Abstract
We report here the genomic sequence of the centromeric portion of HLA class I, extending 424,015 bp from tumor necrosis factor α to a newly identified gene ≈20 kb telomeric of Otf-3. As a source of DNA, we used cosmids centromeric of HLA-B that had been mapped previously with conventional restriction digestion and fingerprinting and previously characterized yeast artificial chromosomes subcloned into cosmids and mapped with multiple complete digest methodologies. The data presented provide a description of the gene content of centromeric HLA class I including new data on intron, promoter and flanking sequences of previously described genes, and a description of putative new genes that remain to be characterized beyond the structural information uncovered. A complete accounting of the repeat structure including abundant di-, tri-, and tetranucleotide microsatellite loci yielded access to precisely localized mapping tools for the major histocompatibility complex. Comparative analysis of a highly polymorphic region between HLA-B and -C was carried out by sequencing over 40 kb of overlapping sequence from two haplotypes. The levels of variation observed were much higher than those seen in other regions of the genome and indeed were higher than those observed between allelic HLA class I loci.
The human major histocompatibility complex (MHC) is located on the short arm of chromosome 6 in the distal portion of the 6p21.3 band and consists of three major linked gene clusters: class I, II, and III genes. Much attention has been paid to this region because of the importance of the class I and II antigens in the immune response. These gene products play fundamental roles by presenting antigen to T lymphocytes as well as interacting with receptors on NK cells to inhibit NK cell-mediated cytotoxicity (1–3). The exceptional polymorphism of these molecules is of considerable interest in clinical transplantation because they are able to elicit strong antibody and cytolytic T lymphocyte responses upon allogeneic stimulation. The class I component is made up of the HLA-A, -B, and -C loci that lie within a 2-Mb region comprising the telomeric half of the MHC.
Further understanding of the molecular genetics of the HLA class I region depends on the identification and characterization of many other resident genes, analogous to the progress made in the class II and III regions (4–6). The susceptibility to a variety of diseases has long been the subject of a plethora of studies on the MHC, primarily a consequence of the immune function of the class I and II loci and their high allelic polymorphism. The portion of HLA class I around HLA-B and -C has received a great deal of attention, both as a region that is associated with disease and as a focal point for analysis in clinical studies of marrow transplants (7). One of the best studied HLA-B-associated diseases include spondyloarthropathies, associated with a series of chronic inflammatory diseases involving the musculoskeletal system (8). Psoriasis vulgaris, a skin disease, is more strongly associated with HLA-C than HLA-B and may be caused by a linked gene rather than the HLA-C locus itself (9). Other disease loci include that associated with: nasopharyngeal carcinoma tightly linked to the HLA region (10), myasthenia gravis, which is associated with HLA-B, possibly located on the telomeric side (11), and the Behcet’s disease locus, which also may be closely linked to HLA-B (12). Most recently, multiple sclerosis was linked to two principal genomic regions, one of which was the MHC (13).
Although numerous studies have mapped and characterized a number of genes within the MHC, a comprehensive analysis of the resident genes within the HLA class I region is incomplete. Analyses of cDNAs homologous to HLA class I region clones have yielded a substantial wealth of new sequences for investigation (14–15). Genomic analysis of the HLA-B and -C region covering over 237 kb has been carried out, yielding a more precise description of the gene content of this region (16). A definitive characterization that can distinguish genes from pseudogenes and other low repeat homologies must include the details of a highly accurate and complete determination of the genomic sequence. A major stumbling block in such analysis is obtaining cloned DNA verified as representative of the genomic DNA from which it was derived. Given this, high throughput sequencing can proceed rapidly when carried out in a well structured facility (17). New methodologies that allow major advances in the mapping of cloned DNA have been developed and promise to provide ready access to verified cloned material (18). Over the last several years, we have established a set of clones spanning HLA class I (19–22) that can be used toward establishing the complete nucleotide sequence of HLA class I.
We report here the complete sequence of a 424,015-bp segment of the centromeric HLA class I region, extending from the tumor necrosis factor α locus to a new gene 130 kb telomeric of HLA-C. These data include a precise description of all of the coding and flanking sequence of previously described loci, reveal potential new coding sequences, and completely define the repeat structure of the segment, including the precise localization of di-, tri-, and tetranucleotide repeats. These data were derived in part by using the new technology of multiple complete digest (MCD) mapping (23) on yeast artificial chromosome (YAC)-derived cosmids and, as such, constitute one of the first tests of the ongoing efforts to advance this technology toward high throughput analysis. To obtain a first relatively large scale view of the extent of nucleotide variation within the MHC exclusive of the class I genes, we obtained over 40 kb of sequence data from different haplotypes spanning a region between HLA-B and HLA-C.
MATERIALS AND METHODS
Source of Cosmids.
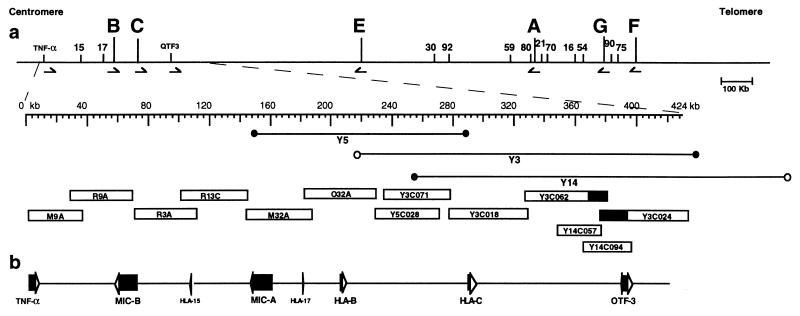
Cosmids M9A, R9A, R3A, R13C, M32A, and O32A were obtained from the study outlined in Spies et al. (24), having been chosen from the available set, to cover the region with minimal overlap between cosmids. As sequencing progressed, two gaps were discovered, one between R9A and R3A and a second between R13C and M32A. Suitable spanning restriction fragments from overlapping cosmids R15A and M17A, respectively, were subcloned and sequenced to bridge the gaps. After cosmid subcloning and MCD mapping of YAC-derived cosmids (23), a cosmid set with 20-fold coverage was available from each of YACs 3, 5, and 14 (19) for selection of suitable cosmids for sequencing. Cosmids derived from YAC 3 (cosmids Y3C071, Y3C018, Y3C062, and Y3C024; Fig. 1) were chosen for minimal overlap. During the course of mapping, it was discovered that Y3 contained an internal deletion within the overlap region with Y14, and cosmids Y14C057 and Y14C094 were chosen to span this gap. Cosmid Y5C028 was chosen both to bridge the gap between Y3CO71 and O32A and to provide maximal overlap between haplotypes. The HLA type of CGM1 (YAC source) is A3, B8, DR3, Dqw2, Drw52 and A29, B14, DR7, Dqw2, Drw53 (19).
Figure 1.
Schematic representation of the region sequenced in centromeric HLA class I. (A) The location of the region sequenced within HLA class I is identified and expanded beneath the HLA class I map. On the larger map, the locations of HLA class I genes are indicated by letters and of pseudogenes by numbers (20). In the expanded map, the locations of cosmids obtained from Spies et al. (24) and of YACs (19) and the cosmids generated from them are indicated beneath. Cosmids derived from YACs are named with the derivative YAC number followed by a designated cosmid number. (B) The positions and transcriptional orientations of known genes within the region are indicated to scale.
Sequencing.
The sequences derived from this study were determined entirely on fluorescent-based sequence-gel readers (Model 373A, Applied Biosystems) by using the enzymatic dideoxy chain-terminating method (16, 17). Sequencing strategies and methods were as described by Janer (M.J., unpublished work). In brief, cosmid DNA was sequenced by the shotgun method. DNA was sonicated, and the ends were made blunt by T4 DNA polymerase. End-repaired DNA was separated on an 1% agarose gel, and fragments of 1.5–3.6 kb were extracted and subcloned into SmaI cut M13. Single-stranded DNA was sequenced by using dye primer cycle sequencing kits and provided protocols (Perkin—Elmer).
Computer Analysis.
Sequence assembly and editing was done with the phred-phrap package developed by Phil Green et al. and with consed (D. Gordon and P. Green). Information about these programs can be found at the web site http://chimera.biotech.washington.edu/UWGC. cross_match with the repeat matrix developed by A. Smit was used to mask repeat sequences (University of Washington Genome Server; RepeatMasker on World Wide Web URL: http://ftp.genome.washington.edu). Various gene analysis programs were tested; however, the results reported here and in the annotated database entry were determined by using xgrail_1.3c (25) and the blastn and blastx-beauty (26, 27) database search tools. The sequence analysis tools developed by T. Smith, Geospiza, Seattle, WA (http://www.geospiza.com) were used to generate repeat maps and to analyze GC content.
RESULTS
This report has focused on the centromeric end of HLA class I extending from tumor necrosis factor α to a distance 130 kb telomeric of HLA-C. As a source of DNA for genomic sequencing, we relied on two distinct sets of clones, each of which provided at least two-fold coverage of the corresponding genomic regions (Fig. 1). The technology of MCD mapping (23) was used to map cosmids subcloned from YACs. Details of the methods used by the MCD mapping approach are described elsewhere (23, 28). In brief, three restriction enzymes, NsiI, HindIII, and BglII, were used in complete digestion of each cosmid DNA followed by gel electrophoresis and translation of the gel image into a computationally understandable format by using a Fluorescent Imager. The gel imaging software of Wong et al. (23) was used to generate highly accurate size measurements of the resultant bands. After identification of the single vector band by Southern blot, maps were assembled by using the mapping software described by Gillett et al. (28). Three YACs were used to construct the maps shown in Fig. 2. Four cosmids from YAC 3 and 2 from YAC 14 were chosen based on minimal overlap and one cosmid from a largely overlapping region from YAC 5 was chosen for comparative sequence analysis of distinct haplotypes.
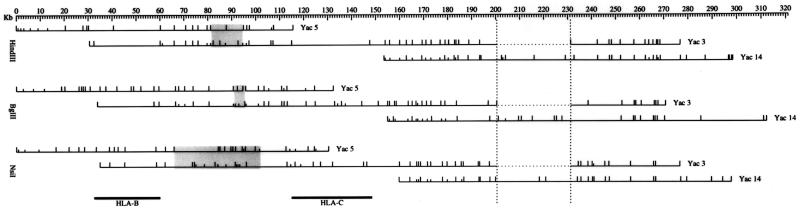
Figure 2.
Multiple complete digest maps of YAC-derived cosmids. Restriction maps for the three indicated enzymes were constructed as described for YACs 3, 5, and 14 (19). The distance spanned is indicated at the top starting with the leftmost mapped restriction site. Only complete fragments verified by more than one cosmid subclone are included in the map, resulting in a portion of the sequences at the ends of the YACs to be excluded. YACs 3 and 14 were derived from the same chromosome, and YAC 5 was derived from the homologous chromosome in CGM1. The constructed maps verified this and identified a region between HLA-B and -C (indicated below) with substantial restriction site variability between YACs 5 and 3 (shaded areas). When comparing the maps derived for YACs 3 and 14, an apparent deletion in YAC 3 was detected as discussed in the text (bounded by vertical dotted lines).
YACs 5 and 3 were derived from homologous chromosomes in heterozygous LCL CGM1 (19) providing access to overlapping DNA from distinct haplotypes. YACs 3 and 14 were derived from the same CGM1 chromosome and were both mapped to provide independent confirmation of the genomic region. After constructing MCD maps of YACs 3 and 14, it was apparent that YAC 3 had suffered a deletion of some 30 kb relative to YAC 14 (Fig. 2). Size measurements of the YAC suggest instead that the deletion was present in the original clone and was not detected in the low resolution mapping previously carried out. Cosmids from YAC 14 spanning the region in question were chosen for sequence analysis, and probes from this region were used in Southern analysis of genomic DNA to verify the integrity of the YAC 14 sequence (data not shown).
Genome-Wide Repeats.
The human genome-wide repeat families are divided into three categories by the cross_match matrices developed by Smit and Green (http://ftp.genome.washington.edu), which includes a highly effective mask for five categories of Alu sequences, non-Alu sequences including MIRs and L1s, and di-, tri-, and tetranucleotide repeats. One complex repeat structure encountered near MICA proved to be a serious test of the sequencing strategy used and presents a potentially interesting example of repeat structure in the human genome. Within the region between HLA-B and tumor necrosis factor α lies a structure of di- and trinucleotide repeats extending some 2,930 bp in length. This structure is a complex mixture of GGA(XY)n and GAA(XY)n, combined among small related repeats. Our sequencing strategy of random shotgun M13 dye primer sequencing failed to cover the region because of early termination of the sequencing reactions. However, dye terminator sequencing of existing templates provided highly accurate data (extending up to 1 kb in some cases) and allowed verified closure of the repeat. This is perhaps a significant tribute to the phred-phrap assembly programs developed by Phil Green and colleagues because some nonoverlapping templates differed by as few as 4 bp and yet were correctly placed into the assembly.
Polymorphism Outside of the HLA Class I Genes.
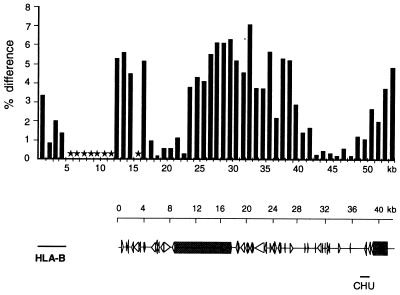
To measure levels of MHC polymorphism outside of the class I loci, we took advantage of the MCD mapping data we had acquired on YACs 5 and 3 covering the region between HLA-B and HLA-C identified by these maps as highly variable (shaded in Fig. 2). Cosmid Y5C028 covered a region largely overlapping with Y3C071 and therefore was chosen for sequence analysis. The polymorphism identified in the MCD maps indeed was reflected in the DNA sequence as illustrated in Fig. 3. The variability in this stretch of sequence is remarkably high when compared with other loci where polymorphism has been measured. For example, in the T cell antigen receptor β locus, an estimate of natural sequence variation of 0.2% was found over a total of 129 kb of overlapping sequence (29). In contrast, variation over this 40-kb stretch was as low as 0.1% but reached up to 7% in the central segment. It appears that there is a gradient of increasing sequence divergence that peaks directly in the center of a retroviral-like sequence. The divergence lowers to baseline briefly proceeding telomeric and rises again to 6% over the final 1-kb block, approximately 20 kb centromeric of HLA-C.
Figure 3.
Sequence comparison of allelic regions between HLA-B and -C. The sequence of cosmid Y5C028 was compared over the first 3 kb with cosmid O32A, which was derived from a non-CGM1 HLA haplotype (24) and over positions 4,000–42,000 with cosmid Y3C071, derived from YAC 3. cross_match analysis in sequential 1-kb blocks of Y5C028 against O32A or Y3C071 was used with the following parameters: minmatch 20, minscore 40, bandwidth 14, indexwordsize 10, masklevel 80. Y5C028 and Y3C071 span the region between HLA-B and -C identified by MCD mapping as highly variable between the two CGM1 chromosomes (shaded region in Fig. 2). Each bar represents the percentage difference over the 1-kb region compared. Included at the left of the bar graph is a similar comparison of the divergence between the HLA-B locus contained in O32A and a genomic HLA-B locus from the database. Below is a depiction of the genetic content including genome wide repeats and other homologies as discussed in the text, aligned to scale with the bar graph. Solid arrows indicate alu repeats, open arrows indicate MIR repeats, and shaded boxes indicate reverse transcriptase homologies. CHU, class I homology unit.
Class I Homology Between HLA-B and -C.
At position 256,865 near HLA-C lies a potentially interesting significant homology to an HLA class I sequence. A cross_match comparison with HLA-B58 genomic DNA showed that the class I homology unit was colinear over the first third of the HLA-B sequence from positions 1 to 211 (72%), 285 to 587 (67%), and 782 to 1090 (67%), after which the homology abruptly ended. In a blastx + beauty (27) comparison with the HLA-B58 protein sequence, homology was limited to the α1-α2 domains of HLA class I. There was ≈40% identity between residues 10 and 28 and 83 and 109 of HLA-B58 followed by 46% identity between positions 112 and 169. The homology to rat or murine class I was essentially similar to that observed in the human comparisons; and nearly all of the identical residues were a subset of those that are highly conserved between the class I antigens of murine and human.
Gene Content.
A schematic diagram of 424,015 kb of genomic sequence depicting predicted genes, pseudogenes, and gene fragments derived from this set of cosmids is shown in Fig. 4. Our analysis consisted of identifying genes by using several available similarity searches (26, 27) and dividing them into two categories, those previously characterized either at the cDNA or genomic DNA level and those not previously identified, but with homology to nucleic acid or protein sequences in the database. The latter group was subdivided further into sequences predicted to be pseudogenes based on apparent interruptions in the available coding sequence and sequences predicted to be genes based on structural criteria and identity or near identity to entries in the EST databases.
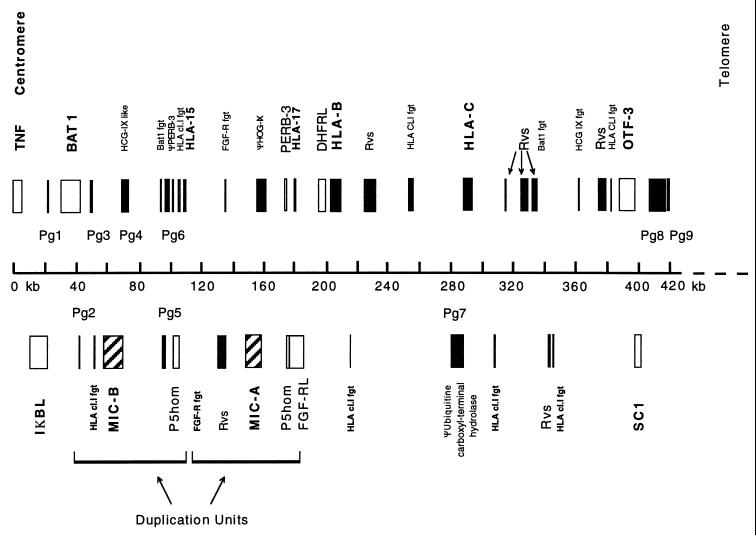
Figure 4.
The gene content of centromeric HLA class I. The positions of genes, pseudogenes, and gene fragments identified by blastn and blastx–beauty analysis are indicated above and below the scale with the relative width of the bar indicating the extent of the gene. The given names of previously described genes (see ref. 5 for references) are indicated above (transcriptional orientation from left to right) and below (transcriptional orientation from right to left) the bars, with the following abbreviations: Rvs, reverse transcriptase homology; HLA CLI fgt, homology to HLA-class I genomic fragment; FGF-R fgt, fibroblast growth factor receptor gene fragment. New putative genes identified as the result of this work are indicated immediately above the scale as Pg1–9 (putative gene) with homologies as described in Table 1.
Among the previously characterized genes, the MICA and MICB loci are included in the sequence centromeric of HLA-B. The MICB genomic sequence is closely homologous to MICA over the entire length of the gene, extending from 16,291 bp 5′ of the MICA ATG start codon to 1,325 bp centromeric of the Poly(A) addition signal. The extent of duplication of the MIC genes and flanking sequence includes a homologue of the P5 family (32) and a portion of a fibroblast growth factor receptor sequence (Fig. 4). Immediately 3′ of the MICB sequence, but not found in the MICA duplication unit, is an HLA class I fragment with homology to the murine H2-M3 α3 domain. The duplication unit defined in this region is repeated as part of a more extensive duplication that gave rise to the MICE pseudogene, located between HLA-G and HLA-F.
To identify potential new coding sequences, the most successful analysis relied on the use of blastn against the expressed sequence tag (EST) and nucleotide sequence databases and the blastx–beauty algorithms (27). Table 1 lists the homologies identified along with their respective locations and identities with proteins in the database. Some of these sequences are clearly pseudogenes, containing evident interruptions of the coding sequence, and others are present as reverse transcribed pseudogenes. The putative genes include a variety of coding sequences including a trichohyalin homology (PG8; ref. 33), an ATPase G subunit-like sequence (PG1; ref. 34), and a putative member of the cyclophilin A family (PG3; ref. 35). We expect that number of genes that have been identified in this analysis is an underestimate because existing databases comprise ≈60% of the human coding sequences.
Table 1.
HLA gene content
| Position from the centromeric start, bp | Protein size, aa | Putative genes | Homology | Identity, % |
|---|---|---|---|---|
| 22,680–23,818 | 117 | Pg1 | Vacuolar ATPase G subunit [Manduca sexta] mRNA (X92805) | 48 |
| 41,101–40,425 | Pg2 | ψ-60S ribosomal protein L15 | 52.7 | |
| 47,857–48,665 | 165 | Pg3 | Peptidyl prolyl cis-trans isomerase A (P05092) | 87.7 |
| 68,960–69,173 | 73 | Pg4 | ψ-HCG-IX | 51 |
| 97,076–95,991 | EST | Pg5 | Homo sapiens (clone 3.8-1) (L29376) | |
| 105,122–104,698 | 143 | Pg6 | P5 homolog | 54 |
| 161,698–161,920 | ψ-HCG-IX | 41 | ||
| 182,023–181,360 | ψ-P5 | |||
| 199,101–199,644 | ψ-DHFR | 84.5 | ||
| 285,987–284,791 | 288 | 60S ribosomal protein L3 TARBP | 91.5 | |
| 290,712–289,553 | Pg7 | ψ-Ubiquitin carboxyl-terminal hydrolase | 74.3 | |
| 409,006–420,319 | Pg8 | Trichohyalin homolog (Q07283) | ||
| 421,996–422,170 | 67 | Pg9 | RNA polymerase II subunit (U37690) | 63.2 |
ψ, predicted pseudogene.
EST, Homology with EST database entry, no protein data available.
DISCUSSION
The 424,015 bp of contiguous genomic sequence reported here represents the largest contiguous MHC-derived sequence determined to date. The 5′ starting point of the sequence was chosen as a centromeric boundary of HLA class I because it includes both active MIC loci and other sequences with homologies found telomeric of this region (e.g., BAT1). In addition, the analysis of >40 kb of DNA between the HLA-B and -C loci from two different haplotypes constitutes the largest contiguous segment of human MHC DNA available for such comparison. All of the data have been verified as representative of the derivative genomic DNA by using only source DNA that was obtained from regions covered by two or more independent clones. The mapping methods used first verified the comparative analysis of clones and second provided a valuable check of the integrity of the assembled sequences. By relying on computational methods for gene identification, we have identified several potential new genes.
As one of the first field test efforts of MCD mapping, this work may provide useful experience contributing to the widespread use of this technology. The absolute necessity of verifying cloned material as representative of the human genome before embarking on a large scale sequencing effort cannot be overstated, and MCD technology directly addresses this need. The relatively small deletion found in YAC 3 was not detected by comparison of the low resolution maps previously constructed for YACs 3 and 14. This example emphasizes the need to have both redundant coverage of the cloned material and high resolution maps for an effective comparison. Sequencing methodologies clearly have matured to the point of allowing megabases of DNA to be sequenced by large genome centers. However, the single major obstacle still remaining is to obtain cloned material with verified integrity. New sources of DNA with an apparently higher level of fidelity than that found in YACs or cosmids are now available in BAC and PAC libraries (36, 37). High resolution MCD maps may be the most efficient means to clearly define the level of rearrangement in these clones.
Of the candidate genes identified by this work, the tricohyalin homologue (PG8) suggests a relationship to a long-standing MHC-related disease association with psoriasis vulgaris. Psoriasis vulgaris is a disease of the skin that has an immunological and genetic basis present in 1–3% of most populations studied (8) and has been consistently associated with HLA-Cw6 and Cw7 (38, 39). The association is clearly stronger with HLA-C than HLA-B, suggesting that either HLA-C is involved directly or that the affected gene lies close to, but telomeric of, HLA-C. The overall structure of PG8 suggests possibilities for involvement as a structural component of skin-related tissues, containing trichohyalin, myosin, and laminin homologies in its coding sequence. Transcripts from this gene have been detected in keratinocyte mRNA and primary skin cell monolayer mRNA via reverse transcriptase PCR (T.G., unpublished work). The homology of PG8 to trichohyalin may be noteworthy because the latter is an intermediate filament-associated protein expressed in the granular layer of the epidermis (33). As potentially the single remaining candidate gene for involvement in psoriasis in this region other than HLA-C itself, PG8 may motivate studies of this disorder.
Although HLA classes I and II include the most polymorphic loci in the human genome, the mechanisms governing the generation of this polymorphism remain the subject of debate. Gene conversion in the generation of local alleles, intra-allelic recombination, and point mutation all may contribute to the generation of new polymorphism (40). However, the extensive sharing of substitutions between nonhuman primate and human class I alleles suggests that point mutation is a relatively infrequent event compared with the rates of recombination mechanisms (41). Our motivation for obtaining comparative sequence data between HLA-B and -C was in part based on a desire to increase our understanding of the mechanisms that generate HLA polymorphism. In this regard, the overall level of sequence divergence in the segment between HLA-B and -C is intriguing when compared with much lower levels detected in the T cell antigen receptor β locus (29) but does not of itself distinguish operative mechanisms. However, the remarkably high allelic variation of this region combined with its location between two of the most ploymorphic loci in the genome does suggest that further analysis of polymorphism within this region and elsewhere in the MHC is warranted.
The class I homology unit between HLA-B and -C provides one intriguing speculation as a source of new material for allelic divergence. Three features of this sequence are relevant: (i) its residence between HLA-B and -C; (ii) the higher degree of homology with HLA-B and -C than with other HLA class I or other class I-related families; and (iii) the conservation of codons encoding conserved residues in the α1 and α2 domains combined with a complete lack of conservation in the variable α2 residues. These features are at least suggestive of a level of interaction between the HLA-B and/or HLA-C genes in the exchange of sequence variation through gene conversion between the homology unit and these HLA loci toward the generation of new allelic variation. These considerations suggest that, although the primary focus of genome annotation is the identification of new genes, further examination of presumably “junk” DNA also may yield useful information.
In summary, this study provides several useful observations. First, within the genomic sequence lies the precise description of the gene content. Some of the genes have been described, and for some of these, new data on intron, promoter, and flanking sequences are now available. Second, some of the genetic information may lead directly to the study of a disease locus by comparative analysis of the new genes among normal and disease samples. Many other disease loci have been mapped to the MHC, examples of which include multiple sclerosis, myasthenia gravis, and nasopharyngeal carcinoma, and more focused mapping efforts are needed to pinpoint the affected genes. The abundant di-, tri-, and tetranucleotide microsatellite loci uncovered provide access to precisely localized mapping tools that could be used very rapidly to rule in or rule out large segments of the MHC (42) or could more finely map a well localized disease like psoriasis vulgaris. Third, the data provide a starting point toward investigations of polymorphism outside of the HLA class I loci. As the structural cataloging of allelic variation at the HLA-A, B, C loci draws to a close, interest can now turn toward better understanding the genetic mechanisms generating that polymorphism. Whether specific mechanisms are operating uniquely on all class I loci or there are recombinational mechanisms specific to only certain class I loci (40) is a question that may find answers in the flanking sequence. Fourth, unraveling the complex events that have given rise to the multigenic content of HLA class I has been a goal of evolutionary biologists for several years. As a paradigm, understanding the evolution of the MHC will help describe evolutionary mechanisms that have led to the divergence of species and in particular to the expansion of the mammalian orders.
Acknowledgments
We thank all of the members of the University of Washington Genome Center for helpful discussion and advice, including Maynard Olson, Phil Green, Arian Smit, Todd Smith, David Gordon, and Chris Abajian among many. The technical help of K. Bubb and Q. Pham was critical for the successful completion of this work. This work was supported by a Department of Energy grant to Maynard Olson.
ABBREVIATIONS
- MHC
major histocompatibility complex
- MCD
multiple complete digest
- YAC
yeast artificial chromosome
Footnotes
References
- 1.Ploegh H L, Orr H T, Strominger J L. Cell. 1981;24:287–299. doi: 10.1016/0092-8674(81)90318-4. [DOI] [PubMed] [Google Scholar]
- 2.Guillet J G, Lai M Z, Briner T J, Buus S, Sette A, Grey H M, Smith J A, Gefter M L. Science. 1987;235:865–870. doi: 10.1126/science.2433769. [DOI] [PubMed] [Google Scholar]
- 3.Lanier L L, Phillips J H. Semin Immunol. 1995;7:75–82. doi: 10.1006/smim.1995.0011. [DOI] [PubMed] [Google Scholar]
- 4.Campbell R D, Trowsdale J. Immunol Today. 1993;14:349–352. doi: 10.1016/0167-5699(93)90234-C. [DOI] [PubMed] [Google Scholar]
- 5.Trowsdale J. Immunogenetics. 1995;41:1–17. doi: 10.1007/BF00188427. [DOI] [PubMed] [Google Scholar]
- 6.Beck S, Abdulla S, Alderton R P, Glynne R J, Gut I G, Hosking L K, Jackson A, Kelly A, Newell W R, Sanseau P, et al. J Mol Biol. 1996;255:1–13. doi: 10.1006/jmbi.1996.0001. [DOI] [PubMed] [Google Scholar]
- 7.Spencer A, Szydlo R M, Brookes P A, Kaminski E, Rule S, van Rhee F, Ward K N, Hale G, Waldmann H, Hows J M, et al. Blood. 1995;86:3590–3597. [PubMed] [Google Scholar]
- 8.Baines M, Ebringer A. Mol Aspects Med. 1992;13:263–378. doi: 10.1016/0098-2997(92)90003-i. [DOI] [PubMed] [Google Scholar]
- 9.Elder J T, Henseler T, Christophers E, Voorhees J J, Nair R P. J Invest Dermatol. 1994;103:150S–153S. doi: 10.1111/1523-1747.ep12399486. [DOI] [PubMed] [Google Scholar]
- 10.Lu S J, Day N E, Degos L, Lepage V, Wang P C, Chan S H, Simons M, McKnight B, Easton D, Zeng Y, et al. Nature (London) 1990;346:470–471. doi: 10.1038/346470a0. [DOI] [PubMed] [Google Scholar]
- 11.Degli-Esposti M A, Andreas A, Christiansen F T, Schalke B, Albert E, Dawkins R L. Immunogenetics. 1992;35:355–364. doi: 10.1007/BF00179791. [DOI] [PubMed] [Google Scholar]
- 12.Mizuki N, Ohno S, Tanaka H, Sugimura K, Seki T, Kera J, Inaba G, Tsuji K, Inoko H. Tissue Antigens. 1992;40:22–30. doi: 10.1111/j.1399-0039.1992.tb01953.x. [DOI] [PubMed] [Google Scholar]
- 13.Sawcer S, Jones H B, Feakes R, Gray J, Smaldon N, Chataway J, Robertson N, Clayton D, Goodfellow P N, Compston A. Nat Genet. 1996;13:464–468. doi: 10.1038/ng0896-464. [DOI] [PubMed] [Google Scholar]
- 14.Gruen J R, Nalabolu S R, Chu T W, Bowlus C, Fan W F, Goei V L, Wei H, Sivakamasundari R, Liu Y C, Xu H X, et al. Genomics. 1996;36:70–85. doi: 10.1006/geno.1996.0427. [DOI] [PubMed] [Google Scholar]
- 15.Wei H, Fan W F, Xu H, Parimoo S, Shukla H, Chaplin D D, Weissman S M. Proc Natl Acad Sci USA. 1993;90:11870–4. doi: 10.1073/pnas.90.24.11870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuki N, Ando H, Kimura M, Ohno S, Miyata S, Yamazaki M, Tashiro H, Watanabe K, Ono A, Taguchi S, et al. Genomics. 1997;42:55–66. doi: 10.1006/geno.1997.4708. [DOI] [PubMed] [Google Scholar]
- 17.Adams M D, Kerlavage A R, Kelley J M, Gocayne J D, Fields C, Fraser C M, Venter J C. Nature (London) 1994;368:474–475. doi: 10.1038/368474a0. [DOI] [PubMed] [Google Scholar]
- 18.Olson M V, Green P. Cold Spring Harbor Symp Quant Biol. 1993;58:349–355. doi: 10.1101/sqb.1993.058.01.041. [DOI] [PubMed] [Google Scholar]
- 19.Bronson S K, Pei J, Taillon-Miller P, Chorney M J, Geraghty D E, Chaplin D D. Proc Natl Acad Sci USA. 1991;88:1676–1680. doi: 10.1073/pnas.88.5.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geraghty D E, Pei J, Lipsky B, Hansen J A, Taillon-Miller P, Bronson SK, Chaplin D D. Proc Natl Acad Sci USA. 1992;89:2669–2673. doi: 10.1073/pnas.89.7.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geraghty D E, Koller B H, Hansen J A, Orr H T. J Immunol. 1992;149:1934–1946. [PubMed] [Google Scholar]
- 22.Janer, M. & Geraghty D. E. (1998) Genomics, in press. [DOI] [PubMed]
- 23.Wong G K-S, Yu J, Olson M V. Proc Natl Acad Sci USA. 1997;94:5225–4230. doi: 10.1073/pnas.94.10.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spies T, Bresnahan M, Strominger J L. Proc Natl Acad Sci USA. 1989;86:8955–8958. doi: 10.1073/pnas.86.22.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uberbacher E C, Xu Y, Mural R J. Methods Enzymol. 1996;266:259–281. doi: 10.1016/s0076-6879(96)66018-2. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 27.Brenner S E. Trends Genet. 1995;11:330–331. doi: 10.1016/s0168-9525(00)89094-0. [DOI] [PubMed] [Google Scholar]
- 28.Gillett W, Hanks L, Wong G K S, Yu J, Lim R, Olson M V. Genomics. 1996;33:389–408. doi: 10.1006/geno.1996.0215. [DOI] [PubMed] [Google Scholar]
- 29.Rowen L, Koop B F, Hood L. Science. 1996;272:1755–1762. doi: 10.1126/science.272.5269.1755. [DOI] [PubMed] [Google Scholar]
- 30.Bahram S, Bresnahan M, Geraghty D E, Spies T. Proc Natl Acad Sci USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Jr, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 32.Vernet C, Ribouchon M T, Chimini G, Jouanolle A M, Sidibe I, Pontarotti P. Immunogenetics. 1993;38:47–53. doi: 10.1007/BF00216390. [DOI] [PubMed] [Google Scholar]
- 33.Lee S C, Kim I G, Marekov L N, O’Keefe E J, Parry D A, Steinert P M. J Biol Chem. 1993;268:12164–12176. [PubMed] [Google Scholar]
- 34.Lepier A, Graf R, Azuma M, Merzendorfer H, Harvey W R, Wieczorek H. J Biol Chem. 1996;271:8502–8508. doi: 10.1074/jbc.271.14.8502. [DOI] [PubMed] [Google Scholar]
- 35.Haendler B, Hofer-Warbinek R, Hofer E. EMBO J. 1987;6:947–950. doi: 10.1002/j.1460-2075.1987.tb04843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ioannou P A, Amemiya C T, Garnes J, Kroisel P M, Shizuya H, Chen C, Batzer M A, de Jong P J. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 37.Shizuya H, Birren B, Kim U J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikaheimo I, Silvennoinen-Kassinen S, Karvonen J, Tiilikainen A. Br J Dermatol. 1994;131:257–259. doi: 10.1111/j.1365-2133.1994.tb08501.x. [DOI] [PubMed] [Google Scholar]
- 39.O’Donnell B F, O’Loughlin S, Codd M B, Powell F C. Irish Med J. 1993;86:65–68. [PubMed] [Google Scholar]
- 40.Parham P, Adams E J, Arnett K L. Immunol Rev. 1995;143:141–180. doi: 10.1111/j.1600-065x.1995.tb00674.x. [DOI] [PubMed] [Google Scholar]
- 41.Lawlor D A, Zemmour J, Ennis P D, Parham P. Annu Rev Immunol. 1990;8:23–63. doi: 10.1146/annurev.iy.08.040190.000323. [DOI] [PubMed] [Google Scholar]
- 42.Epplen J T, Ammer H, Epplen C, Kammerbauer C, Mitreiter R, Roewer L, Schwaiger W, Steimle V, Zischler H, Albert E, et al. EXS. 1991;58:50–69. doi: 10.1007/978-3-0348-7312-3_4. [DOI] [PubMed] [Google Scholar]