biol Chem. 1991;266:19599–19610. [PubMed] [Google Scholar]
- Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Warren G, Davoust J, Cockcroft A. Recycling of transferrin receptors in A431 cells is inhibited during mitosis. EMBO (Eur Mol Biol Organ) J. 1984;3:2217–2225. doi: 10.1002/j.1460-2075.1984.tb02119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman JS, Kim PS. The Pro region of BPTI facilitates folding. Cell. 1992;71:841–851. doi: 10.1016/0092-8674(92)90559-u. [DOI] [PubMed] [Google Scholar]
- Whiteheart SW, Griff IC, Brunner M, Clary DO, Mayer T, Buhrow SA, Rothman JE. SNAP family of NSF attachment proteins includes a brain-specific isoform. Nature (Lond) 1993;362:353–355. doi: 10.1038/362353a0. [DOI] [PubMed] [Google Scholar]
- Wilson DW, Whiteheart SW, Wiedmann M, Brunner M, Rothman JE. A multisubunit particle implicated in membrane fusion. J Cell Biol. 1992;117:531–538. doi: 10.1083/jcb.117.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]