Abstract
Proper positioning of the mitotic spindle is often essential for cell division and differentiation processes. The asymmetric cell division characteristic of budding yeast, Saccharomyces cerevisiae, requires that the spindle be positioned at the mother–bud neck and oriented along the mother–bud axis. The single dynein motor encoded by the S. cerevisiae genome performs an important but nonessential spindle-positioning role. We demonstrate that kinesin-related Kip3p makes a major contribution to spindle positioning in the absence of dynein. The elimination of Kip3p function in dyn1Δ cells severely compromised spindle movement to the mother–bud neck. In dyn1Δ cells that had completed positioning, elimination of Kip3p function caused spindles to mislocalize to distal positions in mother cell bodies. We also demonstrate that the spindle-positioning defects exhibited by dyn1 kip3 cells are caused, to a large extent, by the actions of kinesin- related Kip2p. Microtubules in kip2Δ cells were shorter and more sensitive to benomyl than wild-type, in contrast to the longer and benomyl-resistant microtubules found in dyn1Δ and kip3Δ cells. Most significantly, the deletion of KIP2 greatly suppressed the spindle localization defect and slow growth exhibited by dyn1 kip3 cells. Likewise, induced expression of KIP2 caused spindles to mislocalize in cells deficient for dynein and Kip3p. Our findings indicate that Kip2p participates in normal spindle positioning but antagonizes a positioning mechanism acting in dyn1 kip3 cells. The observation that deletion of KIP2 could also suppress the inviability of dyn1Δ kar3Δ cells suggests that kinesin-related Kar3p also contributes to spindle positioning.
Mitotic spindles commonly segregate chromosomes at specific positions within eukaryotic cells. In a cell type–specific fashion, spindles undergo movements and orient in response to spatial cues originating from cell cortical regions (for reviews see Rhyu and Knoblich, 1995; Gönczy and Hyman, 1996). Spindle-positioning events are often essential for cell propagation or the asymmetric cell divisions required for differentiation. For example, the distinct developmental programs followed by the progeny of the first two cells of the Caenorhabditis elegans embryo require that the cleavage planes of these cells occur perpendicular to each other. In these cells, the plane of cell cleavage is determined by spindle position. The perpendicular cleavage arrangement is achieved by a 90° rotation of the spindle poles, in the posterior cell only, in response to a cortically located structure (Hyman and White, 1987; Hyman, 1989; Waddle et al., 1994). A similar effect has been observed during mammalian brain development. The developmental fate of columnar epithelial progenitor cells is predicted by spindle orientation and the subsequent position of the cleavage plane (Chenn and McConnell, 1995).
The asymmetric cell division characteristic of the budding yeast Saccharomyces cerevisiae makes spindle positioning essential for propagation. In the G1 phase of the cell cycle, the S. cerevisiae spindle is located at a position near the middle of the mother cell body. Proper segregation of progeny nuclei requires that the spindle translocate to the neck separating the mother and bud cell bodies and orient along the long mother–bud cell axis before anaphase. Since this process results in the movement of the entire nuclear contents to the neck region, it is often referred to as nuclear migration. In addition to budding yeast mitotic division, nuclear migration events are essential for gamete fusion processes, insect embryonic development, and the growth of the vegetative mycelium of filamentous fungi (Morris et al., 1995). Nuclear migration events have also been correlated with essential mammalian brain development processes (Book and Morest, 1990; Book et al., 1991; Hager et al., 1995). The mechanisms of many of these processes are not well understood, but all appear to rely upon microtubules and cortically derived signals to guide the movement of nuclei. In addition, nuclear migration processes in S. cerevisiae, Aspergillus nidulans, and Neurospora crassa share in common the participation of the microtubule-based cytoplasmic dynein motor (Eshel, 1993; Li et al., 1993; Plamann et al., 1994; Xiang et al., 1994).
Spindle positioning in S. cerevisiae requires the actions of the cytoplasmic microtubules that extend from the spindle pole bodies out towards the cell cortex (Palmer et al., 1992; Sullivan and Huffaker, 1992). Interactions of the distal ends of the cytoplasmic microtubules with cortical sites have recently been shown to result in spindle movements (Carminati and Stearns, 1997). In mutants specifically defective for cytoplasmic microtubules, nuclei do not migrate to the mother–bud neck, and spindles undergo anaphase chromosome segregation at inappropriate positions in mother cell bodies. The single dynein heavy chain encoded by the S. cerevisiae genome performs an important but nonessential role in spindle positioning (Eshel et al., 1993; Li et al., 1993; Geiser et al., 1997). DYN1 deletion mutants frequently display mislocalized spindles but nonetheless grow at rates indistinguishable from wild-type at room temperature. At lower incubation temperatures, however, the spindle-positioning defect is greatly exacerbated and cell propagation is inhibited. Collected evidence suggests that dynein is contributing to spindle positioning by acting upon the cytoplasmic microtubules from cortical sites (Yeh et al., 1995).
The observation that dynein is not essential for S. cerevisiae spindle positioning indicates that another motor mechanism(s) contributes to this process. In addition to a single dynein heavy chain, the S. cerevisiae genome encodes six kinesin-related proteins (KRPs).1 Studies, including those described here, have demonstrated that none of these seven motors is individually essential for S. cerevisiae viability; all seven motor gene single deletion mutants are viable (Meluh and Rose, 1990; Hoyt et al., 1992; Lillie and Brown, 1992; Roof et al., 1992; Eshel et al., 1993; Li et al., 1993). This is due to overlap in function between the motors such that each essential mitotic spindle movement is accomplished by at least two motors. In this study, we demonstrated that the spindle-positioning function of Dyn1p overlaps with that of Kip3p (encoded by open reading frame YGL216W, also known as RRC805; these sequence data are available from GenBank/EMBL/ DDBJ under accession number Z72739), a KRP revealed by the Saccharomyces Genome Sequencing Project. (A role for Kip3p in spindle positioning was also recently described by DeZwaan et al. [1997].) In the absence of both Dyn1p and Kip3p, spindle positioning was dramatically inhibited by a mechanism that required the Kip2p KRP. dyn1Δ kip2Δ kip3Δ triple mutants were viable and healthy, indicating that yet another motor contributes to spindle positioning. Genetic evidence presented suggests that this other spindle positioning motor is Kar3p.
Materials and Methods
Yeast Strains and Media
The S. cerevisiae strains used in these experiments are derivatives of S288C and are listed in Table I. The cin8::LEU2, dyn1::HIS3, dyn1:: URA3, kar3::LEU2, kip1::HIS3, kip2::URA3, and smy1::LEU2 alleles were described previously (Meluh and Rose, 1990; Hoyt et al., 1992; Lillie and Brown, 1992; Roof et al., 1992; Eshel et al., 1993; Geiser et al., 1997). The kip3::kan allele is a complete disruption of the KIP3 open reading frame (YGL216W) located on chromosome VII and was constructed using the PCR-mediated method described in Wach et al. (1994). Briefly, the kanamycin resistance gene (kanMX) from plasmid pFA6-kanMX4 was amplified with flanking ends homologous to 40 bp upstream and 40 bp downstream from the KIP3 open reading frame. kanMX confers resistance to the antibiotic G418. The PCR product was transformed into strain MAY591, and colonies were selected on rich medium containing 200 mg/liter G418 (Life Technologies, Gaithersburg, MD). PCR primers corresponding to the upstream region of the KIP3 open reading frame and to an internal segment of either KIP3 or kanMX were used to confirm that the kanMX gene had replaced the entire KIP3 open reading frame in G418-resistant cells.
Table I.
Yeast Strains and Plasmids
| Yeast strains | Genotype | |
|---|---|---|
| MAY589 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 | |
| MAY591 | MATα lys2-801 his3-Δ200 leu2-3,112 ura3-52 | |
| MAY2038 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 kip::URA3 | |
| MAY2059 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 cin8::LEU2 | |
| MAY2079 | MATα lys2-801 his3-Δ200 leu2-3,112 ura3-52 kip1::HIS3 | |
| MAY2269 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 kar3::LEU2 | |
| MAY3153 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 dyn1::URA3 | |
| MAY3903 | MAT a leu2-3,112 ura2-52 his4-539 smy1::URA3 | |
| MAY4434 | MAT a ade2-101 his3-Δ200 leu2-3, 112 ura3-52 cyh2 dyn1::HIS3 | |
| MAY4517 | MAT a ade2-101 his3-Δ200 leu2-3, 112 ura3-52 kip3::kan | |
| MAY4544 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3, 112 ura3-52 dyn1::HIS3 kip3::kan | |
| MAY4560 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3, 112 ura3-52 cyh2 dyn1::HIS3 kip3::kan kip2::URA3 | |
| MAY4566 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 chy2 dyn1::HIS3 kip2::URA3 | |
| MAY4619 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3, 112 ura3-52 kar3::LEU2 kip3::kan (pMA1223) | |
| MAY4716 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 kip3::kan kip2::URA3 | |
| MAY4761 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3,112 ura3-52 cyh2 dyn1::HIS3 kar3::LEU2 kip2::URA3 | |
| MAY4816 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 (pFC56) | |
| MAY4817 | MAT a ade2-101 his3-Δ200 leu2-3,112 ura3-52 (p415GALS) | |
| MAY4819 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3,112 ura3-52 cyh2 dyn1::HIS3 kip3::kan kip2::URA3 (pFC56) | |
| MAY4820 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3,112 ura3-52 cyh2 dyn1::HIS3 kip3::kan kip2::URA3 (p415GALS) | |
| MAY4921 | MAT a lys2-801 his3-Δ200 leu2-3,112 ura3-52 dyn1::HIS3 kip3::kan (pFC51kip3-14) | |
| MAY4924 | MAT a lys2-801 his3-Δ200 leu2-3,112 ura3-52 dyn1::HIS3 kip3::kan (pFC51) | |
| MAY5008 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3,112 ura3-52 cyh2 dyn1::HIS3 kip3::kan kip2::URA3 (pFC51) | |
| MAY5009 | MAT a lys2-801 ade2-101 his3-Δ200 leu2-3,112 ura3-52 cyh2 dyn1::HIS3 kip3::kan kip2::URA3 (pFC51kip3-14) | |
| Plasmids | ||
| pFC50 | KIP2 LYS2 (CEN) | |
| pFC51 | KIP3 LYS2 (CEN) | |
| pFC51Kip3-14 | kip3-14 LYS2 (CEN) | |
| pFC56 | Pgals-KIP2 LEU2 (CEN) | |
| p415GALS | Pgals LEU2 (CEN) | |
| pMA1223 | KAR3 URA3 (CEN) |
Rich (YPD) and minimal (SD) media were as described (Sherman et al., 1983). Benomyl (DuPont, Wilmington, DE) was added to solid YPD medium from a 10 mg/ml stock in dimethyl sulfoxide. For G1 synchronization, α-factor (Bachem Bioscience, King of Prussia, PA) was added to 4 μg/ml to log-phase cells in liquid YPD, pH 4.0, and incubated until >80% of cells were unbudded. For arrest in S phase, hydroxyurea (Sigma Chemical Co., St. Louis, MO) was added to 0.1 M to log-phase cells in liquid YPD, pH 5.8, and incubated until >70% of cells were large budded.
DNA Manipulations
The shuttle vectors pRS316, pRS317, and pRS318 are described in Sikorski and Hieter (1989). KIP2 was obtained on a phage λ clone (No. 70186) from the American Type Culture Collection (Rockville, MD). pFC50 (KIP2 LYS2 CEN) is a 3.0-kb subclone containing KIP2 bounded by ClaI and KpnI. To put KIP2 under the control of a galactose-inducible promoter, the KIP2 gene was amplified such that an SpeI site was introduced 6 bp upstream of its open reading frame. The sequence of the 5′ primer is GAATCATCACTAGTGGTATTATGG. The resulting PCR product was subcloned into p415GALS (Mumberg et al., 1994) to create pFC56. KIP3 was obtained as a phage λ clone (No. 70127) from the American Type Culture Collection. pFC51 (KIP3 LYS2 CEN) is a 4.8-kb subclone containing KIP3 bounded by EcoRI and SpeI. pMA1223 (KAR3 URA3 CEN) was constructed by subcloning a 3.4-kb BamHI-ClaI fragment containing KAR3 from pMR1350 (a gift from Mark Rose, Princeton University, Princeton, NJ) into pRS316.
Temperature-sensitive alleles of KIP3 were generated by a mutagenic PCR-based procedure (Staples and Dieckmann, 1993). Primers corresponding to the polylinker region of pRS316 were used to amplify the entire KIP3 gene, plus 1.4 kb of upstream sequence and 1.0 kb of downstream sequence, under mutagenic PCR conditions. We used taq polymerase (Stratagene, La Jolla, CA) according to the manufacturer's instructions, except that Mn2+ and Mg2+ were added to final concentrations of 0.25 and 4.5 mM, respectively. The PCR products were concentrated using QIAquick spin columns (Qiagen, Chatsworth, CA). pFC51 was digested with MscI to create a gapped construct in which all of the KIP3 coding sequence was removed except for the last 18 bp at the 3′ end. The gapped pFC51 construct was gel purified and cotransformed with the concentrated PCR products into strain MAY4619 (kip3Δ kar3Δ lys2 ura3 [pMA1223 = KAR3 URA3]). This strain is unable to survive loss of pMA1223 and is therefore rendered sensitive to 5-fluoro-orotic acid (5-FOA; from US Biological, Swampscott, MA). Approximately 6,000 Lys+ transformants were selected at 26°C and then replica plated to 5-FOA– containing media at 26 and 35°C. 21 colonies were selected that were viable on 5-FOA at 26 but not at 35°C. Plasmids were isolated from the 21 temperature-sensitive transformants and retransformed into MAY 4619. After growth on 5-FOA at 26°C, 18 of the 21 retransformed strains displayed reduced growth on YPD at 35°C. These 18 plasmids were designated pFC51kip3-2 through pFC51kip3-19.
Microscopic Analysis of Cells
To stain for DNA, cells were pelleted out of liquid media, resuspended in 70% ethanol, and stored on ice for 30 min. The fixed cells were washed once with water and then resuspended in 0.32 μg/ml 4,6-diamidino-2-phenylindole (DAPI) plus 1 mg/ml p-phenylenediamine to prevent fading (both from Sigma Chemical Co.). For staining of chitin-containing bud scars (Pringle, 1991), cells were fixed as above, washed once with water, resuspended in 1 mg/ml calcofluor (Sigma Chemical Co.) solution, and stored at room temperature in the dark for 5 min. The cells were washed four times with water and then resuspended in 0.32 μg/ml DAPI plus 1 mg/ml p-phenylenediamine before viewing. For antitubulin immunofluorescence microscopy, cells were fixed by adding formaldehyde directly to the medium to a final concentration of 3.7%. Microtubules were visualized by the procedure described in Pringle et al. (1991) using the rat anti–α-tubulin antibody YOL1/34 (Harlan Bioproducts for Science, Indianapolis, IN) and rhodamine-conjugated goat anti–rat secondary antibodies (Jackson ImmunoResearch, West Grove, PA). Stained cells were examined with an inverted microscope (model Axiovert 135; Carl Zeiss, Inc., Thornwood, NY) equipped with epifluorescent optics using a 100× objective. Digital images were captured with a cooled, slow scan CCD camera.
For microtubule and spindle length measurements, cells were arrested with hydroxyurea at 26°C and stained for microtubule structures as described above. Images of antitubulin-stained cells were captured electronically, and the cursor-based measuring tool of the NIH Image 1.61 software program, calibrated with a stage micrometer, was used to measure the lengths of cytoplasmic microtubules and spindles. Spindle length was determined by measuring the length of the bright bar of antitubulin staining whose ends were coincident with the edges of the DAPI-stained mass. Cytoplasmic microtubules were defined as the less intensely stained fibers emanating from the spindle poles.
To quantitate binucleate cells, cultures were grown to log phase in liquid YPD at 26°C. The cells were either stained with DAPI immediately or shifted to 12°C for 24 h and then stained with DAPI. Binucleate cells were defined as those in which a single cell body contained two (or more) distinguishable DAPI-staining masses. Mother cell bodies were defined as those which contained at least two calcofluor-stained chitin rings on their surface. In our studies of DAPI-stained cells, two aberrant nuclear morphologies were quantitated: (a) Large-budded cells with a single nuclear DNA mass located away from the neck. Nucleus away-from-the-neck cells were defined as those in which the closest distance between the nucleus and the neck was greater than one half of the diameter of the entire nuclear DNA mass, as judged by eye. (b) Large-budded cells with two DAPI-staining masses located within one cell compartment.
Galactose-induced Expression of KIP2
It was discovered that fully induced expression of KIP2 (in 2% galactose) from the GALS promoter on pFC56 resulted in slow growth of wild-type cells. Note that GALS promoter activity has already been weakened by mutation (Mumberg et al., 1994). We empirically determined that a mixture of 2% galactose plus 0.05% glucose reduced the expression of KIP2 sufficiently such that growth of dyn1Δ kip2Δ kip3Δ cells was inhibited but that of wild-type cells was not. Cells to be tested were grown to log phase in liquid synthetic raffinose (2%) media lacking leucine, pH 5.8, and treated with hydroxyurea until >70% of cells were large budded. A sample was removed and fixed with ethanol. The remainder of the culture was transferred to synthetic minus leu media, pH 5.8, containing 0.1 M hydroxyurea plus 2% galactose and 0.05% glucose to induce expression of KIP2. Samples were fixed with ethanol and stained with DAPI, and the percent of large-budded cells with the nucleus away from the neck was determined as described above.
Results
Genetic Interactions between DYN1 and Other Motor Genes
S. cerevisiae cells deleted for the single dynein heavy chain–encoding gene DYN1 exhibit spindle-positioning defects but grow at wild-type rates at 26°C (Eshel et al., 1993; Li et al., 1993). Previous studies from this laboratory and others revealed that five S. cerevisiae KRP genes can also be individually deleted without affecting cell viability (Meluh and Rose, 1990; Hoyt et al., 1992; Lillie and Brown, 1992; Roof et al., 1992). For this study, we created a deletion allele of KIP3, the sixth and final KRP gene revealed by the Saccharomyces Genome Sequencing Project. The entire KIP3 open reading frame was replaced with a bacterial kanamycin resistance gene (see Materials and Methods). Yeast cells deleted for KIP3 were viable and did not exhibit any obvious growth defects at incubation temperatures between 11 and 37°C. Mild microtubule-related phenotypes for kip3Δ cells were observed and are described below.
In an attempt to define the motor(s) that overlap in function with dynein, we created double mutants between dyn1Δ and deletion alleles of the six KRP motor genes. For kip1Δ, kip2Δ, and smy1Δ, viable double mutant cells were recovered that grew at rates comparable to the wild-type at 26°C. In contrast, we were unable to recover viable combinations of dyn1Δ with cin8Δ (Saunders et al., 1995) and kar3Δ. dyn1Δ kip3Δ mutant cells were viable, but grew into colonies at very slow rates (see Figs. 2 C and 5). dyn1Δ kip3Δ cultures also contained high numbers of dead cells detected by both microscopy and by a 48% reduced plating efficiency relative to wild-type (plating efficiency = colony forming units/cells counted in microscope). These findings suggest that Cin8p, Kar3p, and Kip3p overlap with dynein for an essential or important function.
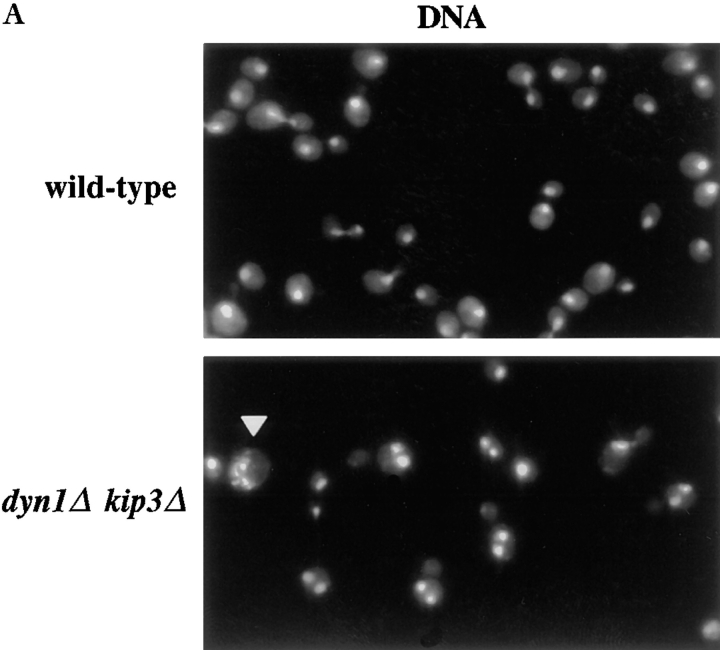
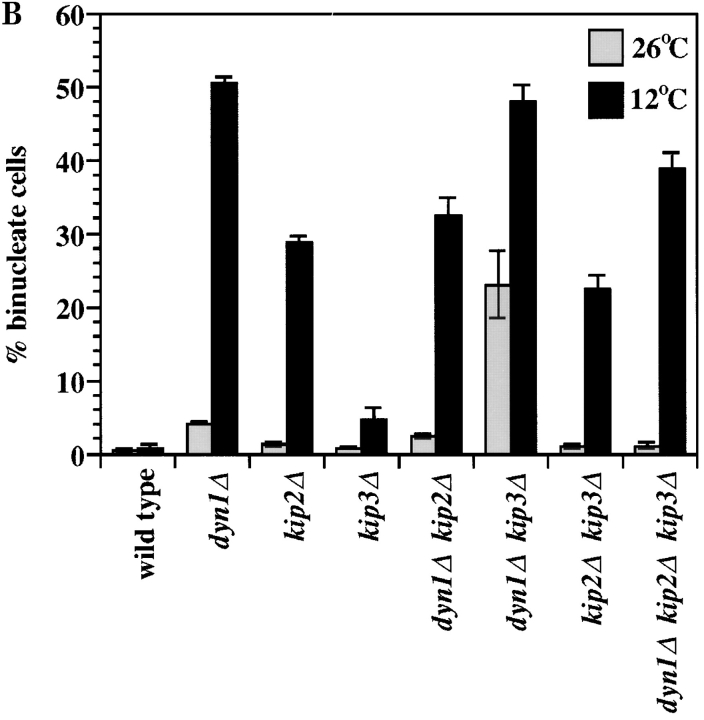
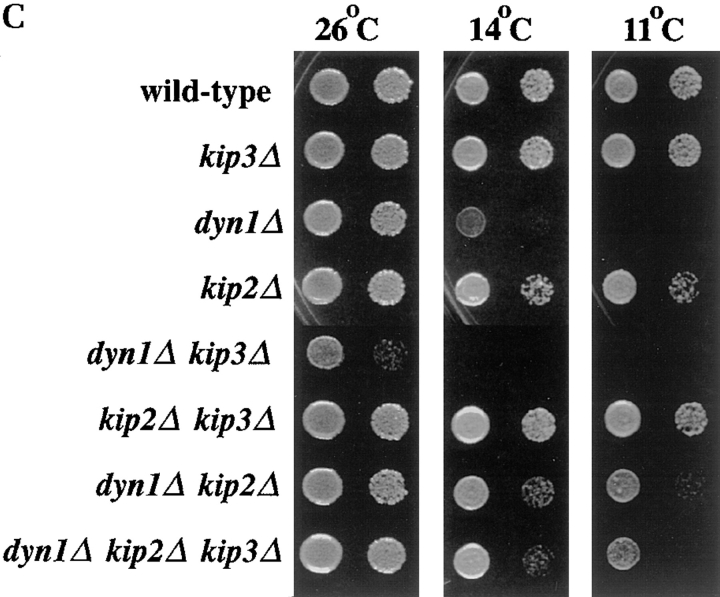
Figure 2.
Binucleate cell production and growth properties of motor mutants. (A) dyn1Δ kip3Δ cells form binucleate cells at 26°C. The arrowhead points to a cell with an aberrant nuclear morphology. (B) Quantitation of binucleate cells produced at 26 and 12°C. For A and B, cells of the indicated genotypes were grown in rich media to log phase at 26°C and stained with DAPI or shifted to 12°C for 24 h before staining. Binucleate cells are those in which two or more nuclear DNA masses could be visualized in one cell body. Each value represents the average of three experiments in which 200 cells were examined. Error bars indicate the SEM. (C) Cells of the indicated genotypes were spotted onto solid media and incubated at the temperature noted. Two spots for each combination are shown, with the second equal to a 40-fold dilution of the first. Yeast strains used: wild-type (MAY589), kip3Δ (MAY4517), dyn1Δ (MAY4434), kip2Δ (MAY2038), dyn1Δ kip3Δ (MAY4544), kip2Δ kip3Δ (MAY4716), dyn1Δ kip2Δ (MAY4566), and dyn1Δ kip2Δ kip3Δ (MAY4560).
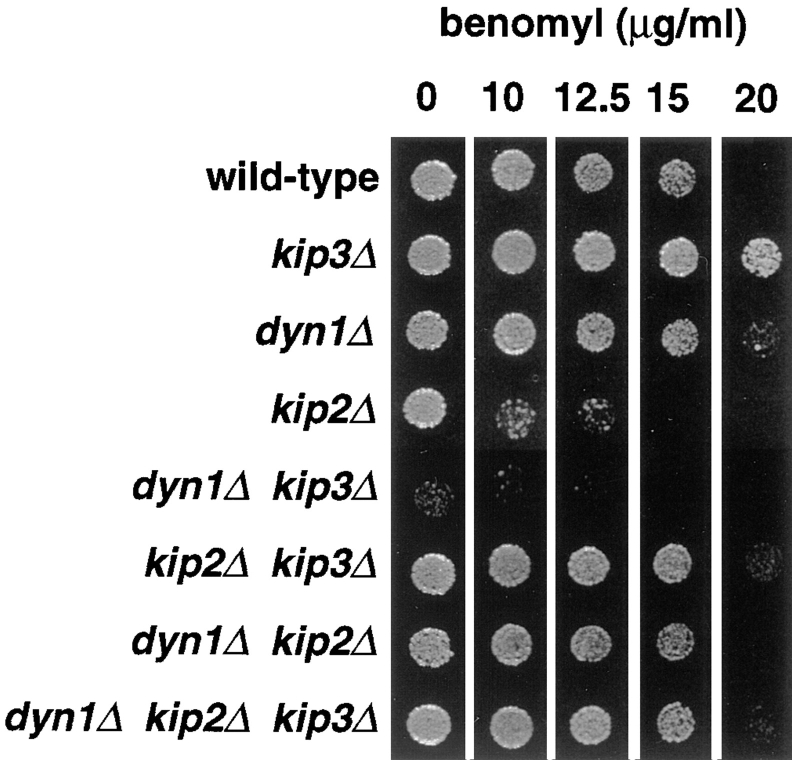
Figure 5.
Benomyl resistance of motor mutants. Cells of the indicated genotypes were spotted to solid rich media containing the concentrations of benomyl indicated. Plates were incubated at 26°C for 3 d. Strains used are the same as in Fig. 2.
In our genetic studies, we noticed an interesting property of the KIP2 deletion allele. kip2Δ single mutant cells exhibited a nuclear migration defect almost as severe as that displayed by dyn1Δ cells (see below). Unexpectedly, the deletion of KIP2 was found to suppress the deleterious effects of the dyn1Δ kip3Δ and dyn1Δ kar3Δ genotypes (Table II). Although dyn1Δ kip3Δ cells exhibited extremely poor colony-forming ability, the dyn1Δ kip3Δ kip2Δ triple mutants formed colonies of wild-type size. dyn1Δ kar3Δ double mutants were inviable. Strikingly, dyn1Δ kar3Δ kip2Δ triple mutants were viable, but formed colonies slightly smaller than wild-type. A demonstration of these antagonistic actions of KIP2 is shown in Fig. 1. A vector plasmid and a plasmid expressing KIP2 were both able to transform wild-type cells with high efficiency. In contrast, dyn1Δ kip3Δ kip2Δ cells and dyn1Δ kar3Δ kip2Δ cells were transformed well by the vector plasmid, but not the KIP2 plasmid. In contrast to the dyn1Δ kip3Δ and dyn1Δ kar3Δ combinations, elimination of KIP2 was unable to suppress the growth defect of kip3Δ kar3Δ cells; both kip3Δ kar3Δ and kip3Δ kar3Δ kip2Δ spores were unable to grow into colonies. Table II summarizes the growth properties of all the dyn1Δ, kip3Δ, kar3Δ, and kip2Δ mutant combinations.
Table II.
Growth Properties of Relevant Motor Mutants
| Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|
| DYN1 | KIP3 | KAR3 | KIP2 | Growth | ||||
| Δ | + | + | + | V | ||||
| + | Δ | + | + | V | ||||
| + | + | Δ | + | V | ||||
| + | + | + | Δ | V | ||||
| Δ | Δ | + | + | V (very slow) | ||||
| Δ | + | Δ | + | I* | ||||
| Δ | + | + | Δ | V | ||||
| + | Δ | Δ | + | I* | ||||
| + | Δ | + | Δ | V | ||||
| + | + | Δ | Δ | V | ||||
| Δ | Δ | Δ | + | I | ||||
| Δ | Δ | + | Δ | V | ||||
| Δ | + | Δ | Δ | V (slightly slow) | ||||
| + | Δ | Δ | Δ | I | ||||
| Δ | Δ | Δ | Δ | I | ||||
V, viable. Indicates that spores of the indicated genotype were able to grow into a colony. I, inviable. Indicates that spores of the indicated genotype were not able to germinate and /or grow into a colony. +, wild-type allele. Δ, deletion mutant allele.
For these genotypes, inviability was also indicated by the inability to survive loss of pMA1223 (KAR3 URA3), rendering these cells sensitive to 5-fluoro-orotic acid.
Figure 1.
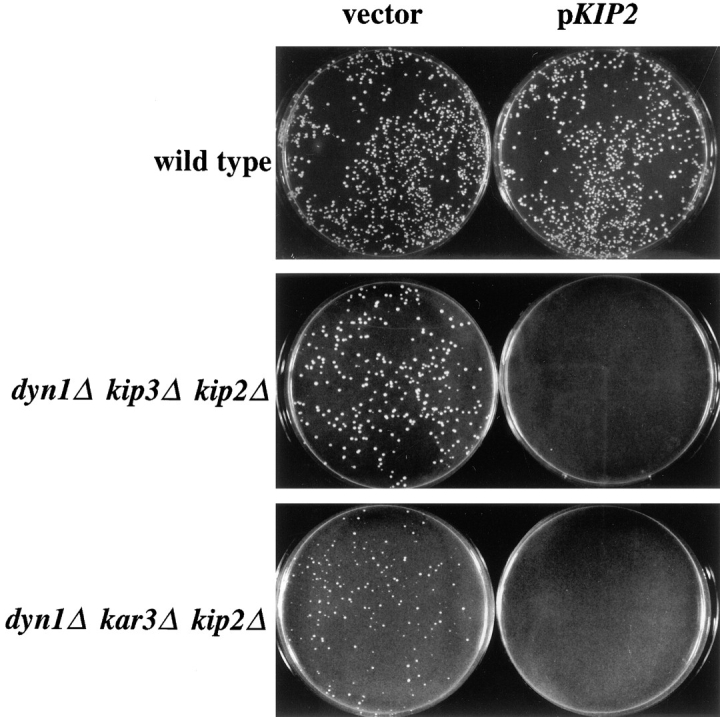
KIP2 is responsible for the growth defects of dyn1Δ kip3Δ and dyn1Δ kar3Δ cells. Wild-type (MAY591), dyn1Δ kip3Δ kip2Δ (MAY4560), and dyn1Δ kar3Δ kip2Δ (MAY4761) yeast strains were transformed with the KIP2-containing plasmid pFC50 and with a vector control (pRS317). Transformation plates were incubated at 26°C for 2 d.
The suppression by kip2Δ of combinations of DYN1, KIP3, and KAR3 alleles functionally links the products of these genes. In contrast, the lethality of dyn1Δ cin8Δ could not be suppressed by deletion of KIP2. In addition, cin8Δ kip2Δ mutants, as well as dyn1Δ cin8Δ and dyn1Δ cin8Δ kip2Δ, were found to be inviable. The cause of the lethality of these cin8Δ combinations is not known. We have previously suggested that the lethality of the Cin8p Dyn1p–deficient combination reflects the contribution of both of these motors to anaphase spindle elongation (Saunders et al., 1995).
In summary, the deleterious effects of dyn1Δ kip3Δ and dyn1Δ kar3Δ, but not kip3Δ kar3Δ, were strongly suppressed by the deletion of KIP2. These findings indicate that the loss of Dyn1p plus either Kip3p or Kar3p can be tolerated, providing that an antagonistically acting function of Kip2p is eliminated. In the remainder of this article, we characterize the spindle positioning and other microtubule phenotypes associated with loss of function of Dyn1p, Kip3p, and Kip2p. The role of Kar3p in spindle positioning has been harder to discern, probably because this motor has more than one mitotic function (see Discussion).
dyn1Δ, kip2Δ, and kip3Δ Cause Spindle-positioning Defects
S. cerevisiae spindle-positioning errors lead to anaphase nuclear division occurring exclusively in the mother cell body. This produces a binucleate mother cell body and an anucleate bud (Fig. 2 A). We examined the dyn1Δ, kip2Δ, and kip3Δ single and multiple mutant combinations for the production of binucleate cell bodies at 26 and 12°C (Fig. 2 B). As previously reported (Eshel et al., 1993; Li et al., 1993; Geiser et al., 1997), dyn1Δ cultures accumulated elevated binucleate cells in a temperature-dependent fashion; very high levels were observed after incubation at 12°C. The kip2Δ mutant behaved similarly, although to a lesser extent. The kip3Δ mutant was only slightly elevated for binucleate cell production at 12°C. However, a role for KIP3 in spindle positioning was indicated by the elevated number of binucleate cells in cultures of the slow-growing dyn1Δ kip3Δ double mutant. This was the only mutant that displayed high levels (>20%) of binucleate cells at 26°C. dyn1Δ kip3Δ double mutant cultures also contained significant numbers of cells with unusual nuclear morphologies (arrow in Fig. 2 A) and anucleate or dead cells. Therefore, the binucleate percentage values determined for this genotype might underrepresent the extent of the defect. The dyn1Δ kip2Δ and kip2Δ kip3Δ double mutants also displayed binucleate cell levels that were greatly elevated over the wild-type, but only at 12°C. Note that the levels exhibited by dyn1Δ kip2Δ and kip2Δ kip3Δ were intermediate to that exhibited by the single mutants. The deletion of KIP2 also partially relieved the cold-sensitive growth defect caused by dyn1Δ (Fig. 2 C). The dyn1Δ kip2Δ kip3Δ triple mutant was reduced for binucleate cell production at 26°C relative to the dyn1Δ kip3Δ double mutant. This reduction corresponded to the greatly improved growth rate of the triple mutant relative to the double mutant (Fig. 2 C).
We examined the mutants to determine if the cell bodies containing two nuclear DNA masses corresponded to the mother or the bud. The mother cell was distinguished from the bud by the presence of chitin-containing bud scars revealed by calcofluor staining. For the dyn1Δ, kip2Δ, and kip3Δ single mutants grown at 12°C, the binucleate bodies were always mother cells (n = 104, 54, and 12 cells, respectively).
Cytoplasmic Microtubules Are Longer than Wild-Type in dyn1Δ and kip3Δ Cells but Shorter in kip2Δ Cells
To examine the causes of the spindle-positioning defects exhibited by the single and multiple mutants, microtubules were visualized by antitubulin immunofluorescence microscopy (Figs. 3 and 4). Before fixation with formaldehyde, the DNA synthesis inhibitor hydroxyurea was added to the cultures to synchronize cells at the preanaphase short spindle stage of the mitotic cycle (Pringle and Hartwell, 1981). Using this technique, stained wild-type spindles appeared as a bright bar of nuclear microtubules with whisker-like cytoplasmic microtubules attached to the ends of the bars (the spindle poles). Striking differences were observed in the length and number of cytoplasmic microtubules in the various mutant genotypes. dyn1Δ and kip3Δ cells exhibited longer cytoplasmic microtubules than wild-type. The slow-growing dyn1Δ kip3Δ double mutant exhibited extremely elongated cytoplasmic microtubules that were on average fourfold longer than the wild-type. In contrast, cytoplasmic microtubules in kip2Δ cells were shorter than wild-type. The kip2Δ culture also exhibited much higher numbers of cells for which we could not visualize a single cytoplasmic microtubule that survived fixation (Fig. 4 A). Combining the short microtubule kip2Δ with either dyn1Δ or kip3Δ caused cytoplasmic microtubule phenotypes resembling those caused by kip2Δ alone (shortened and reduced in number). By both criteria, however, these doubles were less severely affected than the kip2Δ single mutant, consistent with an intermediate phenotype. The elimination of KIP2 in dyn1Δ kip3Δ cells, to create the triple mutant, suppressed the extremely long cytoplasmic microtubule phenotype of this double mutant, resulting in cytoplasmic microtubules intermediate in length between wild-type and kip2Δ. In these experiments, we also detected small but reproducible differences in spindle lengths (pole-to-pole distance; Fig. 4 B). In particular, spindles were longer than wild-type in all strains deleted for KIP3. We note that while formaldehyde fixation may not preserve all yeast microtubule structure (see Carminati and Stearns, 1997), the differences observed here probably reflect actual differences in microtubule lengths and/or stabilities in vivo (see next section).
Figure 3.
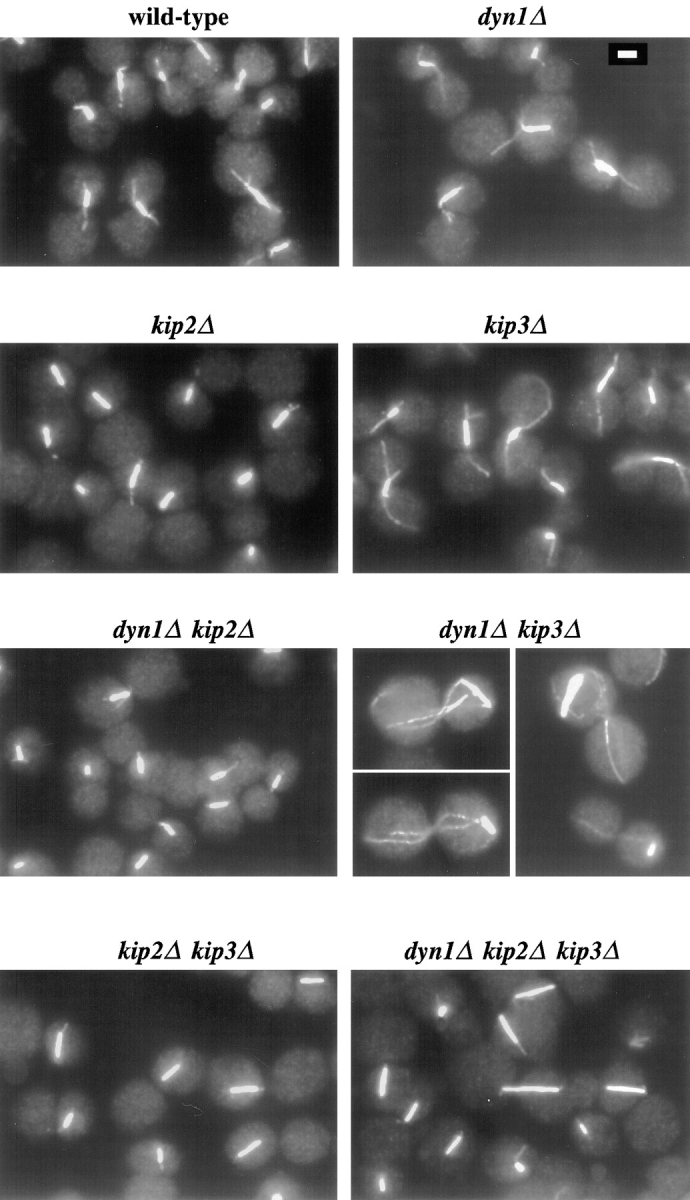
Antitubulin immunofluorescence microscopy of motor mutants. Cells were synchronized with hydroxyurea in rich media at 26°C, fixed with formaldehyde, and processed for antitubulin immunofluorescence microscopy. Strains used are the same as in Fig. 2. Bar (top right), 2 μm.
Figure 4.
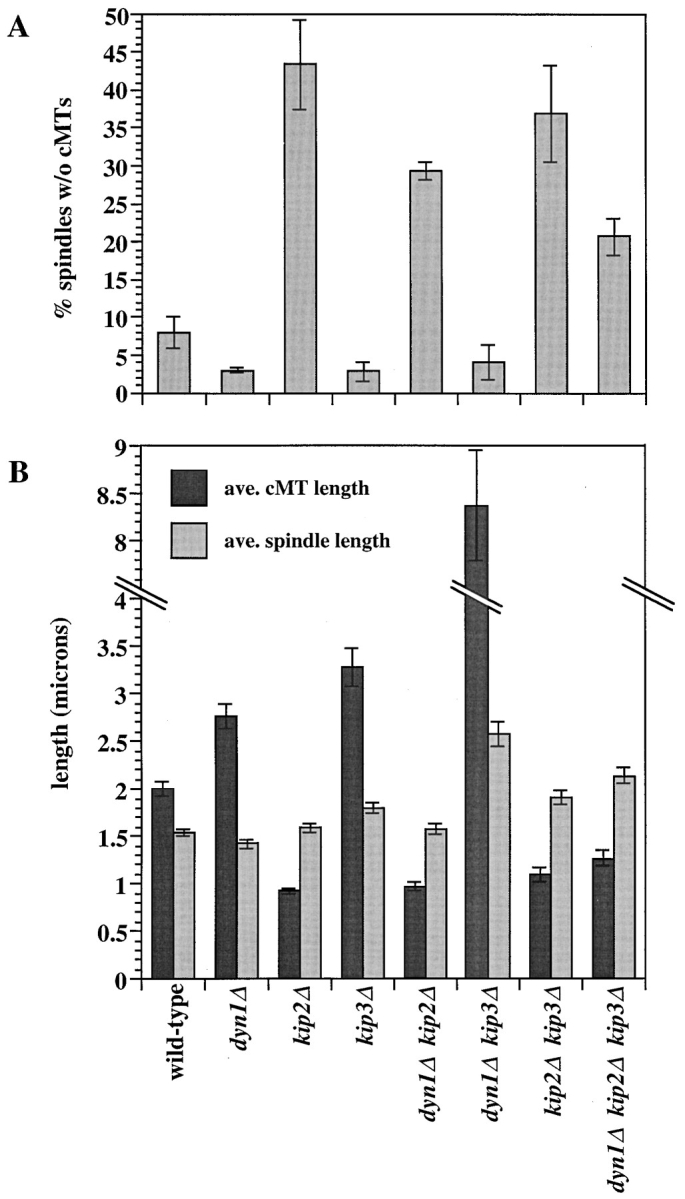
Quantitation of microtubule phenotypes. Digitized images of the cells from Fig. 3 were analyzed for microtubule characteristics. (A) The percent of spindles with no visible cytoplasmic microtubules (cMTs). Values were determined from scoring all cells in four microscope fields (∼40 cells per field) and averaging. (B) The lengths of spindles (pole-to-pole distance) and cMTs were measured (see Materials and Methods). Values equal the average of between 94 to 166 measurements. For both A and B, error bars indicate the SEM.
dyn1Δ and kip3Δ Increase Resistance to Benomyl while kip2Δ Decreases Resistance
The observed differences in cytoplasmic microtubule lengths suggested differences in microtubule stabilities among the various motor mutants. The resistance of yeast cells to compounds that promote microtubule depolymerization, such as benomyl, reflects the intrinsic stability of cellular microtubules. We determined resistance to benomyl by spotting cells onto solid media containing increasing concentrations of the inhibitor (Fig. 5). We found that the dyn1Δ, kip2Δ, and kip3Δ genotypic combinations exhibited opposing and suppressing phenotypes that corresponded to the microtubule length phenotypes described above. kip3Δ cells displayed higher resistance to benomyl than the wild-type. dyn1Δ cells displayed resistance that was only slightly but reproducibly elevated over wild-type. In contrast, kip2Δ cells displayed markedly decreased benomyl resistance. It was not possible to accurately assess the benomyl resistance of the dyn1Δ kip3Δ double mutant because of its extreme slow growth. The two other double mutant combinations, both of which involved kip2Δ, displayed resistances intermediate between those observed for the single mutants. Deletion of either dyn1Δ or kip3Δ was able to restore near wild-type levels of resistance to kip2Δ cells. The dyn1Δ kip2Δ kip3Δ triple mutant also displayed resistance near that of the wild-type.
Dyn1p and Kip3p Are Required to Achieve and Maintain Proper Spindle Positioning in the Presence of Kip2p
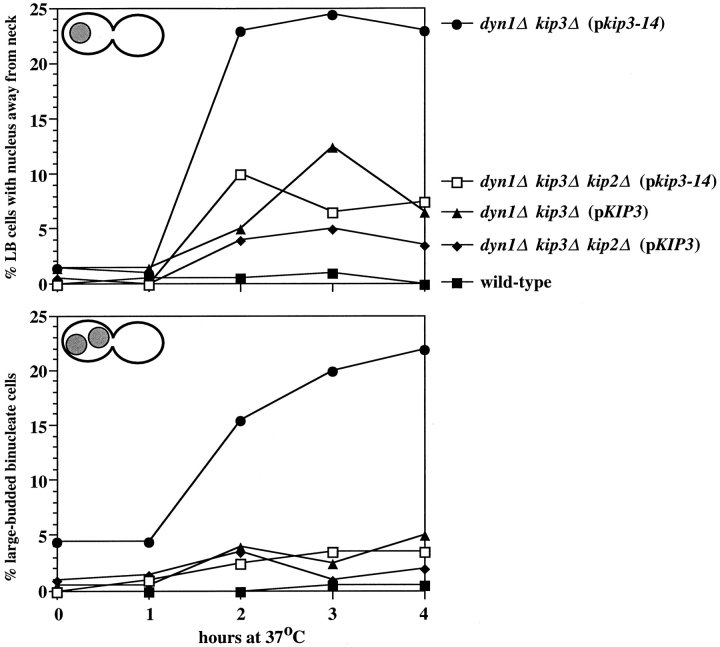
To determine the effects of loss of Kip3p function in various genetic backgrounds, 18 temperature-sensitive alleles of KIP3 were generated using a mutagenic PCR-based approach (see Materials and Methods). A representative temperature-sensitive allele, kip3-14, was selected for further study. Plasmids carrying kip3-14 or KIP3 were introduced into a dyn1Δ kip3Δ double mutant strain, as well as into a dyn1Δ kip2Δ kip3Δ triple mutant strain. The resulting strains, along with a wild-type control, were synchronized in G1 with the α-factor mating pheromone at the permissive temperature of 26°C. The G1 cells were released from the α-factor block, shifted to the nonpermissive temperature (37°C), and assayed for their ability to proceed through mitosis by observation of stained nuclear DNA. As wild-type cells reached a large-budded morphology (bud diameter ∼3/4 that of the mother), nuclear DNA masses were either positioned in the neck or had undergone anaphase division with a single DNA mass positioned in each progeny cell body. For the dyn1Δ kip3Δ (pkip3-14) genotype only, cells with two morphologies that were rarely observed for wild-type accumulated after the shift to 37°C (Fig. 6): large-budded cells with nuclear DNA at a position away from the neck (top) and large-budded cells with two nuclear DNA masses in one cell body (bottom). This finding indicates that Kip3p function is required for efficient spindle positioning in the absence of dynein. The nucleus-away-from-the-neck morphology was somewhat elevated over the wild-type for the three other strains that were deleted for DYN1 (Fig. 6, top). However, as previously described for dyn1Δ mutants (Yeh et al., 1995), these did not lead to the formation of binucleate cell bodies (Fig. 6, bottom), presumably because of the actions of other motors that can resolve dyn1Δ spindle- positioning errors (i.e., Kip3p). Notably, the dyn1Δ kip2Δ kip3Δ (pkip3-14) cells did not accumulate the aberrant forms, indicating that elimination of Kip2p could suppress the spindle-positioning defect exhibited by dyn1Δ kip3Δ (pkip3-14). This also demonstrates that another mechanism acts to position spindles efficiently in the absence of dynein, Kip3p, and Kip2p.
Figure 6.
Loss of Kip3p function in dynein-deficient cells caused spindle-positioning defects. Cell cultures of the indicated genotypes were synchronized with α-factor at 26°C and then shifted to 37°C. At the time points indicated, samples were removed and stained with DAPI. The top panel indicates the percent of cells that were large budded with the nucleus positioned away from the neck (defined as those in which the closest distance between the nucleus and the neck was greater than one half of the diameter of the entire nuclear DNA mass). The bottom panel indicates the percent of cells that were large budded with two DAPI-staining masses located within one cell body. •, dyn1Δ kip3Δ (pkip3-14) (MAY4921); □, dyn1Δ kip2Δ kip3Δ (pkip3-14) (MAY5009); ▴, dyn1Δ kip3Δ (pKIP3) (MAY4924); ♦, dyn1Δ kip2Δ kip3Δ (pKIP3) (MAY5008); ▪, wild-type (MAY589).
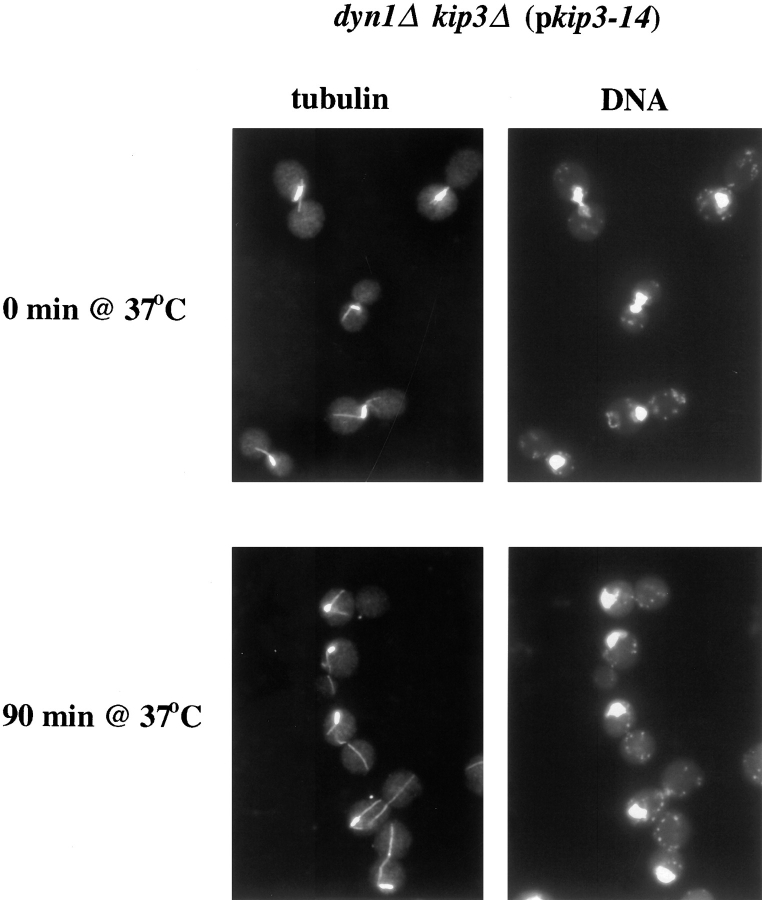
We next examined the consequence of loss of Kip3p function in cells that had completed spindle positioning but had not entered anaphase. Cells of the genotypes described above were grown and treated with hydroxyurea at 26°C. At the hydroxyurea arrest point, the majority of cells had short preanaphase spindles and DNA masses that were positioned in or up against the mother–bud neck (Figs. 7 and 8 A). For all dyn1Δ mutant genotypes, the number of cells with nuclei located away from the neck was slightly elevated over wild-type because of the spindle-positioning defect caused by loss of dynein. Maintaining the presence of hydroxyurea, the cells were then shifted to 37°C. After the shift, the number of nucleus-away-from-neck cells remained fairly constant for all genotypes with the exception of dyn1Δ kip3Δ (pkip3-14). The temperature-induced loss of Kip3p function in the dynein mutant background caused spindles and DNA masses that previously had been positioned at the neck to move to a position distal to the neck. Calcofluor staining of bud scars indicated that the DNA masses had moved exclusively back into the mother cell body (n = 200 cells). The spindles and associated DNA were usually positioned at the peripheral region of the mother cell distal to the neck, with extensive cytoplasmic microtubule arrays directed toward the bud (Fig. 7).
Figure 7.
The spindle and nucleus were drawn away from the mother–bud neck after loss of Kip3p function in the absence of Dyn1p. Cell cycle progression of dyn1Δ kip3Δ (pkip3-14) strain (MAY4921) was arrested by growth in the presence of hydroxyurea at 26°C. The culture was then shifted to 37°C for 90 min. Samples taken before (top row) and after (bottom row) the temperature shift were processed for antitubulin immunofluorescence (left column) and DAPI staining (right column).
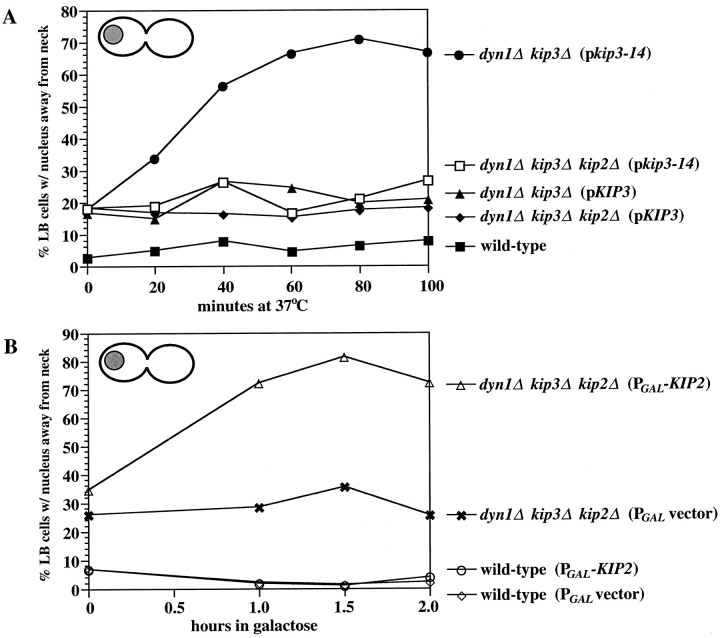
Figure 8.
Kip2p caused nuclear mislocalization in cells deficient for dynein and Kip3p. (A) Quantitation of large-budded cells with mislocalized nuclei after hydroxyurea arrest at 26°C and then a shift to 37°C for the indicated times (see Fig. 7). Strains used were the same as in Fig. 6. (B) Quantitation of large-budded cells with mislocalized nuclei caused by galactose-induced expression of KIP2. Cells were synchronized with hydroxyurea and then switched to media containing galactose to induce expression of KIP2. Samples were analyzed at the indicated times after the addition of galactose. ▵, dyn1Δ kip2Δ kip3Δ (PGAL-KIP2) (MAY4819); ✖, dyn1Δ kip2Δ kip3Δ (PGAL-vector) (MAY4820); ○, wild-type (PGAL-KIP2) (MAY4816); ⋄, wild-type (PGAL-vector) (MAY4817).
The spindle mislocalization phenotype exhibited by dyn1Δ kip3Δ (pkip3-14) was dependent upon Kip2p activity. The deletion of KIP2 suppressed the temperature- induced loss of spindle positioning (see dyn1Δ kip2Δ kip3Δ (pkip3-14) genotype in Fig. 8 A). This strongly suggests that in the absence of Dyn1p and Kip3p activity, Kip2p function directly leads to spindle mislocalization. Additional evidence that Kip2p is responsible for this effect was obtained in an experiment in which KIP2 expression was placed under the control of a galactose-inducible promoter (Fig. 8 B). PGAL-KIP2 or PGAL control plasmids were transformed into wild-type and dyn1Δ kip2Δ kip3Δ strains. After synchronization with hydroxyurea, galactose was added to induce expression from the galactose promoter. Expression of KIP2 in cells deficient for both DYN1 and KIP3 caused properly positioned spindles to mislocalize to sites away from the neck. Neither wild-type cells expressing the same PGAL-KIP2 construct nor cells carrying the PGAL vector displayed nuclear mislocalization when galactose was added.
Discussion
Dynein motor function is important but not essential for spindle positioning in S. cerevisiae. Although spindle-positioning errors can be detected in dyn1Δ cells growing at room temperature, the extent of this defect is not sufficient to reduce the rate of cell division. Presumably, other motor mechanisms contribute to spindle positioning in the absence of dynein. The experiments described here demonstrated that the Kip3p KRP provides an important spindle-positioning function in the absence of dynein. The elimination of Kip3p function in dynein-deficient cells severely compromised spindle movement to the mother–bud neck. Elimination of Kip3p function in dyn1Δ cells that had completed positioning caused spindles to mislocalize into mother cell bodies. We also demonstrated that the spindle-positioning defects exhibited by dyn1 kip3 cells are caused, to a large extent, by the actions of the Kip2p KRP. kip2Δ cells exhibited microtubule phenotypes that were opposite to those exhibited by dyn1Δ and kip3Δ cells. Microtubules in kip2Δ cells were shorter and more sensitive to the action of benomyl than wild-type microtubules, while those of dyn1Δ and kip3Δ cells were longer and more resistant. kip2Δ dyn1Δ and kip2Δ kip3Δ double mutants displayed intermediate microtubule phenotypes. Most significantly, elimination of KIP2 function greatly suppressed the spindle localization defect and slow growth exhibited by dyn1 kip3 cells. Likewise, induced expression of KIP2 caused spindles to mislocalize in cells deficient for dynein and Kip3p.
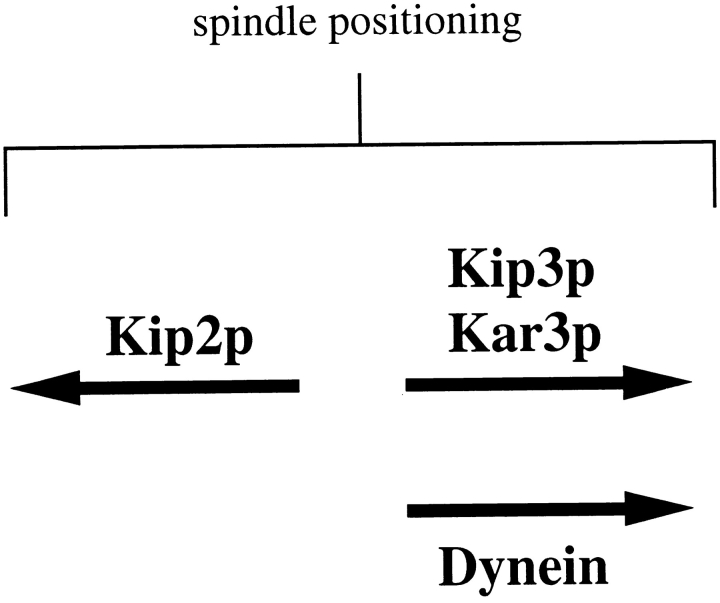
Although Kip2p normally contributes to spindle positioning, its actions can also antagonize a positioning mechanism. In the absence of dynein and Kip3p function, Kip2p activity caused spindles to move to a location in the mother cell distal to the neck. Since kip2Δ suppressed the growth defect of the dyn1Δ kip3Δ double deletion mutant, the antagonistic actions of Kip2p must be directed against whatever is accomplishing spindle positioning in the absence of dynein and Kip3p. Our genetic findings suggest that this activity is provided by the Kar3p KRP. This would be a novel role for the minus end-directed Kar3p motor whose characterized mitotic functions include pulling spindle poles inwardly prior to anaphase and contributing to bipolar spindle structure (Meluh and Rose, 1990; Saunders and Hoyt, 1992; Hoyt et al., 1993; Endow et al., 1994; Saunders et al., 1997b ). Kar3p has also been associated with kinetochore movement along microtubules in vitro (Middleton and Carbon, 1994). Participation of Kar3p in spindle positioning was suggested by the observation that deletion of KIP2 suppressed the lethality of the dyn1Δ kar3Δ combination. A simple hypothesis consistent with these findings is that Kip2p antagonizes the spindle-positioning activities of both Kip3p and Kar3p (Fig. 9). In the absence of dynein, both Kip3p and Kar3p are required to position spindles. If the antagonistic actions of Kip2p are removed, however, then either Kip3p or Kar3p alone is sufficient. kip3Δ kar3Δ cells are inviable and are not suppressed by kip2Δ. This suggests that Kip3p and Kar3p overlap for an essential function that dynein alone cannot provide. In mitotic cells, most Kar3p is concentrated near the spindle poles with a smaller but detectable amount spreading onto the nuclear microtubules (Page et al., 1994; Saunders et al., 1997a ). It is possible that some Kar3p also acts upon the cytoplasmic microtubules to affect spindle positioning. We note that Kar3p is essential for the nuclear migration event that occurs during karyogamy (nuclear fusion during mating). In this capacity, Kar3p acts on the cytoplasmic microtubules to move haploid nuclei towards each other before their fusion (Meluh and Rose, 1990). Kip3p does not appear to overlap with Kar3p for a karyogamy role since Kar3p is essential for this process and kip3Δ cells did not display a defect in karyogamy proficiency (Cottingham, F.R., and M.A. Hoyt, unpublished observations).
Figure 9.
Proposed functional relationship for the Kip2p, Kip3p, Kar3p, and dynein motors. The antagonistic actions of these motors leads to normal spindle positioning. Kip2p is proposed to antagonize the actions of Kip3p and Kar3p because deletion of KIP2 suppressed dyn1Δ kip3Δ and dyn1Δ kar3Δ, but not kip3Δ kar3Δ. Kip3p and Kar3p share a single arrow since they appear to overlap for an essential function that cannot be accomplished by dynein. Dynein is proposed to act in a distinct pathway leading to spindle positioning, although it is possible that it is antagonized by Kip2p as well.
The molecular roles and mechanisms for the four spindle-positioning motors described here are currently not clear. Below, we describe two general models for the roles and interactions of these motors. These models are not mutually exclusive. In the first model, spindle position is determined by directly antagonistic motor activities. The cooperative actions of dynein, Kip3p, and Kar3p exert a force on one spindle pole, pulling it into the bud cell body. At the same time, Kip2p is acting to pull the other pole back towards the mother cell. This antagonism would result in proper positioning and orientation of the spindle along the mother–bud axis. As noted above, we do not view the three bud-directed activities as equivalent. Dynein is unable to perform some activity provided by either Kip3p or Kar3p. Yeh et al. (1995) found that dyn1Δ cells undergo nuclear division that appears different from DYN1 cells. The dynein-deficient cells often performed anaphase at an incorrect position in the mother cell, followed by a phase in which a daughter nucleus was translocated to the bud. It is possible that dynein is primarily responsible for achieving proper spindle orientation before anaphase onset while Kip3p and Kar3p act after anaphase onset in a manner that can correct preanaphase positioning errors. Not all of our findings are easily accommodated by this simple model, however. Although Kip2p was responsible for aberrant spindle movement into the mother in cells deficient for dynein and Kip3p, spindle mislocalization in kip2Δ mutants, similar to dyn1Δ mutants, occurred exclusively in the mother. This finding does not exclude the possibility that the main role of Kip2p is to pull the spindle towards the mother cell, but it does require a more complicated mechanism. Perhaps proper cytoplasmic microtubule connection on the bud side requires that a tension-producing force be exerted from the mother side. In the absence of the mother-directed force supplied by Kip2p, connections in the bud are made improperly, and the spindle remains in the mother cell. A similar tension-based mechanism has been proposed for the attachment of sister kinetochores to spindle fibers (Nicklas, 1997). Spindle fiber–kinetochore connections are unstable in the absence of tension caused by a proper bipolar attachment. A second finding that is difficult to explain by this model is that dyn1 kip3 cells occasionally possessed cytoplasmic microtubules extending towards the bud from both poles of mislocalized spindles (Figs. 3 and 7). This morphology is unexpected if the spindle is being pulled back into the mother cell body by motors operating on a set of cytoplasmic microtubules.
The second model is based upon the opposing microtubule phenotypes exhibited by the motor mutants. dyn1Δ and kip3Δ cells exhibited cytoplasmic microtubules that were longer and more resistant to benomyl treatment than wild-type. Another group has also observed longer and less dynamic microtubules in dyn1Δ cells (Carminati and Stearns, 1997). It has recently been reported that kar3Δ cells also display longer cytoplasmic microtubules (Saunders et al., 1997a ). Coupled with the observation that Kar3p can contribute to microtubule depolymerization in vitro (Endow et al., 1994), it was proposed that Kar3p contributes to microtubule shortening in vivo (Saunders et al., 1997a ). The ability to depolymerize microtubules has also been observed for the XKCM1 KRP motor of Xenopus (Walczak et al., 1996). The observation that dyn1Δ and kip3Δ cells exhibited microtubule phenotypes similar to kar3Δ cells suggests that dynein and Kip3p also contribute to microtubule shortening processes. In contrast, our observation of shorter and benomyl-hypersensitive microtubules in kip2Δ cells suggests that Kip2p actions are required to stabilize microtubules against depolymerization. Perhaps the motor actions of Kip2p contribute in some manner to microtubule polymerization, or the binding of Kip2p to sites on the microtubule lattice increases its stability. For our second model, we propose that the observed spindle-positioning effects were caused by changes in cytoplasmic microtubule polymerization/depolymerization properties. For example, the extremely long cytoplasmic microtubules created in dyn1 kip3 cells may physically mislocalize the spindle. The suppression by kip2Δ may be the result of lowered microtubule stability caused by loss of Kip2p. We note, however, that benomyl treatment did not suppress the dyn1 kip3 growth defect (Fig. 5). Another possibility is that the aberrant polymerization properties of the cytoplasmic microtubules precluded their ability to form the proper cortical connections required to move nuclei. However, it is also possible that the microtubule phenotypes we observed were indirect effects caused by defects in the interactions of microtubules, motors, and the cell cortex.
Both models proposed here require that the spindle-positioning motors act upon the cytoplasmic microtubules. It has been reported that some of the cellular Dyn1p, Kip3p, and Kip2p molecules can be detected in association with cytoplasmic microtubules in mitotic cells (Miller, R.K., and M.D. Rose. 1995. Mol. Biol. Cell. 65:256a; Yeh et al., 1995; DeZwaan et al., 1997). As described above, Kar3p has also been detected on cytoplasmic microtubules, but only in cells undergoing karyogamy (Meluh and Rose, 1990; Page et al., 1994).
Although the mechanisms leading to spindle positioning are currently unclear, we note with interest that both bipolar spindle assembly and spindle positioning are accomplished by antagonistically acting motor proteins. Assembly of the S. cerevisiae preanaphase spindle requires the actions of two KRPs from the BimC family, Cin8p and Kip1p (Hoyt et al., 1992; Roof et al., 1992). The spindle pole–separating actions of the BimC KRPs are antagonized before anaphase by the inwardly directed force produced by Kar3p (Saunders and Hoyt, 1992; Hoyt et al., 1993; Saunders et al., 1997b ). Proper spindle structure is achieved by the balance between the outwardly and inwardly directed forces acting upon the spindle poles. At roughly the same period of mitosis, dynein, Kip3p, and Kar3p, acting antagonistically to Kip2p, are moving and orienting the spindle to permit proper chromosome segregation to progeny cells.
Acknowledgments
The authors wish to thank Mark Rose, and Susan Brown and Sue Lillie (University of Michigan, Ann Arbor, MI) for gifts of mutant strains and Cindy Dougherty, Katie Farr, John Geiser, and Emily Hildebrandt for their comments on the manuscript.
Abbreviations used in this paper
- 5-FOA
5-fluoro-orotic acid
- DAPI
4,6-diamidino-2-phenylindole
- KRP
kinesin-related protein
Footnotes
Address all correspondence to M. Andrew Hoyt, Department of Biology, The Johns Hopkins University, Baltimore, MD 21218. Tel.: (410) 516-7299. Fax: (410) 516-5213. E-mail: hoyt@jhu.edu
These experiments were supported by National Institutes of Health grant GM40714 awarded to M.A. Hoyt.
References
- Book KJ, Howard R, Morest DK. Direct observation in vitro of how neuroblasts migrate: medulla and cochleovestibular ganglion of the chick embryo. Exp Neurol. 1991;111:228–243. doi: 10.1016/0014-4886(91)90011-z. [DOI] [PubMed] [Google Scholar]
- Book KJ, Morest DK. Migration of neuroblasts by perikaryal translocation: role of cellular elongation and axonal outgrowth in the acoustic nuclei of the chick embryo medulla. J Comp Neurol. 1990;297:55–76. doi: 10.1002/cne.902970105. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin- related KIP3 of Saccharomyces cerevisiaeis required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO (Eur Mol Biol Organ) J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. S. cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, Hyman AA. Cortical domains and the mechanisms of asymmetric cell division. Trends Cell Biol. 1996;6:382–387. doi: 10.1016/0962-8924(96)10035-0. [DOI] [PubMed] [Google Scholar]
- Hager G, Dodt H, Zieglgänsberger W, Liesi P. Novel forms of neuronal migration in the rat cerebellum. J Neurosci Res. 1995;40:207–219. doi: 10.1002/jnr.490400209. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiaekinesin-related gene-products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Totis L, Saunders WS. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3motor domain mutants. Genetics. 1993;135:35–44. doi: 10.1093/genetics/135.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:11096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, Brown SS. Suppression of a myosin defect by a kinesin-related gene. Nature (Lond) 1992;356:358–361. doi: 10.1038/356358a0. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Middleton K, Carbon J. KAR3-encoded kinesin is a minus-end- directed motor that functions with centromere binding proteins (CBF3) on an in vitroyeast kinetochore. Proc Natl Acad Sci USA. 1994;91:7212–7216. doi: 10.1073/pnas.91.15.7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR, Xiang X, Beckwith SM. Nuclear migration advances in fungi. Trends Cell Biol. 1995;5:278–282. doi: 10.1016/s0962-8924(00)89039-x. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science (Wash DC) 1997;275:623–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain–related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RE, Sullivan DS, Huffaker TH, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–594. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127:139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 1991;194:732–735. doi: 10.1016/0076-6879(91)94055-h. [DOI] [PubMed] [Google Scholar]
- Pringle, J.R., and L.H. Hartwell. 1981. The Saccharomyces cerevisiae cell cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 97–142.
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Knoblich JA. Spindle orientation and asymmetric cell fate. Cell. 1995;82:523–526. doi: 10.1016/0092-8674(95)90022-5. [DOI] [PubMed] [Google Scholar]
- Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders WS, Koshland D, Eshel D, Gibbons IR, Hoyt MA. Saccharomyces cerevisiaekinesin- and dynein-related proteins required for anaphase chromosome segregation. J Cell Biol. 1995;128:617–624. doi: 10.1083/jcb.128.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hornack D, Lengyel V, Deng D. The Saccharomyces cerevisiaekinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997a;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Lengyel V, Hoyt MA. Mitotic spindle function in Saccharomyces cerevisiaerequires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997b;8:1025–1033. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1983. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 120 pp.
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples RR, Dieckmann CL. Generation of temperature-sensitive cbp1 strains of Saccharomyces cerevisiae by PCR mutagenesis and in vivorecombination: characteristics of the mutant strains imply that CBP1 is involved in stabilization and processing of cytochrome b pre-mRNA. Genetics. 1993;135:981–991. doi: 10.1093/genetics/135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachet A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. . Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Waddle JA, Cooper JA, Waterston RH. Transient localized accumulation of actin in Caenorhabditis elegansblastomeres with oriented asymmetric divisions. Development (Camb) 1994;120:2317–2328. doi: 10.1242/dev.120.8.2317. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: A Xenopuskinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Xiang X, Beckwith SM, Morris NR. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. . Proc Natl Acad Sci USA. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]