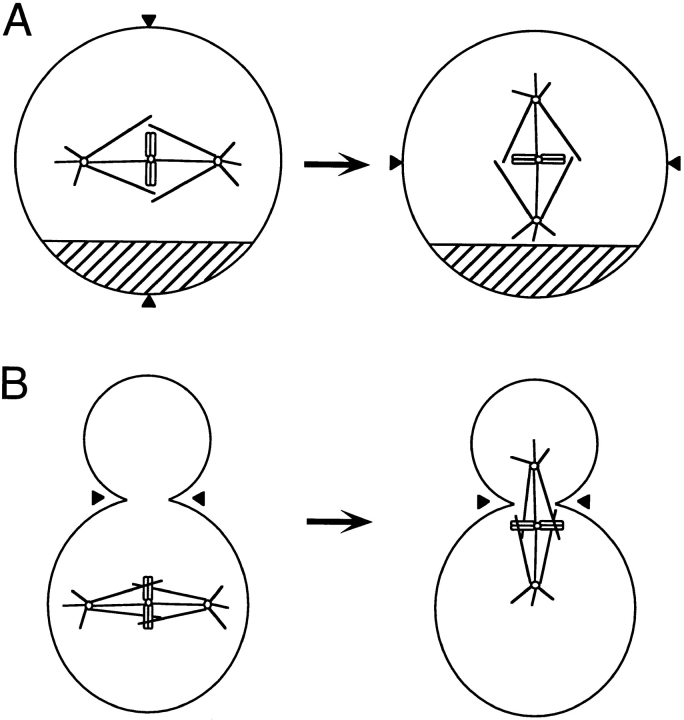
Two papers in this issue of The Journal of Cell Biology address the question of how the mitotic spindle is oriented within a dividing cell (5, 6). In this brief review, these new results will be placed in the context of other work on this topic. Spindle orientation is one of the essential roles of the cytoskeleton in all eukaryotic cells. Two broad classes of mitotic division illustrate the importance of this process: (a) asymmetric division, in which some aspect of the cell is differentially apportioned between the two products of division, and (b) symmetric division within an asymmetric cell, in which a cellular component, such as the nucleus, must be segregated equally to two cells that are unequal in size and/or shape. In the first case, the plane of division is determined by the orientation of the mitotic spindle, as it is in most animal and plant cells (Fig. 1 A). Cytokinesis occurs perpendicular to the spindle axis, at the position of the metaphase plate. Division orientation has been shown to affect cell fate in some cell divisions in animals (3, 11) and is an important part of morphogenesis in plants, where cells are not free to move after division. Thus, spindle orientation in these organisms must respond to both internal and external cues to ensure the proper segregation of developmental determinants or the proper cell division pattern.
Figure 1.
Representations of two situations in which spindle orientation is important. In A, the orientation of the mitotic spindle in a cell with an asymmetrically distributed component must change to segregate the component to only one of the two cells after division. The carets indicate the plane of division, based on spindle position. In B, the orientation of the spindle must change to allow for equal segregation of nuclei in an asymmetric cell, such as for budding yeast. The carets indicate the plane of division, based on the predetermined bud site.
In the second case, the division plane is predetermined, as in the budding yeast Saccharomyces cerevisiae (Fig. 1 B). The division plane in yeast is determined by the position of the bud, and cytokinesis occurs at the narrow neck between the mother and bud cells. The division plane is independent of spindle orientation, and cytokinesis will occur even if the nucleus divides completely within the mother or daughter cell, leaving the other cell without a nucleus. Thus in yeast, proper orientation of the mitotic spindle ensures that mitosis results in the segregation of one nucleus to each cell resulting from the division, and the positional information comes from internal determinants of cell polarity.
There are many common features to suggest that the underlying mechanism for spindle orientation is the same in all cells. The most well-characterized animal system is the spindle pole rotation that occurs in the early development of Caenorhabditis elegans (11). Proper development requires that at the second cleavage, the posterior cell changes its plane of division by 90° from the default plane. This occurs by a rotation of the spindle poles in the posterior cell, in response to a cortical polarity determinant located at the anterior cortex (10). Rotation of the spindle poles is prevented by inhibitors of the actin and microtubule cytoskeletons and by laser irradiation of the region between the centrosomes and the anterior cortex, presumably disrupting the microtubules in that region (10, 11). The anterior cortex of the cell bulges inward during rotation, suggesting that a pulling force is applied between cortex and centrosome, resulting in rotation. Consistent with this hypothesis, both actin and actin-capping protein localize to a site on the anterior cortex at the time of rotation (21); these might mark a cortical site for microtubule attachment.
In yeast cells, the details are different, but the overall picture is quite similar (1). Early in the mitotic cell cycle, the nucleus migrates from a random position in the mother cell to the mother–bud neck, where it remains until the beginning of anaphase. The short intranuclear spindle present in the nucleus at this stage is usually oriented parallel to the mother–bud axis, with one end of the spindle pointing through the neck. At anaphase, the bud-ward spindle pole body of the elongating spindle enters the neck, and further elongation results in a spindle that runs almost the full-length of the cell, with one nuclear mass in the mother cell and one in the bud. Nuclear migration and spindle orientation in yeast are prevented by mutations in actin and tubulin (9, 17, 20) and compromised by mutations in components of the microtubule motor dynein and its associated dynactin complex (4, 8, 13, 15). Cytoplasmic microtubules often appear to extend from the spindle pole body to the cell cortex in fixed cells, and the cytoplasmic microtubules in particular are required for migration and orientation (17). A simple model for migration and orientation, analogous to the one for C. elegans above, is that microtubules extend from the spindle pole body to the cortex, where they generate a dynein-dependent pulling force responsible for the motion. Recent work has shown that transient cortical interactions do occur in living yeast cells, that they require dynein, and that they coincide with directed movement of the spindle (2). However, the two articles in this issue of The Journal of Cell Biology (5, 6) demonstrate that other microtubule motor proteins function in migration and orientation and suggest that the mechanism is more complex than the simple model indicates.
KIP3, the Final Yeast Kinesin
The completion of the yeast genome sequence allows a complete accounting of the members of gene superfamilies. Cottingham and Hoyt (5) and DeZwaan et al. (6) both describe the characterization of the sixth and final kinesin gene in yeast, KIP3. KIP3 had eluded detection by standard genetics and homology-based cloning, but its sequence reveals it to be a card-carrying kinesin, with 39% identity to human kinesin in the conserved motor domain. Kip3p does not have homology to any known proteins outside of the motor domain and, like several of the yeast kinesins, does not fit neatly into any of the established families of kinesins (14). Table I lists all of the yeast kinesin and dynein microtubule motor genes, the class of protein that they encode, and brief functional information.
Table I.
Microtubule Motor Proteins in Yeast
| Gene | Motor type | Functions | ||
|---|---|---|---|---|
| KAR3 | KAR3/ncd kinesin | karyogamy, mitotic spindle (inward force) | ||
| CIN8 | BimC kinesin | mitotic spindle (assembly, outward force) | ||
| KIP1 | BimC kinesin | mitotic spindle (assembly, outward force) | ||
| KIP2 | kinesin-related | spindle orientation | ||
| KIP3 | kinesin-related | nuclear migration, spindle orientation | ||
| SMY1 | kinesin-related (distant) | no known microtubule-related function | ||
| DYN1 | dynein | spindle orientation |
Why are there so many motors in the small, nonmotile yeast cell? The approach to answering this question thus far has been to delete one or more of the relevant genes and observe the phenotypic consequences. These experiments have shown that there is substantial redundancy in motor protein function in most cases. For example, single deletions of any of the kinesin genes or the dynein gene are viable, and in most cases quite healthy. Since it is clear that some of the processes in which these proteins are involved are themselves essential for viability, this result can be interpreted in two ways. Functionally redundant pathways may have evolved to ensure that a given essential event is completed, even if something catastrophic happens to the main pathway for carrying it out. Alternatively, the experimentally observed redundancy may reflect a sort of competition in the wild-type cell: one motor can fill in for another if given the chance in a mutant cell, but it normally does not do so in a wild-type cell. It is also important to realize in these discussions of redundancy that many of the motor protein mutations have phenotypes that would not be insignificant under the selective pressure of life outside the lab.
Both articles conclude that KIP3 is involved in nuclear migration and spindle orientation in a way that is overlapping with but distinct from the role of dynein in those same events. DeZwaan et al. (6) reach this conclusion mostly through a careful phenotypic analysis of the kip3 mutant, and Cottingham and Hoyt (3) reach it mostly through an extensive analysis of double and triple mutant combinations of kip3 with other motor genes.
KIP3 Mutants Are Defective in Nuclear Migration and Early Spindle Orientation
As for the other yeast microtubule motors, a kip3 deletion mutation is viable but has some revealing phenotypes. (Nomenclature note: kip3 refers to the mutant allele, KIP3 to the wild-type allele.) Growth rate, meiosis, chromosome stability, and efficiency of karyogamy were all similar in kip3 and wild-type strains, but nuclear migration was perturbed. DeZwaan et al. (6) noted that none of the previously tested yeast microtubule motor genes had any effect on the earliest step of migration and orientation in which the nucleus migrates from a random position in the mother cell to the mother–bud neck. As described above, dynein is required for proper spindle orientation during mitosis but is not required for nuclear migration before mitosis (23). Nuclear migration was assayed by measuring the distance from the nucleus to the neck in cells with a bud; kip3 cells were found consistently to have a more randomly positioned nucleus than wild-type cells. As might be expected, failure of the nucleus to migrate to the neck resulted in a significant fraction of mitoses in which the spindle elongated within the mother cell, not entering the bud.
Yeast cells are unusual in that they assemble a mitotic spindle early in the cell cycle. In wild-type cells, this spindle is usually located on the mother side of the neck, with one pole pointed into the bud. In kip3 cells, the orientation of the premitotic spindle was almost random, suggesting that migration and orientation before mitosis are both controlled by Kip3p. To examine the kip3 defect in living cells, DeZwaan et al. (6) used time-lapse microscopy of strains expressing a Nuf2–GFP fusion protein to mark the spindle pole bodies (12). Nuclei in wild-type cells migrated to the neck soon after completing anaphase from the previous cell cycle, whereas nuclei in kip3 cells tended to oscillate within the mother cell, occasionally ending up near the neck.
Remarkably, the kip3 phenotypes were almost completely nonoverlapping with the phenotypes of a dynein deletion mutant, dyn1. When DeZwaan et al. (6) ran a dyn1 mutant through the same phenotypic tests described above for the kip3 mutant, they found that the dyn1 mutant was not significantly different from wild-type. Similarly, dyn1 cells were partially defective for insertion of the anaphase spindle through the neck, as reported previously (23), but kip3 cells were not significantly different from wild-type.
Genetic Interactions between KIPs and Dynein Reveal Synergistic and Antagonistic Forces
The phenotypes of the kip3 and dyn1 mutants suggest that the roles of the two motors in nuclear migration and spindle orientation are temporally and functionally distinct. Yet, either gene is sufficient for successful, if inefficient, mitotic divisions, suggesting that their functions are to some extent redundant. This was tested genetically by making dyn1 kip3 double mutants. Both Cottingham and Hoyt (5) and DeZwaan et al. (6) report that the dyn1 kip3 double mutant is inviable, or very nearly so. Both groups also created kip3ts mutants that allowed the phenotype of dyn1 kip3 cells to be studied. The simplest interpretation of these experiments is that the dyn1 kip3 synthetic lethality results from exacerbation of the phenotypes of the single mutants, rather than complete failure of a specific event. This is consistent with the hypothesis that dynein and Kip3p perform overlapping functions that together result in proper migration and orientation.
Cottingham and Hoyt (5) found a striking result when they constructed a dyn1 kip2 kip3 triple mutant: the kip2 mutation was able to suppress the lethality of the dyn1 kip3 double mutant. Indeed, the triple mutant grew almost as well as wild-type. KIP2 is a kinesin gene for which no phenotype had previously been observed (18). This genetic result suggests two things, each of which was a surprise: first, that the nucleus and spindle can be oriented well enough to maintain viability even in the absence of dynein and KIP3, thus there must be yet a third pathway by which this can occur; and second, that the activity of the KIP2 kinesin might normally be opposing the activities of dynein and KIP3 in bringing the nucleus to the neck. This second point was beautifully confirmed by comparing the phenotypes of dyn1 kip3ts and dyn1 kip2 kip3ts strains. Cottingham and Hoyt (5) created an experimental situation in which all cells were arrested in the cell cycle with a short spindle and the nucleus located at the neck. They then shifted the temperature, resulting in loss of Kip3p activity, and observed the position of the nucleus in fixed cells. By 1 h, the nuclei in almost 75% of the dyn1 kip3ts cells were located away from the neck in the mother cell body. In at least some of the cells, the nucleus moved all the way to the far side of the mother cell. In contrast, the nuclei in most of the dyn1 kip2 kip3ts cells remained at the neck after the temperature shift. This result is consistent with Kip2p providing a force that pulls the nucleus away from the neck, antagonizing the forces provided by dynein and Kip3p that pull the nucleus toward the neck. A weakness of these experiments is that the observations were made on fixed cells at time points. Time-lapse analysis of living cells, as done by DeZwaan et al. (6) for the single kip3 mutant, would be very informative here.
What is the phenotype of a kip2 single mutant? If Kip2p acts in opposition to dynein and Kip3p (and whatever else is able to move the nucleus to the neck in their absence), then one might expect that loss of Kip2p activity would leave dynein and Kip3p free to drag the nucleus through the neck, plastering it against the far side of the bud. Unfortunately, kip2 mutant cells have a phenotype that is much like that of dyn1 mutants, although less severe. Why would motors with antagonizing activities have the same phenotype? An interesting possibility suggested by Cottingham and Hoyt (5) is that the tension created by opposing motor activities is important to the mechanism of spindle orientation and that without tension, the orienting apparatus cannot produce directional force. This is based on a similar model by Nicklas (16) for how chromosomes become properly attached to the spindle. Nicklas demonstrated experimentally that tension provided by a microneedle pulling on a chromosome that was only attached to one pole was sufficient to allow spindle function. It might be possible to carry out a similar experiment in yeast by using an optical trap to exert directed force on the nucleus.
Motors and Microtubule Dynamics
Both groups noted that a kip3 mutant is more resistant to benomyl than wild-type. Benomyl is a microtubule-depolymerizing drug, and one interpretation of the increased resistance to benomyl is that the microtubules in a kip3 mutant are more stable to depolymerization than those in wild-type. Interestingly, the cytoplasmic microtubules in the kip3 strain are also longer than in wild-type, which would be consistent with the hypothesis that Kip3p normally acts to destabilize microtubules. In accord with the evidence for antagonistic function, kip2 mutants were more sensitive to benomyl and had shorter cytoplasmic microtubules. The ability of a microtubule motor to alter microtubule dynamics has been demonstrated in vitro for Kar3p (7) and for XKCM1, a frog kinesin (22), and suggested in vivo by the observation that deletion of KAR3 results in longer, more abundant cytoplasmic microtubules (19) in fixed cells. Most recently, analysis of microtubule dynamics in living yeast cells has shown that a dyn1 mutation alters microtubule dynamics in vivo (2). Microtubules were visualized in these experiments with a GFP–tubulin fusion protein and were found to undergo the same sort of dynamic instability behavior observed in animal cells. In dynein mutants, several of the dynamic parameters of microtubule growth were altered, resulting in longer cytoplasmic microtubules (2) that were defective in generating force to orient the spindle. It is possible that this ability of motors to alter microtubule dynamics is as important in some cellular processes as the ability to move along microtubules is in others. For example, abnormally long cytoplasmic microtubules could physically interfere with nuclear migration. Recent findings in which a dynein–GFP fusion protein was used to visualize cytoplasmic microtubules over long periods show that some movements of the nucleus early in the cell cycle appear to result from pushing of growing microtubules against the cell cortex (Shaw, S.I., E. Yeh, E.D. Salmon, and K. Bloom, manuscript submitted for publication), suggesting that microtubule length control will prove to be critical in nuclear migration and spindle orientation.
Conclusions
Like many processes in cell biology, nuclear migration and spindle orientation are not hard wired—each cell follows a unique path to proper division. This flexibility is necessary for the cell but confounding for the scientist because it often reflects functional redundancy that must be teased apart. The Cottingham and Hoyt (5) and DeZwaan et al. (6) papers reviewed here are a big step forward in that they have revealed several layers of redundancy (some not discussed here for brevity) and have suggested some testable models. A comprehensive model for the action of these motors in migration and orientation is not easily derived from these results, however, and will require more analysis, particularly of microtubule function in living cells. In addition to the role of the motors, the central question of how the cytoplasmic microtubules read the polarity of the cell remains unanswered. A recently identified protein, Kar9p, has some of the hallmarks of the necessary link between the determinants of cell polarity and the cytoplasmic microtubules (Miller, R.K., and M.D. Rose, manuscript submitted for publication). Kar9p is localized to the tip of the growing bud at times when microtubules interact with that region, and kar9 mutants exhibit defects in nuclear migration and spindle orientation similar to those for the motor mutants discussed here. It will be particularly interesting to see whether microtubules in kar9 mutant cells fail to make the interactions with the cortex that are associated with nuclear movements (2). The rapidity with which previously unidentified genes can now be assigned to functions in yeast and the ever increasing ability to apply sophisticated microscopic analysis will no doubt result in continued rapid advances in this field.
Footnotes
Address all correspondence to T. Stearns, Department of Biological Sciences, Stanford University, Stanford, CA 94305-5020. Tel.: (415) 725-6934. Fax: (415) 725-8309. e-mail: stearns@stanford.edu
I am grateful to Sid Shaw, Elaine Yeh, Kerry Bloom, Ted Salmon, Rita Miller, and Mark Rose for communication of unpublished results.
References
- 1.Byers, B. 1981. Cytology of the yeast life cycle. In The Molecular Biology of the Yeast Saccharomyces: Life Cycle and Inheritance. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 59–96.
- 2.Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 4.Clark SW, Meyer DI. ACT3: a putative centractin homologue in S. cerevisiaeis required for proper orientation of the mitotic spindle. J Cell Biol. 1994;127:129–138. doi: 10.1083/jcb.127.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiaeis accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiaeis required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO (Eur Mol Biol Organ) J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eshel D, Urrestarazu LA, Vissers S, Jauniaux JC, Van Vliet-Reedijk JC, Planta RJ, Gibbons IR. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc Natl Acad Sci USA. 1993;90:11172–11176. doi: 10.1073/pnas.90.23.11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huffaker TC, Thomas JH, Botstein D. Diverse effects of β-tubulin mutations on microtubule formation and function. J Cell Biol. 1988;106:1997–2010. doi: 10.1083/jcb.106.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyman AA. Centrosome movement in the early divisions of Caenorhabditis elegans: a cortical site determining centrosome position. J Cell Biol. 1989;109:1185–1193. doi: 10.1083/jcb.109.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hyman AA, White TJ. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. . J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahana JA, Schnapp BJ, Silver PA. Kinetics of spindle pole body separation in budding yeast. Proc Natl Acad Sci USA. 1995;92:9707–9711. doi: 10.1073/pnas.92.21.9707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y-Y, Yeh E, Hays T, Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc Natl Acad Sci USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- 15.Muhua L, Karpova TS, Cooper JA. A yeast actin-related protein homologous to that in vertebrate dynactin complex is important for spindle orientation and nuclear migration. Cell. 1994;78:669–679. doi: 10.1016/0092-8674(94)90531-2. [DOI] [PubMed] [Google Scholar]
- 16.Nicklas RB. How cells get the right chromosomes. Science (Wash DC) 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- 17.Palmer RE, Sullivan DS, Huffaker T, Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. . J Cell Biol. 1992;119:583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roof DM, Meluh PB, Rose MD. Kinesin-related proteins required for assembly of the mitotic spindle. J Cell Biol. 1992;118:95–108. doi: 10.1083/jcb.118.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiaekinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waddle JA, Cooper JA, Waterston RH. Transient localized accumulation of actin in Caenorhabditis elegansblastomeres with oriented asymmetric divisions. Development (Camb) 1994;120:2317–2328. doi: 10.1242/dev.120.8.2317. [DOI] [PubMed] [Google Scholar]
- 22.Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopuskinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- 23.Yeh E, Skibbens RV, Cheng JW, Salmon ED, Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast Saccharomyces cerevisiae. . J Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]