Abstract
We have previously shown that Stu2p is a microtubule-binding protein and a component of the Saccharomyces cerevisiae spindle pole body (SPB). Here we report the identification of Spc72p, a protein that interacts with Stu2p. Stu2p and Spc72p associate in the two-hybrid system and can be coimmunoprecipitated from yeast extracts. Stu2p and Spc72p also interact with themselves, suggesting the possibility of a multimeric Stu2p-Spc72p complex. Spc72p is an essential component of the SPB and is able to associate with a preexisting SPB, indicating that there is a dynamic exchange between soluble and SPB forms of Spc72p. Unlike Stu2p, Spc72p does not bind microtubules in vitro, and was not observed to localize along microtubules in vivo. A temperature-sensitive spc72 mutation causes defects in SPB morphology. In addition, most spc72 mutant cells lack cytoplasmic microtubules; the few cytoplasmic microtubules that are observed are excessively long, and some of these are unattached to the SPB. spc72 cells are able to duplicate and separate their SPBs to form a bipolar spindle, but spindle elongation and chromosome segregation rarely occur. The chromosome segregation block does not arrest the cell cycle; instead, spc72 cells undergo cytokinesis, producing aploid cells and polyploid cells that contain multiple SPBs.
In eukaryotic cells, the number, polarity, and organization of cellular microtubules is controlled by the microtubule-organizing center. In the yeast Saccharomyces cerevisiae, the microtubule-organizing center is the spindle pole body (SPB)1 that resides in the nuclear envelope. Cryoelectron microscopy shows that the SPB is composed of six major layers (Bullitt et al., 1997). Although the SPB is an extremely large organelle, estimated to be ∼20 times the size of the yeast nuclear pore complex, each layer may be composed of only one or a few proteins that are present in high copy numbers (Bullitt et al., 1997). Consequently, the SPB may contain a total of ∼20–30 unique proteins. A number of SPB proteins have been identified. These include Cdc31p (Baum et al., 1986; Spang et al., 1993), Kar1p (Rose and Fink, 1987; Spang et al., 1995), Cmd1 (Geiser et al., 1993), Spc42p (Donaldson and Kilmartin, 1996), Spc97p (Knop et al., 1997), Spc98p (Geissler et al., 1996), Spc110p (Kilmartin et al., 1993), and Tub4p (Marschall et al., 1996; Sobel and Snyder, 1995; Spang et al., 1996). Tub4p is the yeast γ-tubulin. It forms a 6S complex with Spc97p and Spc98p (Knop et al., 1997) that has been localized to both the nucleoplasmic and cytoplasmic sides of the SPB (Geissler et al., 1996; Spang et al., 1996). Microtubule nucleation is thought to be accomplished through the action of this γ-tubulin complex (Marschall et al., 1996; Spang et al., 1996). Microtubules that extend from the nucleoplasmic face of the SPB form the mitotic spindle, while microtubules that extend from the cytoplasmic face of the SPB form the cytoplasmic microtubule array that serves to orient the spindle.
Recently, we described an additional component of the yeast SPB, Stu2p (Wang and Huffaker, 1997). Stu2p is unique among the known SPB components because it is a microtubule-binding protein that interacts laterally with microtubules. This property of Stu2p suggests that it may play a role in the attachment, organization, and/or dynamics of microtubules at the SPB. Although Stu2p binds microtubules, its presence at the SPB does not appear to require the presence of microtubules. Therefore, we concluded that Stu2p must also interact with at least one other component of the SPB. In this report, we describe Spc72p, a protein identified by its ability to interact with Stu2p. Like Stu2p, Spc72p is a component of the SPB, but unlike Stu2p, Spc72p is not a microtubule-binding protein. Spc72p is an essential protein, and we describe the effect of an spc72 temperature-sensitive mutation on microtubule assembly and function in vivo.
Materials and Methods
Strains and Media
Yeast strains used in this study are listed in Table I. Yeast extract/peptone/dextrose (YPD) and synthetic dextrose (SD) media were prepared as described by Sherman (1991). YPGal is YPD with 2% galactose substituted for 2% dextrose. Plates containing 5-fluoroorotic acid (5-FOA) were prepared as described by Boeke et al. (1984). To depolymerize microtubules, nocodazole was added to the culture at 10 μg/ml for 1 h at 26°C, and then for 1 h at 4°C. To arrest cells in the cell cycle, 3 μg/ml α-factor or 0.1 M hydroxyurea were added to the culture for 3 h at 26°C.
Table I.
Yeast Strains
| Strain | Genotype | Source | ||
|---|---|---|---|---|
| CUY26 | MATα his3-Δ200 leu2-3, 112 ura3-52 | This study | ||
| CUY546 | MATa/MATα ade2/ADE2 his3-Δ200/his3-Δ200 | This study | ||
| leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 | ||||
| CUY1109 | MATa/MATα spc72-Δ1::URA3/SPC72 ade2/ADE2 | This study | ||
| his3-Δ200/his3-Δ200 leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 | ||||
| CUY1110 | MATa/MATα spc72-Δ1::HIS3/SPC72 ade2/ADE2 | This study | ||
| his3-Δ200/his3-Δ200 leu2-3, 112/leu2-3, 112 ura3-52/ura3-52 | ||||
| CUY1111 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 (pXC255, SPC72 URA3) | This study | ||
| CUY1112 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 (pXC214, SPC72 LEU2) | This study | ||
| CUY1113 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 (pXC226, SPC72-GFP LEU2) | This study | ||
| CUY1114 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 (pXC224, SPC72-GFP LEU2) | This study | ||
| CUY1115 | MATa URA3::Pgal1-SPC72-GFP::ura3-52 his3-Δ200 leu2-3, 112 | This study | ||
| CUY1116 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 (pXC279, spc72-2 LEU2) | This study | ||
| CUY1123 | MATa spc72-2::HIS3 leu2-3, 112 his3-Δ200 ura3-52 | This study | ||
| CUY1124 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 (pXC264, SPC72-myc URA3) | This study | ||
| CUY1125 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 | This study | ||
| (pXC264, SPC72-myc URA3; pXC265, SPC72-HA LEU2) | ||||
| CUY1128 | MATa spc72-Δ1::HIS3 leu2-3, 112 his3-Δ200 ura3-52 (pXC265, SPC72-HA LEU2) | This study | ||
| CUY1130 | MATa leu2-3, 112 his3-Δ200 ura3-52 | This study | ||
| CUY1137 | MATα spc72-2::HIS3 leu2-3, 112 his3-Δ200 ura3-52 | This study | ||
| Y190 | MATa ade2-101 cyhR gal4 gal80 his3 leu2-3, 112 trp1-901 | S. Elledge (Baylor | ||
| ura3-52 URA3::GAL(UAS)-lacZ LYS2::GAL(UAS)-HIS3 | College of Medicine) |
Two-hybrid Screen
The two-hybrid screen was performed as described by Durfee et al. (1993). The pAS2 vector and an S. cerevisiae cDNA library fused to the Gal4p activation domain were obtained from S. Elledge (Baylor College of Medicine, Houston, TX). The bait plasmid, encoding the entire Stu2p fused to the COOH-terminal end of the Gal4p DNA binding domain, was made as follows: STU2 was amplified from yeast genomic DNA by PCR using primers that contain XbaI and SalI sites, respectively. The 2688-bp PCR product was digested with XbaI and SalI, and was ligated into the NheI and SalI sites of pAS2 (Durfee et al., 1993), resulting in plasmid pXC180.
pXC180 and the cDNA library were cotransformed into the host strain Y190. Transformants were selected for growth on SD plates containing 25 mM 3-amino-triazole. A transformation efficiency test indicated that ∼106 Trp+ Leu+ transformants were obtained in this experiment. 63 colonies appeared on the 3-amino-triazole plates. 50 of these colonies showed varying levels of β-galactosidase activity by the filter assay. These 50 colonies were struck on plates containing 10 μg/ml cycloheximide to select cells that had lost the pXC180 plasmid; 42 of these still showed β-galactosidase activity, and were not pursued further. Plasmids were isolated from the remaining 10 strains that had lost pXC180. Each of the 10 plasmids was then introduced into Y190, together with pAS1-SNF1. None of the proteins encoded by the 10 cDNA clones interacted with Snf1p. Partial sequencing was performed to identify genes corresponding to these cDNA clones.
SPC72 Constructs
Selected plasmids used in this study are listed in Table II. A genomic DNA fragment containing SPC72 was amplified from yeast chromosomal DNA by PCR using primers that introduced a BamHI and NheI site at each end, respectively, and was TA-cloned into the pCRII vector (Invitrogen Corp., Carlsbad, CA). The resulting plasmid pXC204 was cut with BamHI and NheI, and the 3.6-kb fragment was cloned into pXC157 to create pXC214. pXC157 is pRS415 (Sikorski and Hieter, 1989) with the SstI site destroyed. A 3.5-kb KpnI-SstII fragment of pXC214 was subcloned into the KpnI and SstII sites of pRS416 to create pXC255.
Table II.
Plasmids
| Plasmid | Class | Relevant markers | Source | |||
|---|---|---|---|---|---|---|
| pACT2 | YEp | Padh-GAL4ad-HA LEU2 | S. Elledge (Baylor College of Medicine) | |||
| pAS2 | YEp | Padh-GAL4bd-HA TRP1 CYH2 | S. Elledge (Baylor College of Medicine) | |||
| pAS1-SNF1 | YEp | Padh-GAL4bd-SNF1 TRP1 | S. Elledge (Baylor College of Medicine) | |||
| pXC180 | YEp | Padh-GAL4bd-STU2 TRP1 CYH2 | This study | |||
| pXC204 | pCRII | SPC72 | This study | |||
| pXC214 | YCp | SPC72 LEU2 | This study | |||
| pXC219 | YEp | Padh-GAL4ad-SPC72 LEU2 | This study | |||
| pXC224 | YCp | SPC72-GFP LEU2 | This study | |||
| pXC226 | YEp | SPC72-GFP LEU2 | This study | |||
| pXC231 | YCp | Pgal1-SPC72 URA3 | This study | |||
| pXC248 | YCp | SPC72-myc LEU2 | This study | |||
| pXC254 | YCp | Pgal1-SPC72 LEU2 | This study | |||
| pXC255 | YCp | SPC72 URA3 | This study | |||
| pXC262 | YEp | SPC72-myc LEU2 | This study | |||
| pXC264 | YEp | SPC72-myc URA3 | This study | |||
| pXC265 | YCp | SPC72-HA LEU2 | This study | |||
| pXC266 | YCp | Pgal1-SPC72-GFP LEU2 | This study | |||
| pXC279 | YCp | spc72-2 LEU2 | This study | |||
| pXC281 | YEp | Padh-GAL4bd-SPC72 TRP1 CYH2 | This study |
SPC72 was also placed under the control of GAL1 promoter. A 0.7-kb EcoRI-BamHI fragment containing GAL1 promoter (Liu, et al., 1992) was cloned into the BamHI site of pRS416 (Sikorski and Hieter, 1989) to create pWP97. The entire SPC72 open reading frame was amplified by PCR and subcloned into pCRII to make pXC215. A 2.0-kb BamHI–SpeI fragment of pXC215 was cloned into the BamHI and XbaI sites of pWP97 to create pXC231. To switch the URA3 marker on pXC231 to LEU2, the 2.8-kb XhoI-SstII fragment of pXC231 was ligated into the XhoI and SstII sites of pRS415 to make pXC254.
SPC72 constructs for use in the two-hybrid screen were created as follows: The SPC72 open reading frame was amplified from yeast chromosomal DNA by PCR using primers that introduced a BamHI and XhoI at the 5′ and 3′ ends, respectively. The 2-kb BamHI-XhoI fragment was cloned into the same sites in pACT2 and pAS2 to create pXC219 and pXC281, respectively.
A SPC72-GFP fusion was created as follows: PCR was used to amplify SPC72 from genomic DNA. One of the primers was designed so that the resulting PCR fragment had a BglII site inserted immediately before the SPC72 stop codon. The PCR product was subcloned into pXC214 (see above) to create pXC223. A BamHI–BamHI fragment containing the S65T version of green fluorescent protein (GFP; Heim et al., 1995) was subcloned into the BglII site of pXC223 to create pXC224 containing the SPC72-GFP fusion construct. pXC224 was transformed into CUY1111. Leu+ transformants were plated on 5-FOA to select cells that had lost the URA3-based plasmid. pXC224 was able to complement the SPC72 disruption. To place the SPC72-GFP construct in a YEp plasmid, a 5.1-kb XhoI– SstII fragment from pXC224 was subcloned into the XhoI/SstII sites of pRS425 (Christianson et al., 1992) to make pXC226. To place SPC72-GFP under the control of GAL1 promoter, the 3.5-kb PstI–SstII fragment from pXC224 was then subcloned into the same sites on pXC254 (see above) to create pXC266. The Pgal1-SPC72-GFP was placed into an integrating vector by cloning the HindIII-SstII fragment from pXC266 into the same sites in pRS406 (Sikorski and Hieter, 1989) to create pXC287. pXC287 was linearized by cutting with StuI, which cuts within the URA3 gene, and was integrated into the URA3 chromosomal locus to create CUY1115.
Epitope-tagged versions of Spc72p were constructed as follows: to make SPC72-HA, a DNA fragment encoding three tandem copies of the hemagglutinin (HA) epitope was amplified by PCR from the plasmid GTEPI (B. Futcher, Cold Spring Harbor Laboratory). The PCR product has a BglII site at each end. It was digested with BglII and ligated into the BglII site on pXC223 to create the YCp plasmid pXC265. To construct SPC72-myc, a DNA fragment encoding two copies of the myc epitope was amplified by PCR from the plasmid pB303 (D. Pellman, Harvard Medical School, Boston, MA). The PCR product contained a BglII site at each end, and was subcloned into the pCRII vector to make pXC242. The BglII fragment containing myc2 was ligated into the BglII site of pXC223 to produce the YCp plasmid pXC248. SPC72-myc was also placed into YEp plasmids. The 2.5-kb XhoI–NotI fragment of pXC248 was subcloned into the XhoI–NotI sites of pRS425 and pRS426 to make pXC262 and pXC264, respectively.
Disruption of SPC72
To disrupt the SPC72 gene, two 69-mers were used to amplify the URA3 gene from pRS416 (Sikorski and Hieter, 1989). The resulting DNA fragment contained the entire URA3 gene and 50 bp of sequence immediately upstream of the SPC72 start codon and 50 bp of sequence immediately downstream of the SPC72 stop codon. This fragment was transformed into a diploid strain CUY546. A Ura+ transformant, CUY1109, containing the correct spc72-Δ1::URA3 deletion was identified by PCR analysis. To convert the URA3 marker at the SPC72 locus to the HIS3 marker, a 3-kb EcoRI fragment from the plasmid B2388 (from G. Fink, Massachusetts Institute of Technology, Cambridge, MA) was transformed into CUY1109. One of these His+ transformants, CUY1110, contains the spc72-Δ1::HIS3 deletion.
Immunoprecipitation Experiments
Mouse monoclonal antibodies 12CA5 and 9E10, which recognize the HA and myc epitopes, respectively, were purchased from Berkeley Antibody Co. (Berkeley, CA). Rabbit anti-Stu2p polyclonal antibody was generated against the 385 carboxy-terminal amino acids of Stu2p. Yeast extracts were prepared essentially as described in Sorger et al. (1994). These extracts were first diluted into PBS (4.3 mM Na2HPO4, 1.4 mM KH2PO4) containing 1% Tween 20. The salt concentration was then adjusted to 50 mM with NaCl. The diluted extracts were incubated with appropriate antibodies at 4°C for 1 h in the presence of protease inhibitors (10 μg/ml leupeptin, 10 μg/ml pepstatin, and 1 mM phenylmethylsulfonylfluoride). The reactions were then spun at 16,000 g for 10 min, and the supernatants were incubated with 10 mg/ml protein G–conjugated agarose beads (Sigma Chemical Co., St. Louis, MO) for 1 h at 4°C. Each reaction was then washed four times with PBS containing 50 mM NaCl and 1% Tween 20. The beads were resuspended in SDS sample buffer, incubated at 100°C for 10 min, and analyzed by SDS-PAGE (Laemmli, 1970) and immunoblotting.
Generation of SPC72 Mutants
SPC72 was amplified by PCR under mutagenic conditions (Muhlrad et al., 1992). A 100-μl PCR reaction contained 40 ng pXC204 template DNA, 7 mM MgCl2, 200 μM dATP, 1 mM dTTP, dGTP, and dCTP, 0.5 μM MnCl2, and 5 U of Taq DNA polymerase. The cycling parameters were as follows: 1 min at 94°C, 1 min at 57°C, and 3 min at 72°C. 30 cycles were performed. PCR products were purified, digested with SstI and BglII, and ligated into pXC214 digested with the same two enzymes so that the 1.4-kb mutagenized SPC72 fragment replaced the wild-type SPC72 fragment. 9,000 bacterial transformants were pooled, 50% of which had the expected insert size based on restriction digests of 10 random clones. To screen for conditionally lethal mutants, the library was transformed into the haploid strain CUY1111. Transformants were replica-plated onto 5-FOA plates at 26°C to select colonies that had lost the URA3 plasmid. The 5-FOA–resistant colonies were replica-plated onto three YPD plates that were then incubated at 14°C, 26°C, and 37°C, respectively. 24 heat-sensitive mutants were identified; no cold-sensitive mutants were found.
The spc72-2 allele was integrated into the yeast chromosome as follows: the plasmid containing spc72-2 was recovered from yeast and called pXC279; a 1.7-kb BamHI–BamHI fragment containing the HIS3 gene was cloned into the BgII site of pXC279, which resides 80 bp downstream of the spc72-2 stop codon; and the resulting plasmid was digested with BamHI and ApaLI to release a 5.2-kb fragment containing spc72-2 and HIS3. This fragment was transformed into diploid strain CUY1109. His+ and Ura− transformants were sporulated and dissected. Tetrads with four viable spores contained two Ts+ and two Ts− progeny. The Ts− spores were always His+, and harbored the spc72-2 allele.
Fluorescence and Electron Microscopy
GFP was visualized in living cells or in cells fixed with formaldehyde as described below. Microtubules and SPBs were visualized by immunofluorescence using β- and γ-tubulin antibodies, respectively, as previously described (Pasqualone and Huffaker, 1994). Cells were fixed for 2 h to visualize microtubules alone. They were fixed for 25 min to visualize both Spc72-GFP and microtubules; the green fluorescence of Spc72p-GFP was sensitive to fixation, but regained fluorescence with reduced intensity after removing formaldehyde. Cells were also fixed for 25 min to visualize SPBs. Rabbit anti-yeast γ-tubulin (Tub4p) antibody was a gift from T. Stearns (Stanford University, Stanford, CA), and was preabsorbed with wild-type fixed yeast spheroplasts overnight at 4°C before use. Rabbit anti-yeast β-tubulin (Tub2p) antibody was a gift from F. Solomon (Massachusetts Institute of Technology, Cambridge, MA). Rhodamine-conjugated goat anti–rabbit and fluorescein-conjugated goat anti–rabbit secondary antibodies were purchased from Organon Teknika-Cappel (Durham, NC). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Electron microscopy was performed as described by Byers and Goetsch (1991).
Results
Two-hybrid Screen Identifies Spc72p as a Protein That Interacts with Stu2p
A yeast two-hybrid screen was carried out to identify proteins that interact with Stu2p. Full-length Stu2p fused to the COOH-terminal end of the Gal4p DNA-binding domain served as the bait. We identified 10 cDNA clones encoding proteins that interact with Stu2p, but not with the unrelated protein, Snf1p. Partial sequencing of each cDNA insert showed that these 10 clones were derived from 6 genes. Two different cDNA clones were from the STU2 gene itself. One clone was from TUB1, and one was from TUB3, the two yeast genes encoding α-tubulins (Schatz et al., 1986). Three different clones were from BIK1; Bik1p has been previously implicated in microtubule function (Berlin et al., 1990; Pellman et al., 1995). Two clones were derived from an open reading frame on chromosome V, YER016w. This open reading frame has recently been identified as encoding a protein that interacts with Tub1p in the two-hybrid screen, and is called BIM1 (Schwartz et al., 1997). The last clone was from an uncharacterized open reading frame on chromosome I, YAL047c, which we call SPC72 for reasons given below.
SPC72 encodes a 622–amino acid protein with a predicted molecular weight of 72 kD. It does not have significant sequence similarity to any proteins in the database. Spc72p is predicted to have an α-helical secondary structure throughout much of its length, and has five predicted coiled-coil regions (Lupas et al., 1991) that together make up 45% of the protein. The cDNA clone obtained from the library encodes residues 110-484 of Spc72p, indicating that this sequence contains the region of Spc72 necessary for its interaction with Stu2p.
SPC72 is Essential for Growth
To determine whether SPC72 is essential for cell viability, we replaced one copy of SPC72 in a diploid strain with the HIS3 gene (See Materials and Methods). The His+ diploid was sporulated, and 30 tetrads were dissected. Most of the tetrads contained two His− spores that were able to form colonies, and two spores that were unable to form colonies after 3 d at 26°C. The two spores that did not form colonies presumably contained the spc72-Δ1::HIS3 disruption. After 5 d at 26°C, ∼30% of these spc72-Δ1::HIS3 spores grew enough to form very small yet visible colonies; the other 70% of spores colonies contained <20 cells. When the small colonies were picked and inoculated into liquid YPD, growth was imperceptible.
A plasmid shuffle experiment (Sikorski and Boeke, 1991) also established that SPC72 is essential for cell growth. We constructed a strain (CUY1111) that has a chromosomal disruption of SPC72 and SPC72 on a URA3-based plasmid. This strain could not grow on plates containing 5-FOA, a drug that is lethal to cells expressing the URA3 gene. In contrast, 5-FOA–resistant colonies were obtained if an additional copy of SPC72 was introduced on a LEU2-based plasmid. Taken together, these data indicate that Spc72p is essential for mitotic growth.
Spc72p Interacts with Stu2p In Vivo
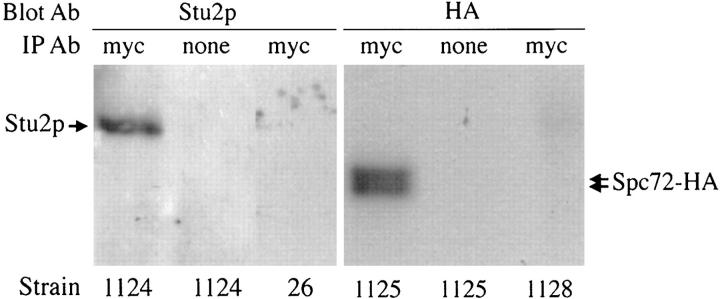
Because Spc72p was identified as a protein that interacts with Stu2p in the two-hybrid system, we investigated whether Spc72p and Stu2p normally interact in yeast cells. We constructed a fusion between SPC72 and the sequence encoding the myc epitope tag. This construct is able to complement a SPC72 deletion, indicating that it is functional. Extracts were made from a yeast strain expressing Spc72-myc, and Spc72-myc was immunoprecipitated using anti-myc antibodies. The precipitated material was separated by SDS-PAGE, and immunoblotting was performed using anti-Stu2p antibodies. As shown in Fig. 1 (lane 1), Stu2p coprecipitates with Spc72-myc. As controls, the immunoprecipitation was performed in the absence of anti-myc antibodies (Fig. 1, lane 2) or using cell extracts lacking Spc72-myc (Fig. 1, lane 3). In either case, no Stu2p was precipitated.
Figure 1.
Spc72p coimmunoprecipitates with Stu2p and itself. Protein extracts from the yeast strains indicated were immunoprecipitated (IP) with anti-myc antibody or were mock-immunoprecipitated without antibody. Immunoprecipitates were subjected to SDS-PAGE, and the gels were immunoblotted with anti-Stu2p or anti-HA antibodies as indicated. CUY1124 contains Spc72-myc, CUY26 contains no epitope-tagged constructs, CUY1125 contains Spc72-myc and Spc72-HA, and CUY1128 contains Spc72-HA.
Spc72p Interacts with Itself In Vivo
We found that Spc72p interacts with itself in the two- hybrid system. Y190 cells containing two plasmids that express Gal4ad-Spc72p and Gal4bd-Spc72p, respectively, grow on 3-amino-triazole plates and produce β-galactosidase activity. To test whether Spc72p normally interacts with itself in vivo, a coimmunoprecipitation experiment was performed. We constructed a yeast strain that expresses two epitope-tagged versions of Spc72p: the Spc72-myc protein mentioned above and Spc72-HA that is also able to substitute functionally for Spc72p in vivo. Spc72-myc was immunoprecipitated from extracts of this strain using the anti-myc antibody, and the immunoprecipitated material was analyzed for the presence of Spc72-HA by immunoblotting with anti-HA antibodies. As shown in Fig. 1 (lane 4), Spc72-HA coimmunoprecipitated with Spc72-myc. Spc72-HA, like Spc72-myc (not shown), migrated as a doublet.
Spc72p Is a Component of the SPB
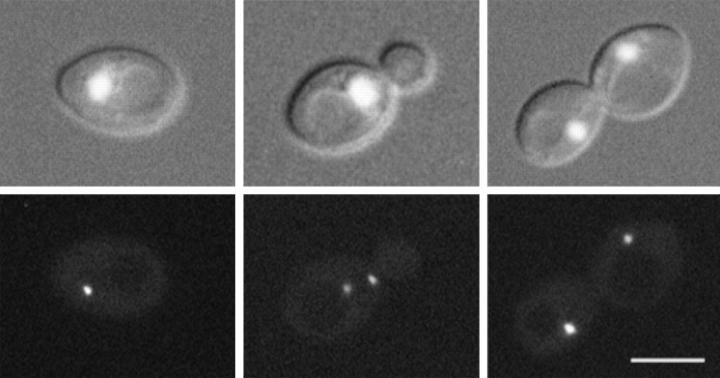
A fusion between SPC72 and the coding region of GFP was constructed to visualize Spc72p in living cells. This SPC72-GFP construct, carried on either a low-copy YCp plasmid or a high-copy YEp plasmid, was able to complement the spc72-Δ1::HIS3 disruption. Spc72-GFP was visualized in spc72-Δ1::HIS3 cells containing these plasmids. The fluorescence intensity was generally stronger when the YEp plasmid was used, but the pattern of staining was similar in cells containing either plasmid (Fig. 2). In living cells, one bright fluorescent dot was observed in unbudded cells, and two dots were generally observed in budded cells. In small-budded cells, the two dots were either adjacent to or separated by ∼1 μm. In large budded cells, the two fluorescent dots were often segregated into each cell body. The intensity of staining did not appear to change over the course of the cell cycle. When cells were grown in the presence of DAPI, we observed that the Spc72-GFP dots resided at the periphery of the nuclear DNA (Fig. 2).
Figure 2.
Localization of Spc72-GFP. GFP fluorescence was visualized in living yeast cells that contain a disruption of the chromosomal SPC72 gene and carry SPC72-GFP on a plasmid (CUY1113). To visualize chromosomal DNA, cells were grown in the presence of 1 μg/ml DAPI for 2 h. (Top) Both DNA staining and differential interference contrast (DIC) image of cells; (bottom) fluorescence of Spc72-GFP. Bar, 5 μm.
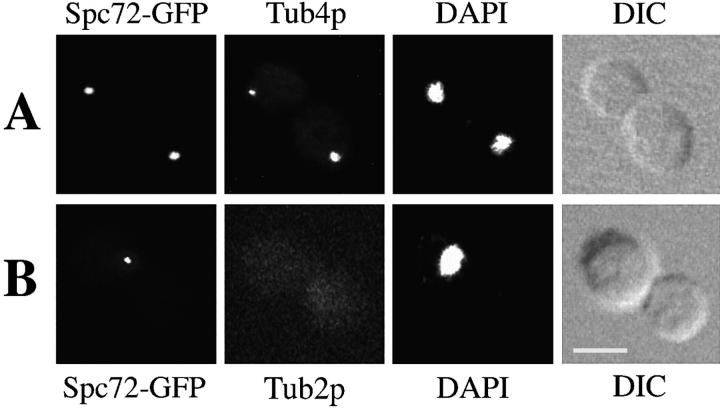
This localization pattern suggested that Spc72p was likely a component of the SPB. To determine directly whether Spc72p localizes to the SPB, we compared Spc72-GFP localization with that of yeast γ-tubulin, Tub4p. Tub4p could be visualized by immunofluorescence after fixation of cells with formaldehyde, but this treatment reduced the Spc72-GFP signal. For this reason, we used another strain in which Spc72-GFP was produced at higher levels from the GAL1 promoter. In >95% of the cells, the Spc72-GFP dots were coincident with the dots stained by anti-Tub4p antibodies (Fig. 3 A). Overexpression of Spc72-GFP did not have a deleterious effect on cell growth, and microtubules appeared normal by immunofluorescence microscopy (not shown).
Figure 3.
Spc72p localization to the SPBs is independent of microtubules. Yeast strain CUY1115, expressing SPC72-GFP under control of the GAL1 promoter, was grown in YPGal at 26°C. (A) Cells were fixed for immunofluorescence and stained with anti-Tub4p antibodies. (B) Cells were treated with nocodazole to depolymerize microtubules, and were stained with anti-β-tubulin antibody. DAPI staining and DIC images are also shown. Bar, 5 μm.
We also tested whether newly synthesized Spc72p is able to associate with an existing SPB. A SPC72 strain containing a plasmid that expresses SPC72-GFP from the GAL1 promoter was grown in glucose-containing medium to repress synthesis of Spc72-GFP. These cells were arrested with α-factor, and then the synthesis of the Spc72p-GFP was induced by galactose in the continued presence of α-factor. Cells arrested with α-factor possess a single SPB (Byers and Goetsch, 1975), so any fluorescence observed at the SPB must be due to association of new Spc72p-GFP with that SPB. After 5 h of induction, Spc72p-GFP was observed at the SPB (data not shown). A similar time course of Spc72p-GFP incorporation into the SPB was observed for mitotically growing cells. Thus, Spc72p can be incorporated into a preexisting SPB.
To determine whether the association of Spc72p with the SPB depends on the presence of microtubules, cells expressing Spc72-GFP were treated with a microtubule- depolymerizing drug nocodazole. After this treatment, microtubules were no longer observed by immunofluorescence (Fig. 3 B). Nocodazole treatment did not affect the number of cells containing the Spc72p-GFP signal, or noticeably change the intensity of staining compared with untreated cells. Thus, the association of Spc72p with the SPB is independent of the presence of detectable microtubules. Although we cannot rule out the possibility that nocodazole-treated cells may contain short microtubules at the SPB that cannot be observed by immunofluorescence, this result suggests that Spc72p is an integral component of the SPB.
Spc72p Does Not Bind Microtubules In Vitro
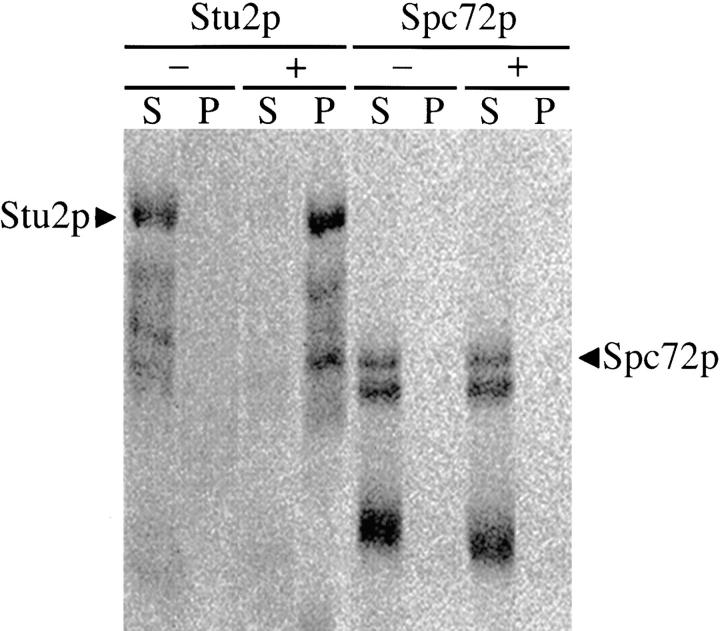
We have previously shown that Stu2p is able to bind microtubules in vitro (Wang and Huffaker, 1997). A similar assay was performed to determine whether Spc72p is able to interact with microtubules as well. We included Stu2p as a positive control for microtubule binding. 35S-labeled Spc72p and Stu2p were synthesized by in vitro transcription and translation, and were incubated separately with 5 μM taxol-stabilized bovine brain microtubules. The microtubules were then pelleted by centrifugation, and the pellet and supernatant fractions were analyzed by SDS-PAGE. Because in vitro transcription and translation often produces truncated proteins, only the upper bands in each lane corresponding to the full-length proteins were quantitated. As expected, most of the Stu2p (95%) pelleted with the microtubules (Fig. 4). We previously showed that the K d for Stu2p binding to microtubules is ∼0.5 μM, so 5 μM tubulin is approximately ten times the concentration of tubulin that is required to cosediment 50% of the Stu2p (Wang and Huffaker, 1997). In contrast, no detectable Spc72p was pelleted with this amount of microtubules.
Figure 4.
Spc72p does not bind microtubules in vitro. 35S-labeled in vitro–translated Stu2p and Spc72p were incubated without microtubules (−) and with 5 μM taxol-stabilized bovine brain microtubules (+). Microtubules were then pelleted by centrifugation, and pellet (P) and supernatant (S) fractions were subjected to SDS-PAGE analysis. Bands were visualized and quantitated using a phosphoimager.
spc72 Mutant Cells are Defective in Nuclear Migration and Division
To determine the role of Spc72p in vivo, we created temperature-sensitive spc72 mutants. The SPC72 gene was mutagenized by error-prone PCR in vitro, and temperature-sensitive mutants were identified by the strategy described in Materials and Methods. The spc72-2 mutant displayed the best conditional growth properties, and was chosen for the phenotypic analysis. spc72-2 cells were grown at their permissive temperature (26°C), and were then shifted to the restrictive temperature (37°C). After 3 h at 37°C, the spc72-2 culture showed a modest increase in the fraction of cells with large buds. More than 40% of the spc72-2 cells were large-budded, compared with ∼20% in the SPC72 population. DAPI staining of chromosomal DNA indicated that this inhibition of cell cycle progression is due to a defect in chromosome segregation (Table III). More than 70% of spc72-2 large-budded cells contained unsegregated chromosomes. In contrast, only ∼30% of the SPC72 cells had failed to segregate chromosomes by the time they had produced large-buds.
Table III.
Cell and Nuclear Morphology of spc72-2 Cells
| Temp | Time |

|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| °C | h | ||||||||||||||||||||||
| SPC72 | 26 | 41 | 0 | 0 | 33 | 0 | 0 | 10 | 16 | 0 | |||||||||||||
| SPC72 | 37 | 3 | 56 | 0 | 0 | 28 | 0 | 0 | 5 | 11 | 0 | ||||||||||||
| spc72-2 | 26 | 35 | 3 | 2 | 28 | 1 | 7 | 5 | 15 | 4 | |||||||||||||
| spc72-2 | 37 | 3 | 21 | 12 | 1 | 21 | 2 | 30 | 1 | 3 | 9 | ||||||||||||
| spc72-2 | 37 | 6 | 14 | 28 | 1 | 13 | 2 | 31 | 4 | 2 | 5 | ||||||||||||
SPC72 (CUY26) and spc72-2 (CUY1123) cells were grown at 26°C, shifted to 37°C for the times indicated, and stained with DAPI. Table shows the percentages of cells with a given cellular morphology (unbudded, small-budded, or large-budded) and chromosomal DNA distribution. For each time point, >100 cells were counted.
Although the nuclear division defect in spc72-2 cells does slow the progression of cells through the cell cycle, it does not produce a cell cycle block. At 37°C, we observed a significant fraction of unbudded cells that lacked nuclear DNA (Table III; see Fig. 6). These cells were presumably generated by cytokinesis of large-budded cells that had failed to segregate chromosomes properly into their bud. As expected, the percentage of anucleate unbudded cells increased with time at 37°C. In addition, flow cytometry showed that spc72-2 cells can undergo a second round of DNA replication in the absence of chromosome segregation (Fig. 5). At 26°C, the spc72-2 culture contained cells with a typical 1C (pre-DNA replication) and 2C (post-DNA replication) DNA content. A small percentage of the cells containing a 4C DNA content were also present, indicating a partial defect in chromosome segregation, even at permissive temperature. This 4C peak became more prominent with increasing time at restrictive temperature, and by 6 h represented about half of the population.
Figure 6.
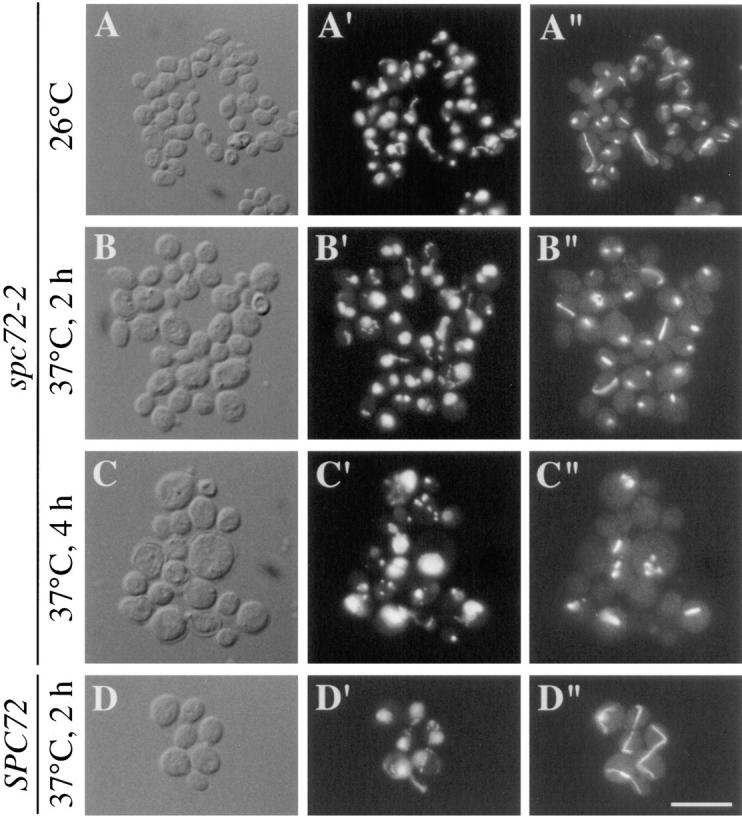
Phenotype of spc72-2 cells. spc72-2 (CUY1123) and SPC72 (CUY26) cells were grown at 26°C, and were then shifted to 37°C for the times indicated. (A) spc72-2 cells at 26°C; (B) spc72-2 cells after 2 h at 37°C; (C) spc72-2 cells after 4 h at 37° C; (D) SPC72 cells after 2 h at 37°C. (A–D) DIC; (A′–D′), DAPI staining; (A′′–D′′), immunofluorescence with Tub2p antibody. Bar, 10 μm.
Figure 5.
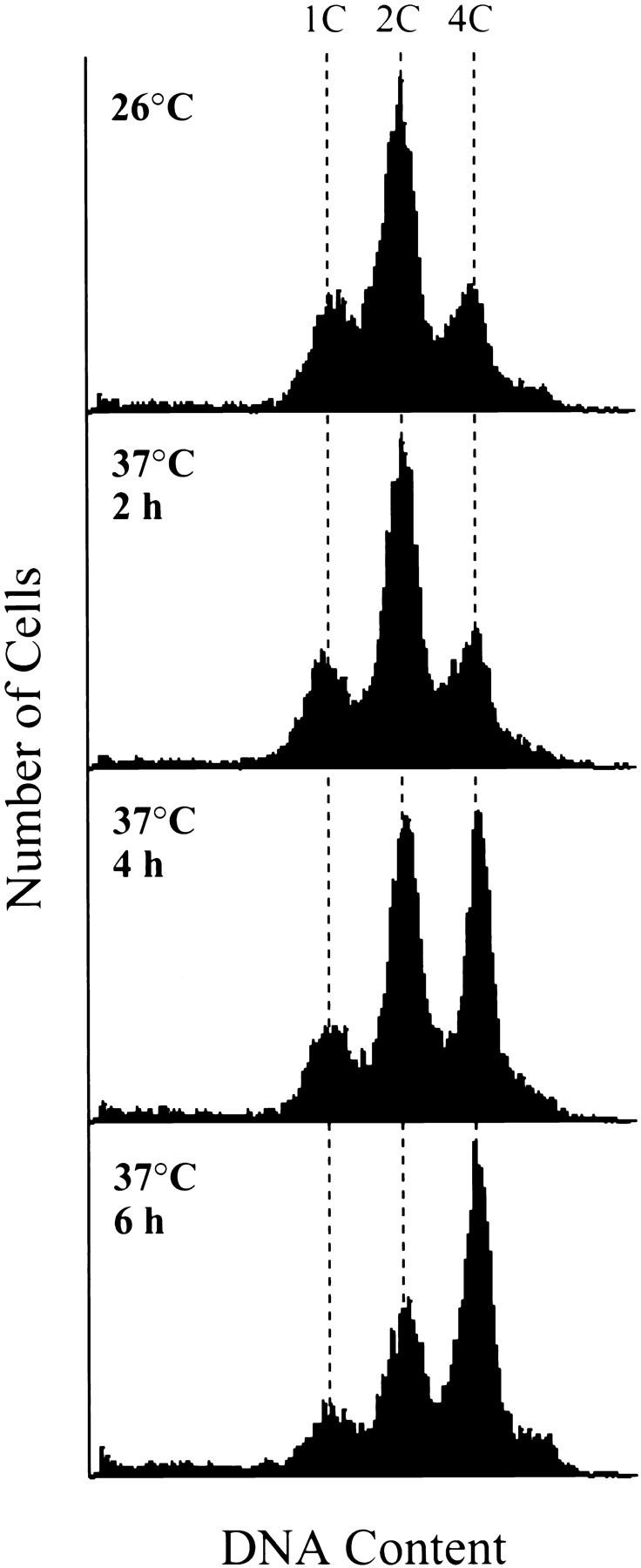
DNA content of spc72-2 cells. CUY1123 cells were grown to logarithmic phase at 26°C, and were then shifted to 37°C. Aliquots of cells were taken 2, 4, and 6 h after the temperature shift. Flow cytometry was performed as described by Hutter and Eipel (1979). The positions of cells with 1C and 2C DNA content were determined by flow cytometry of SPC72 cells (CUY26); the position of cells with a 4C DNA content was extrapolated from this data.
spc72-2 cells also showed defects in nuclear migration at the restrictive temperature (Table III). Before nuclear division, chromosomes in large-budded SPC72 cells were invariably located near the bud neck. In contrast, in nearly all of the spc72-2 cells that were large-budded and contained an undivided nucleus, the chromosomes were located away from the bud neck. In addition, 75% of the spc72-2 cells that did complete chromosome separation contained two nuclei in a single cell body. This latter phenotype also indicates a defect in nuclear migration before mitosis.
To determine whether Spc72p is necessary to maintain the nucleus at the bud neck, we treated spc72-2 cells with hydroxyurea at 26°C. This treatment causes cells to arrest with large buds and a single undivided nucleus. In the spc72-2 population, 85% of these cells contained a nucleus positioned at the bud neck. These cells were then shifted to 37°C for 30 min in the continued presence of hydroxyurea. After this temperature shift, only 40% of the large-budded cells still contained their nuclei at the bud neck. Thus, Spc72p plays a role in maintaining the nucleus at the bud neck.
spc72 Mutant Cells Display Defects in Microtubule Assembly
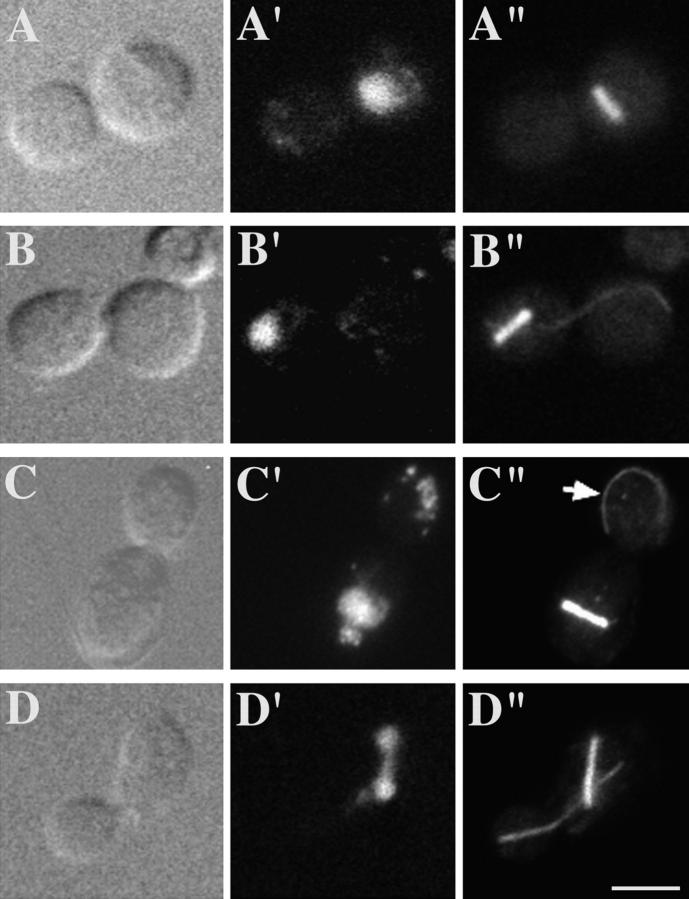
The defects in nuclear migration and division observed in spc72-2 cells suggest that this mutation affects microtubule assembly. The microtubule cytoskeleton in spc72-2 cells was examined by immunofluorescence microscopy. The most dramatic effect of the spc72-2 mutation was on the assembly of cytoplasmic microtubules. After 2 h at 37°, most spc72-2 cells contained only a dot of staining near the periphery of the chromosomal DNA or a bar of staining that stretched across the chromosomal DNA region (Fig. 6 B). The dots presumably indicate the presence of short microtubules at the SPBs, and the bars represent spindle microtubules. Unlike SPC72 cells (Fig. 6 D), spc72-2 cells rarely contained microtubule fibers that extended from the SPBs in the cytoplasm.
The spc72-2 mutation did not block the formation of a short mitotic spindle. After 2 h at 37°C, >75% of the spc72-2 large-budded cells had a mitotic spindle coincident with the chromosomal DNA (Fig. 6 B and Fig. 7). The majority of these contained two separated SPBs, as visualized with γ-tubulin antibody (data not shown), indicating that the spindles observed were in fact bipolar. Most of these spindles (∼75%) were the same length as normal short preanaphase spindles (Fig. 6, B and C, and Fig. 7, A–C). About 15% contained longer spindles that were up to two times the length of a normal short spindle (Fig. 7 D). Relatively few cells (<10%) contained a typical long anaphase spindle that extended the length of the cell, as observed in SPC72 cells (Fig. 6 D). Overall, these results indicate that spindle elongation is substantially inhibited in spc72-2 cells. In cells containing elongated spindles, some degree of chromosome separation was usually observed, but the spindles and chromosomes often remained entirely within the mother cell (Fig. 7 D). As mentioned above, the majority (∼75%) of spc72-2 cells that contained spindles had no detectable cytoplasmic microtubules (Fig. 6, B and C, and Fig. 7 A). However, a few of these cells (∼15%) had an excessively long bundle of cytoplasmic microtubules emanating from only one spindle pole (Fig. 7 B; compare to cytoplasmic microtubules in SPC72 cells in Fig. 6 D). In addition, cytoplasmic microtubules that were not attached to either pole were observed in ∼10% of these cells (Fig. 7 C).
Figure 7.
Phenotype of large-budded spc72-2 cells. spc72-2 cells (CUY1123) were grown at 26°C, and were then shifted to 37°C for 2 h. (A–D), DIC; (A′–D′), DAPI staining; (A′′–D′′), immunofluorescence with Tub2p antibody. Arrow indicates microtubule that is unattached to the SPB. Bar, 5 μm.
The unbudded anucleate spc72-2 cells that accumulated with time at 37°C lacked all microtubules (Fig. 6 C). In addition, there was a corresponding increase in the number of spc72-2 cells with multiple microtubule foci (Fig. 6 C). Both of these classes of cells likely arise from cytokinesis of a large-budded cell that has failed to move a SPB into the bud. In this case, one daughter will lack SPBs. The other will obtain two SPBs that can subsequently duplicate and separate to produce a cell with multiple microtubule-organizing centers.
To assess the primary defect caused by the spc72-2 mutation, we synchronized cells with α-factor at 26°C, and then released them from the α-factor block and incubated them in fresh medium at 37°C. 90 min after release, most cells contained large buds. 90% of these large-budded cells contained a short bipolar spindle, as determined by microtubule and SPB staining, and an undivided nucleus (data not shown). In nearly all of these cells, the nucleus failed to migrate to the bud neck, and was randomly positioned in one cell body. As in asynchronous cultures, very few cytoplasmic microtubules were observed; those few that were observed were excessively long. At later times after release from α-factor, we observed an increasing number of unbudded anucleate cells and cells with multiple SPBs.
These aberrant microtubule phenotypes were also observed in spc72-2 cells grown at 26°C, but to a much lesser extent (Fig. 6 A). Thus, even 26°C is not entirely permissive for microtubule assembly in spc72-2 cells.
SPB Morphology in spc72 Mutant Cells
spc72-2 cells were also examined by thin section electron microscopy. Cells were arrested in α-factor and then released from the block at 37°C for 90 min. As described above, this protocol allowed us to examine a more uniform population than that which could be obtained from asynchronous cultures. Fig. 8 A shows a short bipolar spindle in one of these spc72-2 cells. It is of normal length (1.2 μm) for a preanaphase spindle, and contains organized microtubule arrays typical of those observed in SPC72 cells. We identified about 50 SPBs in both SPC72 and spc72-2 cells by their characteristic dark-staining central plaques. In both cell types, we could also observe a distinct outer plaque in about 20% of these SPBs. The outer plaques of spc72-2 cells displayed a couple of subtle but consistent differences (Fig. 8 B, SPC72; Fig. 8, C and D, spc72-2). First, the distance between the central and outer plaques is about twice as large in spc72-2 cells. Second, the outer plaques of spc72-2 cells appeared to contain discontinuities or small gaps that were not observed in SPC72 outer plaques (Fig. 8, C and D, arrows). Inner plaques could not be observed in sufficient detail to allow any meaningful comparison of these structures.
Figure 8.
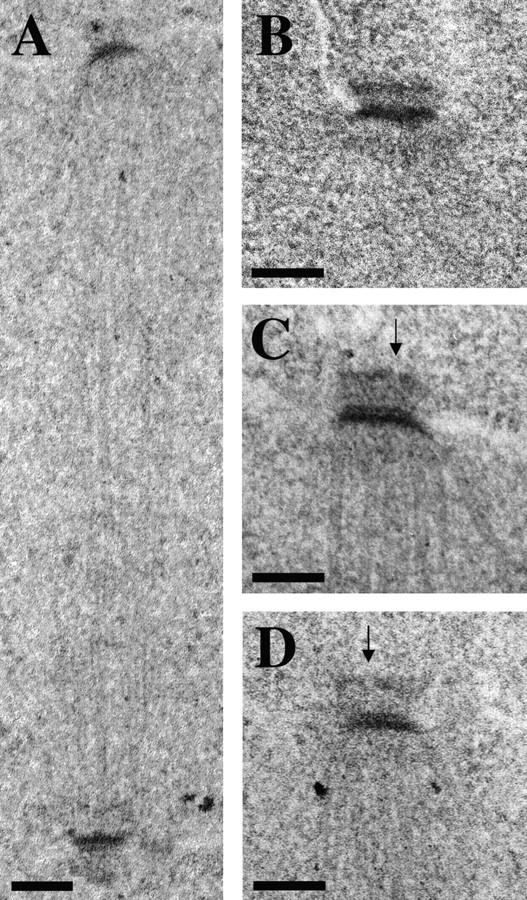
Spindle and SPB morphology in spc72-2 cells. SPC72 (CUY1130) and spc72-2 (CUY1116) were arrested with α-factor at 26°C, and were then released from the arrest at 37°C for 90 min. (A) spc72-2 spindle; (B) SPC72 SPB; (C and D) spc72-2 SPBs. Arrows indicate gaps in the outer plaque of spc72-2 cells. Bars, 0.1 μm.
Discussion
Stu2p Associates with Spc72p In Vivo
We carried out a two-hybrid screen to identify proteins that interact with Stu2p, a microtubule-associated protein that localizes primarily to the SPB. As expected, we identified genes encoding tubulins, specifically the two α-tubulins. We do not believe this result can be taken as evidence that Stu2p interacts directly with α-tubulin, as opposed to β-tubulin, because the Gal4ad-α-tubulin is likely to dimerize with β-tubulin in the cell. Thus, Stu2p could interact exclusively with β-tubulin and still activate transcription through the Gal4ad-α-tubulin-β-tubulin dimer. We did not expect to identify the gene encoding β-tubulin because genes are expressed at high levels in the two-hybrid system, and overexpression of β-tubulin is known to be lethal (Weinstein and Solomon, 1990).
Bik1p and Bim1p were also identified in this screen. Bik1p was originally identified as a protein that is essential for karyogamy (Berlin et al., 1990). Bik1p localizes to the SPB and spindle microtubules in vivo, and cells lacking Bik1p show defects in microtubule assembly. Bim1p was recently identified as a protein that interacts with α-tubulin in a two-hybrid screen (Schwartz et al., 1997). It localizes to both spindle and cytoplasmic microtubules, and cells lacking Bim1p display aberrant spindle morphology. It is possible that the Bik1p and Bim1p are components of a Stu2p complex given their interactions with Stu2p in the two-hybrid assay. However, both Bik1p and Bim1p are believed to be microtubule-binding proteins, so these interactions observed in the two-hybrid assay could be indirect and mediated by tubulin. Further experiments are needed to clarify this issue.
The two-hybrid screen also identified Stu2p and Spc72p. We showed that Spc72p and Stu2p can be coimmunoprecipitated from yeast extracts, indicating that they form a complex. In addition to interacting with itself in the two-hybrid assay, differentially epitope-tagged Stu2p's can be coimmunoprecipitated (K. Kosco, unpublished data). Similarly, Spc72p interacts with itself in the two-hybrid assay, and differentially epitope-tagged Spc72p's can also be coimmunoprecipitated. One interpretation of these results is that the Stu2p–Spc72p complex contains more than one molecule of both Stu2p and Spc72p. Alternatively, cells may contain Stu2p-Stu2p, Spc72p-Spc72p, and Stu2p-Spc72p dimers that do not interact with each other.
Spc72p is a Component of the SPB
A Spc72-GFP fusion protein localizes to the SPB, indicating that Spc72p, like Stu2p, is a component of the yeast SPB. We believe the localization of Spc72-GFP reflects that of endogenous Spc72p because SPC72-GFP is able to rescue the lethality of a spc72 null mutation. Spc72-GFP localization is not affected by treatment of cells with nocodazole, which eliminates all detectable microtubules. Although we cannot rule out the possibility that a small amount of tubulin polymer that is undetectable by immunofluorescence may remain at the SPB after treatment with nocodazole, this result is consistent with the interpretation that Spc72p is an integral component of the SPB. We did not observe even minor amounts of Spc72-GFP along microtubules as we did for Stu2-GFP. This difference may reflect the fact that Stu2p is capable of interacting directly with microtubules, and that Spc72p lacks this ability (see below).
The association of Spc72p with the SPB was examined in more detail by expressing SPC72-GFP under the control of the GAL1 promoter. When Spc72-GFP was produced in α-factor–arrested cells, it localized to the existing SPB. This result indicates that Spc72p can associate with an SPB that is already assembled. A similar result was obtained for the yeast γ-tubulin Tub4p (Marschall et al., 1996). In contrast, another SPB protein, Kar1p, was found to associate only with a new SPB after induction (Vallen et al., 1992). Thus, Spc72p and Tub4p may represent more peripheral components of the SPB that can exchange with their soluble subunits, respectively. This possibility would be consistent with their proposed roles as proteins that interact directly or indirectly with microtubules that contact the inner and outer surfaces of the SPB.
We have shown previously that Stu2p can bind laterally to microtubules in vitro with an affinity that is comparable to that of other known microtubule-associated proteins (Wang and Huffaker, 1997). In contrast, we could not detect any binding of Spc72p to microtubules, even at tubulin concentrations greater than ten times the K d for Stu2p binding. Thus, we conclude that Spc72p does not bind significantly to the sides of microtubules. However, because Spc72p is a component of the SPB, it is reasonable to consider the possibility that it may bind to the ends of microtubules. Microtubules used in the binding assay are on average ∼10 μm in length (Wang and Huffaker, 1997). Assuming 1,700 tubulin dimers per μm microtubule (Mitchison and Kirschner, 1984), the concentration of minus ends in this preparation is ∼3 × 10−10 M. For comparison, the K d for γ-tubulin binding to the minus ends of microtubules is ∼1.5 × 10−10 M. Therefore, our results indicate that Spc72p does not have an affinity for the ends of microtubules comparable to that of γ-tubulin.
Spc72p Plays a Role in Microtubule Assembly and Function
The spc72-2 mutant cells are clearly deficient in cytoplasmic microtubules; the majority of cells contain no cytoplasmic microtubules that can be detected by immunofluorescence microscopy. This phenotype indicates that Spc72p plays a role in the assembly and/or stability of microtubules at the cytoplasmic surface of the SPB or outer plaque. In support of this conclusion, we also observed subtle but consistent defects in the morphology of the outer plaques in spc72-2 cells. Recently, Spc72p has been localized to the outer plaque using immunoelectron microscopy of isolated SPBs (Jensen et al., 1998). This result agrees with our observations of spc72-2 cells.
We observed a single long cytoplasmic microtubule in a minority of the spc72-2 cells. Interestingly, in many cases these cytoplasmic microtubules were not attached to the SPB, indicating that they had been released from the SPB after their assembly. We previously suggested that Stu2p may play a role in tethering microtubules to the SPB through its ability to bind laterally to microtubules. The observation of free cytoplasmic microtubules in spc72-2 cells supports the hypothesis that Stu2p, in conjunction with Spc72p, does play a role in microtubule tethering. We expect that these free microtubules would be relatively unstable, which could account for the fact that they are observed in only a small percentage of cells. Their excessive length may be a secondary consequence of a spc72-2 defect. When fewer microtubules are being assembled, those few that are may be able to draw on a larger pool of soluble tubulin and achieve greater length.
In contrast to their severe defect in cytoplasmic microtubule assembly, most spc72-2 cells are able to assemble bipolar spindles. However, the spc72-2 mutation greatly inhibits spindle elongation. It is unlikely that the spindle elongation defect is a direct consequence of the lack of cytoplasmic microtubules. A conditional lethal mutation in the gene encoding β-tubulin that specifically inhibits cytoplasmic microtubule assembly does not block spindle elongation or chromosome segregation (Sullivan and Huffaker, 1992). In addition, several lines of reasoning argue against the idea that the defect in spindle elongation is due to a cell cycle block. First, although the spc72-2 mutation appears to slow progression through mitosis, it does not cause a cell cycle block. The mutation does not block cytokinesis, as implied by the production of unbudded cells with little or no DNA, additional rounds of DNA replication as shown by flow cytometry, or the SPB duplication cycle as indicated by cells with multiple SPBs. Second, a number of cells do contain slightly elongated spindles, suggesting that they have entered anaphase, but are physically incapable of further spindle elongation. Thus, we conclude that the spc72-2 mutation has a direct effect on spindle elongation.
The mechanisms for spindle pole separation during spindle assembly are likely to be similar to those that increase spindle pole separation during anaphase B elongation. Both processes are thought to be mediated by motor proteins that act to push antiparallel polar microtubules past each other in a direction that forces the poles apart (Ault and Rieder, 1994; Vernos and Karsenti, 1996). In S. cerevisiae, this pushing force is produced by the kinesin-like motors Cin8p and Kip1p (Hoyt et al., 1992). Because the fully elongated yeast spindle is many times the length of its short preanaphase spindle, polar microtubules must lengthen substantially as anaphase separation proceeds. One explanation for the spc72-2 phenotype is that spc72-2 cells are able to assemble the microtubules necessary to form a bipolar spindle, but are unable to assemble spindle microtubules of sufficient length to accomplish spindle elongation. While we do not yet know the mechanism by which Spc72p could influence spindle microtubule length, the fact that the spc72-2 mutation causes a defect in spindle elongation suggests that it affects the nuclear surface of the SPB or inner plaque. Immunoelectron microscopy did not identify Spc72p on the inner plaque (Jensen et al., 1998); however, it is difficult to rule out the possibility that Spc72p is located in this structure while the antigen is masked. Alternatively, Spc72p may be located exclusively in the outer plaque, but the defect caused by the spc72-2 mutation perturbs the SPB sufficiently to affect its function on the intranuclear side as well.
Acknowledgments
We thank Beth Lalonde and Rey-Huei Chen for their comments on the manuscript, and John Kilmartin for sharing results before publication.
This work was supported by a grant from the National Institutes of Health (GM40479).
Abbreviations used in this paper
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- 5-FOA
5-fluoroorotic acid
- GFP
green fluorescent protein
- HA
hemagglutinin
- SPB
spindle pole body
- YPD
yeast extract peptone dextrose
Footnotes
The present address of Xiaoyue Peter Chen is Department of Medicine, College of Physicians and Surgeons, Columbia University, 630 W. 168th St., New York, NY 10032.
Address all correspondence to Tim Huffaker, Section of Biochemistry, Molecular and Cell Biology, Biotechnology Building, Cornell University, Ithaca, NY 14853-2703. Tel.: 607-255-9947. Fax: 670-255-2428. E-mail: tch4@cornell.edu
References
- Ault JG, Rieder CL. Centrosome and kinetochore movement during mitosis. Curr Opin Cell Biol. 1994;6:41–49. doi: 10.1016/0955-0674(94)90114-7. [DOI] [PubMed] [Google Scholar]
- Baum P, Furlong C, Byers B. Yeast gene required for spindle pole body duplication: homology of its product with calcium-binding proteins. Proc Natl Acad Sci USA. 1986;83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin V, Styles C, Fink G. BIK1, a protein required for microtubule function during mating and mitosis in Saccharomyces cerevisiae, colocalizes with tubulin. J Cell Biol. 1990;111:2573–2586. doi: 10.1083/jcb.111.6.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J, LaCroute F, Fink G. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/s0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of saccharomyces cerevisiae. J Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Goetsch L. Preparation of yeast cells for thin-section electron microscopy. Methods Enzymol. 1991;194:602–607. doi: 10.1016/0076-6879(91)94044-d. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV. Spc42p: a phosphorylated component of the S. cerevisiaespindle pole body (SPB) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Geiser JR, Sundberg HA, Chang BH, Muller EGD, Davis TN. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. . Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S, Pereira G, Spang A, Knop M, Soues S, Kilmartin J, Schiebel E. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiaeat the sites of microtubule attachment. EMBO (Eur Mol Biol Organ) J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature. 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiaekinesin-related gene-products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter K-J, Eipel HE. Microbial determinations by flow cytometry. J Gen Microbiol. 1979;113:369–375. doi: 10.1099/00221287-113-2-369. [DOI] [PubMed] [Google Scholar]
- Jensen, O.N., P.A. Wigge, S. Holmes, S. Soues, M. Mann, and J.V. Kilmartin. 1998. Analysis of the Saccharomyces spindle pole by MALDI mass spectrometry. J. Cell Biol. In press. [DOI] [PMC free article] [PubMed]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiaespindle whose transcript is cell cycle regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiaeand functions in microtubule organization and spindle pole body duplication. EMBO (Eur Mol Biol Organ) J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, VanDyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. STU1,a suppressor of a β-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu YH, Fink GR. Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. . J Cell Biology. 1995;130:1373–1385. doi: 10.1083/jcb.130.6.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Fink G. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Schatz P, Pillus L, Grisafi P, Solomon F, Botstein D. Two functional α-tubulin genes of the yeast Saccharomyuces cerevisiaeencode divergent proteins. Mol Cell Biol. 1986;6:3711–3721. doi: 10.1128/mcb.6.11.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz K, Richards K, Botstein D. BIM1encodes a microtubule-binding protein in yeast. Mol Biol Cell. 1997;8:2677–2691. doi: 10.1091/mbc.8.12.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Sikorski R, Boeke J. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent γ-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half-bridge of the spindle pole body. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Grein K, Matzner M, Schiebel E. The Cdc31p-binding protein Kar1p is a component of the half-bridge of the yeast spindle pole body. J Cell Biol. 1995;128:863–877. doi: 10.1083/jcb.128.5.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-tubulin-like Tub4p of Saccharomyces cerevisiaeis associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan DS, Huffaker TC. Astral microtubules are not required for anaphase B in Saccharomyces cerevisiae. . J Cell Biol. 1992;119:379–388. doi: 10.1083/jcb.119.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallen EA, Scherson TY, Roberts T, van Zee K, Rose MD. Asymmetric mitotic segregation of the yeast spindle pole body. Cell. 1992;69:505–515. doi: 10.1016/0092-8674(92)90451-h. [DOI] [PubMed] [Google Scholar]
- Vernos I, Karsenti E. Motors involved in spindle assembly and chromosome segregation. Curr Opin Cell Biol. 1996;8:4–9. doi: 10.1016/s0955-0674(96)80041-x. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Huffaker TC. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein B, Solomon F. Phenotypic consequences of tubulin overproduction In Saccharomyces cerevisiae: differences between alpha-tubulin and beta-tubulin. Mol Cell Biol. 1990;10:5295–5304. doi: 10.1128/mcb.10.10.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]