Abstract
Huntington disease is an autosomal dominant neurodegenerative disorder caused by the pathological expansion of a polyglutamine tract. In this study we directly assess the influence of protein size on the formation and subcellular localization of huntingtin aggregates. We have created numerous deletion constructs expressing successively smaller fragments of huntingtin and show that these smaller proteins containing 128 glutamines form both intranuclear and perinuclear aggregates. In contrast, larger NH2-terminal fragments of huntingtin proteins with 128 glutamines form exclusively perinuclear aggregates. These aggregates can form in the absence of endogenous huntingtin. Furthermore, expression of mutant huntingtin results in increased susceptibility to apoptotic stress that is greater with decreasing protein length and increasing polyglutamine size. As both intranuclear and perinuclear aggregates are clearly associated with increased cellular toxicity, this supports an important role for toxic polyglutamine-containing fragments forming aggregates and playing a key role in the pathogenesis of Huntington disease.
Huntington disease (HD)1 is one of eight neurodegenerative disorders associated with CAG expansion (Ross, 1995; Andrew et al., 1997). Recently, new insights have been gained as to how CAG expansion might be associated with cell death. Intranuclear inclusions have been demonstrated both in vitro and in vivo in spinocerebellar ataxia type I (Skinner et al., 1997), type III (Paulson et al., 1997), Huntington disease (Becher et al., 1997; Davies et al., 1997; DiFiglia et al., 1997; Martindale et al., 1998), and dentatorubropallidoluysian atrophy (DRPLA) (Igarashi et al., 1998). These inclusions contain the respective gene products and only occur in the presence of an expanded polyglutamine tract.
We have previously shown that expression of a truncated form of huntingtin (up to amino acid 548) forms perinuclear aggregates (Martindale et al., 1998). In contrast, a small huntingtin fragment corresponding to exon 1 encoding a protein product of <20 kD and composed almost entirely of the polyglutamine tract clearly enters the nucleus and forms aggregates in vitro (Martindale et al., 1998) and in vivo (Davies et al., 1997). To directly assess the relationship between protein size and nuclear import of huntingtin, we have created progressive truncations of huntingtin and assessed how the length of the protein influences its subcellular localization and aggregate formation.
In mammalian cells, the nucleus is enclosed by an envelope that is intimately associated with the endoplasmic reticulum. The nuclear envelope is punctured at intervals by nuclear pores that are the sites of exchange of molecules between the nucleus and the cytoplasm. Small proteins can diffuse freely through the nuclear pore whereas proteins >40 kD generally have delayed transport (Gorlich and Mattaj, 1996). For proteins >60 kD, passive diffusion is prevented and nuclear import is dependent on active transport (Goldfarb et al., 1986; Newmeyer et al., 1986; Zasloff, 1983; Dingwall and Laskey, 1991).
In an effort to directly explore the relationship between the size of the protein and its subcellular localization, we have created a series of cDNA deletion constructs that express huntingtin proteins of different molecular weights. Additionally, we also have shown that even though wild-type huntingtin is recruited into aggregates (Martindale et al., 1998), aggregates are able to form in the absence of the endogenous huntingtin. Second, the findings of this study reveal that huntingtin molecules of 47 kD and greater are not transported across the nuclear pore. By contrast, smaller NH2-terminal fragments of huntingtin can traverse the nuclear pore complex, which suggests that very small huntingtin fragments are transported into the nucleus mainly via passive diffusion.
The exact relationship between pathology and huntingtin aggregates in vivo is not clear. In an in vitro cell culture model we have demonstrated that the formation of perinuclear aggregates are associated with increased susceptibility to cell death (Martindale et al., 1998). In the present study, we determine the influence of the length of the huntingtin protein on cell death. Here, we show that progressive truncations of huntingtin are correlated with an increasingly severe susceptibility to an apoptotic stress. The findings of intranuclear inclusions (DiFiglia et al., 1997) associated with cytoplasmic perinuclear accumulation of huntingtin (Sapp et al., 1997) in the neurons of patients with Huntington disease, together with findings of this paper, provide significant support for the toxic fragment model for the pathogenesis of HD (Goldberg et al., 1996; Wellington and Hayden, 1997), whereby proteolytic cleavage of huntingtin liberates an NH2-terminal fragment containing the glutamine tract that forms aggregates, which confer increased susceptibility to death from apoptotic stimuli.
Materials and Methods
Vector Construction
Expression constructs containing the full-length huntingtin cDNA (pRcCMV10366-15 and pRcCMV10366-128) or the first 1955 nucleotides of the huntingtin cDNA (pCI1955-15 and pCI1955-128) have been described previously (Goldberg et al., 1996; Martindale et al., 1998). Site- directed mutagenesis was used to create a series of additional NH2-terminal truncations of huntingtin by introducing a translational termination codon at defined positions in the 1955-15 and 1955-128 constructs using the Transformer Mutagenesis kit (CLONTECH, Palo Alto, CA). Termination codons were inserted at nucleotide position 436, 771, 989, and 1597 as defined for huntingtin in GenBank/EMBL/DDBJ under accession number L27350. The following mutagenesis primers were used: 436-15 and 436-128: 5′-CAGCAGCAGCAGCAACAGTGACCACCGCCGCCGCCG-3′; 771-15 and 771-128: 5′-GGCTGACGAATGACTCAACAAAG-3′; 989-15 and 989-128: 5′-GACCCGAAGAATGAGTCCAGGAG-3′; 1597-15 and 1597-128: 5′-GAACTTATAGCTTGAGGGGGTTCC-3′.
The selection primer for each mutagenesis was 5′-CATGGCTCGACACATGTTCAATATTG-3′. Authenticity of the mutagenized constructs was confirmed by DNA sequencing. Also, a huntingtin COOH-terminal construct, encoding amino acids 585–3,144, was constructed by standard subcloning as described (Wellington et al., 1998).
Immunofluorescence and Western Blotting
Human embryonic kidney cells (HEK 293T) were grown on glass coverslips in DME (Gibco Laboratories, Grand Island, NY) with 10% FBS and antibiotics, in 5% CO2 at 37°C. The cells were transfected at 30% confluency with the calcium phosphate protocol by mixing Qiagen-prepared DNA (QIAGEN Inc., Chatsworth, CA) with 2.5 M CaCl2, and then incubating at room temperature for 10 min. 2× Hepes buffer (240 mM NaCl, 3.0 mM Na2HPO4, 100 mM Hepes, pH 7.05) was added to the DNA/calcium mixture, incubated at 37°C for 60 s, and then added to the cells. After 12–18 h, the media was removed, the cells were washed, and fresh media was added. At 36 h after transfection, the cells were treated with 35 μM tamoxifen (Sigma Chemical Co., St. Louis, MO) for 1 h, and then processed for immunofluorescence. The cells were washed with PBS, fixed in 4% paraformaldehyde/PBS solution for 20 min at room temperature, and then permeabilized in 0.5% Triton X-100/PBS for 5 min. After three PBS washes, the cells were incubated with anti-huntingtin antibody mAb 2166 (Chemicon International Inc., Temecula, CA) (1:2,500 dilution) or BKP1 (1:100 dilution) (Kalchman et al., 1997) in 0.4% BSA/PBS for 1 h at room temperature in a humidified container. The primary antibody was removed, the cells were washed, and secondary antibodies conjugated to Texas red or FITC were added at a 1:600–1:800 dilution for 30 min at room temperature. The cells were then washed again, and the coverslips were mounted onto slides with DAPI (4′,6′-diamidino-2-phenylindole; Sigma Chemical Co.) as a nuclear counterstain. Immunofluorescence was viewed using an Axioscope microscope (Carl Zeiss, Inc., Thornwood, NY), digitally captured with a CCD camera (Princeton Instrument Inc., Trenton, NJ) and the images were colorized and overlapped using the Eclipse software program (Empix Imaging Inc., Mississauga, ON). Intranuclear localization was determined by colocalization of huntingtin stain with the DAPI nucleus. The proportion of nuclear and cytoplasmic localization is presented as a percent of the total huntingtin-expressing cells. Appropriate control experiments were performed to determine the specificity of the antibody, including secondary antibody only and mock-transfected cells.
To determine the expression levels of truncated huntingtin, transfected 293T cells were harvested by gentle scraping 36 h after transfection. The cells were pelleted, washed with PBS, and then lysed. Equal amounts of protein were loaded onto standard 7.5% SDS-PAGE gels or 5–20% gradient SDS-PAGE gels, transferred onto polyvinylidene difluoride (PVDF) membrane, Western blotted using anti-huntingtin antibody BKP1 (Kalchman et al., 1997), and then detected using enhanced chemiluminescence (Amersham Corp., Arlington Heights, IL).
Size-exclusion Chromatography
Size exclusion was performed using a 16/60-cm Sephacryl S-300 high resolution column calibrated with a HMW Gel Filtration Calibration Kit (Pharmacia Biotechnology Inc., Piscataway, NJ) using run buffer (5 mM MgCl2, 0.5 mM EDTA, 1 mM PMSF in 1× PBS buffer, pH 7.3) at a flow rate of 0.25 ml/min. The void volume was calculated to be 42 ml using Blue Dextran. A 10-cm plate of 293T cells was transfected with HD 1955-128 and harvested after tamoxifen treatment. The cells were washed with ice-cold PBS, suspended in 200 μl of 1× PBS, and then lysed using 1 ml of 18 mM CHAPS, 5 mM MgCl2, 0.5 mM EDTA in 1× PBS buffer, pH 7.3, containing complete protease inhibitor cocktail (Boehringer Mannheim Corp., Indianapolis, IN) for 30 min on ice. Fractions were collected in run buffer at 0.25 ml/min at 4°C.
Cell Viability Assays
The viability of 293T cells expressing the huntingtin constructs was assessed by a modified MTT assay (Carmichael et al., 1987; Vistica et al., 1991). The cells were seeded at a density of 5 × 104 cells into 96-well plates and transfected with the huntingtin constructs or the control construct lacZ using the calcium phosphate method described above. At 48 h after transfection, a sublethal concentration of tamoxifen (35 μM) (Couldwell et al., 1994) was added to the cells for 3 h. For the modified MTT assay, tamoxifen-treated or untreated cells were incubated for 2 h in a 1:10 dilution of WST-1 reagent (Boehringer Mannheim Corp.) and release of formazan from mitochondria was quantified at 450 nm using an ELISA plate reader (Dynatech Laboratories, Chantilly, VA) (Mosmann, 1983; Carmichael et al., 1987). One-way analysis of variance and Newman-Keuls tests were used for statistical analysis. The transfection efficiency, measured by β-galactosidase staining and immunofluorescence, was ∼50%.
Transfection of Mouse Embryonic Stem Cells
Wild-type and HD−/− embryonic stem (ES) cells were plated onto 25-mm cover glass coated with 0.1% gelatin in H2O in six-well plates (Falcon Plastics, Cockeysville, MD) at a density of 105 cells per well. Cells were cultured in media as described previously (Metzler et al., 1994) and transfected with 5 μg of DNA by lipofection using the cationic compound N-[1-(2,3-Dioleoyloxy)propyl]-N,N, N -trimethylammoniummethylsulfate (DOTAP; Boehringer Mannheim Corp.). The media was changed the following day, and 36 h after transfection cells were incubated in 35 μM tamoxifen for 90 min. Subsequently, cells were washed twice with PBS fixed with 4% paraformaldehyde and immunostained with mAb 2166 as described above.
Results
Small Fragments of Huntingtin Enter the Nucleus
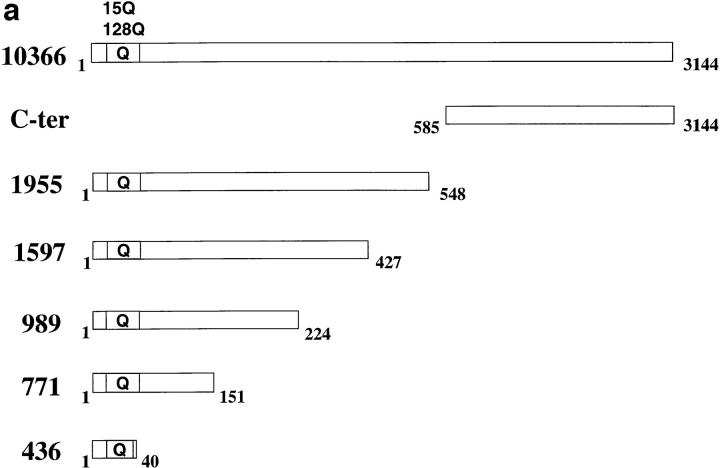
We have demonstrated that huntingtin truncated within exon 1, composed of 40 amino acids and the polyglutamine tract, can enter the nucleus (Martindale et al., 1998). Furthermore, intranuclear aggregates are formed in neurons from transgenic mice expressing truncated huntingtin containing exon 1 with an expanded polyglutamine tract (Davies et al., 1997). In contrast, full-length huntingtin or huntingtin proteins truncated at amino acid 548 (nucleotide 1955) (Fig. 1 a) are not found within the nucleus and accumulate in the cytosol, particularly in the perinuclear region. To demarcate functional domains within huntingtin that may determine its cellular localization, we created a set of constructs expressing various regions of huntingtin and determined the subcellular localization of their expressed products via indirect immunofluorescence of transfected cells.
Figure 1.
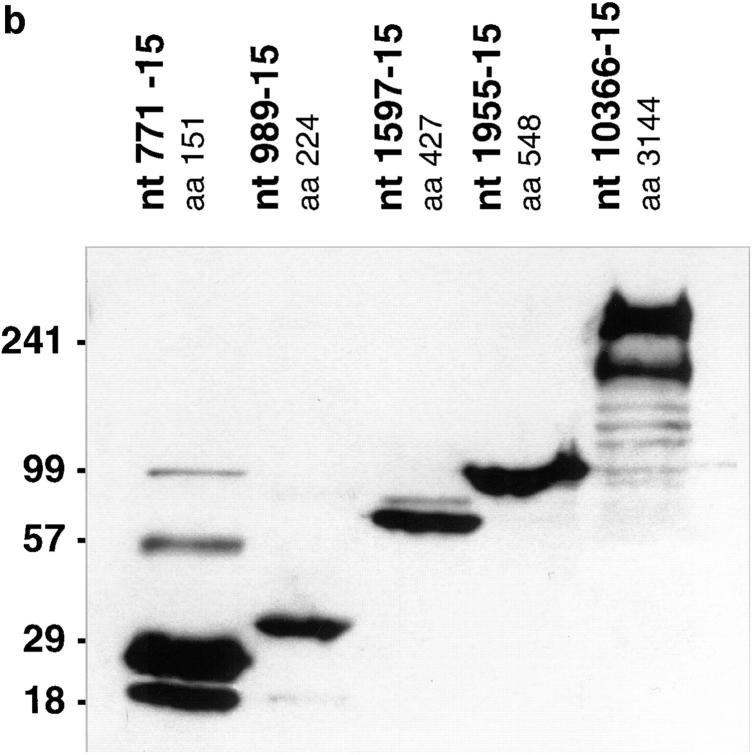
(a) Diagrammatic representation of the cDNA expression constructs used in this study. Bars represent huntingtin protein, with amino acids numbered according to GenBank/EMBL/DDBJ accession number L27350. The locations of the polyglutamine tract (Q) are indicated. The names of the constructs, 436, 771, 989, 1597, 1955, and 10366 correspond to the nucleotide position of the translation termination codon. The amino acid length of each protein is also indicated at the right. (b) Western blot of huntingtin truncation constructs. HEK 293T cells were transfected with the 771, 989, 1597, 1955, and 10366 constructs, containing 15 glutamines. 36 h after transfection, cell lysates were prepared. Equal amounts of protein were loaded on a 5–20% SDS-PAGE gradient gel, and Western blotted using anti-huntingtin antibody BKP1, which recognizes the NH2 terminus. Multiple bands in the lanes represent different conformations of the protein or huntingtin multimers. The amino acid length (aa) of each expressed protein is also indicated.
Subcellular localization of this series of truncated constructs (Fig. 1 a) was evaluated in an in vitro model system that we have described previously (Martindale et al., 1998). In this system, an apoptotic stress is induced in transiently transfected HEK 293T cells with sublethal levels (35 μM) of the apoptotic agent tamoxifen at 36–40 h after transfection. After 60 min of tamoxifen treatment, the cells are processed for indirect immunofluorescence. In 293T cells transfected with 1955 or full-length (10366) huntingtin cDNA constructs containing 128 glutamines, not treated with tamoxifen, huntingtin was diffusely expressed throughout the cytoplasm, with perinuclear aggregates present in only a low frequency of cells (∼1%). However, treatment of these cells with tamoxifen significantly increased the proportion of cells with perinuclear aggregates (Table I).
Table I.
| (a) Frequency of Aggregate Formation in Transfected Cells (%) | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Construct | 436-(128) | 771-(128) | 989-(128) | 1597-(128) | 1955-(128) | 10336-(128) | ||||||||
| Without tamoxifen | 45 | 81 | 70 | 4 | 0-1 | 0-1 | ||||||||
| n = 3 | n = 1 | n = 1 | n = 2 | n = 5 | n = 5 | |||||||||
| With tamoxifen | 43 | 92 | 78 | 13 | 27 | 9 | ||||||||
| n = 4 | n = 3 | n = 2 | n = 2 | n = 4 | n = 4 | |||||||||
| (b) Subcellular Localization of Huntingtin Aggregates with 128 Polyglutamines in 293T Cells (%) | ||||||||||||||
| Construct | 436-(128) | 771-(128) | 989-(128) | 1597-(128) | 1955-(128) | 10336-(128) | ||||||||
| Predicted protein size (kD) | 18 | 31 | 39 | 61 | 73 | 362 | ||||||||
| Estimated* size (kD) | ND | 68 | 75 | 77 | 115 | 530 | ||||||||
| Without tamoxifen | ||||||||||||||
| Nuclear | 21 | 77 | 22 | 0 | 0 | 0 | ||||||||
| Cytoplasmic | 79 | 23 | 78 | 100 | 0-1 | 0-1 | ||||||||
| No. of cells counted | 288 | 176 | 66 | 427 | >1,000 | >1,000 | ||||||||
| n = 3 | n = 3 | n = 2 | n = 2 | n = 5 | n = 5 | |||||||||
| With tamoxifen | ||||||||||||||
| Nuclear | 36 | 71 | 38 | 0 | 0 | 0 | ||||||||
| Cytoplasmic | 64 | 29 | 62 | 100 | 100 | 100 | ||||||||
| No. of cells counted | 314 | 175 | 95 | 320 | >1,500 | >1,000 | ||||||||
| n = 4 | n = 3 | n = 2 | n = 2 | n = 5 | n = 5 | |||||||||
| (c) Subcellular Localization of Huntingtin Protein with 15 Polyglutamines in 293T Cells | ||||||||||||||
| Length/size nucleotides | 436-(15) | 771-(15) | 989-(15) | 1597-(15) | 1955-(15) | 10366-(15) | COOH terminus | |||||||
| Amino acids | 1–40 | 1–151 | 1–224 | 1–427 | 1–548 | 1–3,144 | 585–3,144 | |||||||
| Predicted protein size (kD) | 3 | 16 | 24 | 47 | 59 | 347 | 284 | |||||||
| Estimated size* | ND | 40 | 43 | 72 | 80 | 370 | 280 | |||||||
| Localization without tamoxifen (%) | ||||||||||||||
| Nuclear | 21 | 86 | 11 | 0 | 0 | 0 | 0 | |||||||
| Cytoplasmic | 79 | 14 | 89 | 100 | 100 | 100 | 100 | |||||||
| No. of cells counted | 250 | 185 | 202 | 321 | >1,000 | >1,000 | >200 | |||||||
| n = 2 | n = 3 | n = 2 | n = 2 | n = 5 | n = 6 | n = 1 | ||||||||
| Localization with tamoxifen (%) | ||||||||||||||
| Nuclear | 57 | 82 | 21 | 0 | 0 | 0 | 0 | |||||||
| Cytoplasmic | 43 | 18 | 79 | 100 | 100 | 100 | 100 | |||||||
| No. of cells counted | 181 | 337 | 85 | 319 | >400 | >1,000 | >200 | |||||||
| n = 2 | n = 2 | n = 2 | n = 2 | n = 5 | n = 5 | n = 2 | ||||||||
Numbers in brackets indicate the polyglutamine length. ND, not detected.
Estimated by electrophoretic migration on Western blot. The polyglutamine tract causes aberrant migration.
To assess how protein length alters the subcellular localization of huntingtin, HEK 293T cells were transiently transfected with the 436, 771, 989, 1597, 1955, full-length (10366), and COOH-terminal contructs (Table I; Fig. 1 a), each containing 15 and 128 CAG repeats under the control of the human cytomegalovirus promoter. Protein products of ∼3, 16, 24, 47, 59, and 347 kD are expected from these respective constructs containing 15 repeats (Fig. 1 b), whereas proteins with expanded polyglutamine tracts would migrate as larger protein products (Table I). Indirect immunofluorescence studies were performed before and after the addition of tamoxifen using anti-huntingtin antibodies mAb2166 or BKP1 that recognize the NH2 terminus of huntingtin. Both antibodies gave the same results. Nuclear localization was determined by colocalization with DAPI (Fig. 2 a) or the nuclear protein p53 (data not shown).
Figure 2.
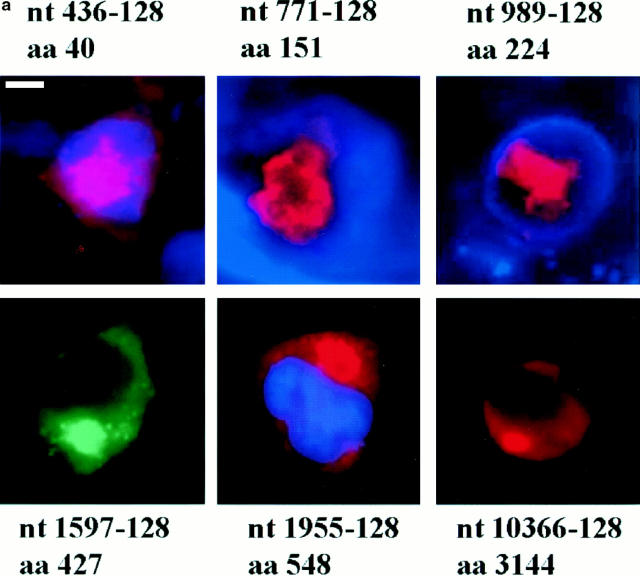
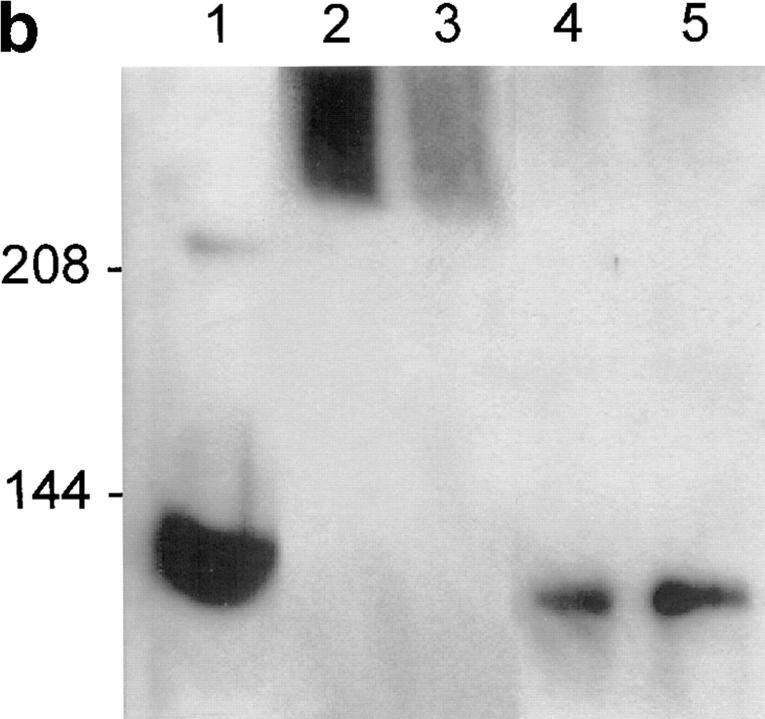
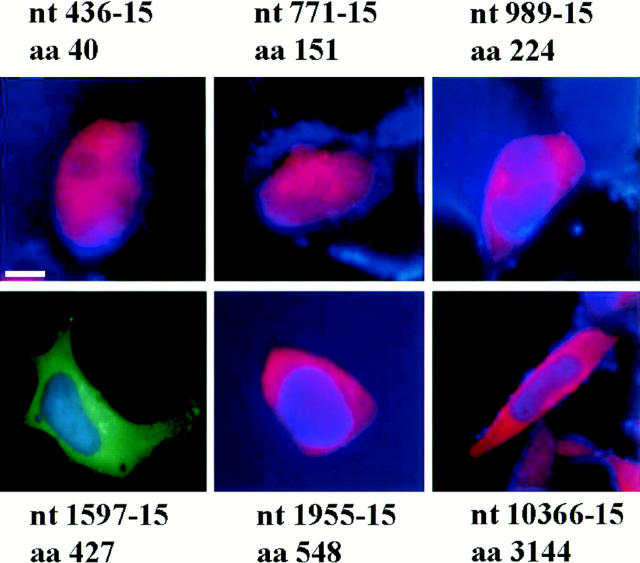
(a) Immunofluorescence of 293T cells transfected with the 436, 771, 989, 1597, 1955, and 10366 constructs, each containing 128 glutamines. 36–40 h after transfection, the cells were processed for immunofluorescence using mAb 2166 (red) or BKP1 (green) to detect huntingtin. The cells were counterstained with the DNA stain DAPI (blue) to label the nucleus (the DAPI stain is not visible in the cells shown for 1597-128 and 10366-128). Huntingtin located in the nucleus appears pink as the red stain is overlaid with blue. Huntingtin from the 436, 771, and 989 constructs (top row) appears as large intranuclear aggregates. Huntingtin from the 1597, 1955, and 10366 constructs (bottom row) show perinuclear aggregates and diffuse cytoplasmic stain. (b) Size-exclusion chromatography of a lysate from 293T cells transfected with HD1955-128 and treated with tamoxifen. 1-ml fractions were collected at 4°C, and samples were immediately analyzed by reducing SDS-PAGE and Western blotting with BKP1 antibody. Large molecular weight complexes were not retained by the column and flowed through into the void volume. Huntingtin aggregates isolated in the void volume were observed as species that did not denature upon SDS-PAGE and did not exit the stacking gel (V o = 42 ml; lanes 2 and 3). Under the same conditions, no aggregation was observed in the whole cell lysate (lane 1) nor in the monomeric huntingtin fractions (V = 72 ml; lanes 4 and 5). Bar in a: (436-128) 11.3 μm; (771-128) 5.31 μm; (989-128) 11.2 μm; (1597-128) 11.97 μm; (1955-128) 12 μm; (10366-128) 11.4 μm.
After transfection of the 436, 771, and 989 constructs, aggregates were present for each construct with and without tamoxifen treatment at similar frequencies, in contrast to the transfection of 1955 and 10366 constructs that resulted in aggregates only after treatment with tamoxifen. These aggregates appear as large, irregularly shaped depositions of huntingtin (Fig. 2 a). Size-exclusion chromatography demonstrated that the huntingtin forms stable high molecular weight complexes that do not denature in SDS-PAGE (Fig. 2 b).
The subcellular localization of different huntingtin fragments varied according to the size of the protein product (Fig. 2 a; Table I) and was similar with and without apoptotic stress. Protein products derived from the three smallest constructs (436, 771, and 989) expressing the most 40, 151, and 224 NH2-terminal residues of huntingtin were each able to enter the nucleus with both 15 and 128 glutamines. However, the frequency of nuclear entry varied among the constructs tested. For example, the localization of aggregates formed by expression of the 436-128 and 989-128 constructs was divided between the nuclear and perinuclear regions (Fig. 2 a), whereas expression of the larger product (31 kD) from the 771-128 construct resulted in more cells with nuclear than perinuclear aggregates (Fig. 2 a; Table I). In contrast, expression of the 1597, 1955, and 10366 constructs with 15 and 128 glutamines resulted in exclusive cytoplasmic localization of the gene products. Aggregates formed after transfection with 1597-128, 1955-128, and 10366-128 constructs were only observed in the perinuclear region (Fig. 2 a; Table I). In addition, cells transfected with a construct containing the COOH-terminal region of huntingtin (amino acids 585–3,144) did not form aggregates, and the COOH-terminal portion of huntingtin was located diffusely throughout the cytoplasm before and after (Fig. 3) tamoxifen treatment.
Figure 3.

Immunofluorescence of 293T cell transfected with the COOH-terminal huntingtin construct (amino acids 585–3,144). Transfected cells were treated with tamoxifen 36–40 h after transfection and processed for immunofluorescence using mAb 2172 to detect huntingtin (red). The protein was localized exclusively throughout the cytoplasm. Bar, 4.42 μm.
The frequency of aggregates varied between constructs of different lengths containing the same number of polyglutamines. An increased frequency of aggregates was seen in cells expressing shorter huntingtin products than in cells transfected with the 1955-128 and 10366-128 constructs (Table I). HEK 293T cells were also transiently transfected with the same series of constructs expressing 15 glutamines (Table I) that did not form aggregates although the subcellular localization of the gene product was similar to that seen after expression of the constructs with 128 repeats (Fig. 4; Table I).
Figure 4.
Immunofluorescence of 293T cells transfected with the 436, 771, 989, 1597, 1955, and 10366 constructs, each containing 15 glutamines. The transfected cells were treated with tamoxifen 36-40 h after transfection and mAb 2166 (red) or BKP1 (green) was used to detect huntingtin. The cells were counterstained with the DNA stain DAPI (blue) (the DAPI stain is not visible in the 1597-15 panel). Huntingtin containing 15 repeats does not form aggregates but can be localized in the nucleus depending on the protein size. Huntingtin is localized entirely in the nucleus in the 436 and 771 constructs in the cells shown. Bar: (436-15) 8.87 μm; (771-15) 11.98 μm; (989-15) 16.18 μm; (1597-15) 16.04 μm; (1955-15) 15.2 μm; (10366-15) 21.37 μm.
Aggregates Can Form in the Absence of Full-Length Endogenous Huntingtin
To further understand which factors contribute to the formation of aggregates, we determined whether endogenous huntingtin was required. Using immunofluorescent confocal microcopy and antibodies directed to epitopes within the NH2- and COOH-terminal portions of huntingtin (Hodgson et al., 1996; Martindale et al., 1998), we have previously shown that aggregates containing truncated huntingtin comprise not only the NH2-terminal fragment, but also contain the COOH terminus of the endogenous protein that was not encoded by the transfected cDNA (Martindale et al., 1998). The fact that COOH-terminal staining was present in aggregates in cells transfected with constructs expressing only truncated NH2-terminal huntingtin suggested that endogenous huntingtin was recruited into these aggregates. The question then arises whether full-length endogenous huntingtin is necessary for the formation of aggregates associated with expression of truncated mutant huntingtin.
HD−/− ES cells are homozygous for a targeted mutation within exon 5 of the HD gene (Nasir et al., 1995) and therefore lack full-length huntingtin expression. In ES cells, we are unable to detect a truncated huntingtin product and therefore if initially present, it is likely to be rapidly degraded in this cell system. Wild-type and HD−/− ES cells were transfected with a truncated form of huntingtin (construct 1955) with 15 or 128 CAG repeats. After tamoxifen treatment, wild-type and HD−/− ES cells transfected with constructs expressing huntingtin with 15 glutamines (1955) had no (wild-type) or very few (1% for HD−/− ES) aggregates. In contrast, the frequency of aggregates was increased in both wild-type (3%) and HD−/− ES cells (6.8%) transfected with huntingtin expressing 128 glutamines (1955-128) (Fig. 5). The aggregates were in the perinuclear region of the ES cells. These results clearly show that the presence of full-length endogenous huntingtin is not necessary or critical for formation of aggregates after transfection of truncated huntingtin with the expanded polyglutamine tract.
Figure 5.
Immunofluorescence of wild-type and null (HD −/−) ES cells transfected with 1955-128. The cells were treated with tamoxifen and immunostained with mAb 2166. Perinuclear aggregates were evident in both cell lines, indicating that the presence of endogenous huntingtin is not required for the formation of aggregates by transfected 1955-128. Bar: (Wild-type) 2.82 μm; (HD −/−) 2.87 μm.
Increased Susceptibility to Cell Death Is Correlated with Aggregate Formation and Inversely Correlated with Length of Huntingtin
We have previously demonstrated that expression of mutant huntingtin results in increased susceptibility to an apoptotic stress in transfected 293T cells induced by sublethal doses of tamoxifen (Martindale et al., 1998). Furthermore, the 1955 constructs resulted in significantly more cell death than the full-length huntingtin constructs (Martindale et al., 1998). To determine whether this observed difference represented a graded effect resulting from protein length, we assessed the change in viability of cells after expression of huntingtin of different lengths.
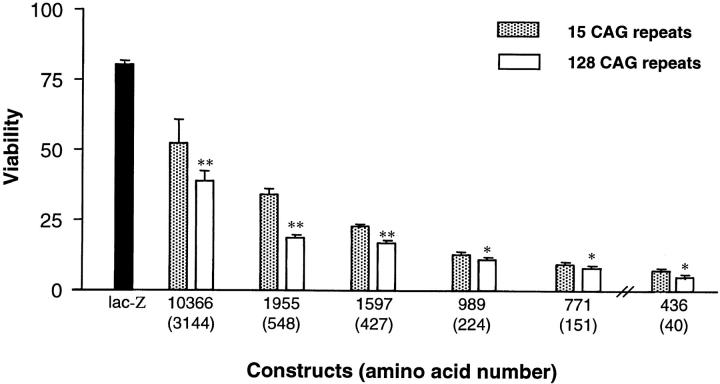
293T cells were transiently transfected with the series of huntingtin truncation and full-length constructs and the viability of the transfected cells was assessed by a modified MTT assay that measures mitochondrial function (Mosmann, 1983; Carmichael et al., 1987; Vistica et al., 1991). Mock- and LacZ-transfected 293T cells treated with tamoxifen served as controls. In each assay, cells transfected with the truncated constructs exhibited significantly higher proportions of injury or death compared with cells that received full-length huntingtin containing the same polyglutamine length (P < 0.01) (Fig. 6). Interestingly, this was seen even with proteins with 15 glutamines indicating that an NH2-terminal fragment of huntingtin may be proapoptotic even in the absence of polyglutamine expansion. However, measurement of cell viability indicated a polyglutamine repeat-dependent increase in cell death (P < 0.01 for all constructs). The differences in toxicity are not due to differences in expression of these constructs in cells as revealed by Western blot (data not shown). Increased susceptibility to cell death for any of these huntingtin gene products were only seen in the presence of apoptotic stress (data not shown). Therefore, the susceptibility to an apoptotic stress is influenced by both the length of the huntingtin protein and the size of the polyglutamine tract.
Figure 6.
Increasing toxicity with decreasing huntingtin length. In the presence of tamoxifen, progressive truncation of huntingtin resulted in increased cellular toxicity (P < 0.01). Interestingly, this trend was seen for proteins containing both 15 and 128 polyglutamines (P < 0.01). 293T cells were transfected with each huntingtin construct, or LacZ control, and cell viability in response to apoptotic stress was measured using a modified MTT assay and presented relative to control (untransfected). All wells were transfected with 0.1 μg DNA except for those transfected with the 436-15 and 128 constructs, where 0.08 μg of DNA was used because all the cells died with 0.1 μg DNA. Results for the 436 construct are normalized, taking into account decreased DNA concentration. Asterisks indicate the level of significance between huntingtin containing 15 and 128 glutamines: * P < 0.01; ** P < 0.001.
Discussion
The factors determining the subcellular localization of huntingtin, and the relationship between its localization and cellular viability, are fundamental to understanding the pathogenesis of HD and may provide new therapeutic targets for delaying or preventing toxicity in vivo. Therefore, we performed experiments designed to investigate factors that may be influencing the nuclear transport of huntingtin. In particular, we wished to dissect how the length of the huntingtin protein was correlated with nuclear localization of huntingtin and its tendency to form aggregates. The findings of this study clearly and directly implicate protein length as a critical factor in influencing the subcellular localization of huntingtin. Whereas nuclear localization is crucially dependent on protein length, the likelihood of formation of aggregates appears to be directly related to the length of the polyglutamine tract.
The fact that these aggregates form even with a huntingtin fragment essentially composed of only 17 amino acids besides the 128-residue polyglutamine stretch is strong evidence for these aggregates forming by expanded polyglutamine tracts joining these molecules together. This also supports the in vitro data generated by assessing interaction between glutathione-S-transferase fusion proteins with expanded polyglutamine tracts (Scherzinger et al., 1997).
Endogenous huntingtin is recruited into aggregates formed by a truncated form of the transfected protein (Martindale et al., 1998). Here we show, using HD−/− ES cells, that aggregates can form in the absence of full-length endogenous huntingtin, demonstrating that in this system, endogenous huntingtin is not necessary for formation of aggregates. Whereas extrapolation from this in vitro model using cell types not normally affected in HD to the in vivo situation must be done with caution, this may suggest that mutant huntingtin can form aggregates alone in vivo providing further evidence for a gain of function resulting from polyglutamine expansion.
Huntingtin derived from three constructs spanning up to 989 nucleotides (224 amino acids) were all seen partially or predominantly within the nucleus. These constructs would be expected to produce predicted proteins of <40 kD. For example, the 989 construct containing 128 repeats would encode a protein of ∼39 kD. By contrast, mutant proteins of 60 kD or greater (derived from the 1597, 1955, and 10366 constructs) were observed in the perinuclear region in the cytosol with no nuclear localization. Furthermore, the expression of the COOH-terminal construct expressing a predicted product of 284 kD also produced a protein exclusively located diffusely throughout the cytoplasm. No aggregates were seen with this construct.
Proteins that traverse from the cytoplasm into the nucleus must go through the nuclear envelope and enter through the nuclear pore. Nuclear pore complexes are composed of many different polypeptides and have a large mass of >100 MD. Transport across the nuclear pore may either be the result of passive diffusion or active transport (Zasloff, 1983; Newmeyer et al., 1986; Gorlich and Mattaj, 1996). Small proteins, generally <40 kD, can diffuse passively through the nuclear pore. For example, cytochrome c (13 kD) diffuses freely through the nuclear pores, whereas ovalbumin (43 kD) has a delayed transport, and BSA (66 kD) is completely prevented from passing through the nuclear pore (Gorlich and Mattaj, 1996). The relationship between size of huntingtin and its localization in the nucleus determined in this study is consistent with transport across the nuclear membrane being size related and resulting from passive diffusion across the nuclear membrane. Thus, a construct expressing a huntingtin product of ∼40 kD has some nuclear but predominantly perinuclear localization (construct 989), and huntingtin of predicted sizes of 60 kD or greater (derived from 1597, 1955, and 10366 constructs) are essentially excluded from the nucleus, similar to that seen with BSA (Gorlich and Mattaj, 1996).
However, size alone is not the only determinant of localization of the expressed product. For example, the gene product of the 771 construct was seen most frequently in the nucleus and seen more often than after expression of a smaller gene product from the 436 construct. This suggests that active transport may influence the localization of this protein.
Active import of proteins requires energy and also a nuclear localizing signal to which different cytosolic receptor complexes bind for transport (Zasloff, 1983; Newmeyer et al., 1986). We have searched the NH2-terminal domain of huntingtin for a sequence motif suggestive of a nuclear localization signal (NLS) (Goldfarb et al., 1986; Dingwall and Laskey, 1991; Chen et al., 1996). Although no stringent consensus sequence for an NLS is defined, a basic hexapeptide sequence and a bipartite sequence are suggestive of this motif (Efthymiadis et al., 1997). Interestingly, within the first 548 amino acids of huntingtin, a single stretch of amino acids (90-PKKELSATKK-99) with some similarity to the NLS of the familial breast cancer BRCA-1 protein (PKKNRLRRKS) has been identified (Chen et al., 1996). Further analysis of the role of this sequence as a functional NLS is necessary in an effort to determine its role in nuclear transport.
We have previously shown that the expression of mutant huntingtin results in increased susceptibility to apoptotic stress in transfected 293T cells (Martindale et al., 1998). This was initially shown in proteins derived from the 1955 compared with the full-length construct. In this study we have extended these findings and now show that constructs with 128 versus 15 repeats were more toxic to cells after apoptotic stress at all lengths of the protein. Furthermore, there is an increase in cell death with shorter constructs, again showing as postulated previously (Martindale et al., 1998), that length of the protein is directly related to the susceptibility to cell death. Interestingly, expression of the truncated form of huntingtin even with a normal sized polyglutamine tract confers increased susceptibility to cell death from apoptotic stimuli demonstrating that this NH2-terminal sequence alone can be toxic to cells in vitro.
This is not inconsistent with the model for this disease whereby cleavage of wild-type huntingtin normally plays a role in influencing cellular viability. Disease could arise when a threshold is reached and inappropriate apoptosis occurs at least influenced by the presence of a truncated product containing an expanded polyglutamine tract. The margin between the effects at any one time between normal and mutant huntingtin may be narrow but cumulative over time resulting eventually in a disease with late onset.
These findings in this manuscript do not explain why each of these polyglutamine expansion diseases result in a selective pattern of neuronal loss, despite the fact that their gene products are expressed in similar regions in the central nervous system (Ross, 1995). One factor that may influence selectivity of neuronal loss is proteolytic cleavage of polyglutamine-containing proteins. For example, our own data suggest that atrophin-1, the androgen receptor, and huntingtin are each cleaved by different caspases with varying efficiencies (Wellington et al., 1998) resulting in the formation of a truncated fragment containing the polyglutamine tract. Because the gene products have differential susceptibility to individual caspases, the particular pattern of cell loss for each disease could be due in part to the altered cellular or subcellular distribution of these particular enzymes, leading to altered interaction with their substrates.
These in vitro data, together with data generated by Martindale et al. (1998) and Cooper et al., (1998), are consistent with a model that proposes that a crucial step in the pathogenesis of this disorder is the progressive truncation of huntingtin resulting in altered localization, aggregate formation, and decreased cellular viability. This model is consistent with data generated from studies of brains from patients with HD which demonstrate both perinuclear accumulation of huntingtin and intranuclear inclusions (DiFiglia et al., 1997; Sapp et al., 1997) suggesting that this in vitro model may significantly recapitulate what occurs in vivo in animals and humans.
Acknowledgments
This work is supported by the Canadian Networks of Centres of Excellence (NCE, Genetics), Medical Research Council (MRC; Canada), and the Huntington Disease Society of America. A. Hackam is an MRC postdoctoral fellow. C.L. Wellington is an Alberta Heritage Foundation for Medical Research postdoctoral fellow. M.A. Kalchman is a MRC (Canada) predoctoral scholar. M.R. Hayden is an established investigator of the British Columbia Children's Hospital.
Abbreviations used in this paper
- ES
embryonic stem
- HD
Huntington disease
- HEK
human embryonic kidney
Footnotes
Address all correspondence to Dr. M.R. Hayden, Department of Medical Genetics, University of British Columbia, 416-2125 East Mall, NCE Bldg., Vancouver, BC V6T 1Z4. Tel.: (604) 822-9240. Fax: (604) 822-9238. E-mail: mrh@ulam.generes.ca
References
- Andrew SE, Goldberg YP, Hayden MR. Commentary: rethinking genotype and phenotype correlations in polyglutamine expansion disorders. Hum Mol Genet. 1997;6:2005–2010. doi: 10.1093/hmg/6.12.2005. [DOI] [PubMed] [Google Scholar]
- Becher, M.W., J.A. Kotzuk, A.H. Sharp, S.W. Davies, G.P. Bates, D.L. Price, and C.A. Ross. 1997. Intranuclear neuronal inclusions in Huntington's disease and dentatorubral and palidoluysian atrophy: correlation between the density of inclusions and IT15 CAG triplet repeat length. Neurobiol. Dis. In press. [DOI] [PubMed]
- Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]
- Chen CF, Li S, Chen Y, Chen PL, Sharp ZD, Lee WH. The nuclear localization sequences of the BRCA1 protein interact with the importin-α subunit of the nuclear transport signal receptor. J Biol Chem. 1996;271:32863–32868. doi: 10.1074/jbc.271.51.32863. [DOI] [PubMed] [Google Scholar]
- Cooper JK, Schilling G, Peters MF, Herring WJ, Sharp AH, Kaminsky Z, Masone J, Khan FA, Delanoy M, Borchelt DR, et al. Truncated N-terminal fragments of huntingtin with expanded glutamine repeats form nuclear and cytoplasmic aggregates in cell culture. Hum Mol Genet. 1998;7:783–790. doi: 10.1093/hmg/7.5.783. [DOI] [PubMed] [Google Scholar]
- Couldwell WT, Hinton DR, He S, Chen TC, Sebat I, Weiss MH, Law RE. Protein kinase C inhibitors induce apoptosis in human malignant glioma cell lines. FEBS (Fed Euro Biochem Soc) Lett. 1994;345:43–46. doi: 10.1016/0014-5793(94)00415-3. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Efthymiadis A, Shao H, Hubner S, Jans DA. Kinetic characterization of the human retinoblastoma protein bipartite nuclear localization sequence (NLS) in vivo and in vitro. A comparison with the SV40 large T-antigen NLS. J Biol Chem. 1997;272:22134–22139. doi: 10.1074/jbc.272.35.22134. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Kalchman MA, Metzler M, Nasir J, Zeisler J, Graham R, Koide HB, O'Kusky J, Sharp AH, Ross CA, et al. Absence of disease phenotype and intergenerational stability of the CAG repeat in transgenic mice expressing the human Huntington disease transcript. Hum Mol Genet. 1996;5:177–185. doi: 10.1093/hmg/5.2.177. [DOI] [PubMed] [Google Scholar]
- Goldfarb DS, Gariepy J, Schoolnik G, Kornberg RD. Synthetic peptides as nuclear localization signals. Nature. 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Smith DJ, McCutcheon K, Koide HB, Nishiyama K, Dinulos MB, Stevens ME, Bissada N, Nasir J, Kanazawa K, et al. Human huntingtin derived from YAC transgenes compensates for loss of murine huntingtin by rescue of the embryonic lethal phenotype. Hum Mol Genet. 1996;5:1875–1885. doi: 10.1093/hmg/5.12.1875. [DOI] [PubMed] [Google Scholar]
- Igarashi S, Koide R, Shimohata T, Yamada M, Hayashi Y, Takano H, Date H, Oyake M, Sato T, Sato A, et al. Suppression of aggregate formation and apoptosis by transglutaminase inhibitors in cells expressing truncated DRPLA protein with an expanded polyglutamine stretch. Nat Genet. 1998;18:111–117. doi: 10.1038/ng0298-111. [DOI] [PubMed] [Google Scholar]
- Kalchman MA, Koide HB, McCutcheon K, Graham RK, Nichol K, Nishiyama K, Lynn FC, Kazemi-Esfarjani P, Wellington CL, Metzler M, et al. HIP1, a human homolog of S. cerevisiae Sla2p, interacts with membrane-associated huntingtin in the brain. Nat Genet. 1997;16:44–53. doi: 10.1038/ng0597-44. [DOI] [PubMed] [Google Scholar]
- Martindale D, Hackam AS, Wieczorek A, Ellerby L, Wellington CL, McCutcheon K, Singaraja R, Kazemi-Esfarjani P, Devon R, Bredesen DE, et al. Length of the protein and polyglutamine tract influence localization and frequency of intracellular aggregates of huntingtin. Nat Genet. 1998;18:150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- Metzler M, Gertz A, Sarkar M, Schachter H, Schrader JW, Marth JD. Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO (Eur Mol Biol Organ) J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O'Kusky JR, Diewert VM, Richman JM, Zeisler J, Borowski A, Marth JD, Phillips AG, Hayden MR. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Lucocq JM, Burglin TR, De Robertis EM. Assembly in vitro of nuclei active in nuclear protein transport: ATP is required for nucleoplasmin accumulation. EMBO (Eur Mol Biol Organ) J. 1986;5:501–510. doi: 10.1002/j.1460-2075.1986.tb04239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Perez MK, Trottier Y, Trojanowski JQ, Subramony SH, Das SS, Vig P, Mandel J-L, Fischbeck KH, Pittman RN. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron. 1997;19:333–344. doi: 10.1016/s0896-6273(00)80943-5. [DOI] [PubMed] [Google Scholar]
- Ross CA. When more is less: pathogenesis of glutamine repeat neurodegenerative diseases. Neuron. 1995;15:493–496. doi: 10.1016/0896-6273(95)90138-8. [DOI] [PubMed] [Google Scholar]
- Sapp E, Schwarz C, Chase K, Bhide PG, Young AB, Penney J, Vonsattel JP, Aronin N, DiFiglia M. Huntingtin localization in brains of normal and Huntington's disease patients. Ann Neurol. 1997;42:604–612. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- Scherzinger E, Lurz R, Turmaine M, Mangiarini L, Hollenbach B, Hasenbank R, Bates GP, Davies SW, Lehrach H, Wanker EE. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 1997;90:549–558. doi: 10.1016/s0092-8674(00)80514-0. [DOI] [PubMed] [Google Scholar]
- Skinner PJ, Koshy BT, Cummings CJ, Klement IA, Helin K, Servadio A, Zoghbi HY, Orr HT. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- Vistica DT, Skehan P, Scudiero D, Monks A, Pittman A, Boyd MR. Tetrazolium-based assays for cellular viability: a critical examination of selected parameters affecting formazan production. Cancer Res. 1991;51:2515–2520. [PubMed] [Google Scholar]
- Wellington CL, Hayden MR. Of molecular interactions, mice and mechanisms: new insights into Huntington's disease. Curr Opin Neurol. 1997;10:291–298. doi: 10.1097/00019052-199708000-00003. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R, McCutcheon K, Salvesen GS, Propp SS, Bromm M, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273:9159–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- Zasloff M. tRNA transport from the nucleus in a eukaryotic cell: carrier-mediated translocation process. Proc Natl Acad Sci USA. 1983;80:6436–6440. doi: 10.1073/pnas.80.21.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]