Abstract
Protein methylation is a posttranslational modification that can potentially regulate signal transduction pathways in a similar manner as protein phosphorylation. The role of protein methylation in NGF signaling was examined by metabolic labeling of PC12 cell proteins with l-[methyl-3H]methionine and by in vitro labeling of cell proteins with l-[methyl-3H]S-adenosylmethionine. Effects of NGF were detected within 15 min. Methyl-labeled proteins were resolved by one and two dimensional SDS-PAGE. NGF affected the methylation of several 68–60-kD proteins (pI 5.8–6.4) and 50-kD proteins (isoelectric point pH 6.7–6.8 and 5.8–6.2). Several NGF-induced changes in methylation increased over several hours and through 4 d. Moreover, methyl labeling of several specific proteins was only detected after NGF treatment, but not in nontreated controls. The effects of NGF on protein methylation were NGF specific since they were not observed with EGF or insulin. A requirement for protein methylation for neurite outgrowth was substantiated with either of two methylation inhibitors: dihydroxycyclopentenyl adenine (DHCA) and homocysteine. DHCA, the more potent of the two, markedly inhibits protein methylation and neurite outgrowth without affecting cell growth, NGF-induced survival, cell flattening, or several protein phosphorylations that are associated with early signaling events. Removal of DHCA leads to rapid protein methylation of several proteins and concurrent neurite outgrowth. The results indicate that NGF regulates the methylation of several specific proteins and that protein methylation is involved in neurite outgrowth from PC12 cells.
Protein methylation is a posttranslational modification that may be used to regulate signal transduction and differentiation pathways by mechanisms that are analogous to regulation by protein phosphorylation (Hrycyna and Clarke, 1993; Rando, 1996). Although a specific role for protein methylation in prokaryote chemotaxis is well established (Shapiro et al., 1995), possible roles for protein methylation in eukaryotic signaling mechanisms have not been extensively explored. Protein carboxyl methylations are reversible, and regulatory roles for carboxyl methylation have been proposed for chemoattractant responses in neutrophils (Philips et al., 1993, 1995), insulin secretion from pancreatic islets (Metz et al., 1993), and photoreceptor signal transduction (Parish et al., 1995). Among signaling proteins that are known to be carboxyl methylated are the Ras and Rho family of small G-proteins (Hrycyna and Clarke, 1993; Rando, 1996), γ subunits of heterotrimeric G-proteins (Philips et al., 1993; Rando, 1996), and the catalytic subunit of protein phosphatase 2A (Lee and Stock, 1993; Favre et al., 1994; Xie and Clarke, 1994). Protein phosphatase 2A is also demethylated by a specific protein carboxyl methylesterase (Lee et al., 1996). Many proteins are known to be N-methylated at arginine, lysine, or histidine residues including cytoskeletal proteins (actin and myosin), nuclear proteins (nucleolin, fibrillarin, histones, heterogeneous nuclear RNPs), the multifunctional calcium binding protein, calmodulin, and FGF-2. The physiological functions of methylation, however, remain largely unknown. Moreover, potential mechanisms for regulating protein methylation within growth factor signaling pathways remain to be explored.
Methylation pathways use S-adenosylmethionine (SAM)1 as the universal methyl donor for methyltransferase-catalyzed methylation of proteins and other methyl acceptors. The role of methylation in NGF signal transduction was previously studied using high concentrations (millimolar) of inhibitors of methyltransferases that use SAM (Seeley et al., 1984; Kujubu et al., 1993). These attempts to examine the role of methylation in cell signaling were somewhat compromised by the lack of specific, nontoxic inhibitors of methylation. In addition to the methylated acceptor, S-adenosylhomocysteine (SAHcy) is a product of all methyltransferase reactions. SAHcy is a strong competitive inhibitor of SAM (Hildeshein et al., 1972) and is normally removed by hydrolysis in a reversible reaction (De la Haba and Cantoni, 1959) catalyzed by S-adenosylhomocysteine hydrolase (SAHH). Thus, inhibition of SAHH offers an alternative means to inhibit methyltransferases. This approach was used in the present work to study protein methylation during NGF signaling. The crucial role of SAHH and methylation in development is evident from the nonagouti (ax) mutation. The lethality of this mutation is due to a deletion of the SAHH gene, and embryonic development is arrested in the preimplantation blastula stage (Miller et al., 1994).
The PC12 clonal cell line was used to examine the role of protein methylation in NGF signal transduction. PC12 cells are derived from a rat pheochromocytoma and serve as a model of neuronal differentiation. When exposed to NGF, PC12 cells assume many of the features of sympathetic neurons including cell cycle arrest, survival in serum-free medium, and elaboration of long neurites (Greene and Tischler, 1976; Tischler and Greene, 1978). Moreover, PC12 cells also respond to other growth factors. For example, in the presence EGF, PC12 cells do not differentiate into neuronlike cells, but do increase their mitotic rate (Huff and Guroff, 1981). Thus, PC12 cells are useful to study specific growth factor signaling mechanisms. The specific signaling events and proteins involved in neurite outgrowth have not been fully elucidated. For example, NGF and EGF both activate several signaling proteins including phospholipase C, phosphoinositide (PI)3-kinase, extracellular signal–related kinase (ERKs) (MAP kinases), and Ras (Kaplan and Stephens, 1994; Marshall, 1995). However, NGF, but not EGF, stimulates neurite outgrowth in PC12 cells. One hypothesis for the specificity of NGF actions involves kinetic differences in the activation of both Ras and ERK (Marshall, 1995). NGF stimulation of PC12 cells results in prolonged activation of both Ras and ERK activity, while EGF causes a transient rise and fall in Ras and ERK activities (Qui and Green, 1992). Specificity may also arise from NGF-specific signaling pathways that are not activated by EGF and other mitogens (Chao, 1992; Kaplan and Stephens, 1994; Peng et al., 1995). The results reported here indicate that signaling pathways involving protein methylation may contribute to NGF specificity.
Previous studies by Seeley et al. (1984) have implied that methylation is necessary for neurite outgrowth in PC12 cells. Kujubu et al. (1993) and Haklai et al. (1993) reported that methylation of small G-proteins is regulated by NGF in PC12 cells. Najbauer and Aswad (1990) have also characterized several methyl-arginine containing proteins in PC12 cells. The effects of NGF on protein methylation and the effect of inhibition of protein methylation on neurite outgrowth are, however, incompletely characterized and have been difficult to interpret due to the lack of specificity of the methylation inhibitors used in previous studies. The present work extends previous findings by examining the methylation of specific cellular proteins and neurite outgrowth before and after inhibition of the protein methylation pathway. 9-(trans-2′, trans-3′-dihydroxycyclopent-4-enyl)-adenine (DHCA), a specific, mechanism-based inhibitor of SAHH, inhibits protein methylation and greatly decreases neurite outgrowth. The results indicate that NGF, but not EGF or insulin, regulates the methylation of several specific proteins and that protein methylation is required for neurite outgrowth from PC12 cells.
Materials and Methods
Reagents
NGF was purified from male mouse submaxillary glands as described previously (Mobley et al., 1976). Insulin, l-homocysteine thiolactone, anisomycin, leupeptin, PMSF, aprotinin, and monoclonal phosphotyrosine antibody (clone PT-66) were purchased from Sigma Chemical Co. (St. Louis, MO). Homocysteine was prepared by incubating l-homocysteine thiolactone with 40 mM NaOH at 37°C for 30 min. EGF was obtained from Upstate Biotechnology Inc. (Lake Placid, NY). DHCA (kindly provided by R.T. Borchardt, University of Kansas, Lawrence, KS) was prepared as 1 and 100 mM stock solutions in DMSO. Erythro-9-(2-hydroxy-3-nonyl) adenine was kindly provided by D. Porter (Burroughs Wellcome, Research Triangle Park, NC). l-[methyl-3H]methionine (71.4 Ci/mmol), l-[methyl- 3H]SAM (70 Ci/mmol), [14C]adenosine (59.8 mCi/mmol), and carrier-free [32P]orthophosphate were purchased from Dupont-NEN (Boston, MA). 125I-donkey anti–rabbit IgG was purchased from Amersham Corp. (Arlington Heights, IL). Protein A–Sepharose (PAS) and ampholines were obtained from Pharmacia Fine Chemicals (Piscataway, NJ). Donor horse serum and fetal bovine serum were from JRH Biosciences (Lenaxa, KS). Cell culture medium, penicillin, and streptomycin were obtained from Gibco Laboratories (Grand Island, NY). Restriction endonuclease MspI was from New England Biolabs Inc. (Beverly, MA), and nuclease P1 from United States Biochemical Corp. (Cleveland, OH).
Cell Culture and Bioassays
Stock cultures of PC12 cells were grown on collagen-coated tissue culture dishes (Falcon Plastics, Cockeysville, MD) in RPMI-1640 medium supplemented with 10% heat-inactivated horse serum and 5% fetal bovine serum at 35°C and 7.5% CO2, as previously described (Greene et al., 1987). For neurite outgrowth studies, PC12 cells (3–5 × 105 cells) were plated onto collagen-coated 35-mm plastic tissue culture dishes and cultured in RPMI-1640 medium containing 1% heat-inactivated horse serum and 50 ng/ml NGF. Neurites >20 μm were counted as neurite-bearing cells. At least 100 cells from each experimental condition were scored from randomly chosen fields.
For the study of proliferation and survival in the presence and absence of DHCA, naive PC12 cells were plated on collagen-coated, 24-well tissue culture dishes at a density of 105 cells per well. 1 d after plating, cells were washed three times in RPMI-1640 medium to remove serum and then placed either in complete medium (RPMI plus 10% horse serum and 5% fetal bovine serum), in RPMI plus 50 ng/ml NGF, or in RPMI alone. Where indicated, 1 μM DHCA was included in the culture medium. The number of viable cells was determined by counting intact nuclei using a hemacytometer (Soto and Sonnenschein, 1985) immediately after washing the cultures with RPMI at time intervals of 1, 3, and 5 d later.
For the study of neurite regeneration, PC12 cells were treated with NGF for 7–14 d. NGF was then thoroughly washed away from the cells, and the cells were detached from the culture dish by trituration. Cells were then replated in the presence or absence of NGF, and neurite outgrowth was scored 24 h later.
Metabolic Radiolabeling of Proteins
To assess endogenous protein methylation under various experimental conditions, PC12 cells were cultured in 1% horse serum for at least 16 h before metabolic radiolabeling for 6 h with 100 μCi/ml l-[methyl-3H]methionine in the presence of 10 μM anisomycin at 35°C in a CO2 incubator. The methionine/cysteine content of RPMI-1640 was reduced by 80% during the labeling period. Growth factors (NGF, EGF, or insulin) and/or 1 μM DHCA were added for the indicated times. At the conclusion of the labeling period, cells were washed three times with 1 ml of PBS (35°C), harvested in 200 μl SDS-PAGE lysis buffer, and held in a boiling water bath for 5 min. Total radiolabeled protein in each lysate was determined by TCA precipitation and liquid scintillation spectrometry. The averages of the determinations are given under Results as the means ± SEM. Aliquots of each cell lysate containing equal TCA-precipitable counts per minute were subjected to SDS-PAGE as described below. For analysis of protein methylation by two-dimensional (2D) IEF × SDS-PAGE, the cells were harvested in IEF lysis buffer (Aletta and Greene, 1987) without heating the sample.
To assess changes in protein phosphorylation, PC12 cell proteins were metabolically radiolabeled as previously described (Aletta, 1996) in a Hepes-buffered Krebs-Ringer solution containing 0.1% d-glucose. When the effect of DHCA was examined, the drug was added to cultures 30 min before addition of the radioisotope. The labeling was carried out with 50 μCi/ml [32P]orthophosphate at 35°C in room air for 2 h. At the end of the labeling period, cells were washed three times with 1 ml of PBS (35°C), harvested in SDS-PAGE lysis buffer, and held in a boiling water bath for 5 min. Total radiolabeled protein in each lysate was determined by TCA precipitation and liquid scintillation spectometry.
In Vitro Protein Methylation
To measure protein methylation in subcellular fractions, cells were washed three times in PBS (4°C) and harvested by scraping with a rubber policeman in ice-cold homogenization buffer (100 mM Tris, pH 8.0, 1 mM EDTA, 2 μM PMSF, and 10 μg/ml leupeptin). After homogenization at 4°C in a Dounce homogenizer, the homogenates were centrifuged at 4°C for 10 min at 500 g to obtain a crude nuclear fraction. The supernatant was centrifuged at 20,000 g for 5 min at 4°C. The resulting supernatant was centrifuged at 100,000 g for 90 min at 4°C to obtain the cytoplasmic fraction. The pellet of the 100,000 g spin was washed three times in homogenization buffer and resuspended in 150 μl of homogenization buffer to obtain the membrane fraction. Protein in each fraction was determined spectrophotometrically (Bradford, 1976). Equal amounts of protein (125 μg) from each fraction were incubated with 4.25 μCi l-[methyl-3H]SAM in a total volume of 50 μl for 1 h at 37°C. The labeling reaction was stopped by adding 12.5 μl of 5× SDS-PAGE sample buffer (0.3 M Tris-HCl, pH 6.8, 45% glycerol, 1.4 M 2-mercaptoethanol, 10% SDS, 0.001% bromophenol blue) and holding the samples in a boiling water bath for 5 min. In vitro–labeled proteins from the cell fractions were then analyzed by 7.5–15% gradient SDS-PAGE as described below.
Gel Electrophoresis
Discontinuous SDS-PAGE (Laemmli, 1970) was performed with 19-cm separating gels composed of polyacrylamide gradients of 6–12, 7.5–15, or 8.5–15%, depending upon the experiment. Gels were fixed, stained with Coomassie blue, and then destained. Gels containing proteins labeled with [32P]orthophosphate were dried and placed in contact with Kodak XAR film to produce an autoradiographic image. Gels containing tritium- labeled proteins were prepared for fluorography by washing the gel for 1 h in three changes of deionized water, followed by immersion in 1 M sodium salicylate for 30 min (Chamberlain, 1979). After drying, the gels were exposed to preflashed Kodak XAR film (Laskey and Mills, 1975) and stored at −70°C (Bonner and Laskey, 1974) with an intensifying screen. Quantitative comparisons of methyl-labeled proteins were obtained by scanning fluorograms into an analysis program (Molecular Analyst; Bio Rad Laboratories, Hercules, CA).
For 2D IEF × SDS-PAGE, PC12 cell proteins were labeled as described above and harvested in a lysis buffer appropriate for IEF (Aletta and Greene, 1987). Equal TCA-precipitable counts per minute of cell lysates were subjected to IEF with pH 5–7, and 3.5–10 ampholines at a ratio of 4:1, respectively. Proteins in IEF gels were further resolved by SDS-PAGE (the second-dimension) using 12-cm separating gels composed of 7.5–15% polyacrylamide gradient. A standard IEF gel containing only lysis solution was used to determine the pH range. Fluorographic images of tritium-labeled proteins were generated as described above.
DNA Methylation
Total methylation of cytosines in DNA was determined by first digesting PC12 cell DNA with MspI, which cleaves methylated or unmethylated CCGG sequences (Bestor et al., 1984). The products are 5′ labeled with [32P]ATP, and digested with nuclease P1. Methyl-cytosines are resolved from unmethylated cytosines by chromatography in isobutyric acid/water/ ammonium hydroxide (66:33:1) and detected by autoradiography (Bestor et al., 1984). Liquid scintillation spectrometry was used to quantify the results.
Immunoprecipitation
Tyrosine phosphorylation of Trk in the presence or absence of DHCA was assessed by immunoprecipitation followed by Western blotting. Cells were treated with 100 ng/ml NGF for 5 min, with or without 1 μM DHCA. Cells were then washed three times in ice-cold PBS and harvested in 1 ml of a lysis buffer containing 1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 25 mM NaF, 5 mM EGTA, 5 mM EDTA, 2 μM PMSF, and 100 U/ml aprotinin. Insoluble material was removed by centrifugation at 4°C for 10 min at 13,000 g. Samples were precleared with 6 mg PAS for 2 h followed by centrifugation for 10 min at 13,000 g. Anti-phosphotyrosine monoclonal antibody (clone PT-66) was added to equal amounts of lysate protein for 2 h on a rotating platform at 4°C followed by addition of 3 mg of PAS and incubation on the rotating platform at 4°C for 1 h. PAS beads were then recovered by centrifugation at 13,000 g for 10 min, and washed three times with 1 ml of 1% Triton X-100 lysis buffer followed by two 1-ml washes in lysis buffer without Triton X-100. The PAS beads were then resuspended in SDS-PAGE sample buffer and held in a boiling water bath for 5 min. The precipitated material was resolved by SDS-PAGE (7.5% acrylamide) followed by transfer to Immobilon P membrane (Millipore Corp., Milford, MA). The blot was probed with 1 μg of rabbit polyclonal Trk antiserum (Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at room temperature followed by incubation with 125I-donkey anti–rabbit IgG. The dried blot was then exposed to Kodak XAR film at −70°C with an intensifying screen.
SAHH Activity
PC12 cells were plated on 150-mm dishes (2 × 107 cells) in the presence of 100 ng/ml NGF plus 10 nM to 3 μM DHCA, or without added DHCA for 7 d. Cells were then washed three times in ice-cold PBS and harvested by scraping in a potassium phosphate buffer, pH 7.0 (25 mM KH2PO4, 25 mM K2HPO4, 1 mM dithiothreitol, 0.5 μM PMSF, and 10 μg/ml leupeptin). Cells were homogenized in a Dounce homogenizer, and the cell nuclei and debris were removed by centrifugation at 13,000 g. Cytosol was then prepared by centrifugation at 100,000 g for 90 min and the protein concentration determined. SAHH activity was determined in the synthesis direction by a TLC method described previously by Hershfield (1979).
Results
NGF-specific Induction of Changes in the Pattern of PC12 Cell Methylated Proteins
Previous studies have implicated regulation of protein methylation in the early events of NGF-mediated signal transduction (Seeley et al., 1984; Kujubu et al., 1993). To examine this possibility in greater detail, protein methylation patterns were assessed after metabolic radiolabeling of cellular proteins with l-[methyl-3H]methionine. Fluorograms of the labeled proteins generated from SDS-PAGE and 2D IEF × SDS-PAGE were used to detect specific changes in protein methylation after NGF treatment. The cellular pool of SAM, the predominant methyl donor in all cells, was radiolabeled using l-[methyl-3H]methionine in control PC12 cells, and PC12 cells treated with either 50 ng/ml NGF, 5 nM EGF or 1 μM insulin. These concentrations and growth factors were chosen based on the biological effects produced by each in PC12 cells. NGF at 50 ng/ ml produces the maximum neurite outgrowth response. EGF at 5 nM enhances cell proliferation (Huff et al., 1981) and 1 μM insulin is commonly used to effect cell survival in serum-free media (Rukenstein et al., 1991). We have verified that receptors for all three of these growth factors are present on the PC12 cells used in these studies, by observation of the aforementioned biological effects at the doses indicated (data not shown). Radiolabeled methionine equilibrates with the cellular SAM pool within 20 min (Chelsky et al., 1985). A protein synthesis inhibitor, anisomycin (10 μM), was present during the metabolic radiolabeling to prevent incorporation of l-[methyl-3H]methionine into newly synthesized proteins.
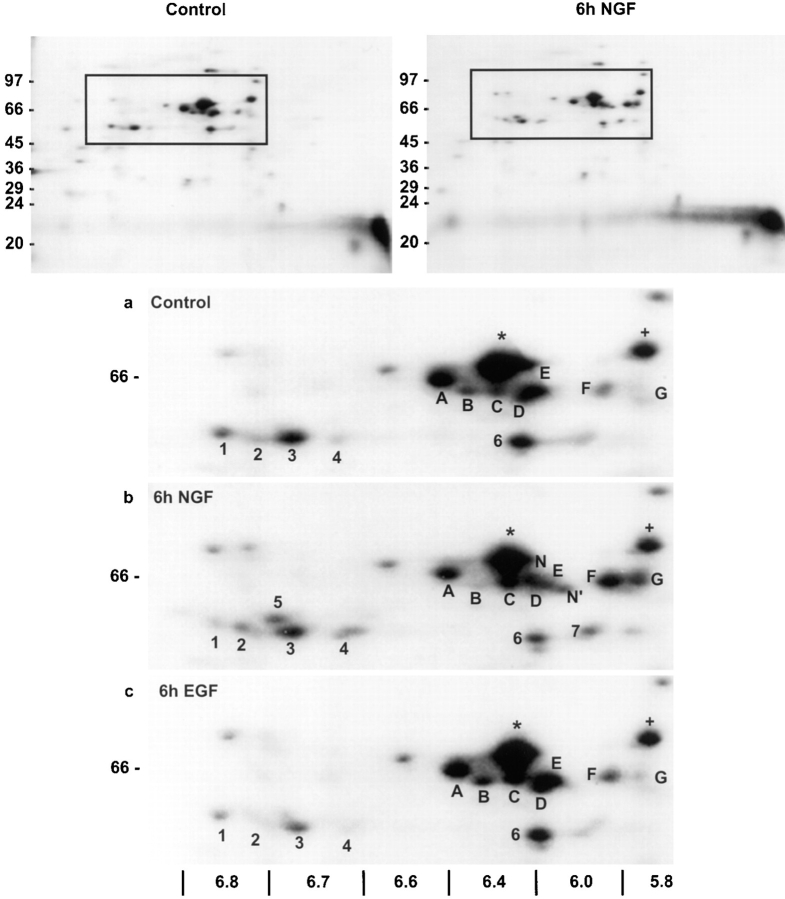
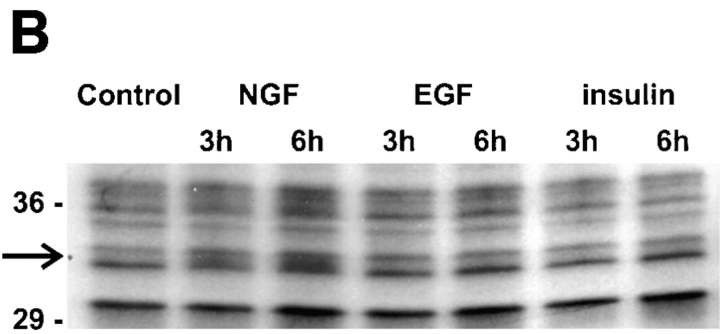
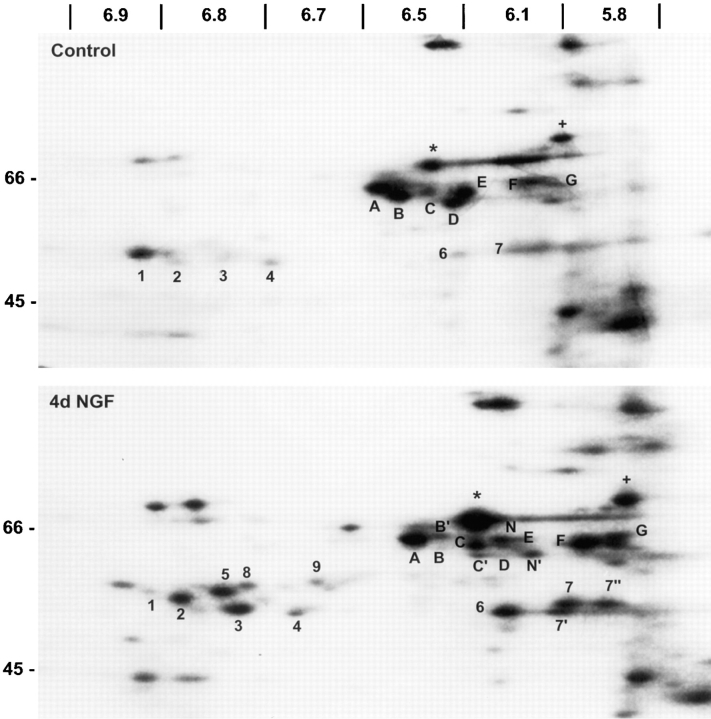
In each of five independent trials, all three trophic factors produced a net increase in methyl group incorporation into PC12 cell protein. After 6 h of treatment, EGF and insulin lead to ∼50% greater incorporation (±0.2, SEM) and NGF to a 31% increase (±0.1, SEM). Despite the slightly larger effects of EGF- and insulin-promoted increases in total methyl-3H–labeled protein, NGF treatment consistently yields protein methylation patterns exhibiting more pronounced changes in specific methylated proteins (Fig. 1). Several, indistinctly resolved proteins that migrated between 24 and 20 kD showed increased methyl labeling relative to total protein methylation after 6 h NGF treatment (Fig. 1, top right). Among the methylated species detected in control cell lysates, were nine 68–60-kD proteins with isoelectric point (pI) values of 5.8–6.4 (Fig. 1, A–G, *, and +). After treatment with 50 ng/ml NGF for 6 h, the proteins labeled A, *, and + in Fig. 1 showed little or no reproducible changes in methylation relative to controls. Although there was a net increase in total protein methylation, both increases and decreases in the methylations of specific proteins relative to total protein methylation were detected in response to NGF. Decreased labeling of proteins B, D, and E was detected with NGF (Fig. 1 b). Methyl labeling of proteins C, F, and G increased markedly, and methyl group incorporation into two proteins (N and N′) was detected only after 6 h in NGF-treated cells, but not in controls (Fig. 1 b).
Figure 1.
NGF induces specific changes in the pattern of in vivo protein methylation observed by 2D IEF × SDS-PAGE. PC12 cell proteins were metabolically labeled with l-[methyl-3H]methionine (100 μCi/ml) in the presence of anisomycin (10 μM) for 6 h. Whole cell lysates containing equal TCA-precipitable cpm (800,000) were separated by IEF. IEF gels were loaded onto 8.5– 15% gradient gels for the separation of methylated proteins by molecular mass. Fluorographic images generated from the control and 6 h NGF (50 ng/ml) conditions are illustrated in the top two panels. Molecular mass standards (in kD) are indicated at the left of each panel. The boxed area in the top panels is enlarged below for control, 6 h NGF and 6 h EGF (5 nM) samples. Fluorograms were generated by exposing x-ray film to the dried gels with an enhancing screen for 10 d at −70°C. The pH gradient of the isoelectric focusing tubes is shown below the enlarged images (a–c). Labels (A, *, and +) indicate unaffected methylations, while the remaining letters and numbers indicate protein methylations affected by NGF. These results are representative of three independent experiments.
Significant NGF-induced effects on protein methylation were also detected at pI 6.7–6.8 and 5.8–6.2 in the 50-kD region (Fig. 1, a and b). Labeling of protein 3 was unaffected by NGF. Decreased methyl labeling of protein 1 was detected after 6 h NGF treatment, while proteins 2, 4, and 7 showed increased methyl labeling, and the [methyl- 3H] incorporation into protein 5 was only detected in response to NGF. The effects of NGF on protein methylation are unlikely to be due to incorporation of l-[methyl-3H]methionine into newly synthesized proteins because protein synthesis was inhibited by 97 ± 0.1% (n = 3) under the conditions of these experiments. In addition, no significant increases or decreases in the amounts of the specific proteins analyzed in Fig. 1 were detected by 2D IEF × SDS-PAGE of [35S]methionine-labeled proteins in whole cell lysates, in the absence of protein synthesis inhibition (data not shown).
The NGF-induced changes in methyl labeling of specific proteins were not observed with the two other trophic factors tested. After EGF treatment for 6 h, the methylation pattern of the 68–60-kD proteins was quite different from the NGF-induced changes relative to the control, nonstimulated cells (Fig. 1). The pattern of protein methylations with EGF was more similar to the pattern with control cells, but small increases in the methylation of proteins in this relative molecular weight range were reproducibly detected (B, C, D, and E). NGF, on the other hand, decreased the methylation of proteins B, D, and E. The changes in protein methylations were less marked with EGF than NGF and no methylated protein was detected with EGF that was not detected with controls. Moreover, no significant effects of EGF were detected in the 50-kD set of methylated proteins. Insulin had no significant effects on the methylation of the 68–60- or 50-kD proteins (data not shown).
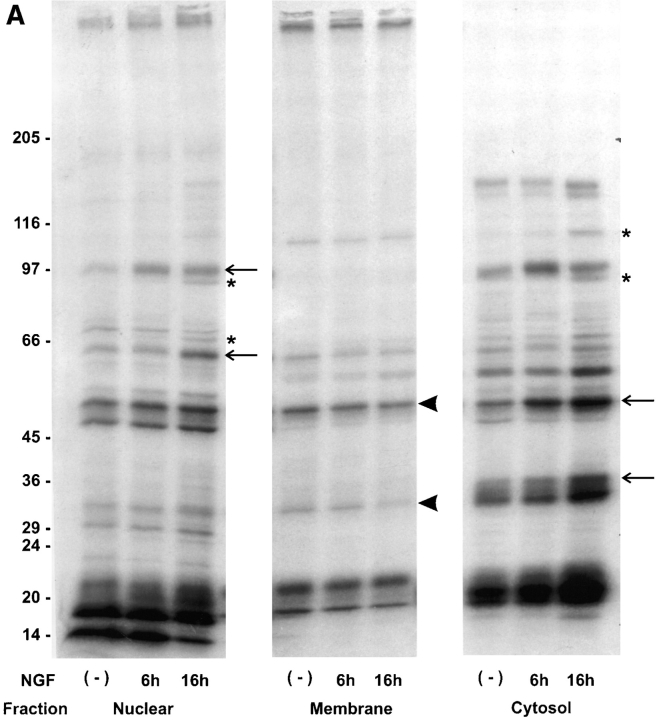
The effects of NGF on the pattern of protein methylation were also examined by in vitro labeling of proteins in subcellular fractions from NGF-treated cells as an independent approach for detecting the protein methylations that are affected by NGF. In vitro labeling provides a complementary method of analysis that does not require a protein synthesis inhibitor. It also indicates the possible cellular locations of protein methylation. PC12 cells were incubated without or with NGF for 15 min, 6 or 16 h. Nuclei (500 g), membranes (100,000 g, pellet), or cytosol (100,000 g, supernatant) were isolated and incubated with [methyl-3H]SAM for 1 h at 37°C, in vitro. Methylated proteins were resolved by 7.5–15% gradient gel SDS-PAGE and detected by fluorography. Changes in protein methylation were detected in nuclei, membranes, and cytosol (Fig. 2). Increases in protein methylation were detected after 15 min of NGF treatment (data not shown) and continued to increase from 6 to 16 h (Fig. 2 A). Moreover, several of the proteins that showed increases in methylation have similar molecular weights as those detected by 2D IEF × SDS-PAGE (Fig. 1) after metabolic radiolabeling of intact cells (e.g., 68–64 kD, and 50 kD). Thus, changes in the methylation of proteins in response to NGF was confirmed by two independent approaches. In the nuclear fraction, time-dependent increases (more than twofold) in protein methylation in response to NGF treatment were detected in proteins migrating at 97, 94, 67, and 64 kD. In the membrane fraction, the methylation of proteins migrating at 50 and 34 kD decreased after 6 or 16 h of NGF. Several proteins in the cytosolic fraction showed increased methylation. Most prominent among these were 114, 94, 50, and 35 kD. Additional NGF-induced effects were observed by the in vitro method. This is most likely due to the higher specific activity of the [methyl-3H]SAM pool in vitro than in the intact cell experiments and the enrichment of proteins by subcellular fractionation. In addition, the in vitro labeling experiments indicate that both the requisite methyltransferase and protein substrate were present at the time of analysis in each of the subcellular fractions that were isolated from NGF-activated cells. Moreover, irrespective of the specific mechanisms responsible for activation of the methylation of specific proteins by NGF, the state of activation was stable during preparation of the fractions.
Figure 2.
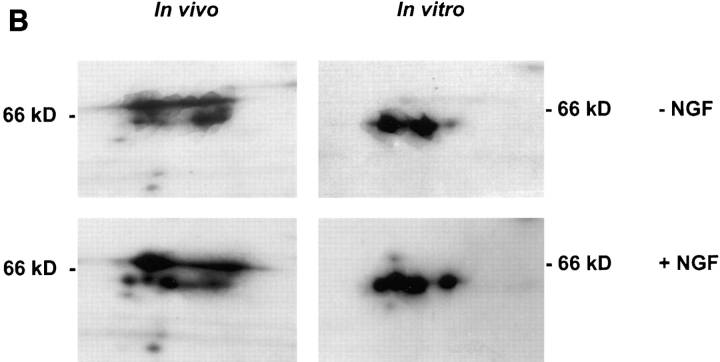
NGF-treatment of intact cells stimulates in vitro protein methylation in cell-free extracts. (A) PC12 cells were incubated in the presence or absence of NGF (50 ng/ml) for 6 or 16 h. Proteins in the nuclear (500 g, pellet), membrane (100,000 g, pellet), or cytosolic (100,000 g, supernatant) fractions were radiolabeled with l-[methyl-3H]SAM for 1 h at 37°C. The labeling reaction was quenched by the addition of 5× SDS-PAGE sample buffer followed by holding the reaction tube in a boiling water bath for 5 min. Labeled proteins were separated by 7.5–15% gradient SDS-PAGE. The image shown is a fluorogram of the dried gel. Asterisks indicate methylated proteins not detectable in extracts from non-NGF-treated control cells. Arrows identify increases of 67% or greater. The arrowheads (membrane fraction) point out decreases of >30%. These results were reproduced in an independent experiment. Specific changes at 16 h were as follows: nuclear proteins at 97 and 64 kD increase 2.4-fold; cytosolic proteins at 50 kD and 35 kD increase 67 and 79%, respectively. (B) Cell proteins were metabolically radiolabeled followed by preparation of cytosol (in vivo) or the cytosol was prepared first, followed by incubation with [methyl-3H]SAM (in vitro). A portion of the 2D IEF × SDS-PAGE fluorograms from control (−NGF) versus 6 h NGF treatment (+NGF) are displayed. There are five individual protein spots migrating at 64 kD, which are most easily discerned in the in vivo +NGF condition. The 64-kD series of spots in each of the other fluorograms (four spots each) are superimposable with those of the in vivo +NGF condition.
To validate this approach further, cytosol was prepared from cells treated with or without NGF after metabolic radiolabeling, and replicate cultures without metabolic radiolabeling were also processed to obtain cytosol for in vitro protein methylation. Comparisons of the 2D IEF × SDS-PAGE fluorograms from each type of preparation indicate that several of the same proteins resolved in the 64 kD range (pI 5.8–6.4) are similarly increased after NGF treatment by either method of analysis (Fig. 2 B). The in vitro data obtained after NGF treatment for 6 h confirm that changes in protein methylation triggered by NGF can occur whether or not protein synthesis has been inhibited. Thus, the in vivo and in vitro experiments independently indicate that NGF produces diverse, marked effects on the regulation of the methylation of several specific proteins.
NGF Affects Protein Methylation during Early and Delayed Signaling
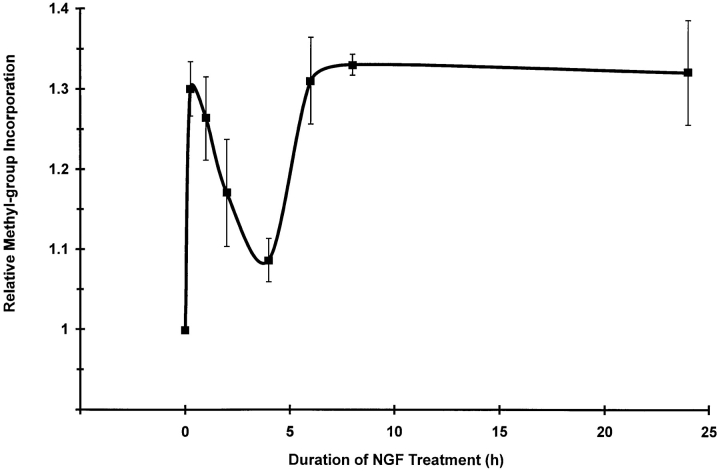
The time dependence of NGF-induced changes in protein methylation was examined to determine the onsets and durations of changes in protein methylation. PC12 cells were incubated with NGF for 15 min to 24 h to examine protein methylation during early stages of NGF signaling and for 4 d to examine changes occurring concurrently with the appearance of neurites. Total protein methylation was assessed by the incorporation of [3H]methyl groups into TCA-precipitable protein during NGF treatment for 24 h (Fig. 3). Significant increases in the total amounts of methyl group incorporation were detected within 15 min after the addition of NGF. The response over the early time course is biphasic with an initial rapid burst of methylation that decays between 1 and 4 h of NGF treatment followed by a second, persistent phase of elevated methylation from 6 to 24 h. The pronounced decrease in methyl group incorporation between 1 and 4 h indicates that the effect of NGF on total protein methylation is likely to be complex and dependent upon multiple factors, including methionine uptake, intracellular compartmentalization, and use of the labeled methyl group in other metabolic pathways. Thus, all comparisons of protein methylation patterns examined by gel electrophoresis were standardized relative to total methyl-3H–labeled protein.
Figure 3.
The time dependence of total methyl group incorporation into proteins in response to NGF. PC12 cell proteins were metabolically labeled with l-[methyl-3H]methionine (100 μCi/ ml) for 6 h in the presence of anisomycin (10 μM). NGF (50 ng/ ml) was added to the cell cultures for the times indicated. Cells were lysed in SDS-PAGE sample buffer. Total methyl labeling of proteins (counts per minute) was measured for each condition after TCA precipitation of an aliquot of the sample. The relative amount of the 3H-labeled methyl groups in lysate proteins at each time point is given with respect to nontreated control cells. The data shown are the means ± SEM for five independent experiments.
Although the resolution of changes in protein methylation in whole cell lysates is reduced on one dimensional SDS-PAGE gels, this method was used to observe the time course of changes in the set of proteins migrating at 64–62 kD. Gradient SDS-PAGE and fluorographic analysis of methyl-3H–labeled proteins in whole cell lysates indicated that there are increases in the methylation of the proteins in this region of the gel, detectable within 1 h after the addition of NGF and persisting for at least 8 h (Fig. 4 A). In addition, when using an 8.5–15% acrylamide gradient, a reproducible NGF-specific change in a 34-kD protein is evident at 3 and 6 h of NGF treatment (Fig. 4 B), but not at times earlier than 2 h (data not shown).
Figure 4.
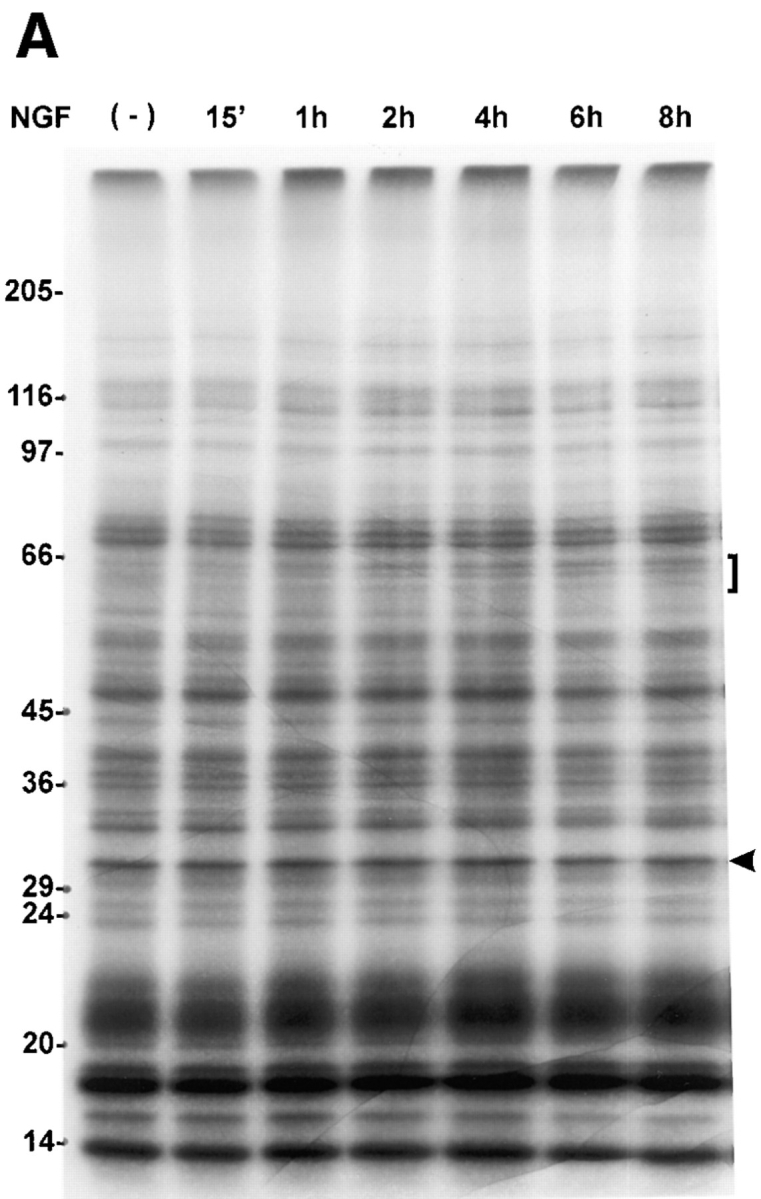
Changes in the pattern of protein methylation are NGF specific and time dependent. (A) PC12 cell proteins were metabolically labeled with l-[methyl-3H]methionine (100 μCi/ml) for 6 h in the presence of anisomycin (10 μM) with or without NGF (50 ng/ml) for the times indicated. Whole cell lysates containing equal TCA-precipitable cpm (300,000) were loaded in each lane. Proteins were separated by 7.5–15% gradient SDS-PAGE. The image shown is a fluorogram of the dried gel after exposure to x-ray film for 3 d at −70°C with an intensifying screen. The bracket indicates the migration position of a 64–62-kD complex of proteins that increase in a time-dependent manner. The arrowhead indicates the migration position of an invariant protein band, the density of which does not change by more than a few percent over the time course. By this measure, the 4-h lane is slightly overloaded, and the 6-h lane, underloaded. Nevertheless, the change in the 64–62-kD protein complex is still evident in the latter. The results illustrated here are representative of five independent experiments. (B) Metabolically radiolabeled methylated proteins (300,000 cpm) were obtained as described above and loaded on 8.5–15% polyacrylamide gradient gels to more favorably resolve the area between 29 and 36 kD. The image shown is a fluorogram derived from the dried gel after exposure to x-ray film for 2 d at −70°C. The arrow indicates the migration position of a regulated 34-kD protein. Cultures were treated with NGF (50 ng/ml), EGF (5 nM), or insulin (1 μM). The results shown are representative of three independent experiments.
To determine if any of the changes in methyl group incorporation persist for several days, the labeling studies were repeated in PC12 cells treated with NGF for 4 d. This time point was chosen because ∼50% of PC12 cells have neurites ⩾20-μm long after 4 d of NGF (Greene et al., 1982). In addition, it was reasoned that some protein methylations may occur during neurite outgrowth that were not detected during early stages of NGF action. Thus, after 4 d of NGF, methylated proteins were analyzed by metabolic radiolabeling of cell proteins followed by 2D IEF × SDS-PAGE (Fig. 5) as described above. The pattern of [3H]methyl incorporation into PC12 cell proteins relative to total methylated protein indicates that many of the changes induced by 6 h of NGF (Fig. 1) persist for several days. Moreover, several of the NGF-induced increases and decreases in protein methylation observed after 6 h of NGF treatment were greater after 4 d of NGF treatment. Methylation of proteins between 68 and 60 kD (C, F, G, N, and N′) and in the 55–50 kD region (2–4, 6, and 7) increased relative to both the control (Fig. 5) and the 6 h NGF result (Fig. 1). Proteins C, F, G, N, and N′ appear to be the same proteins (based on pI and M r) that showed increases in methylation after 6 h of NGF treatment. Also, after 4 d of NGF treatment, the methylation of proteins B, D, and E, as well as protein 1 in the 50-kD region decreased relative to the control or 6-h NGF result. A number of novel methylated proteins were also observed after 4 d of NGF treatment that were not detected after 6 h. These include proteins B′, C′ in the 60-kD region of the gel, and proteins 7′, 7′′, 8, and 9, appearing in the 50-kD region of the gel. The distinctly different time courses for the methylations of specific proteins suggest that NGF may regulate the methylation of proteins involved in multiple signaling pathways. Moreover, most of the NGF- dependent changes in methylated proteins that were detected are not associated with transient phenomena.
Figure 5.
NGF-induced changes in protein methylation persist and progress during prolonged NGF treatment. PC12 cells were incubated with or without NGF (50 ng/ml) for 4 d. Cells were labeled with l-[methyl-3H]methionine (100 μCi/ml) in the presence of anisomycin (10 μM) for 6 h. Equal TCA-precipitable cpm (750,000) of cell lysates were subjected to IEF. IEF gels were loaded on 7.5–15% gradient SDS-PAGE gels to separate methylated proteins by molecular mass. The images shown are fluorograms of the dried gels after exposure of x-ray film for 12 d at −70°C with an intensifying screen. The areas of the gels shown are similar to the boxed areas shown in Fig. 1. The pH gradient of the IEF gels is indicated at the top of the figure. Labels are as in Fig. 1.
Inhibition of Methylation Inhibits Neurite Outgrowth
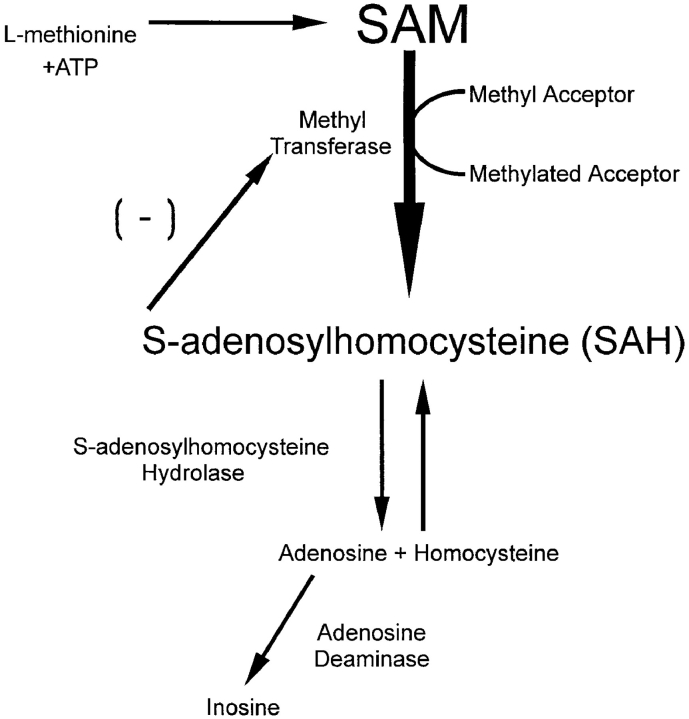
The marked effects of NGF on the methylation of specific proteins calls attention to the possibility that protein methylation is required for NGF signal transduction and neurite outgrowth. This possibility was examined by observing NGF-mediated neurite outgrowth after inhibiting methylation using two approaches based on the biochemical pathway outlined in Fig. 6. One approach was inhibition of SAHH by a mechanism-based, substrate analogue inhibitor, DHCA (Liu et al., 1992). SAHH catalyzes the reversible hydrolysis of SAHcy to homocysteine and adenosine by an NAD-dependent mechanism (De la Haba and Cantoni, 1959; Liu et al., 1992). The reaction is driven in the hydrolysis direction in vivo by the adenosine deaminase–catalyzed conversion of adenosine to inosine. DHCA blocks the methylation pathway by inhibiting SAHH. Like the normal substrate, SAHcy, DHCA is oxidized by SAHH coupled to NAD+ reduction, but the 3-keto DHCA formed cannot be hydrolyzed, thereby trapping SAHH in its inactive, NADH form (Liu et al., 1992). SAHH inhibition results in an accumulation of its substrate, SAHcy, a potent competitive inhibitor of methyltransferases (Hildeshein et al., 1972).
Figure 6.
The methylation pathway. SAHH catalyzes the reversible hydrolysis of SAHcy. Inhibition of SAHH by DHCA or homocysteine leads to an accumulation of SAHcy and inhibition of methyltransferases.
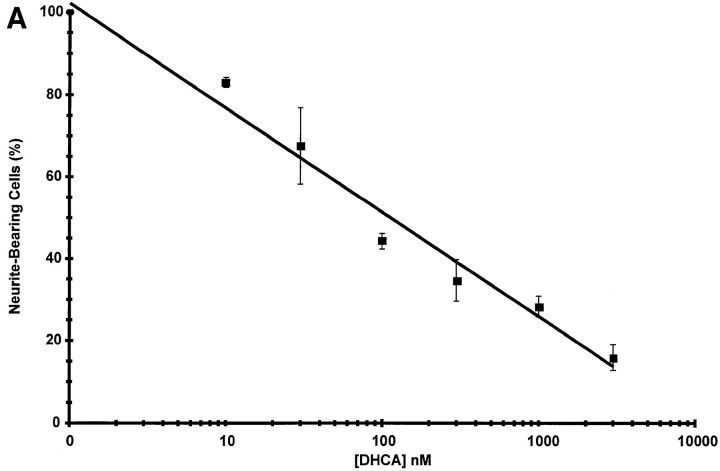
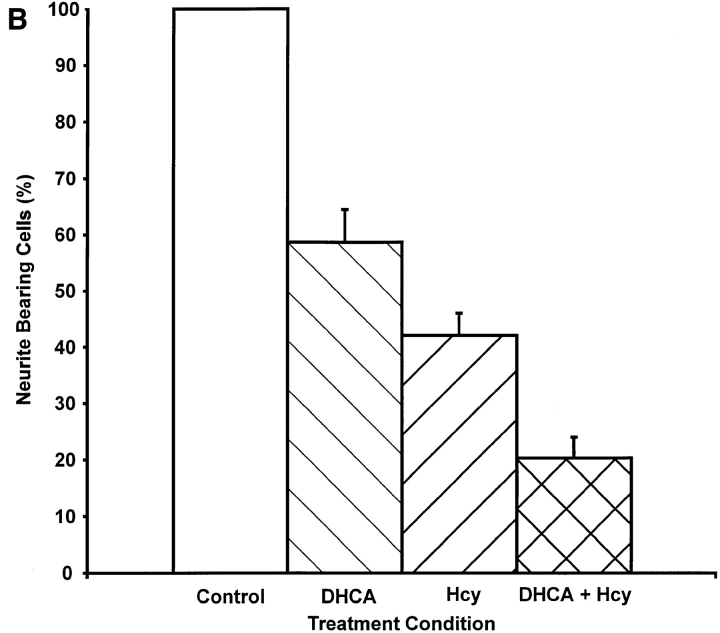
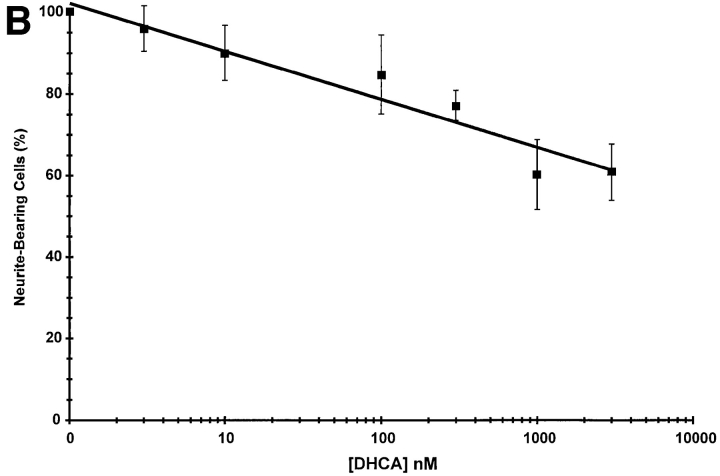
The concentration dependence for inhibition of neurite outgrowth by DHCA (Fig. 7 A) was determined by incubating PC12 cells with NGF for 7 d with or without 10 nM to 3 μM DHCA. Neurite outgrowth was scored after 7 d of NGF treatment as described under Materials and Methods. The concentration of DHCA required to inhibit neurite outgrowth by 50% was ∼100 nM. A second approach that was used to test for a requirement of protein methylation in neurite outgrowth was inhibition of SAHH by addition of extracellular homocysteine, alone or in combination with DHCA. When intracellular levels of homocysteine rise, the SAHH-catalyzed reaction is driven in the synthesis direction (De la Haba and Cantoni, 1959; Hasobe et al., 1989), i.e., towards the formation of SAHcy (Fig. 6). Excess homocysteine acts cooperatively with inhibitors of SAHH in many other cell types (Chiang et al., 1977; Kredich and Martin, 1977; Backlund et al., 1986; Hasobe et al., 1989), and the combination of the two is expected to elicit more inhibition of methylation than either alone. As shown in Fig. 7 B, 100 nM DHCA reduced NGF-induced neurite outgrowth to 59% of control (NGF alone), and 100 μM homocysteine reduced NGF-induced neurite outgrowth to 42%. The combination of 100 nM DHCA plus 100 μM homocysteine inhibited the neurite outgrowth by more than 80%. The findings that homocysteine alone inhibits neurite outgrowth, and that homocysteine enhances the inhibitory effect of a low dose of DHCA on neurite outgrowth indicate that the effect of DHCA on neurite outgrowth is most likely due to inhibition of methylation.
Figure 7.
Concentration-dependent inhibition of neurite outgrowth by inhibitors of methylation. (A) Effect of DHCA. PC12 cells on 35-mm collagen-coated tissue culture dishes (300,000 cells per dish) were treated with NGF (50 ng/ml), or NGF and DHCA (10 nM to 3 μM) for 7 d. Neurite-bearing cells were scored as described under Materials and Methods. Cultures treated with NGF and no DHCA received the DMSO vehicle only, which had no effect on neurite outgrowth. The line was fit by least squares regression analysis. The data shown are the means ± SD for three independent experiments. (B) Effect of homocysteine alone and in combination with DHCA. All cultures were treated with NGF (50 ng/ml) and scored for the presence of neurites after 7 d. The data shown are the means ± SD for three independent experiments. The white bar represents the NGF control condition without DHCA or homocysteine. Results from cultures treated with 100 nM DHCA are represented by the adjacent striped bar (DHCA), and those from cultures treated with 100 μM homocysteine in the next striped bar (Hcy). The cross-hatched bar indicates results from cultures treated with 100 nM DHCA plus 100 μM homocysteine (DHCA + Hcy).
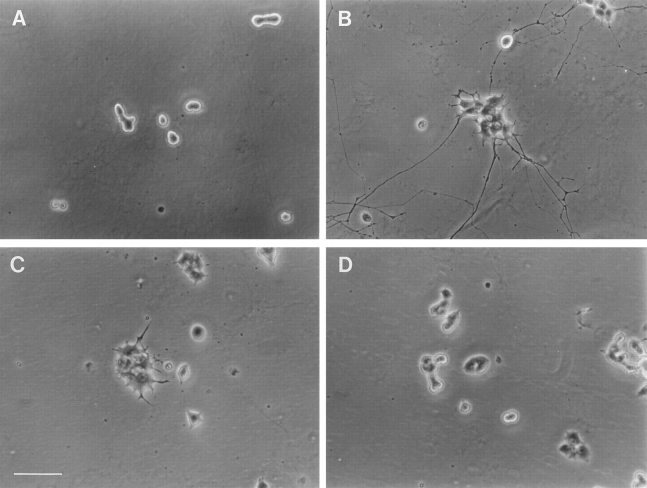
Fig. 8 illustrates the effects of DHCA or DHCA plus homocysteine on cell morphology observed by phase contrast microscopy of NGF-treated PC12 cells. After 7 d of NGF-treatment, PC12 cells flatten, hypertrophy, and elaborate long neurites (Fig. 8 B). In the presence of 1 μM DHCA and NGF for 7 d, the cells continue to respond to NGF by flattening, but neurite outgrowth is greatly attenuated (Fig. 8 C). Short, spike-like processes are observed. In the presence of 100 nM DHCA plus 100 μM homocysteine, the cells appear more flattened than control cells, but little neurite outgrowth is observed (Fig. 8 D). Thus, inhibition of methylation by DHCA, homocysteine, or the cooperative action of the two together suggest that methylation is an important requirement for NGF-induced neurite outgrowth. In addition, the morphological evidence (Fig. 8) indicates that not all NGF actions (e.g., the cell-flattening response) are inhibited by factors that inhibit methylation (see below).
Figure 8.
The effects of DHCA and homocysteine on neurite outgrowth and cell morphology. PC12 cells were plated on 35-mm collagen-coated tissue culture dishes at a density of 300,000 cells per dish. Cells were cultured for 7 d under control conditions without NGF (A), with 50 ng/ml NGF (B), NGF plus 1 μM DHCA (C), or NGF with 100 nM DHCA and 100 μM homocysteine (D). Bar, 50 μm.
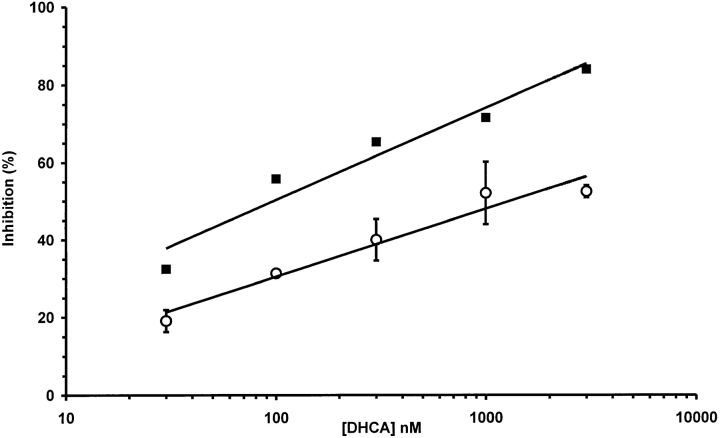
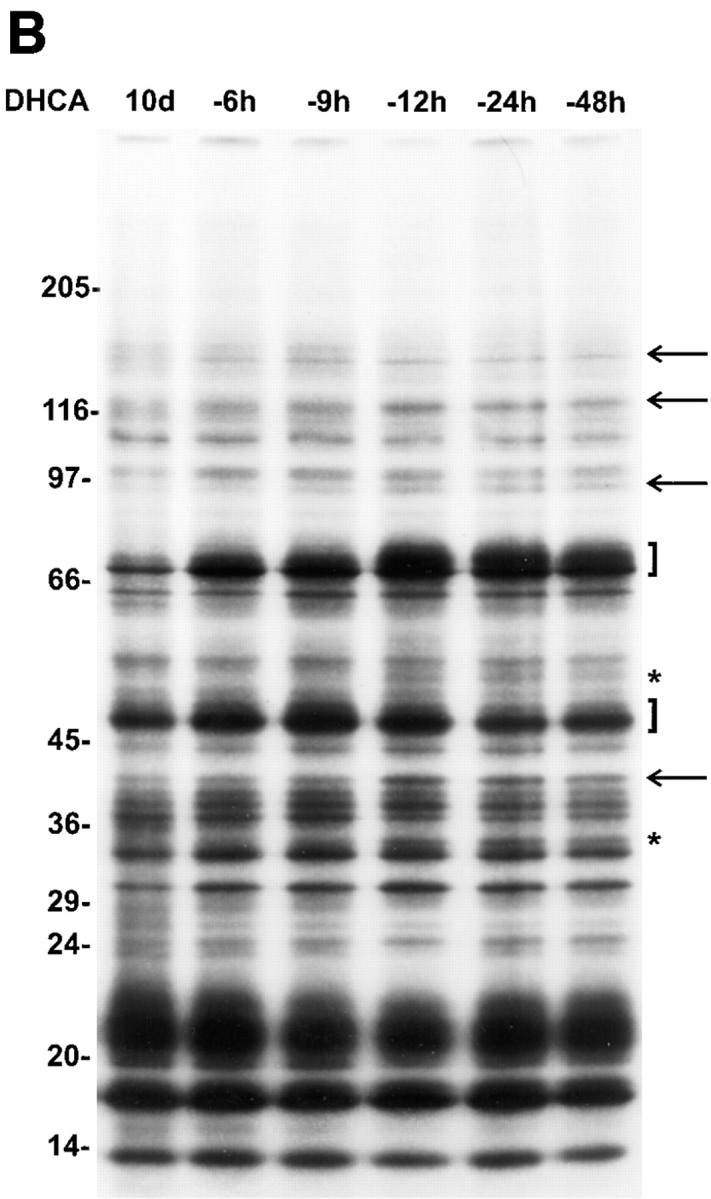
DHCA-induced inhibition of SAHH was verified by direct measurement of SAHH activity. DHCA at 1 nM produced 70% inhibition and >90% inhibition was achieved at 10 nM. The consequence of this inhibition (elevated intracellular level of SAHcy) is expected to inhibit protein methylation (Fig. 6). Thus, the effect of DHCA on protein methylation was directly determined by assessing protein methylation in PC12 cells using metabolic radiolabeling with l-[methyl-3H]methionine in the presence of anisomycin. The dose-dependent inhibition of protein methylation observed is similar to that of the DHCA effect on neurite outgrowth (Fig. 9). DHCA (1 μM) inhibits total methyl group incorporation measured by TCA-precipitation of equal aliquots of whole cell lysates as described under Materials and Methods. The total reduction in radiolabeled, methylated protein was 40% in control, non-NGF-treated cells, and 52% in cells stimulated with NGF for 6 h. Similar differences in the inhibitory effects (±NGF) were observed at all doses of DHCA above 30 nM (data not shown). The inhibitory action of DHCA is thus greater in NGF-treated cells. In conclusion, these experiments indicate that protein methylation is markedly inhibited by DHCA at concentrations that inhibit neurite outgrowth.
Figure 9.
Dose-dependent effect of DHCA on total protein methylation in PC12 cells correlates with the DHCA effect on inhibition of neurite outgrowth. Inhibition of NGF-stimulated neurite outgrowth (▪) by DHCA was compared to the inhibition of total protein methylation (○) at the same doses of DHCA. PC12 cells were treated with DHCA (30 nM to 3 μM) and NGF (50 ng/ ml) for 6 h in the presence of anisomycin to determine total protein methylation as described under Materials and Methods. Inhibition is plotted relative to control cultures treated with NGF only. The neurite outgrowth data are replotted from Fig. 7 A.
In contrast, neither NGF alone nor DHCA with or without NGF have a significant effect on total methylation of cytosines in PC12 cell DNA. The percentage of total cytosines methylated in NGF-treated cells (53.3 ± 1.5) versus that in cells treated with NGF plus DHCA (48.1 ± 0.2) was not statistically significant (n = 3, Student's paired t test; P > 0.05, two tails). All measurements were performed after 24-h treatments.
DHCA Treatment Does Not Interfere with Cell Proliferation or Early NGF-mediated Signaling Events
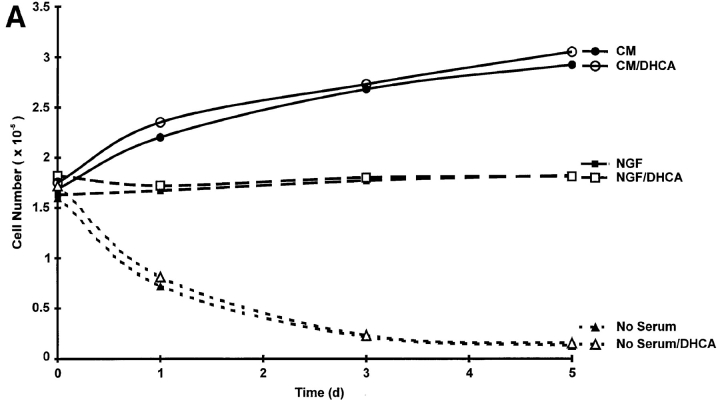
Previous efforts to examine the possible role of methylation in NGF signal transduction (Seely et al., 1984; Haklai et al., 1993; Kujubu et al., 1993) used methylation inhibitors at relatively high concentrations that can produce cytotoxicity (Seeley et al., 1984) and inhibition of receptor tyrosine kinases (Meakin and Shooter, 1991; Maher 1993). The methylation inhibitor used in this study, DHCA, is much less toxic than the related adenosine analogues used previously and is effective at concentrations four orders of magnitude lower than those used previously (Seeley et al., 1984; Meakin and Shooter, 1991; Haklai et al., 1993; Kujubu et al., 1993; Maher, 1993). DHCA does not affect the growth rate of PC12 cells in serum-containing medium (Fig. 10 A). In the absence of serum, PC12 cells degenerate via programmed cell death. DHCA has no effect on the rate that cell death is induced (Fig. 10 A). In the absence of serum, NGF rescues PC12 cells from programmed cell death (Rukenstein et al., 1991). Also shown in Fig. 10 A, DHCA had no effect on the protection afforded by NGF. In addition to confirming the absence of cytotoxicity, these findings suggest that DHCA does not interfere with the ability of NGF to activate its tyrosine kinase receptor (Trk), which prevents programmed cell death (Kaplan and Stephens, 1994). Thus DHCA has no detectable effects on the growth or survival of PC12 cells under any of the conditions examined.
Figure 10.
DHCA treatment does not interfere with cell growth, NGF-mediated survival, or rapid onset NGF-stimulated phosphorylations. (A) PC12 cells were plated on 24-well collagen-coated dishes at a density of 100,000 cells per well. Cells were allowed to attach to the dishes overnight, and then were washed with RPMI-1640 medium to remove serum. Cells were then cultured in complete (serum-containing) medium (CM, ○ and •), plus or minus 1 μM DHCA; RPMI-1640 without serum plus 50 ng/ml NGF, plus or minus 1 μM DHCA (NGF, □ and ▪); RPMI-1640 medium without serum or NGF, plus or minus 1 μM DHCA (No serum, ▵ and ▴). Similar results were observed in two other independent experiments. (B) Effect on Trk phosphorylation. PC12 cells were treated with or without NGF (50 ng/ml) for 5 min. Where indicated, 1 μM DHCA was added to cultures for 1 h before treatment with NGF. Cell lysates were equalized for total protein, immunoprecipitated with anti-phosphotyrosine antibody (as described under Materials and Methods), resolved by 7.5% SDS-PAGE and transferred to Immobilon P membranes. The blot was probed with anti-Trk antibody, followed by donkey anti– rabbit 125I-IgG. The image shown is an autoradiogram of the Western blot after a 5-d exposure at −80°C. This result was verified in a second independent experiment. (C) PC12 cells were treated with or without NGF (50 ng/ml) for 2 h. DHCA (1 μM) was added 1 h before the addition of NGF where indicated. Cells were radiolabeled with [32P]orthophosphate for 2 h as described under Materials and Methods. For long term NGF-induced phosphorylations, PC12 cells were grown in the presence of NGF (50 ng/ml) plus or minus 1 μM DHCA for 10 d. Cells were then radiolabeled with [32P]orthophosphate for 2 h. For both short and long term NGF treatment experiments, cell lysates were equalized for total TCA-precipitable 32P-labeled protein and separated on 6–12% gradient SDS-PAGE. The image shown is an autoradiogram of the dried gel. Exposure time was 16 h. The arrows at the right indicate, in decreasing M r, the positions of the previously identified phosphoproteins, 64-kD chartin, 60-kD tyrosine hydroxylase, and 55-kD β-tubulin.
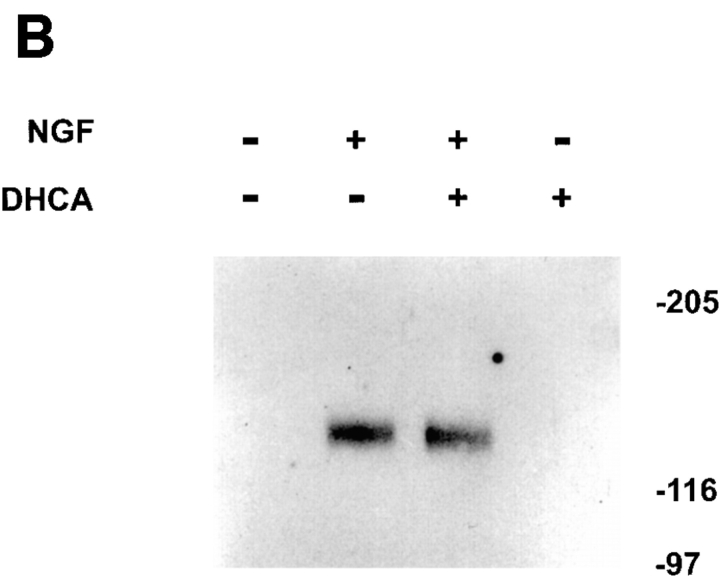
Since other adenosine-analogues, which are putative methylation inhibitors, are known to inhibit phosphorylation and activation of Trk (Meakin and Shooter, 1991; Maher 1993), the effect of DHCA on the tyrosine phosphorylation of Trk was also determined. PC12 cells were incubated with or without 1 μM DHCA for 1 h, and NGF was then added for 5 min. Immunoprecipitation with a phosphotyrosine antibody was followed by SDS-PAGE and immunoblotting with anti-Trk antibody. DHCA had no detectable effect on the Trk tyrosine phosphorylation mediated by NGF (Fig. 10 B), the primary event in NGF signaling.
To determine if DHCA affects other phosphorylation events downstream of Trk activation, total phosphoproteins were analyzed by SDS-PAGE. Several NGF-induced increases in phosphorylation of proteins are resolved by this method (Fig. 10 C). The rapid increase in the phosphorylation of tyrosine hydroxylase (Halegoua and Patrick, 1980) that occurs after NGF treatment is not affected by DHCA. The overall phosphorylation pattern was similarly unaffected by DHCA. Two other NGF-induced increases in phosphorylation, which require long-term (7 d) NGF treatment, are temporally associated with neurite outgrowth. Unlike phosphorylation of tyrosine hydroxylase, the increased 32P incorporation into the 64-kD chartin (Aletta and Greene, 1987) and β-tubulin (Aletta, 1996; Black et al., 1986) is diminished by exposure of the cells to DHCA. The inhibitory effect of DHCA on the phosphorylations of these two proteins is the predicted consequence of DHCA inhibition of neurite outgrowth. Thus, DHCA affects delayed phosphorylation events in NGF signaling but does not inhibit phosphorylations associated with the early signaling events studied here.
DHCA Effects on Neurite Outgrowth Are Diminished by prior NGF Treatment and Are Rapidly Reversible
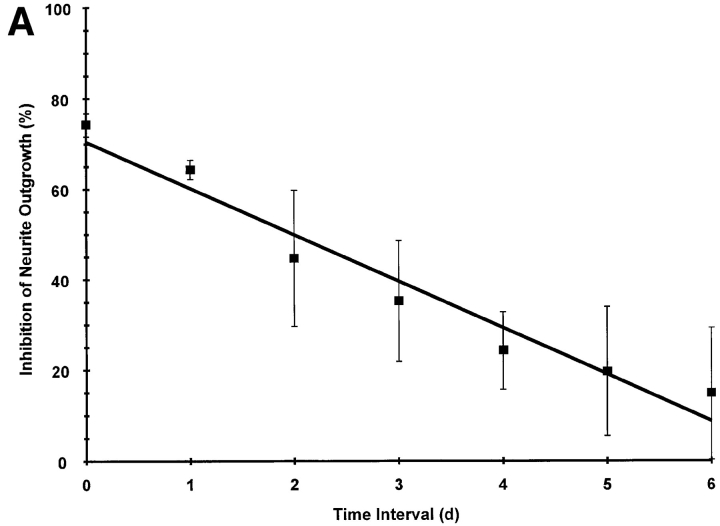
To explore further the functional significance of the time dependence of protein methylation during NGF-mediated neurite outgrowth, DHCA (1 μM) was added to PC12 cells at the same time that NGF was added or 1–6 d after NGF addition. Under these conditions, the later DHCA was added after NGF, the less its inhibitory effect (Fig. 11 A). The decrease in inhibition showed a linear dependence on the time at which DHCA was added to the cells. Once the NGF-signaling cascade begins, neurite outgrowth is progressively less sensitive to inhibition of methylation. This would occur if the rates of demethylation were low for those methylated proteins that are required for neurite outgrowth. Alternatively, neurite outgrowth may be insensitive to inhibiting methylation once neurite outgrowth proceeds past a critical, committed step. While initiation of neurite outgrowth from PC12 cells after exposure to NGF is slow (requiring 7 d to achieve ∼100% neurite bearing cells), PC12 cells can rapidly regenerate after preformed neurites are disrupted as described under Materials and Methods. Fig. 11 B shows the concentration dependence of inhibition of neurite regeneration by DHCA. Neurite regeneration was inhibited 40% by 3 μM DHCA. In contrast, if 3 μM DHCA is used when NGF is first added to the cells, it inhibits neurite outgrowth by 84% (Fig. 7 A). As described above, the difference in the sensitivities of regeneration and de novo neurite outgrowth to inhibition by DHCA is consistent with slow rates of demethylation or decreased sensitivity to DHCA once differentiation has reached a critical stage.
Figure 11.
The effect of prior NGF treatment on the neurite inhibiting action of DHCA. (A) The effect of varying the time interval between NGF treatment and addition of DHCA. PC12 cells were treated with NGF (50 ng/ml) for 7 d. DHCA (1 μM) was added to the cells either at the time NGF was initially added to the cell cultures (time zero), or after a time interval of 1, 2, 3, 4, 5, or 6 d. Cells were scored for the presence of neurites after 7 d of NGF. The data shown are the means ± SD for three independent experiments. (B) The effect of inhibition of methylation on neurite regeneration. PC12 cells were grown in NGF (50 ng/ml) for 7–14 d. NGF was removed from the cultures by an extensive washing procedure described under Materials and Methods. Cells were removed from the culture dishes by trituration and replated without or with DHCA (3 nM to 3 μM). NGF was added to all cultures 1 h after plating. Neurite regeneration was scored 16–24 h later. The line was fit by least squares regression analysis. The data shown are the means ± SD for three independent experiments.
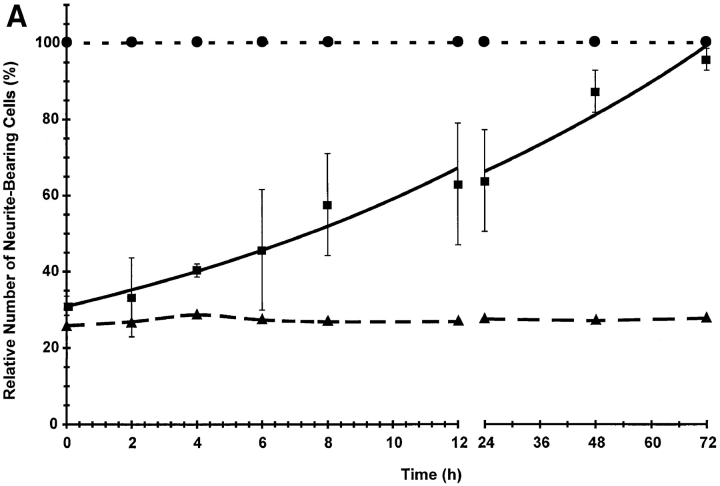
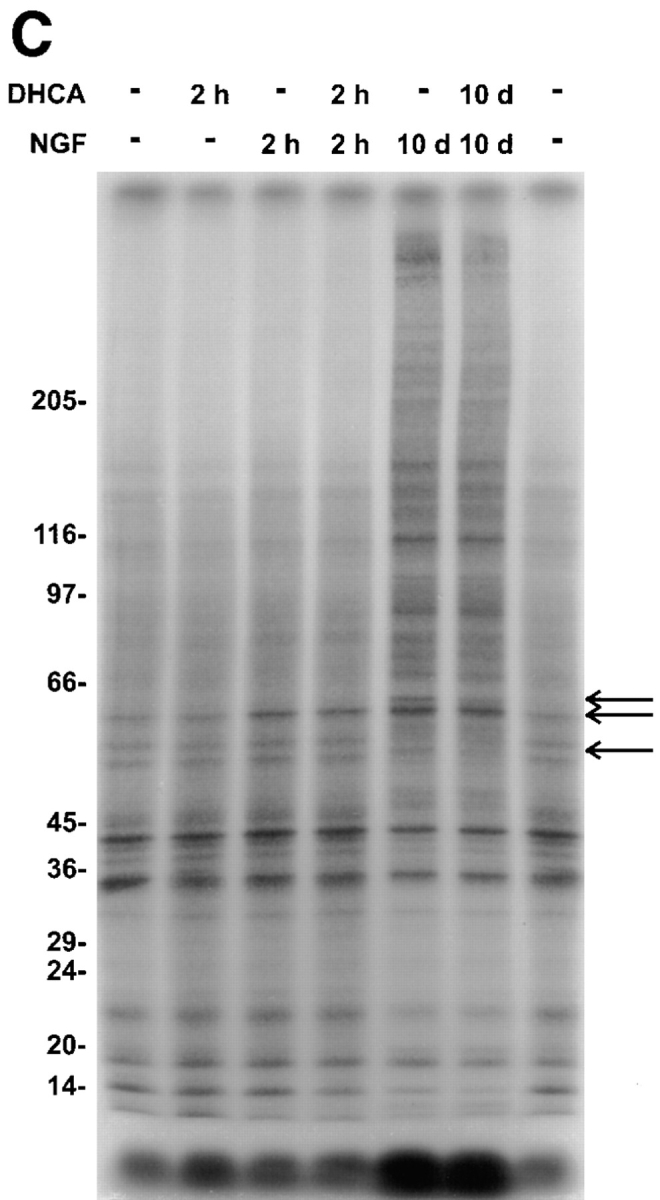
The onset of neurite outgrowth after DHCA is removed was examined to compare the rate of reversing the effect of DHCA on neurite outgrowth to the rate of NGF- induced neurite outgrowth when DHCA had never been present. In addition, removal of DHCA was used to detect protein methylations that are associated with neurite outgrowth during the reversal period. Reversibility of the effects of DHCA on neurite outgrowth and protein methylation was assessed by incubating PC12 cells with NGF and DHCA for 10 d, followed by removal of the DHCA. Interestingly, the inhibitory effect of DHCA on neurite outgrowth was rapidly reversed (Fig. 12 A). No lag was observed before an increase in neurite outgrowth was detected. The t 1/2 for reversal was 12 h (Fig. 12 A). The rate of neurite outgrowth when DHCA is removed is, therefore, about eightfold faster than the normal t 1/2 for neurite outgrowth when NGF is added to naive cells. Thus, the NGF-dependent signaling events that prime the cells (Burstein and Greene, 1978; Greene et al., 1982) for neurite outgrowth were not inhibited greatly by the methylation inhibitor, DHCA.
Figure 12.
Reversal of DHCA-induced inhibition of methyltransferases results in rapid neurite outgrowth and a time-dependent increase in protein methylation. (A) PC12 cells were plated on 35-mm collagen-coated tissue culture dishes at a density of 300,000 cells per dish in NGF (50 ng/ ml) plus or minus 1 μM DHCA for 7 d. The medium was then changed repeatedly to remove DHCA, and neurite outgrowth was scored and plotted over a 3-d period. Controls were maintained in DHCA/NGF (▴) and NGF alone (•). The experimental condition was treatment with DHCA and NGF for 7 d, followed by NGF alone for 3 d (▪). The data are normalized to 100% relative to the number of neurite bearing cells present in cultures treated with NGF alone. The data shown are the means ± SD for three independent experiments. The line shown for the reversal data is the best fit to a single exponential function with a t1/2 of 12 h. After DHCA removal, the protein methylation pattern was also assessed by SDS-PAGE. (B) Cells were harvested after 10 d of DHCA/NGF treatment or after 7 d DHCA/NGF treatment followed by progressively longer times (-h) in the absence of DHCA. Cell lysates were equalized for total TCA-precipitable 3H counts per minute and separated on 7.5–15% gradient SDS-PAGE. The figure shown is a fluorogram of the dried gel. Exposure time was 5 d. The arrows and brackets indicate the positions of proteins that increased in methylation by 25% or more relative to the 10 d DHCA lane. Asterisks mark the position of protein bands not detected in the 10-d DHCA lane. The changes noted during the reversal period were replicated in four independent experiments.
Upon removal of DHCA, the rapid onset of neurite outgrowth was paralleled by rapid reversal of its inhibitory effects on the methylation of several proteins. Proteins that increase by 25% or more in methylation upon removal of DHCA are indicated by the arrows, brackets, and asterisks shown in Fig. 12 B. The molecular weights of proteins that showed increased methylation migrate at 150, 120, 94, 70/68, 50, 48/47, 40, and 35 kD. The time dependencies for methylation were different for different proteins. These results, together with the time course (Figs. 4 and 5), inhibitor studies (Figs. 7–10), and specificity of the NGF- induced effects on protein methylation (Fig. 1) indicate that regulation of protein methylation is required for neurite outgrowth.
Discussion
The Subset of Methylated Proteins that Are NGF-regulated
This work extends previous studies (Seeley et al., 1984; Kujubu et al., 1993) of the role of methylation in growth factor signaling mechanisms and further demonstrates the feasibility of exploring this role by metabolic radiolabeling of specific proteins in intact cells and by in vitro labeling of proteins in cell fractions. The results indicate that protein methylation is involved in the NGF-induced signal transduction that leads to neurite outgrowth. Methylation of numerous proteins was detected by metabolic radiolabeling indicating that protein methylation is an ongoing, active process. NGF specifically affects the methylation of a subset of these proteins. Further evidence for a significant role for protein methylation in neurite outgrowth is the marked concurrent inhibitory effects of DHCA on protein methylation and neurite outgrowth. Moreover, removal of DHCA rapidly leads to concomitant increases in protein methylation and neurite outgrowth. The identities and functions of the NGF-regulated proteins remain to be determined. It is likely that some of the proteins identified by this approach represent previously unidentified players in NGF signal transduction. This is significant because specific protein modifications that are temporally coincident with neuronal differentiation may be useful in the elucidation of the mechanism of action of NGF (Chao, 1992; Kaplan and Stephens, 1994). Thus, signal transduction pathways may be regulated in part by regulating the methylation of specific proteins in much the same manner as protein phosphorylation regulates signaling pathways.
Potential Actions of Protein Methylation During NGF Signal Transduction
The data presented here focus on NGF actions on protein methylations rather than methylation of phospholipids, RNA or DNA. Methylation of genes serves as a general signal for repression of transcription (Razin and Cedar, 1991). Hypomethylation of the genome is associated with cellular differentiation in vitro (Bestor et al., 1984). There were no significant effects of NGF and/or DHCA on total DNA cytosine methylation in PC12 cells. Thus, the potent inhibitory action of DHCA on neurite outgrowth is unlikely to be due to an effect on DNA methylation. Previous work also indicates that NGF does not detectably affect phospholipid methylation (Ferrari and Greene, 1985). These authors further demonstrated that complete inhibition of phospholipid methylation is achieved at concentrations of deaza-adenosine and homocysteine thiolactone that do not block neurite outgrowth.
There are several potential mechanisms through which NGF-induced changes in protein methylation may affect biological responses. Gene regulation may be affected, particularly if the modified proteins are involved in gene expression. For example, many heterogeneous nuclear RNPs that associate with pre-mRNAs are methylated on arginine residues (Liu and Dreyfuss, 1995). Modulation of RNA splicing has been proposed as one potential target for protein methylation in signal transduction (Lin et al., 1996). Consistent with this view, a protein arginine methyltransferase activity appears to be constitutively associated with the type I interferon receptor (Abramovich et al., 1997). A role for protein methylation in cell signaling is further substantiated by the finding that methylation of the γ subunit of a heterotrimeric G-protein enhances activation of PI-specific phospholipase C and PI3-kinase in vitro (Parish et al., 1995). If this biochemical effect occurs in intact cells, protein methylation could play an important general role in the amplification of intracellular signals. Protein methylation may also be involved in altering the subcellular localization of specific proteins, as recently suggested for the cell cycle–dependent methylation of protein phosphatase 2A (Turowski et al., 1995).
Neurite outgrowth requires a fundamental reorganization of the cytoskeleton including formation of intermediate filaments and parallel bundles of microtubules oriented longitudinally in neurites, as well as specialized actin microfilament arrays within motile growth cones. Inhibition of neurite outgrowth by DHCA may be related to inhibition of protein methylation, which affects the cytoskeleton. For example, Ras and Rho proteins, modified by isoprenylation and methylation, are involved in redistribution of actin microfilaments during changes in cell shape (Vojtek and Cooper, 1995). Recently, Luo et al. (1996) presented evidence that constitutively active Rac1 in transgenic mice interferes with the normal formation of Purkinje cell axons and dendrites. Since only the methylation step of small G-protein processing is reversible (Rando, 1996), methylation is more likely than other processing steps to be subject to physiological regulation. NGF-induced increased methyl labeling of G-proteins in PC12 cells (Haklai et al., 1993; Kujubu et al., 1993) and the specific proteins observed in this study indicate that NGF may serve as one mediator of regulatory control in responsive neurons. Furthermore, Klein and colleagues have raised the possibility that protein methylation plays an important role in neurulation (Coelho and Klein, 1990; Moephuli et al., 1997) and neurite outgrowth in rat embryo cultures (Coelho and Klein, 1990). The present work lends additional support to this possibility, especially with regard to neurite outgrowth.
DHCA Is a Potent and Nontoxic Inhibitor of Protein Methylation
DHCA is a promising tool for investigating the possible roles of methylation in signal transduction mechanisms. This inhibitor was developed by Liu and colleagues to obtain a more specific, less toxic inhibitor of SAHH than previously used inhibitors (Liu et al., 1992). The 50% effective dosage (ED50) for the effect of DHCA on neurite outgrowth for PC12 cells (∼100 nM) is four orders of magnitude lower than previously used inhibitors (Seeley et al., 1984). Inhibition of neurite outgrowth by >∼85% is difficult to achieve, even at the highest concentration of the inhibitor. Pretreatment of cells with DHCA for several weeks before NGF stimulation, however, does produce nearly complete inhibition even at 7 d of NGF (Cimato, T.R., M.J. Ettinger, and J.M. Aletta, unpublished observations). These observations provisionally indicate that basal, ongoing protein methylation may serve an auxiliary role in neurite outgrowth in addition to the NGF effects on protein methylation. As expected, DHCA potently inhibits SAHH activity (Hasobe et al., 1989) and consequently leads to decreased total protein methylation. The dose-dependent inhibition of protein methylation observed is quite similar to that for inhibition of neurite outgrowth (Fig. 9). The cooperative effects of homocysteine and DHCA on neurite outgrowth further substantiate a specific effect on methylation. The finding that total protein methylation was inhibited more when NGF was present than when absent, suggests that under the influence of NGF, many of the potential methyltransferases involved in the NGF response may be more sensitive to inhibition due to increased levels of SAHcy. This may occur because, as a result of increased total protein methylation (Fig. 3), intracellular levels of SAHcy are expected to be elevated even before DHCA treatment. NGF has no measureable effect on SAHH activity (Cimato, T.R., M.J. Ettinger, and J.M. Aletta, unpublished results).
DHCA is unique among methylation inhibitors because of its lack of effect on cell flattening, phosphorylation of tyrosine hydroxylase, and tyrosine phosphorylation of Trk. Less specific methylation inhibitors, used to study the role of methylation in PC12 cells previously (Seeley et al., 1986; Kujubu et al., 1993; Maher, 1993; Meakin and Shooter, 1995), blocked each of these actions. The most remarkable property of DHCA in comparison to previously used inhibitors is that DHCA inhibits neurite outgrowth without inhibiting the NGF priming that prepares PC12 cells for neurite outgrowth (Greene et al., 1982). This is best illustrated by the rapid reversal of DHCA effects on neurite outgrowth, which is in marked contrast to that observed with other methylation inhibitors. Reversal of inhibition by 5′-deoxy-S-methyl adenosine requires the same 7 d as required by control PC12 cells when first exposed to NGF (Seeley et al., 1986). Thus, DHCA is clearly more specific than previously used methylation inhibitors. DHCA specifically affects neurite outgrowth without affecting many other signaling effects of NGF, including those associated with priming.
Protein Methylation in Early and Delayed NGF Signaling
Growth factors in general, promote biological responses that are either extremely rapid (within minutes) or require many hours or days to develop fully. The diversity of signaling networks (Pawson, 1985; Hunter, 1995) and the propensity of specific pathways to transmit and/or sustain a given signal (Marshall, 1995) may help to explain the different temporal characteristics of responses. The results of this study and other recent work (Metz et al., 1993; Philips et al., 1993, 1995) indicate that posttranslational modification of proteins by methylation offers another potential mechanism for regulating both early and delayed growth factor signaling.
The importance of early and delayed responses to NGF in PC12 cells, dorsal root ganglion, and sympathetic neurons is well documented (Greene, 1984). One of the most obvious examples of a delayed NGF response is neurite outgrowth. Little neurite outgrowth is evident in PC12 cells treated with NGF for 12–24 h. 50% of the cells extend neurites within 4 d, and nearly all have neurites by 7 d. NGF-mediated neurite outgrowth requires transcription (Burstein and Greene, 1978; Greene et al., 1982). The long latency is apparently due to the progressive accumulation of specific proteins required for neurite assembly during the priming period. This accumulation may depend on protein synthesis (Burstein and Greene, 1978) and/or progressive posttranslational modifications of specific proteins (e.g., as shown here for the methylation of specific proteins). Since regeneration of neurites from PC12 cells previously treated with NGF occurs much more rapidly (within 24 h) than initiation (7 d); the longer time required for neurite outgrowth in the latter is thought to be due to the “priming” events.
When interpreted in light of the priming model of NGF-induced neurite outgrowth, the experiments that used DHCA provide important insights regarding the temporal nature of protein methylation involved in this NGF response. Inhibition of protein methylation by DHCA is most effective in reducing neurite formation when the DHCA is present concomitantly with NGF treatment. There is decreased inhibition of neurite outgrowth as the time interval between NGF treatment and the addition of DHCA increases (Fig. 11 A). DHCA is also less effective at inhibiting neurite regeneration (Fig. 11 B). These results imply progressive accumulation of the methylated protein product(s) necessary for neurite outgrowth, as was detected at 4 d of NGF treatment (Fig. 5). Slow turnover of methylated proteins is consistent with the reduced effect of DHCA when added after NGF signaling has been initiated.
The time dependencies of specific protein methylations, the effects of DHCA on protein methylation, and the lack of effects of DHCA on early protein phosphorylations and priming are consistent with little or no interdependence between protein methylation signaling events and early, priming events. Moreover, the data presented on the phosphorylation of Trk and other proteins that lie downstream of Trk activation (Fig. 10, B and C) further substantiate the view that some cellular signaling events are more susceptible to interference from inhibition of protein methylation than others. Autophosphorylation of Trk and the rapid increase in the phosphorylation of tyrosine hydroxylase (Halegoua and Patrick, 1980) are not affected by DHCA. Two other NGF-induced increases in phosphorylation, which require long term (⩾7 d) NGF treatment for maximal phosphorylation, are temporally associated with neurite outgrowth. In accord with DHCA-mediated inhibition of neurite outgrowth, but unlike the rapid, early phosphorylation events, the increased 32P incorporation into the 64-kD chartin (Aletta and Greene, 1987) and β-tubulin (Black et al., 1986; Aletta, 1996) is diminished by exposure of the cells to DHCA.
It can thus be concluded that DHCA does not disrupt all early NGF signaling events and priming actions, nor does it abrogate many of the biological effects of NGF signaling (e.g., Fig. 10 A, survival; Fig. 9, cell flattening). The effects of DHCA on neurite outgrowth and inhibition of the phosphorylation of neurite-associated proteins (β-tubulin and chartin) are consistent with a selective inhibitory action on an NGF pathway responsible for neurite outgrowth. Based on all of the above results, the protein methylation required is likely to occur downstream from, or parallel to, rapid NGF actions and priming events. This interpretation does not exclude the possibility that protein methylation of rapid onset also plays an important role in neurite outgrowth. Once priming is achieved, NGF stimulation of some of the rapid protein methylations observed in this study may assume added significance, particularly if they occur at the growth cone.
Acknowledgments
We are grateful to R. Borchardt for synthesizing the DHCA that was used for these studies.
This work was supported in part by USDA grant No. 9500773, and by the Multidisciplinary Research Pilot Program from the University at Buffalo, State University of New York.
Abbreviations used in this paper
- DHCA
9-(trans-2′, trans-3′-dihydroxycyclopent-4′-enyl)-adenine
- ERK
extracellular signal-regulated kinase
- PAS
protein-A-Sepharose
- 2D two dimensional; PI
phosphoinositide
- SAHcy
S-adenosylhomocysteine
- SAHH
S-adenosylhomocysteine hydrolase
- SAM
S-adenosylmethionine
- Trk
tyrosine kinase receptor
Footnotes
Please address all correspondence to John M. Aletta, Department of Pharmacology and Toxicology, University at Buffalo School of Medicine and Biomedical Sciences, 3435 Main Street, Buffalo, NY 14214-3000. Tel.: (716) 829-3237. Fax: (716) 829-2801. e-mail: jaletta@ubmed.buffalo.edu
References
- Abramovich C, Yakobson B, Chebath J, Revel M. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO (Eur Mol Biol Organ) J. 1997;16:260–266. doi: 10.1093/emboj/16.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aletta JM. Phosphorylation of type III β-tubulin in PC12 cell neurites during NGF-induced process outgrowth. J Neurobiol. 1996;31:461–475. doi: 10.1002/(SICI)1097-4695(199612)31:4<461::AID-NEU6>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Aletta JM, Greene LA. Sequential phosphorylation of chartin microtubule-associated proteins is regulated by the presence of microtubules. J Cell Biol. 1987;105:277–290. doi: 10.1083/jcb.105.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backlund PS, Jr, Carotti D, Cantoni GL. Effects of the S-adenosylhomocysteine hydrolase inhibitors 3-deazaadenosine and 3-deazaaristeromycin on RNA methylation and synthesis. Eur J Biochem. 1986;160:245–251. doi: 10.1111/j.1432-1033.1986.tb09963.x. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Hellewell SB, Ingram VM. Differentiation of two mouse cell lines is associated with hypomethylation of their genomes. Mol Cell Biol. 1984;4:1800–1806. doi: 10.1128/mcb.4.9.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MM, Aletta JM, Greene LA. Regulation of microtubule composition and stability during nerve growth factor-promoted neurite outgrowth. J Cell Biol. 1986;103:545–557. doi: 10.1083/jcb.103.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritium- labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burstein DE, Greene LA. Evidence for RNA synthesis-dependent and independent pathways in stimulation of neurite outgrowth by nerve growth factor. Proc Natl Acad Sci USA. 1978;75:6059–6063. doi: 10.1073/pnas.75.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JP. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979;98:132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Chao MV. Growth factor signaling: where is the specificity? . Cell. 1992;68:995–997. doi: 10.1016/0092-8674(92)90068-n. [DOI] [PubMed] [Google Scholar]
- Chelsky D, Ruskin B, Koshland DE. Methyl-esterified proteins in a mammalian cell line. Biochemistry. 1985;24:6651–6658. doi: 10.1021/bi00344a053. [DOI] [PubMed] [Google Scholar]
- Chiang PK, Richards HH, Cantoni GL. S-adenosyl-l-homocysteine hydrolase: analogues of S-adenosyl-l-homocysteine as potential inhibitors. Mol Pharmacol. 1977;13:939–947. [PubMed] [Google Scholar]
- Coelho CND, Klein NW. Methionine and neural tube closure in cultured rat embryos: morphological and biochemical analyses. Teratology. 1990;42:437–451. doi: 10.1002/tera.1420420412. [DOI] [PubMed] [Google Scholar]
- De la Haba G, Cantoni GL. The enzymatic synthesis of S-adenosyl-l-homocysteine from adenosine and homocysteine. J Biol Chem. 1959;234:603–608. [PubMed] [Google Scholar]
- Favre B, Zolnierowicz S, Turowski P, Hemmings BA. The catalytic subunit of protein phosphatase 2A is carboxyl-methylated in vivo. J Biol Chem. 1994;269:16311–16317. [PubMed] [Google Scholar]
- Ferrari G, Greene LA. Does phospholipid methylation play a role in the primary mechanism of action of nerve growth factor? . J Neurochem. 1985;45:853–859. doi: 10.1111/j.1471-4159.1985.tb04072.x. [DOI] [PubMed] [Google Scholar]
- Greene LA. The importance of both early and delayed responses in the biological actions of nerve growth factor. Trends Neurosci. 1984;13:91–94. [Google Scholar]
- Greene LA, Tischler AS. Establishment of a noradrenergic clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Proc Natl Acad Sci USA. 1976;73:2424–2428. doi: 10.1073/pnas.73.7.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Burstein DE, Black MM. The role of transcription-dependent priming in nerve growth factor promoted neurite outgrowth. Dev Biol. 1982;91:305–316. doi: 10.1016/0012-1606(82)90037-9. [DOI] [PubMed] [Google Scholar]
- Greene LA, Aletta JM, Rukenstein A, Green SH. PC12 pheochromocytoma cells: culture, nerve growth factor treatment, and experimental exploitation. Methods Enzymol. 1987;147:207–216. doi: 10.1016/0076-6879(87)47111-5. [DOI] [PubMed] [Google Scholar]
- Haklai R, Lerner S, Kloog Y. Nerve growth factor induces a succession of increases in isoprenylated methylated small GTP-binding proteins of PC12-pheochromocytoma cells. Neuropeptides. 1993;24:11–25. doi: 10.1016/0143-4179(93)90036-a. [DOI] [PubMed] [Google Scholar]
- Halegoua S, Patrick J. Nerve growth factor mediates phosphorylation of specific proteins. Cell. 1980;22:571–581. doi: 10.1016/0092-8674(80)90367-0. [DOI] [PubMed] [Google Scholar]
- Hasobe M, McKee JG, Ishii H, Cools M, Borchardt RT, DeClercq E. Elucidation of the mechanism by which homocysteine potentiates the anti-vaccinia virus effects of the S-adenosylhomocysteine hydrolase inhibitor 9-(trans-2′, trans-3′-dihydroxycyclopent-4′-enyl) derivatives of adenine and 3-deazaadenine on the metabolism of S-adenosylhomocysteine in mouse L929 cells. Mol Pharmacol. 1989;36:490–496. [PubMed] [Google Scholar]
- Hershfield MS. Apparent suicide inactivation of human lymphoblast S-adenosylhomocysteine hydrolase by 2′-deoxyadenosine and adenosine arabinoside. J Biol Chem. 1979;254:22–25. [PubMed] [Google Scholar]
- Hildeshein J, Hildeshein R, Lederer E. New syntheses of S-adenosyl homocysteine analogues, potential methyltransferase inhibitors. Biochimie (Paris) 1972;54:989–995. doi: 10.1016/s0300-9084(72)80226-8. [DOI] [PubMed] [Google Scholar]
- Hrycyna CA, Clarke S. Modification of eukaryotic signaling proteins by COOH-terminal methylation reactions. Pharmacol Ther. 1993;59:281–300. doi: 10.1016/0163-7258(93)90071-k. [DOI] [PubMed] [Google Scholar]
- Huff K, End D, Guroff G. Nerve growth factor-induced alteration in the response of PC12 pheochromocytoma cells to epidermal growth factor. J Cell Biol. 1981;88:189–198. doi: 10.1083/jcb.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- Kaplan DR, Stephens RM. Neurotrophin signal transduction by the Trk receptor. J Neurobiol. 1994;25:1404–1417. doi: 10.1002/neu.480251108. [DOI] [PubMed] [Google Scholar]
- Kredich NM, Martin DW., Jr Role of S-adenosylhomocysteine in adenosine mediated toxicity in cultured mouse T lymphoma cells. Cell. 1977;12:931–938. doi: 10.1016/0092-8674(77)90157-x. [DOI] [PubMed] [Google Scholar]
- Kujubu DA, Stimmel JB, Law RE, Herschman HR, Clarke S. Early responses of PC12 cell to NGF and EGF: effect of K252a and 5′-methylthioadenosine on gene expression and membrane protein methylation. J Neurosci Res. 1993;36:58–65. doi: 10.1002/jnr.490360107. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature (Lond) 1970;222:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Mills AD. Quantitative film detection of 3H and 14C in polyacrylamide gels by fluorography. Eur J Biochem. 1975;56:335–341. doi: 10.1111/j.1432-1033.1975.tb02238.x. [DOI] [PubMed] [Google Scholar]
- Lee J, Stock J. Protein phosphatase 2A catalytic subunit is methyl-esterified at its carboxyl terminus by a novel methyltransferase. J Biol Chem. 1993;268:19192–19195. [PubMed] [Google Scholar]
- Lee J, Chen Y, Tolstykh T, Stock J. A specific protein carboxyl methylesterase that demethylates phosphoprotein phosphatase 2A in bovine brain. Proc Natl Acad Sci USA. 1996;93:6043–6047. doi: 10.1073/pnas.93.12.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-J, Gary JD, Yang MC, Clarke S, Herschman HR. The mammalian immediate-early gene TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J Biol Chem. 1996;271:15034–15044. doi: 10.1074/jbc.271.25.15034. [DOI] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wolfe MS, Borchardt RT. Rational approaches to the design of antiviral agents based on S-adenosyl-l-homocysteine hydrolase as a molecular target. Antivir Res. 1992;19:247–265. doi: 10.1016/0166-3542(92)90083-h. [DOI] [PubMed] [Google Scholar]
- Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature (Lond) 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- Maher PA. Inhibition of the tyrosine kinase activity of fibroblast growth factor receptor by the methyltransferase inhibitor 5′-methylthioadenosine. J Biol Chem. 1993;268:4244–4249. [PubMed] [Google Scholar]
- Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- Meakin SO, Shooter EM. Tyrosine kinase activity coupled to the high-affinity nerve growth factor-receptor complex. Proc Natl Acad Sci USA. 1991;88:5862–5866. doi: 10.1073/pnas.88.13.5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz SA, Rubaglia ME, Stock JB, Kowluru A. Modulation of insulin secretion from normal rat islets by inhibitors of the post-translational modifications of GTP binding proteins. Biochem J. 1993;295:31–40. doi: 10.1042/bj2950031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MW, Duhl DMJ, Winkes BM, Arredondo-Vega F, Saxon PJ, Wolff GL, Epstein CJ, Hershfield MS, Barsh GS. The mouse lethal nonagouti (ax) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy)gene. EMBO (Eur Mol Biol Organ) J. 1994;13:1806–1816. doi: 10.1002/j.1460-2075.1994.tb06449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley WC, Schenker A, Shooter EM. Characterization and isolation of proteolytically modified nerve growth factor. Biochemistry. 1976;15:5543–5552. doi: 10.1021/bi00670a019. [DOI] [PubMed] [Google Scholar]
- Moephuli SR, Klein NW, Baldwin MT, Krider HM. Effects of methionine on the cytoplasmic distribution of actin and tubulin during neural tube closure in rat embryos. Proc Natl Acad Sci USA. 1997;94:543–548. doi: 10.1073/pnas.94.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najbauer J, Aswad DW. Diversity of methyl acceptor proteins in rat pheochromocytoma (PC12) cells revealed after treatment with adenosine dialdehyde. J Biol Chem. 1990;265:12717–12721. [PubMed] [Google Scholar]
- Parish CA, Smrcka AV, Rando RR. Functional significance of βγ-subunit carboxyl methylation for the activation of phospholipase C and phosphoinositide 3-kinase. Biochemistry. 1995;34:7722–7727. doi: 10.1021/bi00023a019. [DOI] [PubMed] [Google Scholar]
- Pawson T. Protein modules and signaling networks. Nature (Lond) 1985;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- Peng X, Greene LA, Kaplan DR, Stephens RM. Deletion of a conserved juxtamembrane sequence in trk abolishes NGF-promoted neuritogenesis. Neuron. 1995;15:395–406. doi: 10.1016/0896-6273(95)90043-8. [DOI] [PubMed] [Google Scholar]
- Philips MR, Pillinger MH, Staud R, Volker C, Rosenfeld MG, Weissmann G, Stock JB. Carboxyl methylation of Ras-related proteins during signal transduction in neutrophils. Science (Wash DC) 1993;259:868–872. doi: 10.1126/science.8438158. [DOI] [PubMed] [Google Scholar]
- Philips MR, Staud R, Pillinger M, Feoktistov A, Volker C, Stock JB, Weissmann G. Activation-dependent carboxyl methylation of neutrophil G-protein γ subunit. Proc Natl Acad Sci USA. 1995;92:2283–2287. doi: 10.1073/pnas.92.6.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui M-S, Green SH. PC12 cell neuronal differentiation is associated with prolonged p21rasactivity and consequent prolonged ERK activity. Neuron. 1992;9:705–717. doi: 10.1016/0896-6273(92)90033-a. [DOI] [PubMed] [Google Scholar]
- Rando RR. Chemical biology of protein isoprenylation/methylation. Biochim Biophys Acta. 1996;1300:5–16. doi: 10.1016/0005-2760(95)00233-2. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukenstein A, Rydel RE, Greene LA. Multiple agents rescue PC12 cells from serum-free cell death by translation, and transcription- dependent mechanisms. J Neurosci. 1991;11:2557–2563. doi: 10.1523/JNEUROSCI.11-08-02552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley PJ, Rukenstein A, Connolly JL, Greene LA. Differential inhibition of nerve growth factor and epidermal growth factor on the PC12 pheochromocytoma line. J Cell Biol. 1984;98:417–426. doi: 10.1083/jcb.98.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro MJ, Chakrabarti I, Koshland DE. Contributions made by individual methylation sites of Escherichia coliaspartate receptor to chemotactic behavior. Proc Natl Acad Sci USA. 1995;92:1053–1056. doi: 10.1073/pnas.92.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto AM, Sonnenschein C. The role of estrogens on the proliferation of human breast tumor cells (MCF-7) J Steroid Biochem. 1985;23:87–94. doi: 10.1016/0022-4731(85)90265-1. [DOI] [PubMed] [Google Scholar]
- Tischler AS, Greene LA. Morphologic and cytochemical properties of a clonal line of rat adrenal pheochromocytoma cells which respond to nerve growth factor. Lab Invest. 1978;39:77–89. [PubMed] [Google Scholar]
- Turowski P, Fernandez A, Favre B, Lamb NJC, Hemmings BA. Differential methylation and altered conformation of cytoplasmic and nuclear forms of protein phosphatase 2A during cell cycle progression. J Cell Biol. 1995;129:397–410. doi: 10.1083/jcb.129.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vojtek AB, Cooper JA. Rho family members: activators of MAP kinase cascades. Cell. 1995;82:527–529. doi: 10.1016/0092-8674(95)90023-3. [DOI] [PubMed] [Google Scholar]
- Xie H, Clarke S. Protein phosphatase 2A is reversibly modified by methyl esterification of its C-terminal leucine residue in bovine brain. J Biol Chem. 1994;269:1981–1984. [PubMed] [Google Scholar]