Abstract
Nuclear lamins are thought to play an important role in disassembly and reassembly of the nucleus during mitosis. Here, we describe a Drosophila lamin Dm0 mutant resulting from a P element insertion into the first intron of the Dm0 gene. Homozygous mutant animals showed a severe phenotype including retardation in development, reduced viability, sterility, and impaired locomotion. Immunocytochemical and ultrastructural analysis revealed that reduced lamin Dm0 expression caused an enrichment of nuclear pore complexes in cytoplasmic annulate lamellae and in nuclear envelope clusters. In several cells, particularly the densely packed somata of the central nervous system, defective nuclear envelopes were observed in addition. All aspects of the mutant phenotype were rescued upon P element-mediated germline transformation with a lamin Dm0 transgene. These data constitute the first genetic proof that lamins are essential for the structural organization of the cell nucleus.
Lamins are the major structural proteins of the nuclear lamina, which lines the nucleoplasmic surface of the inner nuclear membrane in higher eukaryotic cells. The nuclear lamina is composed of a meshwork of 10 nm filaments that is thought to provide a skeletal support for the nuclear envelope and to mediate the attachment of the nuclear envelope to interphase chromatin (Aebi et al., 1986; Krohne and Benavente, 1986; Gerace and Burke, 1988; Paddy et al., 1990). Additional functions of the nuclear lamina may include the proper organization and anchoring of nuclear pore complexes (NPCs;1 Aaronson and Blobel, 1975; Aebi et al., 1986; Goldberg and Allen, 1992). During mitosis the lamins also play a crucial role in the disassembly and reassembly of the nuclear envelope (Gerace et al., 1978; Krohne et al., 1978; Gerace and Blobel, 1980).
Sequence comparison and biochemical data indicate that lamin proteins belong to the intermediate filament gene superfamily characterized by a central α-helical rod domain containing heptad repeats (Fisher et al., 1986; McKeon et al., 1986; Franke, 1987; for review see Fuchs and Weber, 1994). A lamin-like protein is thought to constitute the progenitor of the intermediate filament proteins (Weber et al., 1989a ; Dodemont et al., 1990; Döring and Stick, 1990). Highly specific features of lamins include a nuclear localization signal, a COOH-terminal CaaX sequence (C, cysteine; a, aliphatic; X, any amino acid) and characteristic phosphorylation sites in the NH2-terminal head and COOHterminal tail domains. The nuclear localization signal is responsible for rapid transport of lamins into the nucleus, thus preventing cytoplasmic assembly (Loewinger and McKeon, 1988). Modification by isoprenylation and carboxymethylation at the CaaX motif targets lamins to the inner nuclear membrane (Loewinger and McKeon, 1988; Holtz et al., 1989; Krohne et al., 1989; Kitten and Nigg, 1991). The interaction of lamins with the inner nuclear membrane may in addition be supported by integral membrane proteins, e.g., the putative lamin receptor p58 (Senior and Gerace, 1988; Worman et al., 1988; Bailer et al., 1991) or the lamina-associated proteins (LAPs; Foisner and Gerace, 1993). Hyper- and dephosphorylation processes at specific sites close to the lamin's central rod domain are important for regulating the nuclear envelope assembly and disassembly during mitosis (Gerace and Blobel, 1980; Smith and Fisher, 1989; Heald and McKeon, 1990; Peter et al., 1990; for review see Nigg, 1992). The binding of soluble- and/or vesicle-associated lamin to chromosomes is thought to play a critical role during reassembly of the nuclear envelope at the end of mitosis (Gerace and Blobel, 1980; Benavente and Krohne, 1986; Burke, 1990; Glass and Gerace, 1990; Höger et al., 1991) in which the LAPs may also be involved (Foisner and Gerace, 1993). Additionally, lamins seem to be required for DNA replication (Meier et al., 1991; Moir et al., 1994; for review see Hutchison, 1994). However, the precise roles of lamins in nuclear envelope assembly, chromosome condensation, and DNA replication remain to be unraveled.
Vertebrates have two main types of lamins, i.e., A-type and B-type lamins (Gerace et al., 1978; Gerace and Blobel, 1980; Krohne and Benavente, 1986). The developmentally regulated A-type lamins include the lamin A precursor lamin A0 and its alternative splice variant lamin C (Fisher et al., 1986; McKeon et al., 1986; Röber et al., 1989; Lin and Worman, 1993), whereas the constitutively expressed lamins B1 and B2 are products of two distinct genes (Höger et al., 1988, 1990; Zewe et al., 1991; Lin and Worman, 1995). Lamin C, in contrast to lamins A and B, has no COOH-terminal CaaX motif. In case of lamin A, this isoprenylation motif is removed by proteolytic trimming after association with the nuclear lamina (Weber et al., 1989b ; Hennekes and Nigg, 1994). While lamins A and C are soluble during mitosis, the B-type lamins remain membrane associated throughout the cell cycle (Gerace and Blobel, 1980; Burke and Gerace, 1986).
In the invertebrate Drosophila melanogaster, two lamin genes are known. A recently described lamin C gene is expressed only late during embryonic development and thus may constitute an analog of vertebrate A type lamins (Bossie and Sanders, 1993; Riemer et al., 1995). Like the vertebrate lamin C, Drosophila lamin C lacks a COOH-terminal isoprenylation motif. In contrast, the early and ubiquitously expressed Dm0 lamin gene localized at position 25F on the left arm of chromosome 2 encodes a polypeptide precursor of 621 amino acids containing a COOH-terminal CaaX sequence (Gruenbaum et al., 1988; Osman et al., 1990). Both its constitutive expression and the presence of the CaaX motif classify lamin Dm0 as the equivalent of vertebrate lamins B. Proteolytic processing of the Dm0 precursor in the cytoplasm followed by differential phosphorylation in the nucleus generates different mature isoforms (Dm1 and Dm2) that are specifically found in interphase and mitotic nuclei (Smith et al., 1987; Smith and Fisher, 1989).
Because of their central role in nuclear function and cell division, the genetic analysis of lamins has been proven difficult. Here we report the serendipitous isolation and characterization of a Drosophila lamin mutant resulting from a P element insertion into the first intron of the Dm0 gene. Flies homozygous for this mutation show a severe lamin deficiency resulting in impaired viability, fertility, and locomotion. Ultrastructural analysis of the mutant central nervous system indicates that the lamin Dm0 gene product is essential for the structural integrity of the nuclear envelope and the proper integration of NPCs into the nuclear membrane. In addition, annulate lamellae, membranous cisternae containing pore complexes, are enriched in the cytoplasm of the mutant cells.
Materials and Methods
Fly Stocks
All genetic markers used for P element mutagenesis are described in Lindsley and Zimm (1992). The null white allele w1118, the transposaseproviding stock ry506 Sb P(ry + Δ2-3), the inversion In(2LR)Gla used as balancer, and the wild-type (wt) strain Oregon R were obtained through the Bloomington and Umëa stock collections. The mlac strain with four P-lacW elements on the X chromosome was obtained from M. Brandt (Baylor College of Medicine, Houston, TX) via E. Hafen (University of Zürich, Switzerland). The following strains were used throughout this study (synonym in bold letters): w1118: control flies (unbalanced): w1118/ w1118(−); +/+; +/+; In (2LR)Gla: balanced control flies: w1118/w1118 (−); +/In(2LR)Gla, Gla; +/+; lamP: homozygous mutant flies: w1118/w1118(−); lamP-lacW/lamP -lacW; +/+ and heterozygous/balanced mutant flies: w1118/ w1118(−); lamP-lacW/In(2LR)Gla, Gla; +/+; Tw2-lamP: homozygous rescue flies: w1118/w1118(−); lamP-lacW/lamP-lacW; P-lam +/P-lam +, and heterozygous rescue flies: w1118/w1118(−); lamP-lacW/In(2LR)Gla, Gla; P-lam +/+ (P- lam +); wt: Oregon R.
P Element Insertion Screen
P element mutagenesis for autosomal insertions was performed according to standard methods as described (Zinsmaier et al., 1994). P-lacW transposable elements (Bier et al., 1989) were mobilized from the X chromosome with the transposase-providing ry506 Sb P(ry + Δ2-3) element at 99B (Robertson et al., 1988). Jump male progeny of w1118 background and autosomal P-lacW insertions identified by pigmented eyes were crossed in batches of 50 g to 150 w1118 females per batch. For plasmid rescue of genomic fragments flanking the insertion site, genomic DNA prepared from overnight egg lays of the individual batches was restricted with EcoRI, ligated, and transformed into bacteria by electroporation. Plasmid DNA prepared from pooled colonies obtained with the different fly batches was digested with EcoRI and analyzed on Southern blots. Digoxygenin-labeled genomic probes of the glutamate receptor subunits DGluR-I (Ultsch et al., 1992), DGluR-II (Schuster et al., 1991), and DNMDAR-I (Ultsch et al., 1993) were used for hybridization. With the DGluR-II probe localized at 25F1-2 (Schuster et al., 1991), one positive pool was obtained; individual lines were established thereof by single crosses followed by Southern blot analysis. The resulting mutant line carrying a P-lacW element insertion at 25F was balanced with the inversion In(2LR)Gla. Plasmid DNA derived by plasmid rescue from the positive single line was sequenced using a 31mer oligonucleotide primer complementary to the P element's inverted repeat sequence (5′-CGACGGGACCACCTTATGTTATTTCATCATG3′). Sequences were analyzed using the HUSAR program package of the Deutsches Krebsforschungszentrum (Heidelberg, Germany). A European Molecular Biology Laboratory DNA library search using the BLAST program revealed identity to sequences of the lamin Dm0 gene (Osman et al., 1990).
Northern Blot Analysis
Total RNA was isolated from homozygous and heterozygous lamP mutant flies and from w1118 and In(2LR)Gla control flies, as described (Sass et al., 1990). RNA (20 μg/lane) was separated on a 1% agarose/5% (wt/vol) formaldehyde gel and transferred onto a Pall A nylon membrane (Biodyne, Santa Monica, CA). The filter was hybridized to an in vitro transcribed antisense RNA probe of the lamin Dm0 gene (see Fig. 1 A, probe 1) at 65°C in the presence of 50% (wt/vol) formamide as described (Wismar et al., 1995). The probe was 32P labeled using the Riboprobe system (Promega Biotech, Madison, WI) according to the manufacturer's protocol. To normalize for relative amounts of total RNA applied, the blots were rehybridized with a 32P-labeled α-tubulin antisense RNA probe.
Figure 1.
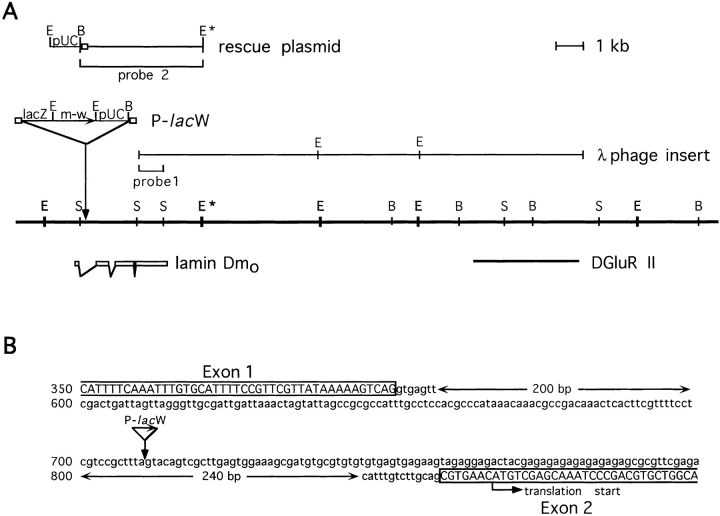
P-lacW insertion into the lamin Dm0 gene. (A) Schematic diagram of the genomic region at position 25F1-2 of the second chromosome covering the lamin Dm0 and the DGluR-II loci. Exonic regions of the lamin gene are indicated by open boxes below the restriction map. The restriction sites given are: E, EcoRI; B, BamHI; S, SalI. E* denotes a polymorphic EcoRI restriction site present in wt but not the w1118 strains. The P-lacW element is inserted into the first intron of the lamin gene (solid arrow). A horizontal arrow indicates the 5′ to 3′ orientation of the β-galactosidase (lacZ) and miniwhite (m-w) genes, and small boxes denote the inverted repeats of the P element. The λ phage insert, originally isolated for DGluR-II, extends into the lamin Dm0 gene region and hybridizes to the rescue plasmid derived from pUC sequences and adjacent genomic regions. Hybridization probes used for Northern (probe 1) and Southern (probe 2) blot analysis are indicated. (B) Precise location of the P-lacW insertion in the Dm0 genomic sequence. The P element is inserted into the center of the first intron of the lamin gene. Open boxes and capital letters indicate exonic regions. The translation start site is found in the second exon.
Western Blot Analysis
Individual flies were homogenized in 100 μl of sample buffer and heated for 10 min at 70°C, and 50 μl of each extract was loaded on a 10% SDS– polyacrylamide gel (Laemmli, 1970). After separation, proteins were transferred electrophoretically onto nitrocellulose. After blocking in 10% (wt/vol) nonfat dry milk in PBS overnight at 4°C, the blot was incubated with hybridoma supernatants of the monoclonal Drosophila lamin Dm0 antibodies U25 (undiluted; kindly provided by H. Saumweber, Humboldt University, Berlin, Germany; Risau et al., 1981) and ADL67 (dilution 1:20; kindly provided by N. Stuurman, Biozentrum, Basel, Switzerland; Riemer et al., 1995) or of the Drosophila lamin C antibody LC28 (dilution 1:20; also provided by N. Stuurmann; Riemer et al., 1995) for 2 h at room temperature. Subsequent incubation with alkaline phosphatase-conjugated anti–mouse antibodies (Promega Biotech) and visualization with NBT/BCIP were performed as described (Morr et al., 1995).
Southern Blot Analysis
Genomic DNA of a single adult fly was restricted with EcoRI, separated on 0.8% agarose gels, and blotted onto Hybond-N+ membrane (Amersham Life Science, Pittsburgh, PA) by alkaline capillary transfer after incubation of the gels in 0.25 M HCl for 10 min. The blot was hybridized with a 32P-labeled probe derived from the rescue plasmid fragment (see Fig. 1 A, probe 2) as described (Ultsch et al., 1992).
Phenotypic Analysis
The righting response of individual flies was tested by tapping Drosophila culture bottles containing a single fly on a desk top, thus forcing the animal onto its dorsal side. The time required for it to return to a standing posture was then recorded.
Light Microscopy
To examine the anatomy of mutant gonads, dissected abdomina of female flies were fixed in Carnoy solution (60% [vol/vol] ethanol/30% [vol/vol] CHCl3/10% [vol/vol] acetic acid) for 4 h, dehydrated, embedded in paraffin, and cut into 15-μm sections. Tissue staining was performed with hematoxilin and eosin for 7 min and 3 s, respectively. The motility of wt and mutant sperm was monitored by visual inspection of slightly squashed, unfixed testis preparations.
Immunofluorescense Microscopy
Anesthetized flies were placed in a fly collar (Ashburner, 1989), covered with Tissue Tek (Miles Inc., Kankakee, IN) and frozen at −70°C. Cryosections (10 μm) of fly heads collected on Superfrost Plus microscopy slides (Menzel, Braunschweig, Germany) were fixed in 4% (wt/vol) formaldehyde for 2 min and washed in PBS, 0.5% (wt/vol) NP-40/PBS, and again in PBS for 5 min each. After blocking with 5% (vol/vol) goat serum in PBS for 20 min, the head sections were incubated overnight at room temperature with 1:50 dilutions in PBS of the monoclonal Drosophila lamin Dm0 antibodies ADL67 (Riemer et al., 1995), U25 or T50 (Risau et al., 1981), or the Drosophila lamin C antibody LC28 (Riemer et al., 1995). After three washes with PBS for 3 min, the slides were incubated with a 1:500 dilution in PBS of secondary mouse antibodies conjugated to the fluorescent dye Cy3 (Dianova GmbH, Hamburg, Germany) for 30 min in the dark. After washing the slides twice with PBS for 15 min, DNA staining was performed with DAPI (1 mg/ml) in the dark. The sections were then rinsed with deionized water and mounted in glycerol/Mowiol (BASF, Ludwigshofen, Germany). Double fluorescence for lamin and DNA was examined in a fluorescence microscope (Axiophot; Zeiss Inc., Oberkochen, Germany).
Electron Microscopy
Drosophila heads were fixed in 2% (wt/vol) OsO4 in 0.1 M sodium cacodylate buffer (pH 7.4) for 2.5 h at 4°C, washed with 0.1 M sodium cacodylate buffer, dehydrated by a series of ethanol dilutions, and incubated subsequently in 1,2-propylene oxide, 1,2-propylene oxide/Epon 812 (1:1; vol/vol), and Epon 812 for 5 min, 30 min, and 16 h, respectively. Epon polymerization was then performed for 24 h at 60°C. Ultramicrotome sections (80 nm) were postfixed with buffered OsO4 and negatively stained with uranyl-acetate and lead citrate for 12 and 3 min, respectively, using the Ultrastainer apparatus (LKB instruments). Electron micrographs were taken with a Zeiss microscope (EM10C; Zeiss Inc.).
For the statistical analysis of cellular differences in morphology, head sections of young adult flies (1 d after eclosion from the pupal envelope) were used. Cells surrounding the medulla, lobula, and lobula plate (including some cells of central brain) were inspected in several sections. Each cell was examined for NPC clusters in tangential and cross sections. In addition, annulate lamellae and defective nuclear envelopes were counted. Only one of these structural features was scored as positive per cell, with defective envelopes and annulate lamellae having higher priorities than nuclear pore clusters. NPC clusters were operationally defined as areas containing more than five NPCs in a regular close distance. Circumferences of cross-sectioned nuclei, NPC densities, and interpore distances were determined from electron micrographs of head sections of two wt and homozygous lamP animals each.
P Element-mediated Rescue Experiments
A genomic fragment containing the entire lamin Dm0 transcription unit was constructed from the P-lacW rescue plasmid and an additional genomic PCR fragment of the 5′ gene region absent from the rescue fragment. The latter was generated on genomic Drosophila DNA using the oligonucleotide primers 5′-AAGGATCCAAAAACAGCGCAGAGCA-3′ (sense) and 5′-CGTGAGATTTTTGTGACTGA-3′ (antisense) at an annealing temperature of 55°C as described (Schuster et al., 1991). Positions 8 of the sense and 1 of the antisense primers correspond to positions 1 and 1026 of the published lamin genomic sequence (Osman et al., 1990; these sequence data are available from GenBank/EMBL/DDBJ under accession No. X16275), respectively. A BamHI recognition sequence was added to the 5′ end of the sense primer. The rescue plasmid and the 1033-bp PCR fragment were restricted with BamHI and MunI and ligated at the MunI recognition site, yielding the intact lamin gene sequence, with additional 5′ and 3′ sequences of ∼0.25 and 0.9 kb, respectively. For P element-mediated germline transformation, the EcoRI site at the 3′ end of this genomic lamin fragment was blunted by fill in reaction with Klenow enzyme, and the fragment cloned into the BamHI and EcoRV sites of the transformation vector pHS85 (Sass, 1990) containing an Hsp82 promotor and a neomycin resistance gene as selection marker. The transformation construct (P-lam +) was coinjected with the transposase-providing helper plasmid pp25.7wc (Karess and Rubin, 1984) into heterozygous lamP preblastoderm embryos according to standard procedures (Rubin and Spradling, 1982). Hatching adults were crossed with heterozygous lamP flies, and the F2 progeny was selected on G418 (Sigma Chemical Co., St. Louis, MO) containing food. Two G418-resistant transformants, Tm1-lamP and Tw2lamP, were obtained. The transformant Tw2-lamP with a P-lam + element insertion on the third chromosome was used throughout these studies.
Results
Identification of a P Element Insertion into the Lamin Gene
A P element insertion into the first intron of the gene encoding the nuclear membrane protein lamin Dm0 of Drosophila melanogaster was discovered in a genetic screen designed to isolate mutants of ionotropic glutamate receptor subunits (Schuster et al., 1991; Ultsch et al., 1992, 1993). The lamin gene insertion was identified by hybridization of a λ phage insert containing DGluR-II genomic sequences (Schuster, C., and B. Schmitt, unpublished results) to plasmid-rescued DNAs isolated from pools of flies that carried random insertions of P-lacW elements in their genomes. Sequence analysis of the hybridizing plasmid containing a 4.7-kb rescue fragment revealed a P element insertion after position 710 of the Dm0 gene sequence (Osman et al., 1990). This position lies within the first of three introns (Fig. 1 A) and is located 350 bp upstream of the translation start site (Fig. 1 B). The genes for both lamin Dm0 (Gruenbaum et al., 1988) and the muscle glutamate receptor subunit DGluR-II (Schuster et al., 1991) have been previously localized to position 25F1-2 on the left arm of the second chromosome. Our analysis showed that the two genes indeed lie closely together, with an intergene distance of only ∼12 kb (Fig. 1 A).
Expression of Lamin Dm0 Is Reduced in Homozygous Mutant Flies
Flies carrying the lamin P-lacW insertion (lamP) were balanced for the second chromosome with the inversion In(2LR)Gla, which contains the easily detectable genetic marker for glaced eyes (Gla). Homozygous and heterozygous lamP adult flies were investigated for lamin gene expression by Northern and Western blot analysis in comparison to w1118 and In(2LR)Gla control flies.
The Dm0 gene encodes two transcripts of 2.8 and 3.0 kb, which are differentially expressed during development and probably originate from alternative polyadenylation (Gruenbaum et al., 1988). Northern hybridizations with an in vitro synthesized RNA probe encompassing most of the last exon of the lamin gene (Fig. 1 A, probe 1) are shown in Fig. 2 A. Both lamin transcripts were barely detectable (<5% of control) in flies homozygous for the P-lacW insertion; only after extensive overexposure of the radioactively labeled blot, faint signals became visible at 2.8 and 3.0 kb (not shown). In lamP flies heterozygous for the insertion, no obvious differences in transcript levels could be detected as compared to control flies.
Figure 2.
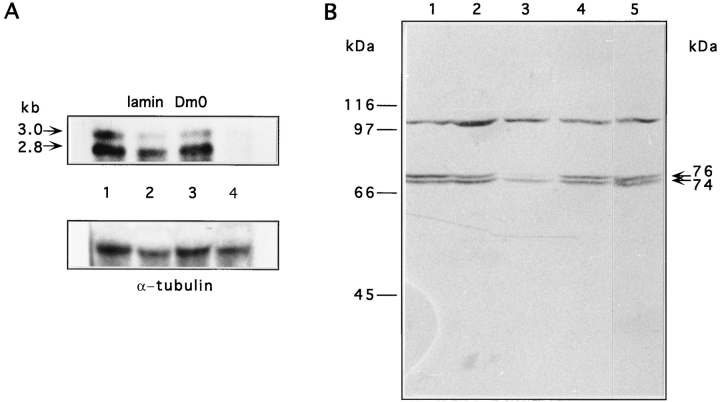
Expression of lamin Dm0 transcripts and proteins. (A) Northern blot of total RNA isolated from adult flies. The top panel shows hybridization with a lamin-specific riboprobe (Fig. 1 A, probe 1) and the lower with an α-tubulin riboprobe as control. Lane 1 contains total RNA of homozygous w1118, lane 2 of In(2LR)Gla, lane 3 of heterozygous lamP, and lane 4 of homozygous lamP flies. Arrows on the left indicate the sizes of the two lamin transcripts derived by alternative polyadenylation. Signals for both transcripts were very faint with the homozygous mutant strain (lane 4), but clearly visible after overexposure (not shown). (B) Western blot analysis of homogenates from single adult flies. The blot was probed with the monoclonal antibody U25 recognizing all Dm isoforms (Risau et al., 1981). Lane 1 contains the homogenate of a wt, lane 2 of a heterozygous lamP, lane 3 of a homozygous lamP, lane 4 of a homozygous Tw2-lamP, and lane 5 of a homozygous w1118 fly. Both lamin protein bands were reduced in the homozygous mutant (lane 3) and restored in the rescue fly (lane 4). A cross reacting protein band of apparent molecular mass of 105 kD is not related to the Dm0 gene product and served as an internal control for protein load. Positions of molecular weight marker proteins are indicated on the left; arrows on the right depict the positions of lamin isoforms.
A single primary translation product (Dm0) of apparent molecular mass of 76 kD is synthesized from both transcripts and constitutes the precursor of multiple posttranslationally modified isoforms (Smith et al., 1987; Smith and Fisher, 1989). During early embryogenesis and in mitosis, a single soluble isoform of 75 kD (Dmmit) is present. In interphase nuclei, however, the two lamin isoforms Dm1 and Dm2 with apparent molecular masses of 74 and 76 kD, respectively, predominate. Western blot analysis (Fig. 2 B) with the monoclonal antibody U25 recognizing all isoforms (Risau et al., 1981; Smith et al., 1987) showed a severe reduction of the Dm1 and Dm2 variants in homozygous lamP flies to <20% of the lamin protein in comparison to wt (Oregon R) or control (w1118) flies. The 75-kD mitotic isoform could not be observed. Notably, the reduction of the 76-kD band seemed to be more pronounced than that of the 74-kD Dm1 form in the homozygous mutant. To rule out a possible crossreaction of the U25 antibody with Drosophila lamin C (Bossie and Sanders, 1993; Riemer et al., 1995), which exhibits 52% amino acid sequence identity to lamin Dm0 and migrates at a similar position as Dm1 in SDS–polyacrylamide gels, we also used the Dm0-specific monoclonal antibody ADL67 (Riemer et al., 1995) for Western blot analysis. A pattern similar to that revealed with the U25 antibody was again obtained with the ADL67 antibody (data not shown). Both antibodies failed to detect a reduction in the total amount of lamin protein in heterozygous lamP animals (Fig. 2 B and data not shown). Parallel Western blots with the lamin C-specific antibody LC28 (Riemer et al., 1995) failed to reveal detectable differences in lamin C expression between wt and both homo- and heterozygous lamP flies (data not shown).
Lamin Insertion Flies Show Delayed Development, Reduced Viability, Impairment of Locomotion, and Sterility
The most obvious phenotypic changes in the homozygous lamP flies concerned locomotion behavior, development and survival, and fertility. After a prolonged developmental time course (delayed by up to 3 d at culture temperature of 24°C), the few hatching homozygous adult mutants were unable to fly; only occasionally, small jumps could be observed. This may reflect differences in penetrance of the insertion in individual animals, resulting in small variations in lamin protein expression. Homozygous lamP mutants moved more slowly than wt or control flies and displayed “spastic” behavior after losing motor coordination. To monitor the locomotion impairment we tested the righting response by measuring the time for returning to a normal upright posture from a dorsal position. Fig. 3 A shows that while heterozygous lamP and wt animals right themselves instantly, the mean righting time of homozygous lamP flies lasted up to several minutes.
Figure 3.

Analysis of locomotor behavior and sterility. (A) Righting responses of wt, homo- and heterozygous lamP, and homozygous Tw2lamP flies. Six animals of each genotype were analyzed in six independent measurements. Mean values ± SEM of the observed righting times are given for each individual. (B and C) Histology of ovaries. Hematoxiline/eosin-stained paraffin sections of the ovaries from a wt (B) and a homozygous lamP (C) female fly are shown. Note shortened oocytes and reduced number of egg chambers in the bottom panel. ec, egg chamber; fc, follicle cell; g, germarium; gt, gut; nc, nurse cell; oc, oocyte. The scale bar in B represents 50 μm and is also valid for C.
Only a small number (5–10%) of the homozygous lamP animals survived until adulthood in the balanced mutant strain. Survival was inversely correlated to the population density in the culture chamber. In overcrowded culture bottles, very few homozygous mutant flies survived, whereas a higher proportion of homozygous individuals was found in less populated cultures. During development, three major periods of lethality were detected in the lamP strain, which included the embryonic stages (lethality of 20–30%), the pupal stages (lethality of 50–60%), and the eclosion of the adult fly (lethality of 5–10%). Mutant adult flies died within 2 wk after eclosion.
Homozygous lamP flies of both sexes were sterile. Mutant female flies showed abnormal ovaries whose anatomy varied among individuals. Comparative sections of wt and homozygous mutant ovaries are presented in Fig. 3, B and C. The number of ovarioles, each consisting of a germarium (g) and the vitellarium containing different egg chamber (ec) stages, was significantly reduced in the mutant, as was the number of individual egg chambers. Late stages of oogenesis were rarely detected in the mutant ovaries, and the ones present showed an abnormal morphology. In contrast to the large oocytes (oc) typically seen in late egg chambers of wt animals, oocytes of comparable stages were shortened in homozygous lamP ovaries. In contrast to the mutant ovaries, the gonads of male mutant flies showed no gross morphological changes but a drastic reduction in or complete loss of sperm motility (data not shown).
Germline Transformation with an Intact Lamin Gene Rescues the Mutant Phenotype
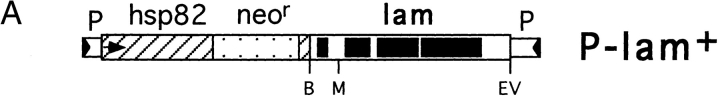
To prove that the P element insertion in the first intron of the lamin Dm0 gene is indeed the cause for the pleiotropic phenotype of homozygous lamP flies, we performed P element-mediated germline transformation with a wt lamin transgene construct, P-lam + (Fig. 4 A). A unique MunI restriction site at the end of the first intron (see Materials and Methods) was exploited to assemble the entire transcription unit from the plasmid rescue fragment and a genomic PCR fragment of the 5′ genomic region. The lamin gene construct begins 251 bp upstream of the transcription start site, includes the putative TATA box located 29 bp upstream from the transcription start site, and ends about 0.9 kb downstream of the polyadenylation site of the 3.0kb lamin transcript (Osman et al., 1990). The lamin gene construct was cloned into the P element transformation vector pHS85 that has a neomycin resistance as selection marker and provides the hsp82 promoter, which is constitutively active (Sass, 1990). After germline transformation, two neomycin-resistant transformant lines, Tm1-lamP and Tw2-lamP, were obtained, in which the mutant phenotype of the homozygous lamP fly was rescued. The transformant line Tw2-lamP was used throughout the experiments described below.
Figure 4.
Germline transformation with a lamin transgene. (A) Schematic diagram of the P element construct used for transformation. The Dm0 lamin transgene was inserted into the P transposable vector pHS85. Exonic regions of the lamin Dm0 gene (lam) are shown as black boxes. Restriction sites used for lamin transgene construction (see Materials and Methods) are indicated below: B, BamHI; M, MunI; EV, EcoRV. The P element transfer vector pHS85 contains a fusion of hsp82 protein (hatched) and neomycin phosphotransferase (stippled) gene. The hsp82 promoter is indicated by an arrow. Flanking P element sequences (P) are shown with their inverted repeats (triangles). (B) Southern blot analysis of the transformed rescue strain. EcoRI-restricted genomic DNA isolated from single flies was hybridized to a radioactively labeled plasmid rescue fragment (Fig. 1 A, probe 2). DNA samples were prepared from the following flies: hetero- and (lane 1), homozygous lamP (lane 2), heterozygous Tw2-lamP (one transgene copy, lane 3; two transgene copies, lane 4), homozygous Tw2-lamP (lane 5), and homozygous w1118 (lane 6). Arrows on the right indicate the control (9.8 kb), transgene (9.3 kb), and lamP (6.7 kb) specific hybridization bands.
Southern blot analysis with probe 2 (Fig. 1 A) showed that the original P-lacW insertion was still present in the Tw2-lamP flies (Fig. 4 B). Control flies (w1118), in which a polymorphic EcoRI restriction site (Fig. 1 A, E*) is absent, showed a single hybridization signal at ∼9.8 kb (Fig. 4 B, lane 6). Heterozygous lamP flies gave the same 9.8-kb signal but, in addition, a band at ∼6.7 kb resulting from the P-lacW insertion (Fig. 4 B, lane 1). The 9.8-kb band was absent in homozygous lamP animals (Fig. 4 B, lane 2). In the Tw2-lamP transformant strain, a new signal was seen at ∼9.3 kb (Fig. 4 B, lane 3); this indicates the insertion of one transgene copy in the transformant strain. The intensity of this rescue signal increased in both male and female animals homozygous for the transgene, indicating a location of the transgene on the third or fourth chromosome (Fig. 4 B, lane 4, and not shown). The band of 9.8 kb characteristic of control flies was absent in rescue flies homozygous for the lamP mutation (Fig. 4 B, lane 5).
Western blot analysis of homozygous rescue flies (Fig. 2 B) showed that the Dm lamin isoforms of apparent molecular masses of 74 and 76 kD were present in similar amounts as in wt, control, and heterozygous mutant strains. Thus, the defect was indeed rescued at the molecular level. Consistent with this recovery of protein expression, the homozygous rescue animals developed normally, were fully viable, fertile in both sexes, and showed wt locomotion behavior (Fig. 3 A). No obvious phenotypic differences could be found between animals heterozygous and homozygous for the transgenic P-lam + insertion (not shown).
Lamin Dmo Immunoreactivity Is Reduced in Perikarya-rich Regions of the Homozygous lamP Mutant Brain
To examine whether the reduction of lamin immunoreactivity found by Western blot analysis was due to organspecific deficits in Dm0 gene expression, heads and thoraces of adult homozygous lamP mutants were inspected by immunofluorescence microscopy using the Dm0 specific monoclonal antibody ADL67 (Riemer et al., 1995) and compared to tissue from wt flies (Fig. 5). In heads from the homozygous mutant, lamin Dm0 immunoreactivity was significantly decreased in nuclei of the densely packed cell bodies (perikarya-rich region) of the central nervous system (Fig. 5 B) as compared to heads from wt flies (Fig. 5 A), whereas the intensities of nuclear DNA staining, as revealed by DAPI fluorescence, were comparable. Notably, lamin C immunoreactivities of the same perikarya-rich regions were not significantly different between wt (Fig. 5 C) and the lamP (Fig. 5 D) flies. Moreover, the majority of the neuronal cells strongly immunoreactive with ADL67 showed no detectable lamin C immunofluorescence in either wt or mutant animals. Similar differences in lamin Dm0 staining were also seen in muscle cells of the thoracic region which express, however, high levels of lamin C in both wt and lamP mutant. The lamin C expression was comparable to that of Dm0 seen in wt (data not shown).
Figure 5.
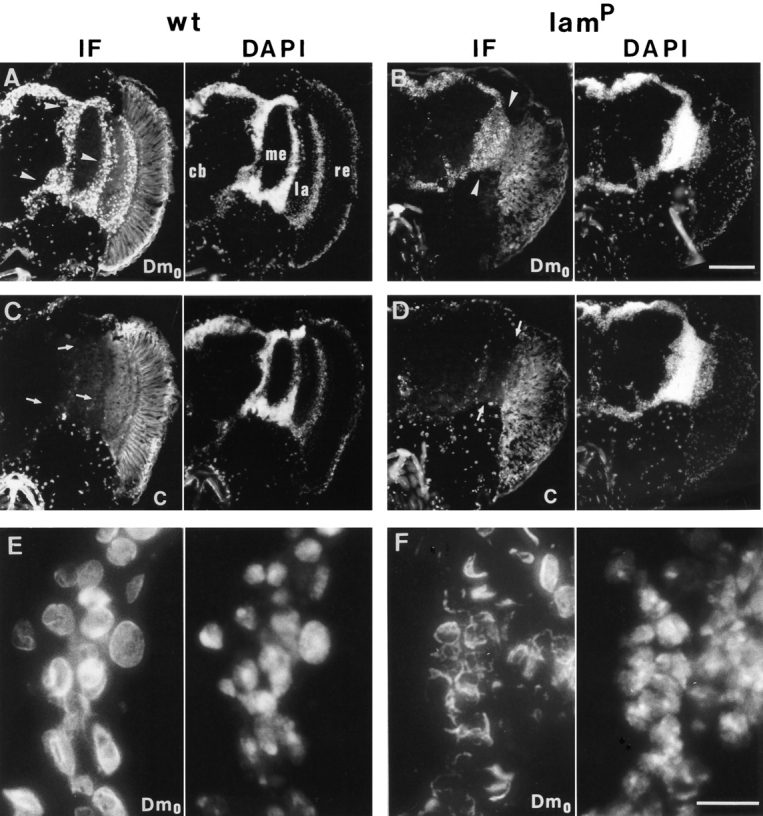
Indirect immunofluorescence staining of head cryosections by lamin Dm0- and lamin C-specific monoclonal antibodies. Simultaneously processed head sections of a wt (left column) and a homozygous lamP (right column) fly are depicted. The left half column of each depicts the lamin antibody staining detected by indirect immunofluorescense (IF) and the right half the corresponding DNA staining by DAPI of the same section. A and B show lamin Dm0-specific staining by antibody ADL67 and C and D lamin C-specific staining by antibody LC28, of a total head hemisection each, with A and C as well as B and D, representing consecutive sections. E and F show magnifications of selected areas stained with antibody ADL67. Note the altered lamin Dm0 nuclear staining in lamP flies (B and F) as compared to wt (A and E). Arrowheads in A and B indicate the medulla and lobula/lobula plate cell body regions inspected in the electron-microscopic analysis. Arrows in C and D indicate the same areas displaying low lamin C expression. Anatomical structures of the fly's central nervous system are indicated in A: cb, central brain; me, medulla; la, lamina; re, retina. The retina displays strong autofluorescence, which is more obvious in wt. Bars: (A–D) 100 μm; (E and F) 10 μm.
Analysis at higher magnification revealed an altered nuclear distribution of lamin Dm0 in neuronal cells of homozygous lamP mutant flies. While the ADL67 immunofluorescence nicely delineated the surface of wt nuclei (Fig. 5 E), the residual lamin Dm0 staining of homozygous mutant nuclei was often fuzzy and irregular (Fig. 5 F). Similar results (not shown) were also obtained with the monoclonal lamin Dm0 antibodies U25 and T50 (Risau et al., 1981). High resolution micrographs in addition suggested alterations in the distribution of DNA staining in lamP flies. Whereas wt nuclei showed rather homogeneous DAPI fluorescence, the DNA staining of mutant nuclei sometimes appeared decompacted and irregular (compare Fig. 5, E and F).
No differences in lamin Dm0 immunoreactivity were seen in animals heterozygous for the lamP mutation (data not shown). Similarly, in Tw2-lamP rescue animals, lamin expression in the perikaryal region of the central nervous system seemed to be restored to that of wt flies (not shown). In addition, the morphology of the nuclei stained by lamin Dm0 antibodies or DAPI was indistinguishable from that found in wt preparations (data not shown).
Ultrastructural Analysis Reveals Incomplete Nuclear Envelopes, Clustering of NPCs in the Nuclear Envelope, and an Increased Number of Annulate Lamellae
To further characterize any morphological changes that might underlie the altered lamin immunofluorescence pattern seen in the visual system of homozygous lamP heads we performed electron microscopy. The high density of neuronal cell bodies in this region facilitated the simultaneous inspection of many nuclei. This ultrastructural analysis of cross sections through the heads of young adult flies revealed striking abnormalities in the homozygous mutant. These included (a) the clustering of NPCs in the nuclear membrane, (b) a high incidence of annulate lamellae, and (c) a partial loss or even total absence of the nuclear envelope. Fig. 6 shows representative images obtained from sections of the optic lobe region around the medulla, lobula, and lobula plate of adult wt, homo- and heterozygous lamP mutants, and homozygous Tw2-lamP rescue flies. Already at low magnification, a high frequency of NPC clusters and annulate lamellae was routinely seen in homozygous lamP flies (Fig. 6 B) but not in wt (Fig. 6 A), heterozygous mutant (Fig. 6 C), or rescue (Fig. 6 D) animals. In addition, incomplete nuclear envelopes were often found in the homozygous mutant. A quantitative summary of the morphological characteristics observed in the medulla and lobula/lobula plate regions of different wt, homo- and heterozygous lamP mutants, and homozygous Tw2-lamP rescue flies is given in Table I. The incidence of NPC clusters, annulate lamellae, and incomplete nuclear envelopes was increased in the homozygous mutant flies, with ∼67% of the mutant nuclei displaying one or more of these abnormal structures. It should be emphasized that due to the mode of analysis, i.e., inspection of distant cross- sections, the extent of the morphological changes observed might even represent an underestimate of the existing alterations. A slightly increased incidence of NPC clusters, but not of annulate lamellae or incomplete nuclear envelopes, was observed in the optic lobe cells of heterozygous lamP flies. No obvious differences as compared to wt flies could be detected in the rescued Tw2-lamP animals. The changes produced by the mutation in the homozygous lamP animals were not accompanied by gross alterations in nuclear size; the average nuclear circumference (±SD) in cross sections of wt nuclei was 7.92 ± 1.80 μm (n = 366), whereas nuclei of lamP flies had an average circumference of 8.97 ± 2.22 μm (n = 301).
Figure 6.

Thin section, electron-microscopic analysis of cell bodies surrounding the medulla and lobula/lobula plate in wt, homo- and heterozygous lamP mutant, and in Tw2-lamP rescue flies. Electron micrographs of negatively stained sections are shown at lower magnification. A section through several optic lobe cell bodies of the homozygous (ho) lamP mutant (B) reveals striking differences to wt (A). Arrowheads in B point to examples of annulate lamellae (lower left cell) and NPC clusters in tangential (middle right cell) and transversal (lower right cell) nuclear sections. Similar sections from heterozygous (he) lamP (C) and homozygous Tw2-lamP (D) flies show no obvious differences to wt (A). Bar, 2 μm.
Table I.
NPC Clusters, Annulate Lamellae, and Incomplete Nuclear Envelopes in Cells Surrounding the Medulla and Lobula/ Lobula Plate*
| Strain | No. of cells analyzed | Cells with abnormal nuclear morphology | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | NPC clusters | Annulate lamellae | Incomplete nuclear envelopes | |||||||
| % ± SD | ||||||||||
| Homozygous lamP (n = 3) | 594 | 67.2 ± 2.7 | 45.6 ± 2.0 | 12.7 ± 4.9 | 8.9 ± 2.0 | |||||
| Oregon R (n = 2) | 519 | 1.5 ± 1.0 | 1.2 ± 0.7 | 0.2 ± 0.2 | 0.3 ± 0.3 | |||||
| Heterozygous lamP (n = 2) | 769 | 9.1 ± 0.1 | 7.6 ± 0.6 | 1.0 ± 0.5 | 0.4 ± 0.1 | |||||
| Homozygous Tw2-lamP (n = 2) | 682 | 1.9 ± 1.0 | 1.0 ± 0.4 | 0.6 ± 0.6 | 0.3 ± 0.1 | |||||
NPC clusters, annulate lamellae, and incomplete nuclear envelopes were identified by electron microscopy and counted as described in Materials and Methods. The total cell values as percentage include all cells displaying one or more of these phenotypes. n, number of animals analyzed.
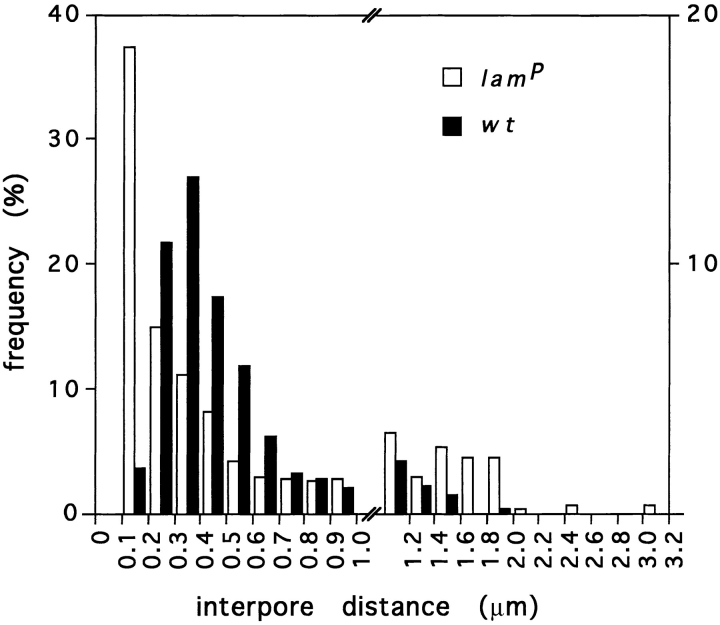
The most obvious consequence of the lamP genotype was a high incidence of NPC clusters. Cross sections of wt nuclei displayed few randomly dispersed NPCs detectable by a narrowing of the intermembrane space of the nuclear envelope (Fig. 7, A and E). In homozygous mutants, however, such cross sections frequently contained clustered NPCs at distinct regions of the nuclear envelope (Fig. 7, B and F). This clustering of NPCs in the homozygous mutant cells was particularly obvious in tangential sections (Fig. 7 D), while similar sections of nuclei from wt (Fig. 7 C), heterozygous mutant, and rescue (data not shown) animals carried only a few pore complexes. High resolution images showed that the NPCs in the homozygous mutant nuclei were often very densely packed and showed tetragonal symmetry (Fig. 7 H), a feature never found in wt nuclei (Fig. 7 G). This could not be attributed to major changes in NPC density resulting from the mutation, since the mean pore complex density (±SD) was only modestly increased to 2.73 ± 0.60 NPCs/μm nuclear envelope for lamP (nuclei, n = 296) as compared to a wt value of 2.15 ± 0.67 NPCs/μm nuclear envelope (nuclei, n = 195). Frequency histograms of the interpore distance between individual NPCs confirmed that the major fraction (37%) of the NPCs in the mutant cells was located within about one pore diameter distance (0.1–0.2 μm from center to center) from a neighboring NPC (Fig. 8). Consistently, a significant portion of NPCs is separated by distances >1.0 μm. In contrast, interpore distances in wt nuclei showed the highest incidence between 0.3–0.4 μm, and distances ⩾1.0 μm were found to a lesser extent than in mutant nuclei.
Figure 7.
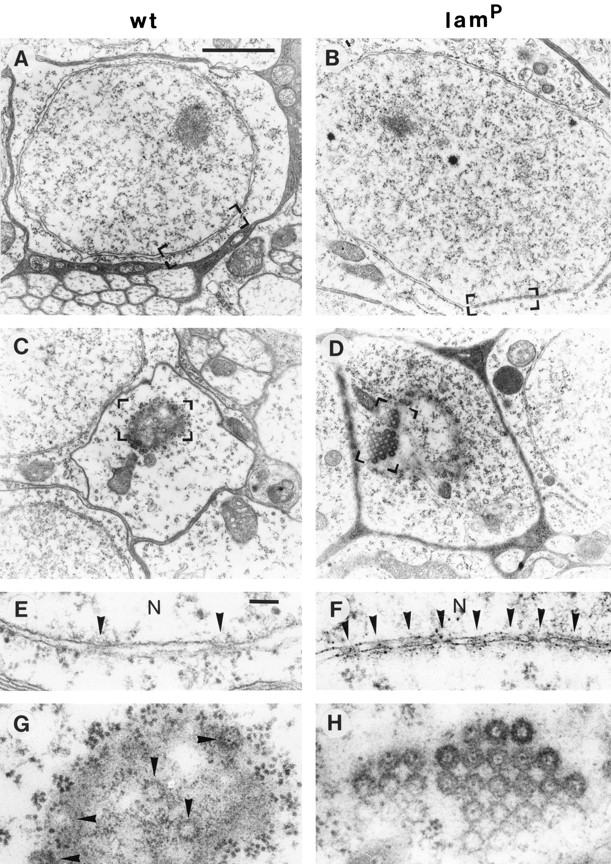
Distribution of NPCs in wt and homozygous lamP mutant cells. Electron micrographs representing thin sections from selected cells of the optic lobe region show NPCs in transversal (A and B) and tangential (C and D) nuclear sections. Bracketed areas (A–D) are shown at higher magnification in E–H, respectively. Arrowheads indicate NPCs in E–G. Note the high packing density of NPCs (NPC clusters) in B, D, F, and H with an apparent tetragonal symmetry in D and H. N, nucleoplasm. Bars: (A–D) 2 μm; (E–H) 100 nm.
Figure 8.
Frequency of interpore distances of NPCs in wt and homozygous lamP mutant nuclei. Interpore distances between the NPCs of 30 cross sectioned nuclei, each of wt and lamP mutant cells determined from electron micrographs and classified into bin sizes of 0.1 μm (for distances ⩽1.0 μm) and 0.2 μm (for distances >1.0 μm), respectively. The graph shows the histogram of frequency (as a percentage) of wt (solid bars) and mutant (open bars) interpore distances (wt, n = 527; lamP, n = 529); scales differ on both sides of the figure, with the lamP values on the left and wt on the right. Note high frequency of short (<0.2 μm) interpore distances as well as an increased occurrence of long (>1.0 μm) interpore distances in the mutant nuclei.
The second striking feature of cells from homozygous lamP mutants was an abundance of annulate lamellae. Annulate lamellae are stacked sheets of membranes in the cytoplasm, which contain a high density of pore complexes and are often continuous with rough endoplasmic reticulum cisternae (for review see Kessel, 1992). The structure of pore complexes in annulate lamellae is similar, if not identical, to that of NPCs in the nuclear envelope. In ∼13% of the mutant cells in the perikarya region (Table I), annulate lamellae were found as parallel cisternae apposed to NPCs of the nuclear envelope (Fig. 9 A) but also as independent curvilinear structures in the cytoplasm (Fig. 9 C). The high resolution micrograph shown in Fig. 9 E demonstrates the high packing density of NPCs in these cytoplasmic membranes.
Figure 9.
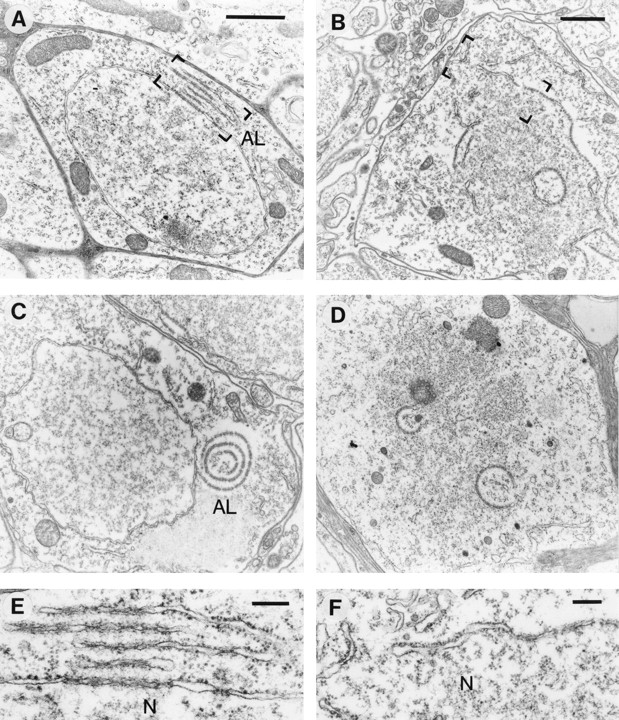
Annulate lamellae and defective nuclear envelopes in homozygous lamP mutant cells. Thin-section electron micrographs show annulate lamellae (left column) and defective nuclear envelope (right column) structures typically found in mutant cells of the optic lobe region. A depicts a parallel stack of annulate lamellae (AL) close to an NPC cluster and (C) a circular annulate lamellae structure. B shows a cell with a partially fragmented nuclear membrane and D one without nuclear envelope. Bracketed areas of A and B are shown at higher magnification in E and F, respectively. N, Nucleoplasm. Bars: (A and B) 1 μm; (E and F) 200 nm; Bar in A is also valid for C and D.
While nearly all cell nuclei from wt, heterozygous mutant, and rescue flies had complete nuclear envelopes, those of homozygous mutants appeared incomplete in ∼9% of the total cell population (Table I). Frequently, a fragmentation of the nuclear envelope coincided with the presence of annulate lamellae (Fig. 9 B). A higher magnification of the fragmented envelope is shown in Fig. 9 F. (Due to the absence of nuclear pores, the residual membrane compartments could not always be classified with certainty as nuclear; however, in many cases the membraneous structures surrounded electron-dense material, most probably chromatin). In several cells, a nearly complete loss of the nuclear envelope was found, leaving only a few annulate lamellae-like structures (Fig. 9 D). Changes in the homozygous mutant similar to those described above for the cells surrounding the medulla and lobula/lobula plate were also occasionally seen in retina, lamina, central brain, and muscle (not shown).
Discussion
The identification of a P element insertion into the first intron of the Dm0 lamin gene reported here constitutes the first successful isolation of a mutation in a gene of the nuclear lamin family. Our data show that reduced Dm0 expression results in a severe mutant phenotype that strongly affects viability, fertility, and locomotion. The observed cellular changes, including NPC clusters, cytoplasmic annulate lamellae, and incomplete nuclear envelopes, indicate that lamin Dm0 is required for normal nuclear envelope assembly and nuclear pore distribution. These data provide clear genetic evidence for the important role of this lamin in the structural organization of the nuclear envelope.
Northern and Western blot analysis of homozygous lamP flies showed that the P element insertion into the first intron of the lamin Dm0 gene caused a marked reduction in the respective transcript and protein levels. Random insertions of P elements into the genome often occur within regulatory regions and are used in enhancer trap screens to search for cis-acting elements conferring tissue- or stagespecific expression (O'Kane and Gehring, 1987; Cooley et al., 1988; Bier et al., 1989). It is presently unclear whether such a regulatory function exists in the first intron of the lamin gene. Alternatively, the P element insertion may affect the efficiency of pre-mRNA splicing, as reported for transposon insertions in mouse (Pattanakitsakul et al., 1992; Mülhardt et al., 1994). Only low levels of correctly processed Dm0 transcripts of 2.8 and 3.0 kb were found here in homozygous lamP animals. We did not observe, however, a preferential expression of the 3.0-kb transcript as reported for adult flies by Gruenbaum et al. (1988).
In contrast to the developmentally regulated lamin C (Riemer et al., 1995), the lamin Dm0 gene is constitutively expressed from early stages of development onwards; some decrease in transcript levels is seen during the larval stages (Gruenbaum et al., 1988; Riemer at al., 1995). This is consistent with marked effects, in particular high lethality and delayed morphogenetic maturation, of the insertional mutation at different stages of development. Only a low percentage of homozygous mutant animals reached the adult stage. This may reflect variations in mutational penetrance, with comparatively low levels of lamin Dm0 expression being sufficient for survival and/or partial compensation by lamin C (see below). The lack of full lethality at early developmental stages, in which no expression of lamin C occurs, may be due to maternal transmission from the heterozygous mothers. Indeed, lamin Dm0 protein is highly enriched inside the oocyte nucleus, which may serve as a storage compartment for lamin required during the early nuclear divisions in the embryo (Frasch et al., 1988).
Deficits in the oocyte's lamin pool may also explain another characteristic of the lamP phenotype, female sterility. While sufficient lamin may be provided by heterozygous mutant mothers, the low amount of Dm0 protein produced in homozygous mutant females most likely cannot support oocyte development. Together with the observed abnormalities in mutant ovary anatomy, this may be a major cause of female sterility. Interestingly, the Drosophila lamin Dm0 gene seems to be ubiquitously expressed in all cells analyzed so far with one exception: mature Drosophila sperm lack lamin Dm0 (Riemer et al., 1995). Male infertility in homozygous lamP flies thus seems to be caused not by low sperm lamin contents but rather by an effect of the lamin Dm0 deficiency on spermatogenesis.
The third phenotypic characteristic of the homozygous lamP mutant, a severe deficiency in locomotion with delayed righting response and complete loss of flying behavior, suggested a critical role of lamin Dm0 in the neuromuscular system. Immunocytochemistry of homozygous lamP mutants disclosed a significant reduction of lamin Dm0 immunofluorescence in both the perikarya region of the central nervous system, which contains many densely packed neuronal somata, and muscle cells. Importantly, a lamin C-specific antibody failed to reveal detectable levels of lamin C in most Dm0-deficient neuronal nuclei, whereas muscle nuclei showed significant lamin C immunoreactivity in both mutant and wt flies. This suggests that lamin C may be able to compensate for lamin Dm0 function and is consistent with the data of Riemer et al. (1995), who reported a high accumulation of lamin Dm0, but not C, transcripts and protein in the central nervous system of late Drosophila embryos. The same authors also showed that low lamin C expression persists in the larval eye-antennal discs that give rise to several structures of the visual system. We interpret the striking manifestation of the lamP phenotype in neurons of the visual system (and other regions of the central nervous system) as a consequence of the combined effects of both a mutation-induced reduction in lamin Dm0 expression and low lamin C gene activity in these cells. Our high resolution analysis of lamin Dm0 immunofluorescence indicates that under the latter conditions not only a reduction in lamin Dm0 content but also a severe distortion of the regular geometry of the nuclear lamina results.
Additional changes in the structural organization of neuronal nuclei in homozygous lamP animals were disclosed by electron microscopy. First, we frequently observed incomplete or even missing nuclear envelopes. The nuclear envelope is disassembled at the onset of open mitosis. During prophase, the nuclear membrane is fragmented into vesicles, and the lamina is depolymerized into soluble and membrane-bound lamin. Nuclear envelope reassembly takes place in late anaphase and telophase, when membranes, pore complexes, and lamins reassociate with chromatin. Several studies have suggested a role of lamins in targeting nuclear envelope precursor vesicles to chromatin (for review see Lourim and Krohne, 1994). For example, both inclusion into nuclear assembly-competent, cell-free extracts (Burke and Gerace, 1986; Dabauvalle et al., 1991; Ulitzur et al., 1992) and microinjection in cultured cells during mitosis (Benavente and Krohne, 1986) of antilamin antibodies prevent assembly of the nuclear envelope. The observations made here are consistent with these data and extend the evidence for a critical role of lamins in envelope formation to the intact organism. It should, however, be emphasized that in homozygous lamP flies only a fraction of the inspected cells displayed a partial or total absence of the nuclear envelope membrane (Table I); in most cells, a closed nuclear membrane was seen. Earlier studies with cell-free extracts from Xenopus eggs and antibodies specific for the Xenopus B-type lamin XB3, which was initially thought to be the only lamin expressed during early developmental stages, have reported formation of a complete membrane including pores around the nucleus without a lamina present (Newport et al., 1990; Jenkins et al., 1993). Therefore, a lamin-independent nuclear membrane assembly pathway was proposed from these experiments. However, minor amounts (∼5–10% of those of lamin XB3) of another lamin of the B2 type were subsequently found to be present in these Xenopus extracts (Lourim and Krohne, 1993). This residual lamin might have been sufficient to promote nuclear membrane assembly. Similarly, the small amount of lamin Dm0 still expressed in homozygous lamP flies might suffice for complete nuclear membrane assembly even in cells that do not (yet) express lamin C.
The most prominent feature revealed upon ultrastructural analysis was a high abundance of NPC clusters in the nuclear envelope of homozygous lamP neurons. In 67% of the nuclei in the perikarya-rich region of the mutant visual system, NPC clusters were detected, and 37% of all NPCs were located within 0.1 to 0.2 μm distance from the next pore complex. The nuclear lamina is thought to organize and anchor NPCs (Aebi et al., 1986; Stewart and Whytock, 1988; Goldberg and Allen, 1992), which are located in “holes” of the lamina's fibrillar meshwork (Belmont et al., 1993). Our data constitute a convincing demonstration that lamins are indeed essential for the proper spatial organization of NPCs in the nuclear membrane. The latter might involve either interactions of lamin Dm0 with components of the NPC or its binding to specific proteins in the inner nuclear membrane. A recent study with lamin XB3-, but not XB2-depleted, cell-free extract from Xenopus eggs reported an increased formation of regions containing a high density of NPCs on the in vitro assembled nuclei (Goldberg et al., 1995). Nuclear pore clustering has also been observed in mutants of yeast nucleoporin genes including NUP120, NUP133, NUP145, and NUP159 (Doye et al., 1994; Wente and Blobel, 1994; Aitchison et al., 1995; Gorsch et al., 1995; Pemberton et al., 1995). The nucleoporin proteins are components of the NPC, and some of them have been implicated in positioning of the NPCs. The high incidence of NPC clusters in the homozygous lamP mutant is consistent with a direct interaction between nuclear pore proteins and the lamins. It should be noted that an uneven distribution of NPCs is often observed in nuclei at early stages of nuclear assembly (Burke and Gerace, 1986; Goldberg and Allen, 1992). Thus, the NPC clusters found in the lamin-deficient flies may also be indicative of the disturbed assembly process highlighted by the more severely affected nuclei with incomplete nuclear envelopes.
The third striking feature found in homozygous lamP mutant cells was an accumulation of annulate lamellae. These may also be a reflection of early nuclear assembly stages. Annulate lamellae, membraneous cisternae–containing pore complexes, are frequently found in germ cells and rapidly dividing somatic cells, like embryonic and tumor cells (for review see Kessel, 1992). They are usually located in the cytoplasm but occasionally also in the nucleoplasm. Viral infection and chemical treatment can enhance annulate lamellae formation. It has been shown that annulate lamellae do not contain lamins; however, their disassembly and reassembly behavior during mitosis follows closely that of the nuclear envelope (Cordes et al., 1996). Formation of annulate lamellae has been proposed to represent a default pathway, in which pore complexes and other nuclear membrane components can be stored upon saturation or absence of chromatin templates (Stafstrom and Staehelin, 1984; Meier et al., 1995). Xenopus cell-free extracts form annulate lamellae instead of a nuclear envelope when anti-lamin antibodies are added during incubation with external DNA or chromatin (Dabauvalle et al., 1991). We interpret, therefore, the high incidence of annulate lamellae in homozygous lamP mutant flies as an accumulation of pore complexes resulting from impaired assembly of the nuclear envelope.
We frequently observed annulate lamellae apposed to pore clusters in the nuclear envelope; this may be indicative of cytosolic regions specialized for NPC assembly from soluble components. Another interesting finding of our ultrastructural analysis is that clusters of NPCs at the nuclear surface were often densely packed into crystal-like structures of tetragonal symmetry, which differs from the hexagonal symmetry of dense NPC packing most commonly found in annulate lamellae (Scheer and Franke, 1969; Stafstrom and Staehelin, 1984; Kessel, 1992). The reason for this different packing geometry is presently unclear but may reflect differences in the membrane composition of annulate lamellae and the nuclear envelope.
The causal relationship between the observed phenotypic and ultrastructural changes and the insertional disruption of the Dm0 gene was demonstrated here by gene rescue. Introduction of a Dm0 transgene harboring the entire transcription unit into lamP flies not only restored all phenotypic features of the wt, but also reversed the ultrastructural changes seen in the mutant. In particular, normalized lamin Dm0 protein levels were paralleled by the reappearance of an intact nuclear membrane, recovery of locomotion and flight behavior, and normal fertility. In addition, whereas eclosion of homozygous lamP pupae was delayed by up to 3 d as compared to control flies, the time course of development was normal in the case of the rescued mutant. Since it appears highly unlikely that the short flanking and intronic sequences contained in the rescue construct in addition to the Dm0 open reading frame correspond to another functional gene, we confidently conclude that the lamP phenotype indeed resulted from reduced lamin Dm0 expression.
The availability of the lamP mutant strain described in this study should foster further genetic and cellular approaches to lamin function. In particular, mobilization of the inserted P element should allow the isolation of novel mutant alleles, including severe deficiencies and null phenotypes. The detailed biochemical and ultrastructural analysis of such mutants may crucially contribute to further deciphering the role(s) of lamin Dm0 proteins in nuclear organization and dynamics. In addition, such mutants may provide a tool to dissect genetically the interactions of lamins with other nuclear envelope and chromatin proteins implicated in the disassembly and reassembly of the nucleus during mitosis.
Acknowledgments
We thank Dr. H. Saumweber for kindly providing the monoclonal antibodies U25 and T50, Drs. A. Püschel and C. Morgans for critical reading of the manuscript, and Ms. M. Baier, Ms. H. Reitz, and Ms. S. Wartha for secretarial assistance. We also are indebted to Dr. N. Stuurman for kindly providing the monoclonal antibodies ADL67 and LC28 and advice on their use. We are particularly grateful to Dr. G. Krohne (Theodor-BoveriInstitut, University of Würzburg) for help with the interpretation of the ultrastructural data, to Dr. B. Klagges for participating in the mutagenesis screen, to W. Hofer for performing electron microscopy, and to Dr. J.H. Brandstätter for help with ultrastructural analysis and preparation of figures.
This work was supported by Deutsche Forschungsgemeinschaft (Schm 726/4-1) and Fonds der Chemischen Industrie.
Footnotes
1. Abbreviations used in this paper: LAP, lamina-associated proteins; NPC, nuclear pore complex; wt, wild type.
Please address all correspondence to B. Schmitt, Max-Planck-Institut für Hirnforschung, Abteilung Neurochemie, Deutschordenstrasse 46, D-60528 Frankfurt am Main, Germany. Tel.: (49) 69-96769-261; Fax: (49) 69-96769-441.
R. Reifegerste's current address is the Department of Biological Sciences, University of Southern California, Los Angeles, CA 90084-7340.
References
- Aaronson RP, Blobel G. Isolation of pore complexes in association with a lamin. Proc Natl Acad Sci USA. 1975;72:1007–1011. doi: 10.1073/pnas.72.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U, Cohn JB, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature (Lond) 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Nup120p: a yeast nucleoporin required for NPC distribution and RNA transport. J Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner, M. 1989. Drosophila. A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- Bailer SM, Eppenberger HM, Griffiths G, Nigg EA. Characterization of a 54-kD protein of the inner nuclear membrane: evidence for cell cycle–dependent interaction with the nuclear lamina. J Cell Biol. 1991;114:389–400. doi: 10.1083/jcb.114.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavente R, Krohne G. Involvement of nuclear lamina in postmitotic reorganization of chromatin as demonstrated by microinjection of lamin antibodies. J Cell Biol. 1986;103:1847–1854. doi: 10.1083/jcb.103.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, et al. Searching for pattern and mutation in the Drosophilagenome with a P-lacZ vector. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Bossie CA, Sanders MM. A cDNA from Drosophila melanogasterencodes a lamin C-like intermediate filament protein. J Cell Sci. 1993;104:1263–1272. doi: 10.1242/jcs.104.4.1263. [DOI] [PubMed] [Google Scholar]
- Burke B. On the cell-free association of lamins A and C with metaphase chromosomes. Exp Cell Res. 1990;186:169–176. doi: 10.1016/0014-4827(90)90223-w. [DOI] [PubMed] [Google Scholar]
- Burke B, Gerace L. A cell-free system to study reassembly of the nuclear envelope at the end of mitosis. Cell. 1986;44:639–652. doi: 10.1016/0092-8674(86)90273-4. [DOI] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. Insertion mutagenesis of the Drosophilagenome with single P-elements. Science (Wash DC) 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Franke WW. Cytoplasmic annulate lamellae in cultured cells: composition, distribution, and mitotic behavior. Cell Tissue Res. 1996;284:177–191. doi: 10.1007/s004410050578. [DOI] [PubMed] [Google Scholar]
- Dabauvalle MC, Loos K, Merkert H, Scheer U. Spontaneous assembly of pore complex–containing membranes (“annulate lamellae”) in Xenopusegg extract in the absence of chromatin. J Cell Biol. 1991;112:1073–1082. doi: 10.1083/jcb.112.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodemont H, Riemer D, Weber K. Structure of an invertebrate gene encoding cytoplasmic intermediate filament (IF) proteins: implications for the origin and the diversification of IF proteins. EMBO (Eur Mol Biol Organ) J. 1990;9:4083–4094. doi: 10.1002/j.1460-2075.1990.tb07630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring V, Stick R. Gene structure for nuclear lamin Liii of Xenopus laevis: a model for the evolution of IF proteins from a lamin-like ancestor. EMBO (Eur Mol Biol Organ) J. 1990;9:4073–4081. doi: 10.1002/j.1460-2075.1990.tb07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V, Wepf R, Hurt EC. A novel nuclear pore protein Nup133p with distinct roles in poly(A)+RNA transport and nuclear pore distribution. EMBO (Eur Mol Biol Organ) J. 1994;13:6062–6075. doi: 10.1002/j.1460-2075.1994.tb06953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filaments. Proc Natl Acad Sci USA. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the inner nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Franke WW. Nuclear lamins and the cytoplasmic intermediate filament proteins: a growing multigene family. Cell. 1987;48:3–4. doi: 10.1016/0092-8674(87)90345-x. [DOI] [PubMed] [Google Scholar]
- Frasch M, Paddy M, Saumweber H. Developmental and mitotic behavior of two novel groups of nuclear envelope antigens of Drosophila melanogaster. . J Cell Sci. 1988;90:247–263. doi: 10.1242/jcs.90.2.247. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Weber K. Intermediate filaments: structure, dynamics, function, and disease. Annu Rev Biochem. 1994;63:345–382. doi: 10.1146/annurev.bi.63.070194.002021. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- Gerace L, Burke B. Functional organization of the nuclear envelope. Annu Rev Cell Biol. 1988;4:335–374. doi: 10.1146/annurev.cb.04.110188.002003. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blum A, Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex–lamina fraction. Interphase and mitotic distribution. J Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JR, Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J Cell Biol. 1990;111:1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD. High resolution scanning electron microscopy of the nuclear envelope: the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol. 1992;119:1429–1440. doi: 10.1083/jcb.119.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Jenkins H, Allen TD, Whitfield WGF, Hutchison CJ. Xenopus lamin B3has a direct role in the assembly of a replication competent nucleus: evidence from cell-free egg extracts. J Cell Sci. 1995;108:3451–3461. doi: 10.1242/jcs.108.11.3451. [DOI] [PubMed] [Google Scholar]
- Gorsch LC, Dockendorff TC, Cole CN. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J Cell Biol. 1995;129:939–955. doi: 10.1083/jcb.129.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Landesman Y, Drees B, Bare JW, Saumweber H, Paddy MR, Sedat JW, Smith DE, Benton BM, Fisher PA. Drosophila nuclear lamin precursor Dm0is translated from either of two developmentally regulated mRNA species apparently encoded by a single gene. J Cell Biol. 1988;106:585–596. doi: 10.1083/jcb.106.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-y. [DOI] [PubMed] [Google Scholar]
- Hennekes H, Nigg EA. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J Cell Sci. 1994;107:1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- Höger TH, Krohne G, Franke WW. Amino acid sequence and molecular characterization of murin lamin B as deduced from cDNA clones. Eur J Cell Biol. 1988;47:283–290. [PubMed] [Google Scholar]
- Höger TH, Zatloukal K, Waizenegger I, Krohne G. Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma. 1990;99:379–390. doi: 10.1007/BF01726689. [DOI] [PubMed] [Google Scholar]
- Höger TH, Krohne G, Kleinschmidt JA. Interaction of Xenopus lamins A and LII with chromatin in vitromediated by a sequence element in the carboxyterminal domain. Exp Cell Res. 1991;197:280–289. doi: 10.1016/0014-4827(91)90434-v. [DOI] [PubMed] [Google Scholar]
- Holtz D, Tanaka RA, Hartwig J, McKeon FD. The CaaX-motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell. 1989;59:969–977. doi: 10.1016/0092-8674(89)90753-8. [DOI] [PubMed] [Google Scholar]
- Hutchison CJ, Bridger JM, Cox LS, Kill IR. Weaving a pattern from disparate threads: lamin function in nuclear assembly and DNA replication. J Cell Sci. 1994;107:3259–3269. doi: 10.1242/jcs.107.12.3259. [DOI] [PubMed] [Google Scholar]
- Jenkins HE, Holman T, Lyon C, Lane EB, Stick R, Hutchison CJ. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci. 1993;106:275–285. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- Karess RE, Rubin GM. Analysis of P transposable element functions in Drosophila. . Cell. 1984;38:135–146. doi: 10.1016/0092-8674(84)90534-8. [DOI] [PubMed] [Google Scholar]
- Kessel RG. Annulate lamellae: a last frontier in cellular organelles. Int Rev Cytol. 1992;133:43–120. doi: 10.1016/s0074-7696(08)61858-6. [DOI] [PubMed] [Google Scholar]
- Kitten GT, Nigg EA. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J Cell Biol. 1991;113:13–23. doi: 10.1083/jcb.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohne G, Benavente R. The nuclear lamins: a multigene family of proteins in evolution and differentiation. Exp Cell Res. 1986;162:1–20. doi: 10.1016/0014-4827(86)90421-0. [DOI] [PubMed] [Google Scholar]
- Krohne G, Franke WW, Ely S, D'Arcy A, Jost E. Localization of a nuclear envelope-associated protein by indirect immunofluorescence microscopy using antibodies against a major polypeptide from rat liver fractions enriched in nuclear envelope-associated material. Cytobiologie. 1978;18:22–38. [PubMed] [Google Scholar]
- Krohne G, Waizenegger I, Höger TH. The conserved carboxy-terminal cysteine of nuclear lamins is essential for lamin association with the nuclear envelope. J Cell Biol. 1989;109:2003–2011. doi: 10.1083/jcb.109.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J Biol Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27:230–236. doi: 10.1006/geno.1995.1036. [DOI] [PubMed] [Google Scholar]
- Lindsley, D.L., and G.G. Zimm. 1992. The genome of Drosophila melanogaster. Academic Press Inc., San Diego, CA.
- Loewinger I, McKeon FD. Mutations in the nuclear lamin proteins resulting in their aberrant assembly in the cytoplasm. EMBO (Eur Mol Biol Organ) J. 1988;7:2301–2309. doi: 10.1002/j.1460-2075.1988.tb03073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourim D, Krohne G. Membrane-associated lamins in Xenopusegg extracts: identification of two vesicle populations. J Cell Biol. 1993;123:501–512. doi: 10.1083/jcb.123.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourim D, Krohne G. Lamin-dependent nuclear envelope reassembly following mitosis: an argument. Trends Cell Biol. 1994;4:314–318. doi: 10.1016/0962-8924(94)90228-3. [DOI] [PubMed] [Google Scholar]
- McKeon FD, Kirschner MW, Caput D. Primary and secondary structural homologies between the major nuclear envelope and cytoplasmic intermediate filament proteins. Nature (Lond) 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- Meier J, Campbell KH, Ford CC, Stick R, Hutchison CJ. The role of lamin LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopuseggs. J Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Meier E, Miller BR, Forbes DJ. Nuclear pore complex assembly studied with a biochemical assay for annulate lamellae formation. J Cell Biol. 1995;129:1459–1472. doi: 10.1083/jcb.129.6.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Montag-Lowy M, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morr J, Rundström N, Betz H, Langosch D, Schmitt B. Baculovirus-driven expression and purification of glycine receptor α1 homooligomers. FEBS (Fed Eur Biochem Soc) Lett. 1995;368:495–499. doi: 10.1016/0014-5793(95)00721-k. [DOI] [PubMed] [Google Scholar]
- Mülhardt C, Fischer M, Gass P, Simon-Chazottes D, Guénet J-L, Kuhse J, Betz H, Becker C-M. The spastic mouse: aberrant splicing of glycine receptor β subunit mRNA caused by intronic insertion of a L1 element. Neuron. 1994;13:1003–1015. doi: 10.1016/0896-6273(94)90265-8. [DOI] [PubMed] [Google Scholar]
- Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Assembly-disassembly of the nuclear lamina. Curr Opin Cell Biol. 1992;4:105–109. doi: 10.1016/0955-0674(92)90066-l. [DOI] [PubMed] [Google Scholar]
- O'Kane C, Gehring W. Detection in situ of genomic regulatory elements in Drosophila. . Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman M, Paz M, Landesman Y, Fainsod A, Gruenbaum Y. Molecular analysis of the Drosophilanuclear lamin gene. Genomics. 1990;8:217–224. doi: 10.1016/0888-7543(90)90274-x. [DOI] [PubMed] [Google Scholar]
- Paddy MR, Belmont AS, Saumweber H, Agard DA, Sedat JW. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell. 1990;62:89–106. doi: 10.1016/0092-8674(90)90243-8. [DOI] [PubMed] [Google Scholar]
- Pattanakitsakul S, Zheng JH, Natsuume-Sakai S, Takahashi M, Nonaka M. Aberrant splicing caused by the insertion of the B2 sequence into an intron of the complement C4 gene is the basis for low C4 production in H-2k mice. J Biol Chem. 1992;267:7814–7820. [PubMed] [Google Scholar]
- Pemberton LF, Rout MP, Blobel G. Disruption of the nucleoporin gene NUP133 results in clustering of nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:1187–1191. doi: 10.1073/pnas.92.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990;61:591–602. doi: 10.1016/0092-8674(90)90471-p. [DOI] [PubMed] [Google Scholar]
- Riemer D, Stuurman N, Berrios M, Hunter C, Fisher PA, Weber K. Expression of Drosophilalamin C is developmentally regulated: analogies with vertebrate A-type lamins. J Cell Sci. 1995;108:3189–3198. doi: 10.1242/jcs.108.10.3189. [DOI] [PubMed] [Google Scholar]
- Risau W, Saumweber H, Symmons P. Monoclonal antibodies against a nuclear membrane protein of Drosophila. . Exp Cell Res. 1981;133:47–54. doi: 10.1016/0014-4827(81)90355-4. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz D, Benz WK, Engels WR. A stable source of P-element transposase in Drosophila melanogaster. . Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röber RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development (Camb) 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophilawith transposable element vectors. Science (Wash DC) 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sass H. P-transposable vectors expressing a constitutive and thermoinducible hsp82-neo fusion gene for Drosophilagermline transformation and tissue-culture transfection. Gene (Amst) 1990;89:179–186. doi: 10.1016/0378-1119(90)90004-b. [DOI] [PubMed] [Google Scholar]
- Scheer U, Franke WW. Negative staining and adenosine triphosphatase activity of annulate lamellae of newt oocytes. J Cell Biol. 1969;42:519–533. doi: 10.1083/jcb.42.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster CM, Ultsch A, Schloss P, Cox JA, Schmitt B, Betz H. Molecular cloning of an invertebrate glutamate receptor subunit expressed in Drosophilamuscle. Science (Wash DC) 1991;254:112–114. doi: 10.1126/science.1681587. [DOI] [PubMed] [Google Scholar]
- Senior A, Gerace L. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamin. J Cell Biol. 1988;107:2029–2036. doi: 10.1083/jcb.107.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Fisher PA. Interconversion of Drosophilanuclear lamin isoforms during oogenesis, early embryogenesis and upon entry of cultured cells into mitosis. J Cell Biol. 1989;108:255–265. doi: 10.1083/jcb.108.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Gruenbaum Y, Berrios M, Fisher PA. Biosynthesis and interconversion of Drosophilanuclear lamin isoforms during normal growth and in response to heat shock. J Cell Biol. 1987;105:771–790. doi: 10.1083/jcb.105.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafstrom JP, Staehelin LA. Are annulate lamellae in the Drosophila embryo the result of overproduction of nuclear pore components? . J Cell Biol. 1984;98:699–708. doi: 10.1083/jcb.98.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M, Whytock S. The structure and interactions of components of nuclear envelopes from Xenopusoocyte germinal vesicles observed by heavy metal shadowing. J Cell Sci. 1988;90:409–423. [Google Scholar]
- Ulitzur N, Harel A, Feinstein N, Gruenbaum Y. Lamin activity is essential for nuclear envelope assembly in a Drosophilaembryo cell-free system. J Cell Biol. 1992;119:17–25. doi: 10.1083/jcb.119.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A, Schuster CM, Laube B, Schloss P, Schmitt B, Betz H. Glutamate receptors of Drosophila melanogaster.Cloning of a kainateselective subunit expressed in the central nervous system. Proc Natl Acad Sci USA. 1992;89:10484–10488. doi: 10.1073/pnas.89.21.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A, Schuster CM, Laube B, Betz H, Schmitt B. Glutamate receptors of Drosophila melanogaster.Primary structure of a putative NMDA receptor protein expressed in the head of the adult fly. FEBS (Fed Eur Biochem Soc) Lett. 1993;324:171–177. doi: 10.1016/0014-5793(93)81387-f. [DOI] [PubMed] [Google Scholar]
- Weber K, Plessmann U, Ulrich W. Cytoplasmic intermediate filament proteins of invertebrates are closer to nuclear lamins than are vertebrate intermediate filament proteins; sequence characterization of two muscle proteins of a nematode. EMBO (Eur Mol Biol Organ) J. 1989a;8:3221–3227. doi: 10.1002/j.1460-2075.1989.tb08481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K, Plessmann U, Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor: implications for the structure of the nuclear lamina. FEBS (Fed Eur Biochem Soc) Lett. 1989b;257:411–414. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wismar J, Löffler T, Habtemichael N, Vef O, Geißen M, Zirwes R, Altemeyer W, Sass H, Gateff E. The Drosophilatumor suppressor gene lethal(3)malignant brain tumor encodes a proline-rich protein with a novel zinc finger. Mech Dev. 1995;53:141–152. doi: 10.1016/0925-4773(95)00431-9. [DOI] [PubMed] [Google Scholar]
- Worman HJ, Juan J, Blobel G, Georgatos SD. A lamin B receptor in the nuclear envelope. Proc Natl Acad Sci USA. 1988;85:8531–8534. doi: 10.1073/pnas.85.22.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinsmaier KE, Eberle KK, Buchner E, Walter N, Benzer S. Paralysis and early death in cysteine string protein mutants of Drosophila. . Science (Wash DC) 1994;263:977–980. doi: 10.1126/science.8310297. [DOI] [PubMed] [Google Scholar]
- Zewe M, Höger TH, Fink T, Lichter P, Krohne G, Franke WW. Gene structure and chromosomal localization of the murine lamin B2 gene. Eur J Cell Biol. 1991;56:342–350. [PubMed] [Google Scholar]