Abstract
One of the fundamental goals of lipidomics research is to identify the linkage of an individual gene with a given lipidome, thereby revealing the role of that gene in lipid metabolism, transport, and homeostasis. In this study, we have identified four apolipoprotein E (apoE)-induced alterations in the lipidome of mouse dorsal root ganglia (DRG) through utilizing the technology of shotgun lipidomics. First, apoE mediates sulfatide mass content in mouse DRG, which is comparable to its role in the CNS. Second, apoE contributes to galactosylceramide and ceramide homeostasis in mouse DRG. Third, apoE significantly modulates cholesterol levels in mouse DRG. The latter two functions of apoE are distinct from those in the CNS. Finally, mice null for apoE have dramatically less triacylglycerol mass content in DRG which are opposite to the effects observed in the peripheral organs and vascular system. Collectively, this study identifies the specific alterations in the DRG lipidome induced by apoE knockout and suggests the potential roles of apoE in lipid transport and homeostasis in a tissue specific manner, thereby providing insights into the biochemical mechanisms underlying the functions of apoE in the PNS.
Keywords: apolipoprotein E, dorsal root ganglion, electrospray ionization mass spectrometry, lipidomics, peripheral nervous system, shotgun lipidomics, sphingolipid metabolism, sulfatide
Lipidomics, the large-scale study of organic solvent-soluble lipids by integration of many different modern techniques such as mass spectrometry (MS), is a rapidly expanding field following the tremendous progress made in genomics and proteomics (Han and Gross 2003; Lagarde et al. 2003). The recent rapid development of lipidomics has been driven by intense interest in lipid-related disease states, lipid-regulated protein expression, and defining alterations in the lipid profiles of the cells resulting from their metabolic, environmental, or nutritional influences (Han et al. 2001, 2004a). Such information can provide insight into the biophysical states of cellular membranes (Han et al. 2001), differences in lipid pools and turnover rates (Delong et al. 1999; Boumann et al. 2003, 2004; Su et al. 2004), alterations in cellular energy supply (Han et al. 2004a), and changes in lipid second messenger levels reflecting cellular metabolic responses and transcriptional programs (Listenberger et al. 2003). Therefore, lipidomics plays many essential roles in identifying the biochemical mechanisms of lipid metabolism, investigating the functions of an individual gene of interest, identifying novel biomarkers, and evaluating drug efficacy. Lipidomics has been greatly advanced by the development and application of MS, particularly electrospray ionization (ESI) MS (see Griffiths 2003; Han and Gross 2003; Pulfer and Murphy 2003; Welti and Wang 2004; Han and Gross 2005a,b; Merrill et al. 2005; Wenk 2005 for recent reviews).
Apolipoprotein E (apoE), highly expressed in the liver (Lin et al. 1986) and other tissues, plays a prominent role in the transport and metabolism of plasma cholesterol, triacylglycerols (TG), and phospholipids among various cells of the body (Mahley 1988). Therefore, apoE depletion leads to substantial accumulations of cholesterol and TG in plasma and many peripheral organs (Mahley 1988; Kypreos et al. 2001; Mensenkamp et al. 2004). ApoE is the major apolipoprotein in the CNS (Elshourbagy et al. 1985; Demattos et al. 2001; Koch et al. 2001) and thus, plays a key role in neurobiology (see Horsburgh et al. 2000; Mahley and Rall 2000; Laws et al. 2003 for recent reviews) by mediating the transport of cholesterol, phospholipids, and sulfatides (Weisgraber et al. 1994; Han et al. 2003a; Han 2004). However, in contrast to its role in peripheral organs, apoE modulates sulfatide mass content without disturbing the homeostasis of cholesterol, phospholipid, and other examined sphingolipid pools in the CNS (Han 2004). ApoE may also function in the development, remodeling, and regeneration of the nervous system independent of lipid transport (Boyles et al. 1990; Poirier et al. 1993; Masliah et al. 1995; Posse De Chaves et al. 2000), but the biochemical mechanism(s) underlying these activities are not entirely clear.
ApoE has attracted intense interest because of its association with Alzheimer's disease (AD), Parkinson's disease, stroke, brain injury, and possibly longevity (see recent review Han 2004 for detailed references). Among the researches, we have hypothesized that apoE-mediated sulfatide transport and alterations in lipid metabolism in the CNS are linked to the pathogenesis of AD. Notably, the apoE allele 4 is a primary risk factor for development of AD (Strittmatter and Roses 1996; Bales et al. 2002). Furthermore, sulfatide mass content has recently been shown to be substantially and specifically depleted in the brain of AD patients at the earliest clinically recognizable stage of AD (Han et al. 2002, 2003b; Cheng et al. 2003). In addition, apoE specifically modulates overall sulfatide mass content in the CNS (Han et al. 2003a).
However, it is unknown whether apoE also affects lipid transport and metabolism in the PNS as it does in the CNS or in the peripheral organs, and if so, to what degree does apoE affect the PNS lipidome? It has been shown that apoE knockout (KO) mice have abnormal morphology of unmyelinated axons in their peripheral nerves, severe peripheral nerve conduction defects, and impaired blood–nerve barriers, among others (Fullerton et al. 1998, 2001; Pola et al. 2003). Surprisingly, virtually nothing is known regarding the relationship between apoE-mediated lipid alterations and these apoE KO-induced PNS defects. Therefore, we hypothesized that apoE plays an important and specific role in lipid transport and homeostasis in the PNS, which might provide insights into the biochemical mechanisms underlying these apoE KO-induced PNS defects. In this report, we determined the specific changes of the PNS lipidome induced by apoE depletion using mouse cervical dorsal root ganglion (DRG) as a model utilizing our newly developed shotgun lipidomics technology (Han et al. 2004b; Han and Gross 2005a,b). Through a systematic examination of the DRG lipidomes of wild-type (WT) and apoE KO mice, we demonstrated multiple specific changes induced by apoE depletion including substantial increases in sulfatide, galactosylceramide (GalCer), ceramide (Cer), and cholesterol mass contents and a dramatic decrease in TG mass content. These changes are very different from those observed in either the CNS or the peripheral organs and may partially explain several apoE KO-induced PNS defects. Accordingly, this study identifies the specific alterations in the DRG lipidome induced by apoE depletion. The results suggest apoE participates in tissue-specific lipid transport and homeostasis and may implicate apoE dysfunction as a contributing factor to multiple disease states such as peripheral neuropathy.
Materials and methods
Materials
Synthetic phospholipids including 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (14:1−14:1 GPCho), 1,2-dilinoleoyl-sn-glycero-3-phosphoethanolamine (18:2−18:2 GPEtn), 1,2-dipentadecanoyl-sn-glycero-3-phosphoglycerol (15:0−15:0 GPGro), 1,2-dimyristoyl-sn-glycero-3-phosphoserine (14:0−14:0 GPSer), N-lauroryl sphingomyelin (N12:0 SM), and N-heptadecanoyl ceramide (N17:0 Cer) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Semisynthetic N-palmitoyl sulfatide (N16:0 sulfatide) and perdeuterated N-stearoyl GalCer (d35-N18:0 GalCer) were obtained from Matreya, Inc. (Pleasant Gap, PA, USA). Deuterated cholesterol (d6-chol) was purchased from Cambridge Isotope Laboratories, Inc. (Cambridge, MA, USA). The Amplex Red cholesterol assay kit was obtained from Molecular Probe, Inc. (Eugene, OR, USA). All the solvents were obtained from Burdick and Jackson (Honeywell International Inc., Burdick and Jackson, Muskegon, MI, USA). All other chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA).
Preparation of lipid extracts from mouse cervical DRG
Both WT and apoE KO mice (male, C57 BL/J background, at the indicated ages) were either purchased from the Jackson Laboratory (Bar Harbor, ME, USA) and/or housed in a full barrier facility with a 12-h light/dark cycle and maintained on standard chow (Diet 5053; Purina Inc., St Louis, MO, USA). All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (National Academy of Science, 1996) and were approved by the Animals Studies Committee at Washington University. Mice were killed by asphyxiation with carbon dioxide.
The cervical DRG along with associated nerve bundles (DRG, ∼2 mg) of each individual mouse were dissected quickly and perfused with ice-cooled, 10× diluted phosphate-buffered saline. Each piece of mouse DRG was cut into tiny segments prior to homogenization in 1 mL of ice-cold LiCl aqueous solution (50 mmol/L) with a Potter–Elvehjem tissue grinder. Protein assays on each individual homogenate were performed. After a certain amount of homogenate (∼1 mL) from each DRG sample was transferred to a disposable culture borosilicate glass tube (16 × 100 mm), internal standards including 14:0−14:0 GPSer (20.0 nmol/mg protein), 15:0−15:0 GPGro (8.0 nmol/mg protein), 18:2−18:2 GPEtn (35.0 nmol/mg protein), 14:1−14:1 GPCho (40.0 nmol/mg protein), T17:1 TG (10 nmol/mg protein), d35-N18:0 GalCer (20.0 nmol/mg protein), N12:0 SM (20 nmol/mg protein), N16:0 sulfatide (8.0 nmol/mg protein), d6-chol (250.0 nmol/mg protein), and N17:0 Cer (300 pmol/mg protein) were added to each DRG homogenate based on protein concentration, thereby allowing the final quantitated lipid content to be normalized to the protein content and the elimination of a potential loss from the incomplete recovery. These internal standards were selected because they only represent of the endogenous cellular lipid mass as demonstrated by ESI/MS lipid analysis.
Lipids from each homogenate were extracted by a modified Bligh and Dyer (1959) procedure as described previously (Cheng et al. 2006). Each lipid extract was reconstituted with a volume of 500 μL/mg of protein (which was based on the original protein content of the samples as determined from protein assays) in CHCl3/MeOH (1:1, v/v). The lipid extracts were finally flushed with nitrogen, capped, and stored at −20°C for ESI/MS analyses (typically within 1 week). Each lipid solution was diluted approximately 50-fold immediately prior to infusion and lipid analysis.
Mass spectrometric analysis of DRG lipids
A triple-quadrupole mass spectrometer (ThermoElectron TSQ Quantum Ultra, San Jose, CA, USA) equipped with an electrospray ion source and Xcalibur system software was utilized in the study under conditions as previously described (Han et al. 2004b). The first and third quadrupoles serve as independent mass analyzers using a mass resolution setting of peak width 0.7 Th while the second quadrupole serves as a collision cell for tandem MS. The diluted lipid extract was directly infused into the ESI source at a flow rate of 4 μL/min with a syringe pump. Typically, a 2-min period of signal averaging in the profile mode was employed for each MS spectrum. For tandem MS, a collision gas pressure was set at 1.0 mTorr, but the collision energy varied with the classes of lipids as described previously (Han et al. 2004b; Han and Gross 2005a). Typically, a 2- to 5-min period of signal averaging in the profile mode was employed for each tandem MS spectrum. All the MS spectra and tandem MS spectra were automatically acquired by a customized sequence subroutine operated under Xcalibur software. Individual GPEtn molecular species were identified and low-abundant GPEtn molecular species were quantitated or refined after derivatization with fluorenylmethoxycarbonyl (Fmoc) (Han et al. 2005a). Data processing of 2D MS analyses including ion peak selection, data transferring, peak intensity comparison, and quantitation was conducted using self-programmed MicroSoft Excel macros (Han et al. 2004b).
Quantitation of cholesterol and its esters by ESI MS
An aliquot (10 μL) of a stock solution of mixed dimethyl aminopyridine (1 mol/L) and methoxyacetic acid (1 mol/L) in CH2Cl2 and 10 μL of a stock solution of 1-(3-dimethyl aminopropyl)-3-ethylcarbodiimide in CH2Cl2 (0.142 mol/L) were sequentially added to a DRG lipid extract (containing ∼30 nmol of cholesterol in 25 μL of an individual lipid extract) with a cholesterol internal standard (i.e. d6-chol). The mixed solution (make to a final volume of 100 μL with CH2Cl2) was stirred at 23°C overnight (approximately 14 h). To each reaction mixture, 1 mL of milli-Q purified water was added prior to extraction of derivatized cholesterol with ethyl ether (2 mL × 3 times). The combined organic extract was filtered through a 0.2-μm polytetrafluoroethylene syringe filter (Millex®-FG, Millipore Corporation, Bedford, MA, USA) and evaporated under a nitrogen stream. Each individual residue was resuspended in 1 mL of CHCl3/MeOH (1:1, v/v) which was diluted 10-fold prior to MS analysis. To 1 mL of this diluted solution was added a LiOH solution in methanol (10 μL of LiOH solution in MeOH which was diluted 100-fold from a saturated LiOH stock solution in methanol). This LiOH-added solution was directly infused to the ESI mass spectrometer by a Harvard syringe pump at a flow rate of 4 μL/min for quantitative analysis. Specifically, a precursor ion (PI) scan of m/z 97.1 (corresponding to a fragment ion of [methoxyacetate + MeOH + Li]+, resulting from both cholesterol and d6-chol derivatives) was performed at a collisional energy of 22 eV with a collisional gas pressure of 1 mTorr in the positive-ion mode. Quantitative analysis was achieved by direct comparison of the peak intensity of cholesterol derivative to that of the internal standard derivative from the PI scanning. A detection limit of 1 fmol/μL, a linear dynamic concentration range from 1 fmol/μL to 100 pmol/μL, and a linear dynamic range of the relative ratio of cholesterol versus d6-chol from 0.05 to 20 (i.e. ∼400-fold) were achieved in the technique. These results were obtained through analyses of mixtures of cholesterol and d6-chol at various ratios and concentrations by MS as described above.
For quantitation of total esterified cholesterols, cholesterol esters were first converted to free cholesterol in the presence of the selected internal standard (d6-chol). Specifically, a DRG lipid extract (25 μL of the prepared lipid extract) containing an internal standard was dissolved in tetrahydrofuran (25 μL) followed by addition of working buffer [0.5% (w/v) taurocholate in phosphate buffer, pH 7.0, 1 mL]. The mixture was sonicated for 10 min followed by addition of cholesterol esterase stock solution (200 U/mL working buffer, 10 μL). The resulting mixture was incubated at 37°C overnight (about 16 h), cooled to 23°C, and extracted with ethyl ether (2 mL × 3 times). The combined organic extract was evaporated under a nitrogen stream, and the residue containing cholesterol and d6-chol was directly derivatized with methoxyacetic acid and quantitated as described above. The mass content of total esterified cholesterol in DRG lipid extracts was obtained by subtraction of the mass content of free cholesterol from that of total cholesterol in the sample.
Miscellaneous
Protein concentration was determined with a bicinchroninic acid protein assay kit (Pierce, Rockford, IL, USA) using bovine serum albumin as a standard. The cholesterol and cholesterol ester contents of DRG lipid extracts were also determined by using an enzymatic methodology with an Amplex Red cholesterol assay kit. Data from biological samples were normalized to the protein content and all data are presented as the means ± SD of six separate animals. Statistical differences between mean values were determined by nested anova analysis.
Results
Analyses of the mouse cervical DRG lipidome
Intrasource separation and selective ionization of the mouse DRG lipidome
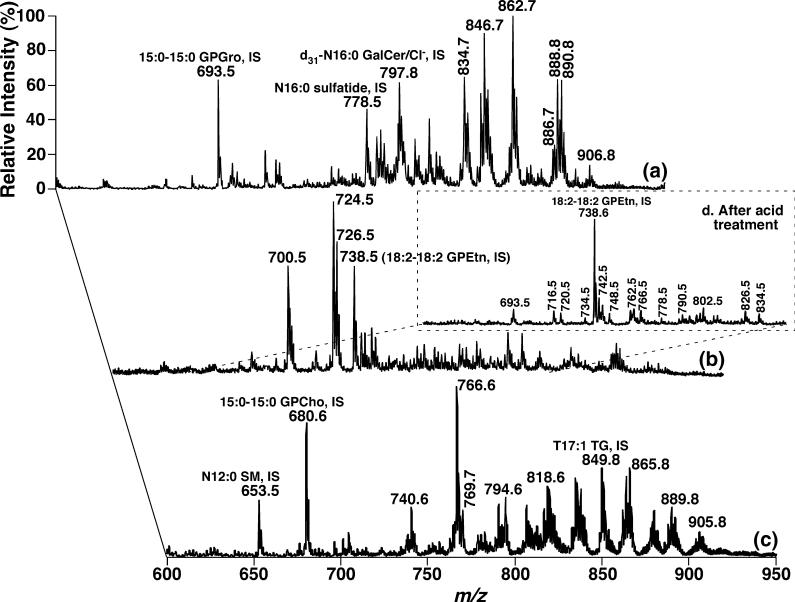
By employing different ionization conditions, different lipid classes can be resolved and selectively ionized in the ion source (i.e. intrasource separation) (Han et al. 2004b, 2006). By using this technique, many of the lipid classes in DRG lipid extracts of WT mice at 4 months of age were profiled under three ionization conditions in the absence or presence of a low concentration of LiOH (Fig. 1). Trace a was acquired in the negative-ion mode after direct infusion of a diluted DRG lipid extract. The displayed abundant ion peaks correspond to molecular species of anionic lipids (e.g. GPGro and sulfatide) and chlorinated neutral but polar lipids (e.g. GalCer) (Fig. 1a). Trace b was acquired in the negative-ion mode as trace a; however, a small amount of LiOH was added to the diluted DRG lipid extract before infusion. The displayed abundant ion peaks correspond to GPEtn molecular species (Fig. 1b). Trace c was acquired in the positive-ion mode after infusion of the diluted DRG lipid extract in the presence of LiOH. The displayed abundant ion peaks correspond to lithiated neutral, polar lipids (such as GPCho, GalCer, SM, and TG) (Fig. 1c). A detailed description of the identification and quantitation of these molecular species using 2D ESI/MS array analyses is presented in the following subsections.
Fig. 1.
Electrospray ionization/mass spectrometry analyses of the mouse dorsal root ganglia lipidome after intrasource separation and selective ionization. Spectrum (a) was acquired in the negative-ion mode directly from a lipid extract that was diluted to less than 50 pmol of total lipids/μL. Spectrum (b) was acquired again in the negative-ion mode from the diluted lipid solution as used for spectrum (a) after addition of approximately 25 pmol LiOH/μL to the lipid solution. Spectrum (c) was acquired in the positive-ion mode from the identical diluted lipid solution as used in spectrum (b) after direct infusion. Spectrum (d) (inset) was taken in the negative-ion mode after the diluted lipid solution used in spectrum (a) was treated with acid vapor and a small amount of LiOH (approximately 25 pmol LiOH/μL) was added to the infused solution. The spectrum (d) is displayed after normalization of the ion peak of the glycerophosphoethanolamine (GPEtn) internal standard to that in spectrum (b). ‘IS’ denotes internal standard. All mass spectral traces are displayed after normalization to the base peak in each individual spectrum.
Identification and quantitation of individual anionic lipid molecular species of the mouse DRG lipidome
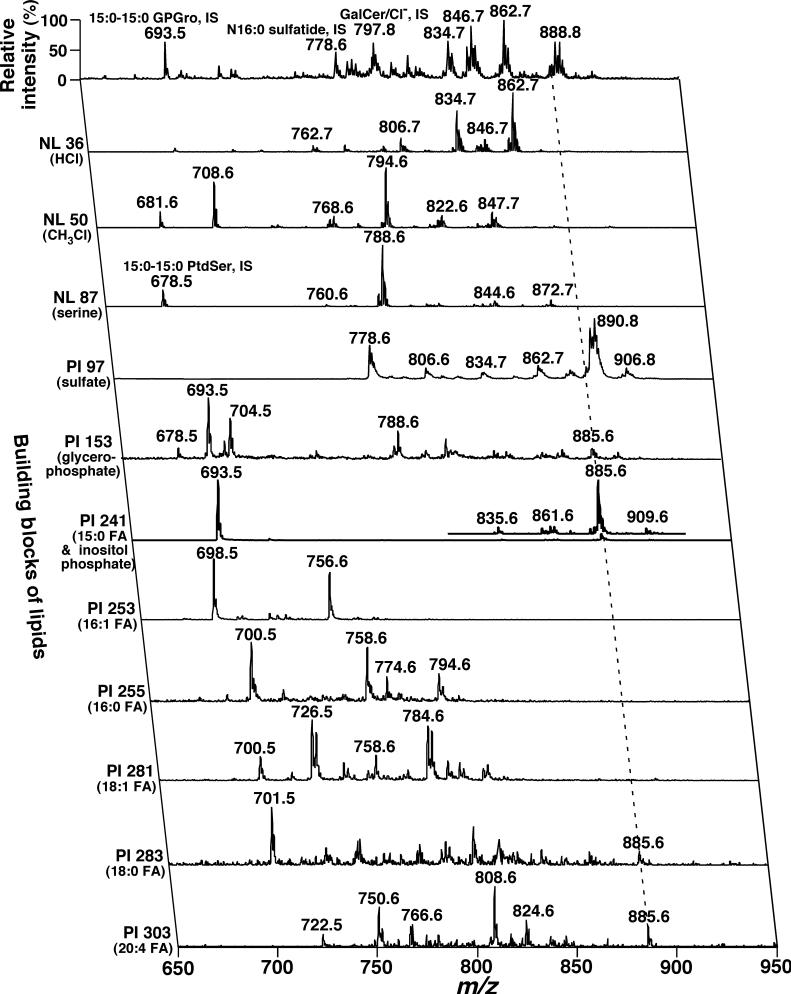
The individual molecular species corresponding to the ion peaks displayed in trace a of Fig. 1 were identified by 2D ESI/MS array analyses (Fig. 2). The second dimension of this mass spectrum represents the building blocks of lipid classes shown in the trace. These building blocks represent either a specific component of the head group moiety of each class or the acyl chain(s) of each molecular species. For example, neutral loss of 36.0 U (NL 36.0) represents the NL of HCl from chlorinated GalCer molecular species. In the scanning, the peak intensity ratios of the hydroxy to non-hydroxy GalCer molecular species are enhanced three times in comparison to those in the first dimension (Han and Cheng 2005). Therefore, analysis of this building block not only identifies all of GalCer molecular species but also distinguishes hydroxy GalCer from non-hydroxy GalCer. For example, the ion clusters at m/z 806.7, 834.7, and 862.7 are hydroxy GalCer molecular species, whereas other ion clusters are non-hydroxy ones (Table 1).
Fig. 2.
Representative 2D electrospray ionization/mass spectrometry (ESI/MS) analyses of anionic lipids and neutral lipid chlorine-adducts in a lipid extract of mouse dorsal root ganglia. The diluted lipid extract from a wild-type mouse at 4 months of age used for acquisition of spectrum (a) in Fig. 1 was employed. Each MS or MS/MS trace of the 2D ESI mass spectrum was acquired by sequentially programmed customized scans operating utilizing Xcalibur software. ‘PI’ denotes precursor-ion scanning, ‘NL’ stands for neutral loss scanning, and ‘IS’ represents internal standard. All mass spectral traces are displayed after normalization to the most intense peak (base peak) in each individual trace.
Table 1.
Mass contents of sphingolipid molecular species in the dorsal root ganglia of wild-type and apolipoprotein E knockout mice at 4 months of agea
| |
|
Molecular species |
WT mice |
ApoE KO mice |
|---|---|---|---|---|
| [M - H]− | Sulfatide | |||
| 804.7 | N18:1 sulfatide | 0.3 ± 0.1 | 0.4 ± 0.1 | |
| 806.7 | N18:0 sulfatide | 2.3 ± 0.3 | 3.3 ± 0.1 | |
| 820.7 | OH N18:1 sulfatide | 0.9 ± 0.3 | 1.1 ± 0.3 | |
| 822.7 | OH N18:0 sulfatide | 1.9 ± 0.2 | 2.4 ± 0.3 | |
| 832.7 | N20:1 sulfatide | 0.5 ± 0.1 | 0.6 ± 0.2 | |
| 834.7 | N20:0 sulfatide | 1.6 ± 0.1 | 2.4 ± 0.1 | |
| 848.7 | OH N20:1 sulfatide | 0.5 ± 0.1 | 0.6 ± 0.1 | |
| 850.7 | OH N20:0 sulfatide | 1.1 ± 0.1 | 1.4 ± 0.1 | |
| 860.7 | N22:1 sulfatide | 1.3 ± 0.1 | 1.8 ± 0.2 | |
| 862.7 | N22:0 sulfatide | 4.6 ± 0.4 | 6.6 ± 0.5 | |
| 876.7 | OH N22:1 sulfatide | 1.7 ± 0.2 | 2.5 ± 0.2 | |
| 878.7 | OH N22:0 sulfatide | 3.0 ± 0.2 | 3.8 ± 0.2 | |
| 886.8 | N24:2 sulfatide | 4.4 ± 0.3 | 5.8 ± 0.1 | |
| 888.8 | N24:1 sulfatide | 17.8 ± 1.2 | 25.7 ± 0.6 | |
| 890.8 | N24:0 sulfatide | 13.1 ± 1.1 | 23.6 ± 0.6 | |
| 902.8 | OH N24:2 sulfatide | 0.9 ± 0.1 | 1.0 ± 0.1 | |
| 904.8 | OH N24:1 sulfatide | 1.6 ± 0.2 | 2.0 ± 0.1 | |
| 906.8 | OH N24:0 sulfatide | 3.3 ± 0.3 | 4.7 ± 0.2 | |
| 916.8 | N26:1 sulfatide | 0.7 ± 0.1 | 0.7 ± 0.1 | |
| 918.8 | N26:0 sulfatide | 0.5 ± 0.1 | 0.4 ± 0.1 | |
| 932.8 | OH N26:1 sulfatide | 0.3 ± 0.1 | 0.4 ± 0.1 | |
| 934.8 | OH N26:0 sulfatide | 0.5 ± 0.1 | 0.6 ± 0.1 | |
| Total sulfatide | 62.8 ± 3.1 | 91.4 ± 1.5** | ||
| Hydroxy sulfatide | 15.8 ± 1.0 | 20.3 ± 0.7** |
| [M + CI]− | [M + Li]+ | Galactosylceramide (GalCer) | ||
|---|---|---|---|---|
| 732.6 | 704.6 | N16:1 GalCer | DA | DA |
| 734.6 | 706.6 | N16:0 GalCer | DA | DA |
| 760.6 | 732.6 | N18:1 GalCer | 0.3 ± 0.1 | 0.3 ± 0.1 |
| 762.6 | 734.6 | N18:0 GalCer | 4.0 ± 0.2 | 4.3 ± 0.2 |
| 776.6 | 748.6 | OH N18:1 GalCer | 0.5 ± 0.1 | 0.9 ± 0.2 |
| 778.6 | 750.6 | OH N18:0 GalCer | 3.2 ± 0.2 | 3.6 ± 0.3 |
| 788.6 | 760.6 | N20:1 GalCer | DA | DA |
| 790.6 | 762.6 | N20:0 GalCer | 7.2 ± 0.8 | 7.9 ± 0.5 |
| 804.6 | 776.6 | N21:0/OH N20:1 GalCer | 1.6 ± 0.2 | 1.7 ± 0.3 |
| 806.6 | 778.6 | OH N20:0 GalCer | 8.7 ± 0.7 | 9.7 ± 0.4 |
| 816.6 | 788.6 | N22:1 GalCer | 0.4 ± 0.1 | 0.9 ± 0.1 |
| 818.7 | 790.7 | N22:0 GalCer | 5.3 ± 0.6 | 7.1 ± 0.1 |
| 820.7 | 792.7 | OH N21:0 GalCer | 3.5 ± 0.3 | 3.7 ± 0.5 |
| 832.7 | 804.7 | N23:0/OH N22:1 GalCer | 7.0 ± 0.5 | 7.2 ± 0.4 |
| 834.7 | 806.7 | OH N22:0 GalCer | 19.0 ± 1.1 | 23.7 ± 1.7 |
| 842.7 | 814.7 | N24:2 GalCer | 2.6 ± 0.2 | 2.9 ± 0.2 |
| 844.7 | 816.7 | N24:1 GalCer | 14.0 ± 1.2 | 17.3 ± 0.4 |
| 846.7 | 818.7 | N24:0 GalCer | 19.8 ± 1.9 | 25.2 ± 0.7 |
| 848.7 | 820.7 | OH N23:0 GalCer | 4.4 ± 0.7 | 5.8 ± 0.6 |
| 858.7 | 830.7 | OH N24:2 GalCer | 4.1 ± 0.5 | 3.8 ± 0.4 |
| 860.8 | 832.8 | N25:0/OH N24:1 GalCer | 11.6 ± 0.6 | 13.2 ± 0.4 |
| 862.8 | 834.8 | OH N24:0 GalCer | 24.3 ± 1.9 | 29.2 ± 1.5 |
| 872.8 | 844.8 | N26:1 GalCer | 0.3 ± 0.1 | 0.4 ± 0.1 |
| 874.8 | 846.8 | N26:0 GalCer | 0.2 ± 0.1 | 0.5 ± 0.1 |
| 876.8 | 848.8 | OH N25:0 GalCer | 1.8 ± 0.2 | 2.8 ± 0.4 |
| 888.8 | 860.8 | OH N26:1 GalCer | 0.7 ± 0.1 | 0.6 ± 0.2 |
| 890.8 | 862.8 | OH N26:0 GalCer | 1.9 ± 0.6 | 1.4 ± 0.3 |
| Total GalCer | 146.8 ± 10.5 | 174.1 ± 5.6* | ||
| Hydroxy GalCer | 92.2 ± 4.7 | 107.3 ± 3.8* |
| [M + Li]+ | Sphingomyelin (SM) | |||
|---|---|---|---|---|
| 707.5 | N16:1 SM | 0.3 ± 0.1 | 0.4 ± 0.1 | |
| 709.5 | N16:0 SM | 3.2 ± 0.4 | 3.8 ± 0.1 | |
| 735.5 | N18:1 SM | 0.4 ± 0.1 | 0.5 ± 0.1 | |
| 737.5 | N18:0 SM | 6.0 ± 0.3 | 6.1 ± 0.3 | |
| 765.6 | N20:0 SM | 3.8 ± 0.4 | 3.3 ± 0.3 | |
| 791.6 | N22:1 SM | 0.5 ± 0.2 | 0.5 ± 0.1 | |
| 793.6 | N22:0 SM | 5.2 ± 0.3 | 5.6 ± 0.3 | |
| 807.6 | N23:0 SM | 1.5 ± 0.2 | 1.2 ± 0.2 | |
| 817.6 | N24:2 SM | 2.1 ± 0.2 | 2.9 ± 0.3 | |
| 819.6 | N24:1 SM | 14.0 ± 0.9 | 14.9 ± 0.6 | |
| 821.6 | N24:0 SM | 7.2 ± 0.3 | 7.4 ± 0.3 | |
| 833.6 | N25:1 SM | 0.3 ± 0.1 | 0.3 ± 0.1 | |
| 835.6 | N25:0 SM | 0.3 ± 0.1 | 0.5 ± 0.1 | |
| Total SM | 44.7 ± 1.6 | 47.4 ± 1.1 |
| [M - H]− | Ceramide (Cer) | |||
|---|---|---|---|---|
| 536.5 | N16:0 Cer | 39 ± 3 | 51 ± 6 | |
| 564.5 | N18:0 Cer | 120 ± 3 | 133 ± 7 | |
| 592.5 | N20:0 Cer | 62 ± 5 | 84 ± 4 | |
| 606.5 | N21:0 Cer | 7 ± 1 | 6 ± 1 | |
| 608.5 | OH N20:0 Cer | 11 ± 1 | 15 ± 1 | |
| 620.5 | N22:0 Cer | 60 ± 3 | 80 ± 2 | |
| 632.5 | N23:1 Cer | 2 ± 1 | 4 ± 2 | |
| 634.5 | N23:0 Cer | 25 ± 4 | 27 ± 3 | |
| 636.5 | OH N22:0 Cer | 25 ± 5 | 29 ± 5 | |
| 644.5 | N24:2 Cer | 21 ± 1 | 30 ± 2 | |
| 646.5 | N24:1Cer | 120 ± 6 | 177 ± 5 | |
| 648.5 | N24:0 Cer | 78 ± 7 | 93 ± 6 | |
| 664.5 | OH N24:0 Cer | 36 ± 7 | 42 ± 3 | |
| Total Cer | 593 ± 26 | 769 ± 27** |
Mouse DRG lipids were extracted by a modified Bligh and Dyer procedure and the sphingolipid molecular species in the lipid extracts were identified and quantified using a 2D ESI/MS technique as described under Materials and methods. The results are expressed in nmol/mg of protein (except for ceramide in which data are expressed in pmol/mg of protein) and represent X ± SD of six different animals.
DRG, dorsal root ganglia; WT, wild type; ApoE, apolipoprotein E; KO, knockout; DA, presence of a detectable ion peak; ESI, electrospray ionization; MS, mass spectrometry.
p < 0.01.
p < 0.001.
Similarly, building blocks of NL 50.0 (NL of chloromethane from molecular species containing a phosphocholine head group), NL 87.0 (NL of serine from GPSer molecular species), PI 97.0 (a sulfate ion fragmented from those containing a sulfate), PI 153.1 (a glycerophosphate derived from anionic phospholipids), and PI 241.2 [a phosphoinositol derivative ion arising from glycerophosphoinositol (GPIns) molecular species] (Han and Gross 2005a) identified individual molecular species of GPCho (and SM), serine glycerophospholipids (GPSer), sulfatide, anionic phospholipids, and GPIns, respectively. The acyl chain moieties of these molecular species were identified by precursor-ion scanning (e.g. PI 241.2, PI 253.2, etc.) of their fatty acyl carboxylates (e.g. 15:0, 16:1, etc., respectively). Therefore, each individual lipid molecular species including both its head group and acyl moieties were identified through analyses of this 2D spectrum. For example, the ion peak at m/z 885.6 in the first dimension is crossed with the building blocks of PI 153.1, PI 241.2, PI 283.3, and PI 303.3 (the broken line in Fig. 2) in addition to the higher ion intensity of m/z 885.6 in PI 303.3 than in PI 283.3, thereby identifying this species as 18:0−20:4 GPIns. Several minor GPSer molecular species containing either vinyl ether- or alkyl ether-linked aliphatic chains were also present in the lipid extracts of mouse DRG (Table 2).
Table 2.
Mass contents of phospholipid molecular species in the DRG of wild-type and apolipoprotein E knockout mice at 4 months of agea
| |
Molecular species |
WT mice |
ApoE KO mice |
|---|---|---|---|
| [M - H]− | Phosphatidic acid (GPA) | ||
| 671.5 | 16:0−18:2 GPA | 0.3 ± 0.1 | 0.3 ± 0.1 |
| 673.5 | 16:0−18:1 GPA | 0.2 ± 0.1 | 0.2 ± 0.1 |
| 699.5 | 18:1−18:1 GPA | 0.6 ± 0.1 | 0.5 ± 0.1 |
| 701.5 | 18:0−18:1 GPA | 2.2 ± 0.2 | 1.9 ± 0.2 |
| Total GPA | 3.2 ± 0.2 | 3.0 ± 0.3 |
| [M - H]− | Phosphatidylglycerol (GPGro) | ||
|---|---|---|---|
| 745.5 | 16:0−18:2 GPGro | 0.1 ± 0.1 | 0.1 ± 0.1 |
| 747.5 | 16:0−18:1 GPGro | 0.5 ± 0.1 | 0.5 ± 0.1 |
| 771.5 | 18:1−18:2 GPGro | 0.3 ± 0.1 | 0.2 ± 0.1 |
| 773.5 | 18:1−18:1 GPGro | 0.2 ± 0.1 | 0.2 ± 0.1 |
| 775.5 | 18:0−18:2 GPGro | DA | DA |
| Total GPGro | 1.0 ± 0.1 | 1.0 ± 0.1 |
| [M - H]− | Serine glycerophospholipid (GPSer) | ||
|---|---|---|---|
| 758.5 | 16:0−18:2 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| 760.5 | 16:0−18:1 | 1.8 ± 0.2 | 2.2 ± 0.1 |
| 770.6 | p18:1−18:1 | 0.7 ± 0.2 | 0.9 ± 0.2 |
| 772.6 | p18:0−18:1 | 2.8 ± 0.2 | 2.9 ± 0.1 |
| 774.6 | p18:0−18:0 | 1.8 ± 0.2 | 1.9 ± 0.1 |
| 786.6 | 18:1−18:1 | 16.5 ± 0.3 | 17.2 ± 0.5 |
| 788.6 | 18:0−18:1 | 74.1 ± 0.8 | 71.8 ± 2.8 |
| 810.6 | 18:0−20:4 | 2.5 ± 0.3 | 2.7 ± 0.2 |
| 812.6 | 18:0−20:3 | 1.4 ± 0.1 | 1.5 ± 0.1 |
| 814.6 | 18:0−20:2 | 1.0 ± 0.1 | 1.2 ± 0.2 |
| 816.6 | 18:1−20:0 | 5.6 ± 0.2 | 5.5 ± 0.2 |
| 834.6 | 18:0−22:6 | 2.8 ± 0.3 | 2.4 ± 0.2 |
| 836.6 | 18:0−22:5 | 0.6 ± 0.2 | 0.4 ± 0.1 |
| 842.7 | 18:1−22:1 | 3.4 ± 0.2 | 3.4 ± 0.1 |
| 844.7 | 18:0−22:1 | 11.2 ± 0.4 | 11.4 ± 0.2 |
| 858.7 | a18:0−24:1 | 2.6 ± 0.2 | 2.6 ± 0.1 |
| 870.7 | 18:1−24:1 | 4.2 ± 0.2 | 4.1 ± 0.1 |
| 872.7 | 18:0−24:1 | 15.4 ± 0.5 | 16.4 ± 0.6 |
| Total GPSer | 148.5 ± 1.6 | 148.8 ± 4.5 |
| [M - H]− | Phosphatidylinositol (GPIns) | ||
|---|---|---|---|
| 833.6 | 16:0−18:2 GPIns | 1.5 ± 0.1 | 0.7 ± 0.1 |
| 835.6 | 16:0−18:1 GPIns | 1.2 ± 0.1 | 1.3 ± 0.1 |
| 857.6 | 16:0−20:4 GPIns | 0.8 ± 0.1 | 0.9 ± 0.1 |
| 859.6 | 18:1−18:2 GPIns | 0.8 ± 0.1 | 0.8 ± 0.1 |
| 861.6 | 18:1−18:1 GPIns | 3.2 ± 0.1 | 3.4 ± 0.2 |
| 863.6 | 18:0−18:1 GPIns | 1.8 ± 0.2 | 2.0 ± 0.1 |
| 883.6 | 18:1−20:4 GPIns | 0.4 ± 0.1 | 0.4 ± 0.1 |
| 885.6 | 18:0−20:4 GPIns | 4.7 ± 0.6 | 5.7 ± 0.2 |
| 887.6 | 18:0−20:3 GPIns | 2.8 ± 0.3 | 3.4 ± 0.1 |
| 889.6 | 18:0−20:2 GPIns | 2.5 ± 0.2 | 2.8 ± 0.2 |
| 909.6 | 18:0−22:6 GPIns | 0.4 ± 0.1 | 0.4 ± 0.1 |
| 911.6 | 18:0−22:5 GPIns | 0.1 ± 0.1 | 0.1 ± 0.1 |
| Total GPIns | 20.1 ± 1.4 | 22.8 ± 0.5 |
| [M - H]− | Ethanolamine glycerophospholipid (GPEtn) | ||
|---|---|---|---|
| 646.5 | p16:0−14:0 | 0.1 ± 0.1 | 0.1 ± 0.1 |
| 672.5 | p14:0−18:1/p18:1−14:0/p16:0−16:1 | 1.0 ± 0.1 | 1.0 ± 1.0 |
| 674.5 | p16:0−16:0/p18:0−14:0 | 0.4 ± 0.1 | 0.4 ± 0.0.1 |
| 698.5 | p16:0−18:2/p18:2−16:0/p18:1−16:1 | 4.0 ± 0.2 | 4.0 ± 0.1 |
| 700.5 | p16:0−18:1/p18:1−16:0/p18:0−16:1 | 29.1 ± 1.7 | 28.7 ± 0.7 |
| 714.5 | d16:0−18:2/d16:1−18:1 | 5.5 ± 0.6 | 4.8 ± 0.1 |
| 716.5 | d16:0−18:1 | 7.9 ± 0.5 | 7.7 ± 0.1 |
| 722.5 | p16:0−20:4 | 2.4 ± 0.1 | 2.4 ± 0.1 |
| 724.5 | p18:1−18:2 | 9.9 ± 0.1 | 10.4 ± 0.8 |
| 726.5 | p18:1−18:1/p18:0−18:2/p20:1−16:1 | 93.6 ± 5.5 | 98.3 ± 3.7 |
| 728.5 | p18:0−18:1/p18:1−18:0/p20:1−16:0/p16:0−20:1/p20:0−16:1 | 73.8 ± 2.7 | 72.2 ± 2.1 |
| 738.5 | d16:0−20:4/d18:2−18:2 | 17.5 ± 0.8 | 18.2 ± 0.5 |
| 740.5 | d18:1−18:2/p19:1−18:1/p18:1−19:1 | 4.8 ± 1.0 | 3.7 ± 0.6 |
| 742.5 | d18:1−18:1/p19:0−18:1/p18:1−19:0 | 18.8 ± 1.1 | 19.9 ± 0.6 |
| 744.5 | d18:0−18:1 | 10.4 ± 0.5 | 10.1 ± 0.2 |
| 746.5 | p16:0−22:6 | 1.7 ± 0.1 | 1.6 ± 0.1 |
| 748.5 | p18:1−20:4/p16:0−22:5 | 10.7 ± 0.3 | 10.2 ± 0.7 |
| 750.5 | p18:0−20:4/p16:0−22:4 | 9.6 ± 0.3 | 9.2 ± 0.5 |
| 752.5 | p20:1−18:2 | 2.1 ± 0.1 | 2.2 ± 0.1 |
| 754.5 | p20:1−18:1/p20:0−18:2/p18:0−20:2/p16:0−22:2/d19:1−18:2/D19:2−18:1 | 4.1 ± 0.2 | 4.2 ± 0.2 |
| 756.5 | p20:0−18:1/d19:1−18:1/p18:0−20:1/p20:1−18:0 | 2.2 ± 0.1 | 2.2 ± 0.1 |
| 760.5 | d16:1−22:6 | 0.8 ± 0.1 | 0.8 ± 0.1 |
| 762.5 | d16:0−22:6/d16:1−22:5 | 3.3 ± 0.2 | 3.3 ± 0.2 |
| 764.5 | d16:0−22:5/d18:1−20:4 | 3.2 ± 0.1 | 3.1 ± 0.2 |
| 766.5 | d18:0−20:4 | 8.0 ± 1.2 | 7.3 ± 0.3 |
| 770.5 | p18:2−22:6 | 1.8 ± 0.1 | 1.9 ± 0.1 |
| 772.5 | p18:1−22:6 | 4.6 ± 0.1 | 4.5 ± 0.2 |
| 774.5 | p18:0−22:6 | 11.1 ± 0.8 | 10.2 ± 0.4 |
| 776.5 | p18:0−22:5/p18:1−22:4/p20:1−20:4 | 8.6 ± 0.2 | 8.6 ± 0.4 |
| 778.5 | p18:0−22:4/p20:0−20:4 | 8.5 ± 0.2 | 8.3 ± 0.3 |
| 780.5 | a20:0−20:4/p20:0−20:3/p18:0−22:3/d19:2−20:2 | 1.8 ± 0.2 | 1.8 ± 0.1 |
| 782.5 | p22:1−18:1/p18:1−22:1/d19:1−20:2/d19:2−20:1 | 2.7 ± 0.2 | 2.8 ± 0.1 |
| 784.5 | a18:1−22:1/p18:0−22:1/d19:1−20:1 | 1.8 ± 0.1 | 1.9 ± 0.1 |
| 788.5 | d18:1−22:6 | 1.6 ± 0.1 | 1.4 ± 0.1 |
| 790.5 | d18:0−22:6 | 9.6 ± 1.3 | 7.9 ± 0.6 |
| 792.5 | d18:0−22:5 | 1.4 ± 0.1 | 1.4 ± 0.1 |
| 794.5 | d20:0−20:4/d20:2−20:2/d18:0−22:4 | 2.1 ± 0.2 | 2.2 ± 0.1 |
| 796.5 | d20:1−20:2 | 0.7 ± 0.1 | 0.8 ± 0.1 |
| 798.5 | d20:1−20:1/d20:0−20:2/d18:1−22:1 | 2.0 ± 0.2 | 2.1 ± 0.1 |
| 800.5 | d20:1−20:0 | 1.5 ± 0.1 | 1.4 ± 0.1 |
| 802.5 | d18:0−22:0/p20:0−22:6 | 4.4 ± 0.2 | 4.5 ± 0.2 |
| 804.5 | p20:0−22:5 | 0.6 ± 0.1 | 0.7 ± 0.1 |
| 806.5 | p20:0−22:4 | 0.7 ± 0.1 | 0.7 ± 0.1 |
| Total GPEtn | 390.1 ± 17.6 | 388.6 ± 11.4 | |
| Total plasmalogen | 284.9 ± 11.4 | 286.0 ± 10.6 | |
| Plasmalogen (%) | 73.0 ± 2.2 | 73.6 ± 1.6 |
| [M + Li]+ | Choline glycerophospholipid (GPCho) | ||
|---|---|---|---|
| 614.5 | a12:0−12:0 | 0.5 ± 0.2 | 0.6 ± 0.1 |
| 712.5 | d14:0−16:0 | 3.3 ± 0.2 | 3.5 ± 0.3 |
| 724.5 | p16:0−16:0 | 0.7 ± 0.1 | 1.0 ± 0.3 |
| 726.5 | a16:0−16:0 | 1.0 ± 0.3 | 1.1 ± 0.2 |
| 738.5 | d16:1−16:0 | 8.2 ± 1.0 | 6.3 ± 0.2 |
| 740.5 | d16:0−16:0 | 18.7 ± 1.9 | 16.0 ± 0.3 |
| 750.5 | p16:0−18:1 | 0.6 ± 0.2 | 0.9 ± 0.1 |
| 752.5 | p16:0−18:0 | 2.5 ± 0.5 | 2.9 ± 0.2 |
| 754.5 | a16:0−18:0 | 0.7 ± 0.1 | 0.5 ± 0.1 |
| 764.6 | d16:0−18:2 | 8.9 ± 1.7 | 10.1 ± 0.1 |
| 766.6 | d16:0−18:1 | 61.5 ± 4.6 | 59.2 ± 1.2 |
| 776.6 | p18:1−18:1 | 0.4 ± 0.3 | 0.6 ± 0.3 |
| 778.6 | p18:0−18:1 | 0.9 ± 0.2 | 0.9 ± 0.1 |
| 780.6 | p18:0−18:0 | 2.2 ± 0.2 | 1.9 ± 0.2 |
| 788.6 | d16:0−20:4/d16:1−20:3 | 6.5 ± 0.4 | 5.9 ± 0.1 |
| 790.6 | d18:1−18:2/d16:0−20:3 | 3.2 ± 0.5 | 3.4 ± 0.1 |
| 792.6 | d18:0−18:2/d18:1−18:1 | 8.9 ± 0.8 | 9.4 ± 0.1 |
| 794.6 | d18:0−18:1 | 19.3 ± 1.3 | 19.3 ± 0.2 |
| 812.6 | d16:0−22:6 | 12.6 ± 1.3 | 12.5 ± 0.5 |
| 814.6 | d18:1−20:4/d18:2−20:3 | 2.1 ± 0.4 | 1.4 ± 0.2 |
| 816.6 | d18:0−20:4/d18:2−20:2 | 5.5 ± 0.4 | 5.1 ± 0.3 |
| 818.6 | d18:0−20:3 | 1.2 ± 0.3 | 1.1 ± 0.4 |
| 830.6 | d19:0−20:4 | 0.5 ± 0.1 | 0.5 ± 0.1 |
| 836.6 | d18:2−22:6 | 0.6 ± 0.1 | 0.5 ± 0.1 |
| 838.6 | d18:1−22:6 | 1.4 ± 0.3 | 1.7 ± 0.1 |
| 840.6 | d18:0−22:6 | 3.2 ± 0.3 | 2.6 ± 0.1 |
| 848.6 | d20:1−20:1/d18:0−22:2 | 0.6 ± 0.3 | 0.8 ± 0.1 |
| 850.6 | d20:0−20:1/p20:1−22:6 | 3.4 ± 0.3 | 3.2 ± 0.1 |
| 864.6 | d20:2−22:6 | 0.3 ± 0.1 | 0.3 ± 0.1 |
| 876.6 | p22:3−22:5/d20:1−22:1 | 1.2 ± 0.1 | 1.0 ± 0.2 |
| 878.6 | p22:1−22:6/p22:6−22:1/d20:0−22:1 | 4.6 ± 0.6 | 6.1 ± 0.2 |
| Total GPCho | 184.4 ± 5.5 | 180.1 ± 1.5 |
Mouse DRG lipids were extracted by a modified Bligh and Dyer procedure and the phospholipid molecular species in the lipid extracts were identified and quantified using a 2D ESI/MS technique as described under Materials and methods. The results are expressed in nmol/mg of protein and represent X ± SD of six different animals.
‘d’ (diacyl), ‘p’ (plasmenyl), and ‘a’ (alkyl) denote the three subclasses (i.e. phosphatidyl-, plasmenyl-, and plasmanyl-), respectively, in the classes of GPCho, GPEtn, and GPSer. DRG, dorsal root ganglia; WT, wild type; ApoE, apolipoprotein E; KO, knockout; DA, presence of a detectable ion peak.
Once each individual molecular species in a lipid class is identified, quantitation of these molecular species can be performed by a two-step procedure. First, the abundant molecular species in each lipid class are quantitated by ratiometric comparison of each of the ion peak intensities of the molecular species in the class of interest with that of the internal standard that is pre-selected for this lipid class after de-isotoping (Han and Gross 2001, 2005a). Next, all of the quantitated molecular species are used as standards along with the pre-selected internal standard to quantitate low-abundance or overlapping molecular species in the first dimensional spectrum using a tandem MS trace that is employed for analysis of a specific building block of the lipid class. For example, sulfatide molecular species at m/z 806.7, 888.8, 890.8, and 906.8 were determined from the first dimensional mass spectrum whereas other molecular species of the sulfatide class were assessed from the trace of PI 97.0 using these quantitated molecular species as standards (Fig. 2 and Table 1). GPIns molecular species at m/z 885.6 and GPSer molecular species at m/z 788.6 were quantitated from the first dimensional mass spectrum whereas other molecular species of these classes were determined from the traces of PI 241.1 and NL 87.0 using these quantitated molecular species as standards, respectively (Fig. 2 and Table 2). A total mass content of 20.1 ± 1.4 nmol GPIns/mg of protein or 148.5 ± 1.6 nmol GPSer/mg of protein was present in the WT mouse DRG lipidome at 4 months of age (Table 2).
GalCer molecular species were quantitated in the positive-ion mode as described below due to the overlap of GalCer molecular species with sulfatide molecular species in the negative-ion mode. Similarly, owing to the low intensities of ion peaks corresponding to the chlorinated GPCho or SM in the first dimensional mass spectrum in Fig. 1, it was typically required that the first step of quantitation was performed in the positive-ion mode. However, NL 50.0 was redundantly used to quantitate minor molecular species of GPCho and SM.
Quantitative analyses of individual GPEtn molecular species in the mouse DRG lipidome
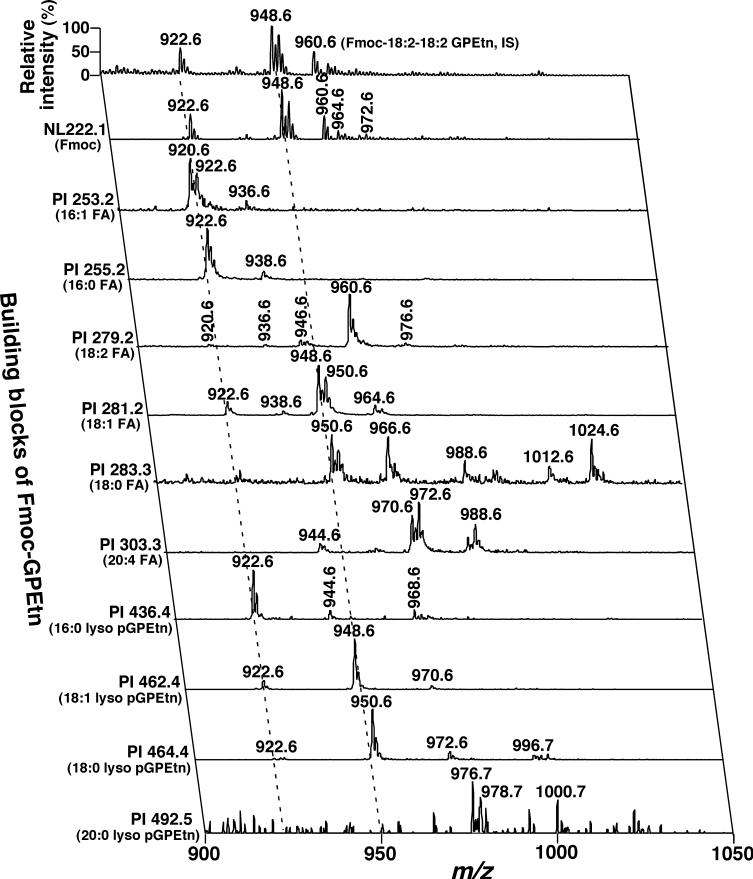
To clearly identify all individual GPEtn molecular species in the mouse DRG lipidome as shown in trace b of Fig. 1, we employed the newly developed methodology of Fmoc derivatization (Han et al. 2005a). After derivatization and MS analysis, NL 222.1 (NL of the Fmoc moiety from Fmoc–GPEtn molecular species) unambiguously demonstrates the presence of all GPEtn molecular ions (Fig. 3) with a dynamic range over four orders of magnitude (Han et al. 2005a). The identities of the aliphatic chains of these molecular species were efficiently identified through a 2D ESI/MS array analysis (Fig. 3). The building blocks of the GPEtn molecular ions included fatty acyl carboxylates and lysoplasmenylethanolamine (lyso pGPEtn) derivatives (Fig. 3). From these 2D MS array analyses, many isobaric GPEtn molecular species were readily identified. For example, the Fmoc–GPEtn ion peak at m/z 922.6 (i.e. de-protonated GPEtn molecular species at m/z 700.5) was comprised of at least four pGPEtn molecular species (i.e. 16:0−18:1, 16:1−18:0, 18:0−16:1, and 18:1−16:0 pGPEtn) (see the left broken line in Fig. 3). To further confirm that these identified pGPEtn molecular species did not overlap with any aGPEtn molecular species, a small portion of each DRG lipid extract was treated with acidic vapor. Each residue was analyzed again in the negative-ion mode in the presence of LiOH (Inset of Fig. 1b). This mass spectrum demonstrates that the major ion peaks at m/z 700.5, 724.5, and 726.5 in Fig. 1b are all absent, suggesting that these ions correspond to pGPEtn molecular species (Table 2).
Fig. 3.
Representative negative-ion 2D electrospray ionization mass spectrum of a diluted chloroform extract of mouse dorsal root ganglia after treatment in situ with Fmoc chloride. A conventional electrospray ionization mass spectrum of Fmoc-glycerophosphoethanolamine (GPEtn) was acquired in the negative-ion mode directly from a diluted mouse dorsal root ganglia lipid extract after derivatization with Fmoc chloride (Han et al. 2005a). Analyses of Fmoc–GPEtn building blocks in the second dimension including the Fmoc moiety, fatty acyl carboxylaytes, and lyso plasmenylethanolamine (pGPEtn) ions by precursor-ion (PI) scanning and neutral loss (NL) scanning were performed. ‘IS’ denotes internal standard; (m:n) indicates an acyl chain containing m carbons and n double bonds; FA stands for fatty acyl chain. Each of the broken lines indicates the crossing peaks (fragmental ions) of a molecular ion with Fmoc–GPEtn building blocks. All mass spectral traces were displayed after normalization to the base peak in each individual spectrum.
Again, quantitation of individual GPEtn molecular species was performed through the aforementioned two-step procedure. The mass contents of molecular ions at m/z 700.5, 724.5, 726.5, 742.5, and 766.5 were determined by ratiometric comparison of these ion peak intensities with that of the selected internal standard (i.e. 18:2−18:2 GPEtn at m/z 738.5) after de-isotoping. Other minor ions displayed in NL 222.1 of Fig. 3 were quantitated by using these quantitated GPEtn molecular species as standards. It was found that the mouse DRG lipidome at 4 months of age was enriched with pGPEtn molecular species which account for 73 mol% of the total GPEtn mass content (Table 2). Intriguingly, pGPEtn molecular species predominantly contain monounsaturated fatty acids at sn-2 position.
Identification and quantitation of individual GPCho and GalCer molecular species in the mouse DRG lipidome
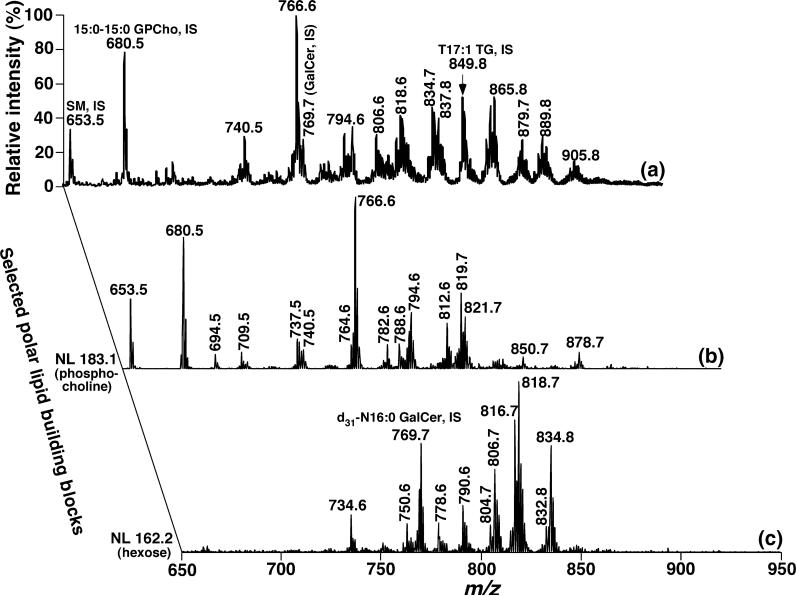
The molecular ion peaks in Fig. 1c which correspond to lithiated GPCho, lithiated SM, lithated GalCer, and lithiated TG species were identified by 2D MS array analyses. For example, the presence of GPCho, SM, and GalCer molecular species was established through the characterization of the building blocks corresponding to GPCho and SM head groups via NL of 183.1 U (i.e. phosphocholine) (Han and Gross 2005a) and NL of 162.2 (i.e. the galactose moiety of GalCer) (Han and Cheng 2005) (Fig. 4). Identification of the fatty acyl chains of GPCho molecular species were repeatedly performed through NL of all potential fatty acids (spectra not shown) in addition to the 2D MS array analyses of chlorinated GPCho species in Fig. 2. As NL 162.2 (corresponding to the loss of hexose) was not specific to GalCer molecular species, but to all cerebrosides containing mono-hexose (Han and Cheng 2005), we attempted to identify the coexistence of glucosylceramide (GlcCer) with GalCer as previously described (Han and Cheng 2005). Under these conditions, the mass levels of GlcCer in DRG lipidome were found to be below the detection limitation of the technique. In addition, the acyl amide moieties were directly derived from the m/z of GalCer molecular species after identification of hydroxyl GalCer species with the assumption that sphingosine is the predominant sphingoid base in GalCer molecular species. Identification and quantitation of TG molecular species in the DRG lipidome has recently been reported in detail (Cheng et al. 2006) and will be further described in a later section.
Fig. 4.
Representative 2D electrospray ionization mass spectrum of a diluted lipid extract of mouse dorsal root ganglia in the positive-ion mode. A conventional positive-ion electrospray ionization mass spectrum was acquired from a diluted mouse dorsal root ganglia lipid extract after direct infusion in the presence of LiOH. Analyses of the building blocks corresponding to glycerophosphocholine (GPCho) and galactosylceramide or galactocerebroside (GalCer) molecular species in the second dimension by neutral loss (NL) scanning were performed. ‘IS’ denotes internal standard. All mass spectral traces were displayed after normalization to the base peak in each individual spectrum.
Lithiated GPCho molecular species at m/z 740.6, 766.6, 794.6, and 812.6 in the first dimensional mass spectrum (Fig. 4a) were quantitated by comparison of these ion intensities with that of the GPCho internal standard at m/z 680.6 after de-isotoping. The mass contents of lithiated SM molecular species at m/z 709.5, 737.5, and 819.6 (Fig. 4a) were similarly determined by comparison with the SM internal standard at m/z 653.5. Then, other minor or overlapping molecular species of GPCho and SM were repeatedly assessed from the traces of NL 50.0 (Fig. 2) and NL 183.1 (Fig. 4b) by using all quantitated GPCho and SM molecular species as standards, respectively (Tables 1 and 2). The GPCho molecular species in the mouse DRG lipidome at 4 months of age are dominated by the presence of a single major species of 16:0−18:1 GPCho, accounting for 33.4 mol% of the total GPCho class, and the presence of molecular species containing very long and polyunsaturated fatty acyl chains such as p22:1−22:6 GPCho (m/z 878.6) (Table 2).
Quantitation of major lithiated GalCer molecular species was performed by comparison of the peak intensities of ions at m/z 790.7, 806.7, 816.7, 818.7, and 834.8 to that of an internal standard at m/z 769.6 from the first dimension of the 2D MS spectrum after de-isotoping (Fig. 4a). The mass contents of other GalCer molecular species were determined from the trace of the NL 162.2 (Fig. 4c) by ratiometric comparison of each of their peak intensities to those of the quantitated GalCer molecular species (Table 1). Hydroxy and non-hydroxy GalCer subclasses account for 62.8 and 31.2 mol%, respectively, and the molecular species containing very long fatty acyl chains such as 24:0 and 24:1 are dominant in this class (Table 1).
Quantitation of mass contents of cholesterol and cholesterol esters in the mouse DRG lipidome by MS
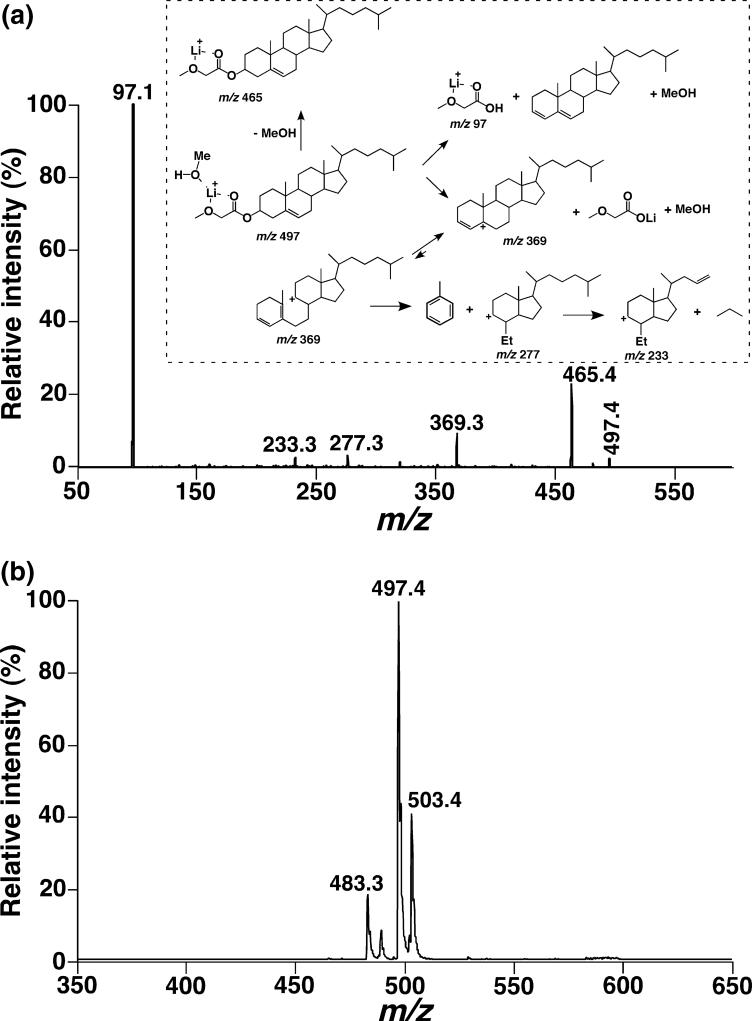
To determine the mass contents of cholesterol and cholesterol esters, a new methodology based on ESI/MS after in situ derivatization of diluted lipid extracts was developed in the study. ESI/MS analysis in the positive-ion mode in the presence of LiOH displayed an abundant ion at m/z 497.4, corresponding to lithiated cholesteryl methoxyacetate plus a methanol molecule, and a low-intensity ion at m/z 483.3, corresponding to the lithiated cholesteryl methoxyacetate plus a water molecule (spectrum not shown). Product-ion analysis of the ion at m/z 497.4 shows informative fragments of this molecular ion such as m/z 369.3, 277.3, 233.3, and 97.1. The possible structures of these fragment ions are proposed (Inset of Fig. 5a). The generated fragment at m/z 97.1 is very intense, representing the facile fragmentation of the C3−O bond arising from the formation of a very stable conjugated sterol compound. This unique fragment ion was employed to detect lithiated cholesteryl methoxyacetate at high sensitivity (a detection limit of 1 fmol/μL was obtained) and to quantitate the mass content of cholesterol in the DRG lipidome by comparison of the peak intensity with that of the one at m/z 503.4 resulting from lithiated d6-cholesteryl methoxyacetate (Fig. 5b). The mass contents of cholesterol in the DRG lipidomes of both WT and apoE KO mice at 4 months of age were determined as 579.6 ± 67.1 and 680.6 ± 54.2 nmol/mg of protein (p < 0.01, n = 6), respectively. These data were consistent with those obtained using a classic fluorometric assay method. The mass content of esterified cholesterol was also determined as 46.5 ± 24.9 and 37.7 ± 24.9 nmol/mg protein, respectively.
Fig. 5.
Characterization and quantitation of cholesterol in mouse dorsal root ganglia (DRG) lipidomes by electrospray ionization/mass spectrometry (ESI)/MS analyses. Mouse DRG lipid extracts were prepared and each of the diluted lipid extract solutions was individually modified with methoxyacetic acid. The reaction-workup solution of each lipid extract was analyzed in the positive-ion mode in the presence of LiOH. Panel (a), a product-ion ESI mass spectrum of a selected ion at m/z 497.4 that was shown in the ESI/MS analysis of the lipid extract (after derivatization with methoxyacetyl acid) was used to identify the lithiated cholesteryl methoxyacetate plus a methanol molecule. The inset of panel (a) represents the proposed fragmentation pathways and fragments of lithiated cholesteryl methoxyacetate plus a methanol corresponding to the fragmental ions in panel (a). Panel (b), a precursor-ion ESI mass spectrum of m/z 97.1 which was acquired (from the derivatized DRG lipid extract with methoxyacetic acid in the presence of LiOH) in the positive-ion mode and used for quantitation of cholesterol in the lipid extract.
Alterations in the mouse DRG lipidome induced by apoE depletion
The effects of apoE on the mass contents of sphingolipid classes
Shotgun lipidomics analysis of the DRG lipidome of apoE KO mice at 4 months of age demonstrated substantial alterations in mass contents of sulfatide, GalCer, and Cer molecular species in comparison to age-matched WT mice. Specifically, comparison of the negative-ion ESI mass spectrum of the DRG lipidome of apoE KO mice to that of WT mice shows substantial increases in the peak intensities of ions at m/z 834.8 or greater, representing major sulfatide and GalCer molecular species (Fig. 1 and Table 1). As some of the sulfatide and GalCer molecular species were overlapped in Fig. 1, MS/MS analysis using NL of 162.2 U in the positive-ion mode was performed to further document the increases in the mass content of GalCer molecular species in mouse DRG induced by apoE depletion (spectra not shown). Repetitive sample analyses demonstrated substantial increases in sulfatide mass content from 62.8 ± 3.1 nmol/mg of protein in WT mice to 91.4 ± 1.5 nmol/mg of protein in apoE KO mice at 4 months of age (p < 0.001, n = 6). GalCer mass content increased from 146.8 ± 10.5 to 174.1 ± 5.6 nmol/mg of protein (p < 0.01, n = 6) in apoE KO mice at 4 months of age (Table 1).
To examine the effects of apoE depletion on other sphingolipid classes within the detection limits of current shotgun lipidomics techniques, alterations in Cer molecular species in the DRG lipidome of apoE KO mice at 4 months of age relative to those of age-matched WT mice were also determined. MS/MS analyses in the neutral-loss mode demonstrated substantial increases induced by apoE depletion in many Cer molecular species containing long fatty amide chains but not in those containing relatively short fatty amide chains (Table 1). An increase in total Cer mass content from 593 ± 26 pmol/mg of protein in WT mice to 769 ± 27 pmol/mg of protein in apoE KO mice at 4 months of age (p < 0.001, n = 6) was obtained from repetitive sample analyses (Table 1).
The effects of apoE on the mass contents of TG and other lipid classes
The effects of apoE depletion on other lipid classes in mouse DRG including GPCho, GPEtn, GPGro, GPSer, GPIns, glycerophosphate, cholesterol, and TG were also examined by shotgun lipidomics. There were no significant differences in the total amounts of all examined phospholipid classes in the DRG lipidomes between WT and apoE KO mice at 4 months of age (Table 2). The increase in cholesterol mass induced by apoE depletion was significant as described previous subsection. However, the pool of TG molecular species in the DRG lipidome of apoE KO mice at 4 months of age was quite different from that of age-matched WT mice in several aspects (Table 3 and Fig. 6). First, positive-ion ESI/MS analyses demonstrated substantially lower mass levels of TG molecular species from apoE KO mouse cervical DRG in comparison to those of WT mice (Table 3). Second, 2D MS analyses show that TG molecular species in each cluster in apoE KO cervical DRG possessed predominantly molecular species with lower molecular weight relative to those in WT mice (Fig. 6).
Table 3.
Mass content of triacylglycerol molecular species in the DRG of wild-type and apolipoprotein E knockout mice at 4 months of agea
| [M + Li]+ | Major molecular species | WT mice | ApoE KO mice |
|---|---|---|---|
| 783.7 | 14:0/16:0/16:1 | 0.4 ± 0.1 (0.7 ± 0.1) | 0.1 ± 0.1 (0.3 ± 0.1) |
| 807.7 | 14:0/16:1/18:2, 16:1/16:1/16:1 | 0.6 ± 0.2 (0.8 ± 0.3) | 0.2 ± 0.1 (0.4 ± 0.1) |
| 809.7 | 14:0/16:0/18:2, 16:0/16:1/16:1 | 2.0 ± 0.2 (2.9 ± 0.2) | 0.8 ± 0.1 (1.7 ± 0.2) |
| 811.7 | 14:0/16:0/18:1, 16:0/16:0/16:1 | 2.2 ± 0.1 (3.3 ± 0.1) | 1.0 ± 0.2 (2.2 ± 0.3) |
| 813.7 | 16:0/16:0/16:0 | 0.8 ± 0.1 (1.2 ± 0.2) | 0.8 ± 0.2 (1.7 ± 0.2) |
| 833.7 | 16:1/16:1/18:2 | 1.0 ± 0.3 (1.5 ± 0.5) | 0.5 ± 0.1 (1.1 ± 0.2) |
| 835.7 | 14:0/18:1/18:2, 16:0/16:1/18:2, 14:0/18:0/18:2 | 4.2 ± 0.6 (6.2 ± 0.5) | 1.7 ± 0.2 (3.9 ± 0.3) |
| 837.7 | 14:0/18:0/18:2, 16:0/16:0/18:2, 16:0/16:1/18:1 | 7.2 ± 0.1 (10.5 ± 0.4) | 3.8 ± 0.3 (8.6 ± 0.7) |
| 839.7 | 16:0/16:0/18:1,16:0/16:1/18:0 | 3.9 ± 0.1 (5.7 ± 0.1) | 2.5 ± 0.3 (5.5 ± 0.3) |
| 851.7 | 16:1/17:0/18:1 | 1.0 ± 0.1 (1.4 ± 0.1) | 0.7 ± 0.1 (1.6 ± 0.2) |
| 853.7 | 16:0/17:0/18:1 | 0.6 ± 0.1 (0.9 ± 0.1) | 0.3 ± 0.1 (0.6 ± 0.1) |
| 859.8 | 16:0/16:1/20:4, 16:1/18:2/18:2 | 1.4 ± 0.1 (2.0 ± 0.1) | 0.8 ± 0.1 (1.9 ± 0.3) |
| 861.8 | 16:0/18:2/18:2, 16:1/18:1/18:2 | 5.3 ± 0.3 (7.8 ± 0.3) | 3.3 ± 0.3 (7.6 ± 1.4) |
| 863.8 | 16:0/18:1/18:2, 16:1/18:0/18:2, 16:1/18:1/18:1 | 9.4 ± 0.3 (13.7 ± 0.2) | 5.6 ± 0.3 (12.6 ± 0.4) |
| 865.8 | 16:0/18:0/18:2, 16:0/18:1/18:1 | 6.1 ± 0.4 (9.0 ± 0.4) | 3.8 ± 0.5 (8.6 ± 1.1) |
| 867.8 | 16:0/18:0/18:1 | 0.6 ± 0.1 (0.9 ± 0.1) | 0.6 ± 0.1 (1.4 ± 0.1) |
| 873.8 | 17:1/18:2/18:2 | 0.5 ± 0.1 (0.7 ± 0.1) | 0.6 ± 0.1 (1.4 ± 0.1) |
| 877.8 | 17:0/18:1/18:2 | 0.4 ± 0.1 (0.5 ± 0.1) | 0.2 ± 0.1 (0.6 ± 0.2) |
| 885.8 | 16:0/18:2/20:4, 18:2/18:2/18:2 | 1.2 ± 0.2 (1.8 ± 0.3) | 1.1 ± 0.2 (2.5 ± 0.2) |
| 887.8 | 16:0/18:1/20:4, 18:0/18:2/18:3, 18:1/18:2/18:2 | 3.0 ± 0.2 (4.5 ± 0.2) | 2.7 ± 0.1 (6.1 ± 0.3) |
| 889.8 | 18:0/18:2/18:2, 18:1/18:1/18:2 | 5.1 ± 0.3 (7.5 ± 0.3) | 3.4 ± 0.2 (7.7 ± 0.2) |
| 891.8 | 16:0/18:2/20:1, 18:0/18:1/18:2, 18:1/18:1/18:1 | 3.8 ± 0.3 (5.6 ± 0.4) | 3.2 ± 0.4 (7.4 ± 0.6) |
| 893.8 | 16:0/18:1/20:1, 18:0/18:1/18:1 | 1.0 ± 0.1 (1.4 ± 0.1) | 0.8 ± 0.1 (1.8 ± 0.2) |
| 911.8 | 16:0/18:1/22:6 | 0.5 ± 0.1 (0.7 ± 0.1) | 0.4 ± 0.1 (0.8 ± 0.1) |
| 913.8 | 18:1/18:1/20:4, 18:0/18:2/20:4 | 0.5 ± 0.1 (0.7 ± 0.1) | 0.4 ± 0.1 (0.8 ± 0.1) |
| 915.8 | 18:0/18:1/20:4 | 0.5 ± 0.1 (0.7 ± 0.1) | 0.5 ± 0.1 (1.1 ± 0.1) |
| 917.8 | 18:1/18:2/20:1 | 0.6 ± 0.1 (0.8 ± 0.1) | 0.4 ± 0.1 (1.0 ± 0.1) |
| 919.8 | 18:1/18:1/20:1 | 0.5 ± 0.1 (0.7 ± 0.1) | 0.3 ± 0.1 (0.8 ± 0.1) |
| ...... | ...... | ...... | ...... |
| Total TG | 68.2 ± 3.5 | 44.1 ± 2.7** | |
| FA containing odd-numbered carbons | 7.0 ± 0.8 (3.5 ± 0.7) | 7.7 ± 0.5 (5.7 ± 0.4) | |
| FA containing 2 or more double bonds | 42.0 ± 4.0 (20.5 ± 1.1) | 35.1 ± 2.0 (26.0 ± 1.3) | |
| FA containing 20 or 22 carbons | 8.1 ± 0.5 (4.0 ± 0.2) | 6.6 ± 0.4 (4.9 ± 0.1) |
Mouse DRG lipids were extracted by a modified Bligh and Dyer procedure and the TG molecular species in the lipid extracts were identified and quantified using a 2D ESI/MS technique as described under Materials and methods. The results are expressed in nmol/mg of protein and represent X ± SD of six different animals whereas the data in parentheses are the percentages of each TG molecular species or the acyl chains in the pool.
p < 0.001 relative to TG mass content in wild-type mouse DRG.
FA, fatty acyl chains; DRG, dorsal root ganglia; WT, wild type; ApoE, apolipoprotein E; KO, knockout; DA, presence of a detectable ion peak; TG, triacylglycerol.
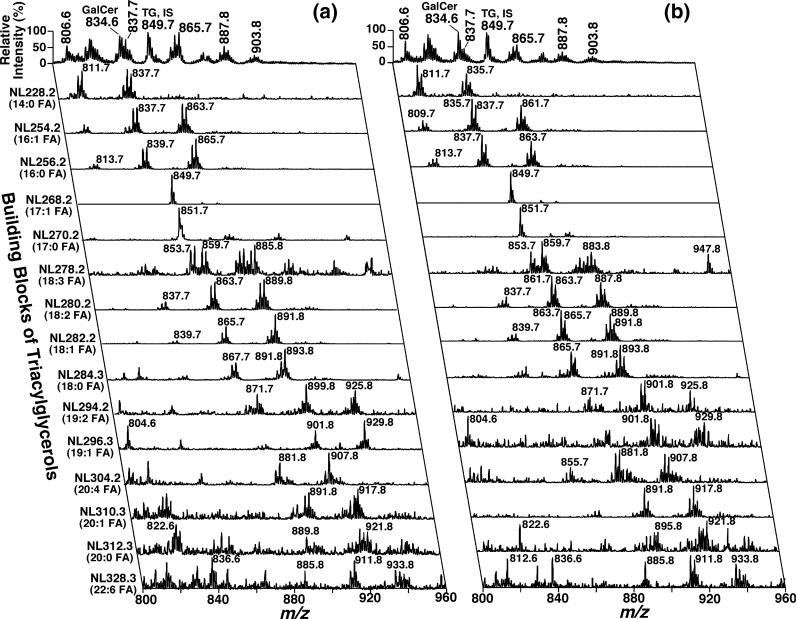
Fig. 6.
Two-dimensional electrospray ionization mass/spectrometry fingerprint and quantitation of triacylglycerol (TG) molecular species in the dorsal root ganglia lipid extracts of wild-type and apoE knockout mice. Dorsal root ganglia lipids of wild-type (Panel a) and apolipoprotein E knockout (Panel b) mice at 4 months of age were extracted by a modified Bligh and Dyer method. Two-dimensional electrospray ionization/mass spectrometry analyses were performed by neutral loss scanning of all naturally occurring fatty acids (building blocks of TG molecular species) in the positive-ion mode in the presence of LiOH as described under Materials and methods.
For example, the base peaks in NL228.2, NL254.2, NL256.2, NL280.2, and NL 282.2 were at m/z 809.7, 835.7, 837.7, 861.7, and 863.7 in the 2D mass spectrum of apoE KO mouse DRG lipids (Fig. 6b) whereas at m/z 835.7, 861.7, 865.7, 889.8, and 891.8 in the 2D mass spectrum of WT mouse DRG lipid extract (Fig. 6a), respectively. Third, although the total TG mass content in apoE KO mouse DRG was much less than that in WT mouse DRG, the compositions of fatty acyl chains containing either an odd number of carbons, two or more double bonds, or 20 or more carbons were not affected within experimental error by apoE depletion (Table 3).
The alterations in mouse DRG lipidome induced by apoE depletion are age dependent
To determine whether the observed changes in the mouse DRG lipidome of apoE KO mice relative to WT mice at 4 months of age is truly due to the deficiency of apoE which leads to defects in lipid transport and homeostasis, we have also examined the apoE KO mouse DRG lipidomes at 2 and 10 months of age relative to those of WT mice. Shotgun lipidomics analyses unambiguously confirmed the alterations in mouse DRG lipidome induced by apoE deficiency in an age-dependent manner (Fig. 7). Specifically, the mass levels of DRG sulfatide and GalCer of WT mice are essentially identical at the different ages examined (Fig. 7a). As anticipated, the mass levels of DRG sulfatide and GalCer of apoE KO mice are significantly higher in comparison to those of the age-matched WT mice. Furthermore, the differences in the alterations of DRG sulfatide and GalCer between apoE KO and WT mice are increased with advancing age (Fig. 7a). The alterations in mouse DRG Cer mass content induced by apoE KO at different ages are intriguing. The mass levels of mouse DRG Cer in both apoE and WT mice are reproducibly higher at the youngest age examined (i.e. 2 months) in comparison to those of older ages (e.g. 4 and 10 months) (Fig. 7b). However, the age-dependent mass level differences of DRG Cer mass content is larger among apoE KO mice relative to WT mice. Again, the mass levels of mouse DRG Cer in apoE KO mice are always higher than those of the age matched WT mice (Fig. 7b).
Fig. 7.
The mass levels of selected lipid classes in the dorsal root ganglia (DRG) lipidome of apolipoprotein E knockout and wild-type mice at different ages. DRG lipid extracts from apolipoprotein E knockout (closed symbols) and wild-type (open symbols) mice were prepared by a modified Bligh and Dyer procedure. The mass levels of lipid classes including sulfatide (panel a, circle), galactosylceramide or galactocerebroside (GalCer) (panel a, square), ceramide (panel b), triacylglycerol (panel c), and cholesterol (panel d) at the indicated ages were quantitated by shotgun lipidomics as described under Materials and methods. Each data point represents mean ± SD of six different animals and some error bars are within the symbols.
Age-dependent changes in DRG TG mass levels and composition in WT mice have recently been determined (Cheng et al. 2006) and have been reproduced for this study (Fig. 7c). It should be pointed out that the different TG mass levels in mouse DRG in these two studies represent the differences between sample preparations (i.e. the DRG dissected in this study contains associated nerve bundles which were not present in our early study). We also found that the mass levels of DRG TG in apoE KO mice were consistently and substantially lower (i.e. 15 ± 2, 33 ± 3, and 40 ± 5 mol% at 2, 4, and 10 months of age) than those in age-matched WT mice (Fig. 7c). In contrast to the depletion of DRG TG mass levels induced by apoE KO, cholesterol mass levels in the DRG lipidome dramatically accumulate in apoE KO mice in comparison to those in age-matched WT mice (Fig. 7d). The mass levels of other components of DRG lipidome (e.g. glycerophosphate, GPGro, GPSer, GPIns, GPEtn, GPCho, and SM) in both apoE KO and WT mice at 2 and 10 months of age were also determined by shotgun lipidomics. It was found that the mass levels of these lipid classes in DRG are not significantly different between apoE KO and WT mice at the different ages examined.
Discussion
The current study systematically determined the alterations in mouse DRG lipidome induced by apoE depletion at different ages using a lipidomics approach. We identified four specific alterations in mouse DRG lipidome resulting from apoE KO including (i) accumulation of the sulfatide mass content, (ii) increases in the mass levels of both GalCer and Cer classes, (iii) elevation of mass content in cholesterol pool, and (iv) depletion of the TG storage. These specific alterations in the mouse DRG lipidome as induced by apoE deficiency suggest the involvement of apoE in DRG lipid metabolism, transport, and/or homeostasis in a tissue-specific manner as these alterations are very different from those observed in the circulatory system, other peripheral organs (e.g. liver and heart), and the CNS. It should be specifically stated that although we have identified four specific lipid mass level alterations in the mouse DRG induced by apoE deficiency, the composition of the individual molecular species (i.e. the lipid profile) of each lipid class examined is essentially unaffected. This result indicates that apoE deficiency primarily affects the homeostasis and transport of entire lipid classes and not individual lipid molecular species. In addition, our study also indicates that the lipid profiles of the WT mouse DRG lipidome display little variability at the ages examined.
Our recent studies have demonstrated that apoE can modulate sulfatide mass content and metabolism in the CNS through the same transport and metabolic pathways that regulate levels of apoE-containing CNS lipoproteins (Han et al. 2003a; Han 2004). Following this line of reasoning, we anticipate that apoE depletion induces a substantial increase in the sulfatide mass content in mouse cervical DRG. We demonstrated a dramatic mass increase in sulfatide in apoE KO mouse DRG in comparison to that in WT mouse DRG at 4 months of age or older, suggesting that a parallel role of apoE in modulating sulfatide mass and metabolism in the PNS to that in the CNS is likely present. However, this apoE-depletion induced increase in sulfatide levels in the DRG is smaller than that in the CNS (Han et al. 2003a), suggesting that some other alternate metabolic pathways of sulfatide in mouse DRG might compensate against sulfatide accumulation.
A reduction in accumulated sulfatide can occur either through an acceleration of hydrolysis or an inhibition of biosynthesis. Both pathways would alter the homeostasis of sulfatide metabolites or biosynthetic precursors, resulting in an accumulation of GalCer and Cer in the apoE KO mouse DRG. Indeed, we demonstrated a significant increase in the mass content of the GalCer pool and a substantial elevation of the Cer level in apoE KO mouse DRG relative to those in WT mouse DRG at all examined ages (Fig. 7). Moreover, these results explain why sulfatide in apoE KO mouse DRG accumulates to lesser extent in comparison to the CNS where both GalCer and Cer pools are not significantly affected by apoE depletion (Han et al. 2003a). Why apoE depletion differentially affects the GalCer and Cer homeostasis in mouse DRG and in the CNS is unclear. It should be specifically emphasized that many other sphingolipid classes such as gangliosides, lactosylceramide, and lysosphingolipids are present in the DRG lipidome and that apoE depletion may also lead to alterations in these lipid classes. Although we are able to identify some of these sphingolipid classes by shotgun lipidomics at its current stage, quantitation of the individual molecular species of these sphingolipids has not yet been achieved due to the lack of suitable internal standards for analysis of these lipid classes by shotgun lipidomics.
Impaired catabolism of Cer, GalCer, or sulfatide is associated with Farber's disease (Moser 1995), Krabbe's disease (Suzuki et al. 1995), or metachromatic leukodystrophy (Von Figura et al. 2001), respectively. High levels of Cer can result in diverse cellular consequences including up-regulation of cytokines, generation of reactive oxygen species, uncoupling of the mitochondrial respiratory chain, and apoptosis (see Sawai et al. 2005 for a recent review). On the other hand, multiple lines of evidence demonstrate the presence of abnormal electrophysiology and pathology in the PNS of apoE KO mice. For example, apoE KO mice have peripheral sensory nerve defects, reduced and delayed responses to noxious thermal stimuli, and an abnormal morphology of unmyelinated fibers (Fullerton et al. 1998; Pola et al. 2003). These observations indicate the disturbed packing of the myelin sheath in the PNS of apoE KO mice, which is directly linked to the accumulation of Cer, GalCer, and sulfatide. Accordingly, our results provide important clues to understanding the biochemical mechanisms underlying these neurological observations.
It is intriguing that apoE modulates cholesterol homeostasis in mouse DRG as although this role of apoE is identical to that present in peripheral organs and in the circulatory system, it is different from that in the CNS where no apparent apoE-induced alterations in cholesterol homeostasis is present (Han et al. 2003a). This role of apoE in mouse DRG may reflect its peripheral location. As apoE in mouse DRG is isolated from the circulatory system by the presence of the blood–nerve barrier, the degree of the alteration in cholesterol level induced by apoE depletion is considerably smaller than that in peripheral organs and in the circulatory system (Mahley 1988). We have recently demonstrated that abundant TG is present in DRG of several examined species including mouse, rat, and rabbit (Cheng et al. 2006). We have also found that the mass content of DRG TG is substantially depleted in diabetic mice relative to controls (Cheng et al. 2006). Intriguingly, apoE depletion also induced a dramatic reduction of the TG pool in mouse DRG (Table 3). The true biochemical mechanism underlying this decrease remains to be elucidated.
In addition to the dramatic TG depletion in DRG induced by apoE depletion, which is similar to that induced by diabetes, two other alterations in the TG pool are also quite similar in both types of mice (apoE KO and diabetic). First, TG molecular species containing polyunsaturated fatty acyl chains are preserved in both apoE KO (Table 3) and diabetic mouse DRG (Cheng et al. 2006) relative to WT controls. Second, there exists a substantial TG mass accumulation in peripheral organs or in the circulatory systems of both mice. For example, heart and muscle of diabetic mice accumulate large amounts of TG (Finck et al. 2002, 2003; Unger 2002; Han et al. 2005b) whereas dramatic elevations in serum TG (over twofold, unpublished results) and hepatic TG (Kypreos et al. 2001; Mensenkamp et al. 2004) are also found in apoE KO mice. TG accumulation in diabetic mouse heart is lipotoxic and associated with diabetic cardiomyopathy (Finck et al. 2002, 2003; Unger 2002; Han et al. 2005b) whereas the substantial elevation of TG in apoE KO mouse serum may contribute to atherosclerotic plaque development. The paradoxical depletion of TG in the DRG accompanying an accumulation of TG in peripheral organs or the circulatory system may imply that TG is redistributed during the pathogenesis of these diseases by a yet unknown mechanism. Understanding the regulation underlying the distribution of TG in normal and diabetic individuals will likely provide insights into the disease pathogenesis as well as provide a gateway for potential pharmaceutical treatment.
In addition to the identification of the roles of apoE in modulating lipid homeostasis, this study also presented a largely complete determination of the mouse cervical DRG lipidome at the molecular species level in comparison to any previous studies by various techniques or methodologies (Linington et al. 1980; Yao et al. 1981; Fressinaud et al. 1986, 1987a,b; Ganser et al. 1988). Shotgun lipidomics analyses demonstrated that cholesterol is the most abundant lipid in the mouse DRG lipidome, accounting for ∼35 mol% of total lipid, followed by GPEtn, accounting for 23 mol% of total lipid mass (Table 4). The GPCho, GalCer, and GPSer classes are also very abundant in the mouse cervical DRG lipidome, each accounting for ∼10 mol% of the total lipid mass (Table 4). As sphingolipids are prominent constituents in the CNS, it is not surprising that the mouse DRG lipidome is enriched in sphingolipids including GalCer, sulfatide, and SM, particularly GalCer (Tables 1 and 4). It is interesting that the molar ratio of hydroxy versus non-hydroxy GalCer present in mouse DRG (Tables 1 and 4) is much higher than that found in the CNS (Han et al. 2003a; Han and Cheng 2005). This difference likely result from the different cellular functions of Schwann cells in the PNS and of oligodendrocytes in the CNS.
Table 4.
Summary of the mass contents of lipid classes in the dorsal root ganglia lipidome of wild-type and apolipoprotein E knockout mice at 4 months of agea
| Lipid class | WT mice | ApoE KO mice |
|---|---|---|
| Phosphatidic acid | 3.2 ± 0.2 (0.2) | 3.0 ± 0.3 (0.2) |
| Phosphatidylglycerol | 1.0 ± 0.1 (0.1) | 1.0 ± 0.1 (0.1) |
| Serine glycerophospholipid | 148.5 ± 1.6 (8.8) | 148.8 ± 4.5 (8.2) |
| (Phosphatidylserine)b | (140.6 ± 1.3) | (140.5 ± 4.3) |
| (Plasmenylserine)b | (5.3 ± 0.5) | (5.7 ± 0.3) |
| Phosphatidylinositol | 20.1 ± 1.4 (1.2) | 22.8 ± 0.5 (1.2) |
| Ethanolamine glycerophospholipid | 390.1 ± 17.6 (23.0) | 388.6 ± 11.4 (22.0) |
| (Phosphatidylethanolamine)b | (104.1 ± 6.4) | (101.4 ± 2.5) |
| (Plasmenylethanolamine)b | (284.9 ± 11.4) | (286.0 ± 10.6) |
| Choline glycerophospholipid | 184.4 ± 5.5 (10.9) | 180.1 ± 1.5 (9.9) |
| (Phosphatidylcholine)b | (171.4 ± 4.6) | (165.6 ± 1.1) |
| (Plasmenylcholine)b | (10.8 ± 0.8) | (11.8 ± 0.8) |
| Galactosylceramide | 146.8 ± 10.5 (8.7) | 174.1 ± 5.6 (9.6) |
| (Hydroxy galactosylceramide)b | (92.2 ± 4.7) | (107.3 ± 3.8) |
| Sphingomyelin | 44.7 ± 1.6 (2.6) | 47.4 ± 1.1 (2.6) |
| Sulfatide | 62.8 ± 3.1 (3.7) | 91.4 ± 1.5 (5.0) |
| (Hydroxy sulfatide)b | (15.8 ± 1.0) | (20.3 ± 0.7) |
| Ceramide | 0.59 ± 0.03 (0.1) | 0.77 ± 0.03 (0.1) |
| Free cholesterol | 579.4 ± 67.1 (34.2) | 680.6 ± 54.2 (37.4) |
| Cholesterol esters | 46.5 ± 24.9 (2.7) | 37.7 ± 24.9 (2.1) |
| Triacylglycerol | 68.2 ± 3.5 (4.0) | 44.1 ± 2.7 (2.4) |
| Total | 1696.3 ± 137.5 (100) | 1820.4 ± 110.4 (100) |
The results are summarized from Tables 1-3. The data are expressed in nmol/mg of protein and represent X ± SD of six different animals whereas the data in parentheses represent the relative percentage of each lipid class in the entire examined lipidome.
These rows summarize the mass contents of the subclass(es) of each indicated class.
WT, wild type; ApoE, apolipoprotein E; KO, knockout.
Ethanolamine plasmalogen molecular species are predominant in the GPEtn class in mouse DRG, accounting for ∼73 mol% of the total GPEtn (Table 2). It is known that the presence of plasmalogens makes membrane bilayers more compact (Han and Gross 1990), thereby contributing to the increased packing efficiency of the myelin sheath. Moreover, DRG plasmalogen molecular species largely contain monounsaturated fatty acids at the sn-2 position (Table 2), accounting for ∼40 mol% of the total GPEtn. This result is quite different from the GPEtn profiles in the CNS where molecular species containing polyunsaturated fatty acyl chains in GPEtn lipid class are quite abundant, although plasmalogen molecular species containing monounsaturated fatty acids at the sn-2 position are also enriched in the white matter of the brain (Han et al. 2001).
The presence of a high mass content of GPSer molecular species in the nervous system has been reported by several investigators (Jungalwala et al. 1984; Kevala and Kim 2001; Larsen et al. 2002). It is believed that the abundant GPSer molecular species containing docosahexanate are involved in signal transduction, cell growth, cell survival, and apoptosis, thereby serving many important roles in neuronal functions (Pepeu et al. 1996; Hamilton et al. 2000; Wen and Kim 2004). However, GPSer molecular species containing docosahexanate in mouse DRG accounted for only ∼2 mol% of the total GPSer. The majority of GPSer molecular species contain monounsaturated fatty acyl chains (i.e. 18:1, 22:1, and 24:1, a total of 90 mol%) at the sn-2 position, particularly 18:1, which accounts for ∼65 mol% of the total GPSer (Table 2). These results suggest that 18:1-enriched GPSer molecular species in the mouse DRG lipidome likely contribute to the packing and/or function of the myelin sheath and are not affected by apoE depletion. Collectively, the current study not only identifies multiple specific roles of apoE in a tissue-specific manner, but also presents a detailed mouse DRG lipidome that provides abundant information about lipid chemistry and its possible functions in the PNS.
Acknowledgements
This work was supported by NIA Grant R01 AG23168 and the Neurosciences Education and Research Foundation. The authors are grateful to Dr Christopher M. Jenkins for his comments and to Dr Kui Yang for her technical help.
Abbreviation used
- AD
Alzheimer's disease
- apoE
apolipoprotein E
- Cer
ceramide
- d6-chol
deuterated cholesterol
- DRG
dorsal root ganglion
- ESI
electrospray ionization
- GalCer
galactosylceramide or galactocerebroside
- GPCho
glycerophosphocholine(s)
- GPEtn
glycerophosphoethanolamine(s)
- GPGro
glycerophosphoglycerol(s)
- GPIns
glycerophosphoinositol(s)
- GPSer
glycerophosphoserine(s)
- KO
knockout
- MS
mass spectrometry
- NL
neutral loss
- pGPEtn
alkenylacyl GPEtn (i.e. plasmenylethanolamine)
- PI
precursor ion
- SM
sphingomyelin
- TG
triacylglycerol(s)
- WT
wild type
References
- Bales KR, Dodart JC, Demattos RB, Holtzman DM, Paul SM. Apolipoprotein E, amyloid, and Alzheimer disease. Mol. Interv. 2002;2:363–375. 379. doi: 10.1124/mi.2.6.363. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Boumann HA, Damen MJA, Versluis C, Heck AJR, De Kruijff B, De Kroon AIPM. The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species. Evidence for lipid remodeling. Biochemistry. 2003;42:3054–3059. doi: 10.1021/bi026801r. [DOI] [PubMed] [Google Scholar]
- Boumann HA, Chin PTK, Heck AJR, De Kruijff B, De Kroon AIPM. The yeast phospholipid N-methyltransferases catalyzing the synthesis of phosphatidylcholine preferentially convert di-C16:1 substrates both in vivo and in vitro. J. Biol. Chem. 2004;279:40, 314–40, 319. doi: 10.1074/jbc.M406517200. [DOI] [PubMed] [Google Scholar]
- Boyles JK, Notterpek LM, Anderson LJ. Accumulation of apolipoproteins in the regenerating and remyelinating mammalian peripheral nerve. Identification of apolipoprotein D, apolipoprotein A-IV, apolipoprotein E, and apolipoprotein A-I. J. Biol. Chem. 1990;265:17, 805–17, 815. [PubMed] [Google Scholar]
- Cheng H, Xu J, Mckeel DW, Jr, Han X. Specificity and potential mechanism of sulfatide deficiency in Alzheimer's disease: an electrospray ionization mass spectrometric study. Cell. Mol. Biol. 2003;49:809–818. [PubMed] [Google Scholar]
- Cheng H, Guan S, Han X. Abundance of triacylglycerols in ganglia and their depletion in diabetic mice: implications for the role of altered triacylglycerols in diabetic neuropathy. J. Neurochem. 2006;97:1288–1300. doi: 10.1111/j.1471-4159.2006.03794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong CJ, Shen Y-J, Thomas MJ, Cui Z. Molecular distinction of phosphatidylcholine synthesis between the CDP-choline pathway and phosphatidylethanolamine methylation pathway. J. Biol. Chem. 1999;274:29, 683–29, 688. doi: 10.1074/jbc.274.42.29683. [DOI] [PubMed] [Google Scholar]
- Demattos RB, Brendza RP, Heuser JE, Kierson M, Cirrito JR, Fryer J, Sullivan PM, Fagan AM, Han X, Holtzman DM. Purification and characterization of astrocyte-secreted apolipoprotein E and J-containing lipoproteins from wild-type and human apoE transgenic mice. Neurochem. Int. 2001;39:415–425. doi: 10.1016/s0197-0186(01)00049-3. [DOI] [PubMed] [Google Scholar]
- Elshourbagy NA, Liao WS, Mahley RW, Taylor JM. Apolipoprotein E mRNA is abundant in the brain and adrenals, as well as in the liver, and is present in other peripheral tissues of rats and marmosets. Proc. Natl Acad. Sci. USA. 1985;82:203–207. doi: 10.1073/pnas.82.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Lehman JJ, Leone TC, et al. The cardiac phenotype induced by PPARα overexpression mimics that caused by diabetes mellitus. J. Clin. Invest. 2002;109:121–130. doi: 10.1172/JCI14080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Han X, Courtois M, Aimond F, Nerbonne JM, Kovacs A, Gross RW, Kelly DP. A critical role for PPARα-mediated lipotoxicity in the pathogenesis of diabetic cardiomyopathy: modulation by dietary fat content. Proc. Natl Acad. Sci. USA. 2003;100:1226–1231. doi: 10.1073/pnas.0336724100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fressinaud C, Rigaud M, Vallat JM. Fatty acid composition of endoneurium and perineurium from adult rat sciatic nerve. J. Neurochem. 1986;46:1549–1554. doi: 10.1111/j.1471-4159.1986.tb01775.x. [DOI] [PubMed] [Google Scholar]
- Fressinaud C, Vallat JM, Rigaud M, Leboutet MJ. Analysis of unsaturated fatty acids of endoneurium and perineurium from normal and degenerating rat sciatic nerve. Morphological correlations. J. Neurol. Sci. 1987a;81:85–92. doi: 10.1016/0022-510x(87)90186-9. [DOI] [PubMed] [Google Scholar]
- Fressinaud C, Vallat JM, Durand J, Archambeaud-Mouveroux F, Rigaud M. Changes in composition of endoneurial and perineurial fatty acids during glycerol-induced Wallerian degeneration and regeneration in the sciatic nerve of the adult rat. J. Neurochem. 1987b;49:797–801. doi: 10.1111/j.1471-4159.1987.tb00963.x. [DOI] [PubMed] [Google Scholar]
- Fullerton SM, Strittmatter WJ, Matthew WD. Peripheral sensory nerve defects in apolipoprotein E knockout mice. Exp. Neurol. 1998;153:156–163. doi: 10.1006/exnr.1998.6872. [DOI] [PubMed] [Google Scholar]
- Fullerton SM, Shirman GA, Strittmatter WJ, Matthew WD. Impairment of the blood–nerve and blood–brain barriers in apolipoprotein E knockout mice. Exp. Neurol. 2001;169:13–22. doi: 10.1006/exnr.2001.7631. [DOI] [PubMed] [Google Scholar]
- Ganser AL, Kerner AL, Brown BJ, Davisson MT, Kirschner DA. A survey of neurological mutant mice. I. Lipid composition of myelinated tissue in known myelin mutants. Dev. Neurosci. 1988;10:99–122. doi: 10.1159/000111961. [DOI] [PubMed] [Google Scholar]
- Griffiths WJ. Tandem mass spectrometry in the study of fatty acids, bile acids, and steroids. Mass Spectrom. Rev. 2003;22:81–152. doi: 10.1002/mas.10046. [DOI] [PubMed] [Google Scholar]
- Hamilton L, Greiner R, Salem N, Jr, Kim HY. n-3 fatty acid deficiency decreases phosphatidylserine accumulation selectively in neuronal tissues. Lipids. 2000;35:863–869. doi: 10.1007/s11745-000-0595-x. [DOI] [PubMed] [Google Scholar]
- Han X. The role of apolipoprotein E in lipid metabolism in the central nervous system. Cell. Mol. Life Sci. 2004;61:1896–1906. doi: 10.1007/s00018-004-4009-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Cheng H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J. Lipid Res. 2005;46:163–175. doi: 10.1194/jlr.D400022-JLR200. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Plasmenylcholine and phosphatidylcholine membrane bilayers possess distinct conformational motifs. Biochemistry. 1990;29:4992–4996. doi: 10.1021/bi00472a032. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of the cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005a;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: multi-dimensional mass spectrometric analysis of cellular lipidomes. Expert Rev. Proteomics. 2005b;2:253–264. doi: 10.1586/14789450.2.2.253. [DOI] [PubMed] [Google Scholar]
- Han X, Holtzman DM, Mckeel DW., Jr Plasmalogen deficiency in early Alzheimer's disease subjects and in animal models: molecular characterization using electrospray ionization mass spectrometry. J. Neurochem. 2001;77:1168–1180. doi: 10.1046/j.1471-4159.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- Han X, Holtzman DM, Mckeel DW, Jr, Kelley J, Morris JC. Substantial sulfatide deficiency and ceramide elevation in very early Alzheimer's disease: potential role in disease pathogenesis. J. Neurochem. 2002;82:809–818. doi: 10.1046/j.1471-4159.2002.00997.x. [DOI] [PubMed] [Google Scholar]
- Han X, Cheng H, Fryer JD, Fagan AM, Holtzman DM. Novel role for apolipoprotein E in the central nervous system: modulation of sulfatide content. J. Biol. Chem. 2003a;278:8043–8051. doi: 10.1074/jbc.M212340200. [DOI] [PubMed] [Google Scholar]
- Han X, Fagan AM, Cheng H, Morris JC, Xiong C, Holtzman DM. Cerebrospinal fluid sulfatide is decreased in subjects with incipient dementia. Ann. Neurol. 2003b;54:115–119. doi: 10.1002/ana.10618. [DOI] [PubMed] [Google Scholar]
- Han X, Cheng H, Mancuso DJ, Gross RW. Caloric restriction results in phospholipid depletion, membrane remodeling and triacylglycerol accumulation in murine myocardium. Biochemistry. 2004a;43:15, 584–15, 594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Cheng H, Ye H, Gross RW. Towards fingerprinting cellular lipidomes directly from biological samples by two-dimensional electrospray ionization mass spectrometry. Anal. Biochem. 2004b;330:317–331. doi: 10.1016/j.ab.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Han X, Yang K, Cheng H, Fikes KN, Gross RW. Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J. Lipid Res. 2005a;46:1548–1560. doi: 10.1194/jlr.D500007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang J, Cheng H, Yang K, Abendschein DR, Gross RW. Shotgun lipidomics identifies cardiolipin depletion in diabetic myocardium linking altered substrate utilization with mitochondrial dysfunction. Biochemistry. 2005b;44:16, 684–16, 694. doi: 10.1021/bi051908a. [DOI] [PubMed] [Google Scholar]
- Han X, Yang K, Yang J, Fikes KN, Cheng H, Gross RW. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J. Am. Soc. Mass Spectrom. 2006;17:264–274. doi: 10.1016/j.jasms.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Horsburgh K, Mccarron MO, White F, Nicoll JA. The role of apolipoprotein E in Alzheimer's disease, acute brain injury and cerebrovascular disease: evidence of common mechanisms and utility of animal models. Neurobiol. Aging. 2000;21:245–255. doi: 10.1016/s0197-4580(00)00097-x. [DOI] [PubMed] [Google Scholar]
- Jungalwala FB, Evans JE, Mccluer RH. Compositional and molecular species analysis of phospholipids by high performance liquid chromatography coupled with chemical ionization mass spectrometry. J. Lipid Res. 1984;25:738–749. [PubMed] [Google Scholar]
- Kevala JH, Kim HY. Determination of substrate preference in phosphatidylserine decarboxylation by liquid chromatography–electrospray ionization mass spectrometry. Anal. Biochem. 2001;292:130–138. doi: 10.1006/abio.2001.5076. [DOI] [PubMed] [Google Scholar]
- Koch S, Donarski N, Goetze K, Kreckel M, Stuerenburg HJ, Buhmann C, Beisiegel U. Characterization of four lipoprotein classes in human cerebrospinal fluid. J. Lipid Res. 2001;42:1143–1151. [PubMed] [Google Scholar]
- Kypreos KE, Van Dijk KW, Van Der Zee A, Havekes LM, Zannis VI. Domains of apolipoprotein E contributing to triglyceride and cholesterol homeostasis in vivo: carboxyl-terminal region 203−299 promotes hepatic very low density lipoprotein–triglyceride secretion. J. Biol. Chem. 2001;276:19, 778–19, 786. doi: 10.1074/jbc.M100418200. [DOI] [PubMed] [Google Scholar]
- Lagarde M, Geloen A, Record M, Vance D, Spener F. Lipidomics is emerging. Biochim. Biophys. Acta. 2003;1634:61. doi: 10.1016/j.bbalip.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Larsen A, Mokastet E, Lundanes E, Hvattum E. Separation and identification of phosphatidylserine molecular species using reversed-phase high-performance liquid chromatography with evaporative light scattering and mass spectrometric detection. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2002;774:115–120. doi: 10.1016/s1570-0232(02)00184-8. [DOI] [PubMed] [Google Scholar]
- Laws SM, Hone E, Gandy S, Martins RN. Expanding the association between the APOE gene and the risk of Alzheimer's disease: possible roles for APOE promoter polymorphisms and alterations in APOE transcription. J. Neurochem. 2003;84:1215–1236. doi: 10.1046/j.1471-4159.2003.01615.x. [DOI] [PubMed] [Google Scholar]
- Lin CT, Xu YF, Wu JY, Chan L. Immunoreactive apolipoprotein E is a widely distributed cellular protein. Immunohistochemical localization of apolipoprotein E in baboon tissues. J. Clin. Invest. 1986;78:947–958. doi: 10.1172/JCI112685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linington C, Neuhoff V, Waehneldt TV. The lipid and glycoprotein composition of peripheral-nervous-system myelin subfractions isolated by zonal centrifugation. Biochem. Soc. Trans. 1980;8:69–70. [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Jr, Ory DS, Schaffer JE. Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl Acad. Sci. USA. 2003;100:3077–3082. doi: 10.1073/pnas.0630588100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- Mahley RW, Rall SC., Jr Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- Masliah E, Mallory M, Ge N, Alford M, Veinbergs I, Roses AD. Neurodegeneration in the central nervous system of apoE-deficient mice. Exp. Neurol. 1995;136:107–122. doi: 10.1006/exnr.1995.1088. [DOI] [PubMed] [Google Scholar]
- Mensenkamp AR, Van Luyn MJA, Havinga R, et al. The transport of triglycerides through the secretory pathway of hepatocytes is impaired in apolipoprotein E deficient mice. J. Hepatol. 2004;40:599–606. doi: 10.1016/j.jhep.2003.12.011. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Moser HW. Farber lipogranulomatosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Diseases. 7th edn McGraw-Hill; New York: 1995. pp. 2589–2599. [Google Scholar]
- Pepeu G, Pepeu IM, Amaducci L. A review of phosphatidylserine pharmacological and clinical effects. Is phosphatidylserine a drug for the aging brain? Pharmacol. Res. 1996;33:73–80. doi: 10.1006/phrs.1996.0013. [DOI] [PubMed] [Google Scholar]
- Poirier J, Baccichet A, Dea D, Gauthier S. Cholesterol synthesis and lipoprotein reuptake during synaptic remodelling in hippocampus in adult rats. Neuroscience. 1993;55:81–90. doi: 10.1016/0306-4522(93)90456-p. [DOI] [PubMed] [Google Scholar]
- Pola R, Gaetani E, Flex A, Aprahamian T, Proia Anna S, Bosch-Marce M, Smith Roy C, Pola P. Peripheral nerve ischemia: apolipoprotein E deficiency results in impaired functional recovery and reduction of associated intraneural angiogenic response. Exp. Neurol. 2003;184:264–273. doi: 10.1016/s0014-4886(03)00260-7. [DOI] [PubMed] [Google Scholar]
- Posse De Chaves EI, Vance DE, Campenot RB, Kiss RS, Vance JE. Uptake of lipoproteins for axonal growth of sympathetic neurons. J. Biol. Chem. 2000;275:19, 883–19, 890. doi: 10.1074/jbc.275.26.19883. [DOI] [PubMed] [Google Scholar]
- Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- Sawai H, Domae N, Okazaki T. Current status and perspectives in ceramide-targeting molecular medicine. Curr. Pharm. Des. 2005;11:2479–2487. doi: 10.2174/1381612054367463. [DOI] [PubMed] [Google Scholar]
- Strittmatter WJ, Roses AD. Apolipoprotein E and Alzheimer's disease. Annu. Rev. Neurosci. 1996;19:53–77. doi: 10.1146/annurev.ne.19.030196.000413. [DOI] [PubMed] [Google Scholar]
- Su X, Han X, Yang J, Mancuso DJ, Chen J, Bickel PE, Gross RW. Sequential ordered fatty acid α oxidation and Δ9 desaturation are major determinants of lipid storage and utilization in differentiating adipocytes. Biochemistry. 2004;43:5033–5044. doi: 10.1021/bi035867z. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Suzuki Y, Suzuki K. Galactosylceramide lipidosis: globoid-cell leukodystrophy (Krabbe disease) In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Diseases. 7th edn McGraw-Hill; New York: 1995. pp. 2671–2692. [Google Scholar]
- Unger RH. Lipotoxic diseases. Annu. Rev. Med. 2002;53:319–336. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- Von Figura K, Gieselmann V, Jaeken J. Metachromatic leukodystrophy: lysosomal disorders. In: Sachdev HS, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Diseases. 8th edn McGraw-Hill; New York: 2001. pp. 3695–3724. [Google Scholar]
- Weisgraber KH, Roses AD, Strittmatter WJ. The role of apolipoprotein E in the nervous system. Curr. Opin. Lipidol. 1994;5:110–116. doi: 10.1097/00041433-199404000-00007. [DOI] [PubMed] [Google Scholar]
- Welti R, Wang X. Lipid species profiling: a high-throughput approach to identify lipid compositional changes and determine the function of genes involved in lipid metabolism and signaling. Curr. Opin. Plant Biol. 2004;7:337–344. doi: 10.1016/j.pbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Wen Z, Kim HY. Alterations in hippocampal phospholipid profile by prenatal exposure to ethanol. J. Neurochem. 2004;89:1368–1377. doi: 10.1111/j.1471-4159.2004.02433.x. [DOI] [PubMed] [Google Scholar]
- Wenk MR. The emerging field of lipidomics. Nat. Rev. Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- Yao JK, Dyck PJ, Vanloon JA, Moyer TP. Free fatty acid composition of human and rat peripheral nerve. J. Neurochem. 1981;36:1211–1218. doi: 10.1111/j.1471-4159.1981.tb01720.x. [DOI] [PubMed] [Google Scholar]