Abstract
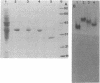
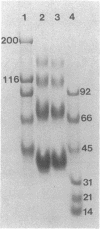
The citric acid cycle enzyme malate dehydrogenase was purified to homogeneity from the nonsulfur purple bacteria Rhodobacter capsulatus, Rhodospirillum rubrum, Rhodomicrobium vannielii, and Rhodocyclus purpureus. Malate dehydrogenase was purified from each species by either a single- or a two-step protocol: triazine dye affinity chromatography was the key step in purification of malate dehydrogenase in all cases. Purification of malate dehydrogenase resulted in a 130- to 240-fold increase in malate dehydrogenase specific activity, depending on the species, with recoveries ranging from 30 to 70%. Homogeneity of malate dehydrogenase preparations from the four organisms was determined by sodium dodecyl sulfate and nondenaturing polyacrylamide gel electrophoresis; a single protein band was observed in purified preparations by both techniques. The molecular weight of native malate dehydrogenases was determined by four independent methods and estimated to be in the range of 130,000 to 140,000 for the enzyme from R. capsulatus, R. rubrum, and R. vannielii and 57,000 for that from R. purpureus. It is concluded that malate dehydrogenase from R. capsulatus, R. rubrum, and R. vannielii is a tetramer composed of four identical subunits, while the enzyme from R. purpureus is a dimer composed of two identical subunits.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews P. The gel-filtration behaviour of proteins related to their molecular weights over a wide range. Biochem J. 1965 Sep;96(3):595–606. doi: 10.1042/bj0960595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter F. H., Harrington K. T. Intermolecular cross-linking of monomeric proteins and cross-linking of oligomeric proteins as a probe of quaternary structure. Application to leucine aminopeptidase (bovine lens). J Biol Chem. 1972 Sep 10;247(17):5580–5586. [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Davies G. E., Stark G. R. Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc Natl Acad Sci U S A. 1970 Jul;66(3):651–656. doi: 10.1073/pnas.66.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossebüter W., Hartl T., Görisch H., Stezowski J. J. Purification and properties of malate dehydrogenase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Biol Chem Hoppe Seyler. 1986 Jun;367(6):457–463. doi: 10.1515/bchm3.1986.367.1.457. [DOI] [PubMed] [Google Scholar]
- Kristjansson H., Ponnamperuma C. Purification and properties of malate dehydrogenase from the extreme thermophile Bacillus caldolyticus. Orig Life. 1980 Jun;10(2):185–192. doi: 10.1007/BF00928668. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- Mevarech M., Eisenberg H., Neumann E. Malate dehydrogenase isolated from extremely halophilic bacteria of the Dead Sea. 1. Purification and molecular characterization. Biochemistry. 1977 Aug 23;16(17):3781–3785. doi: 10.1021/bi00636a009. [DOI] [PubMed] [Google Scholar]
- Murphey W. H., Barnaby C., Lin F. J., Kaplan N. O. Malate dehydrogenases. II. Purification and properties of Bacillus subtilis, Bacillus stearothermophilus, and Escherichia coli malate dehydrogenases. J Biol Chem. 1967 Apr 10;242(7):1548–1559. [PubMed] [Google Scholar]
- Murphey W. H., Kitto G. B., Everse J., Kaplan N. Malate dehydrogenases. I. A survey of molecular size measured by gel filtration. Biochemistry. 1967 Feb;6(2):603–610. doi: 10.1021/bi00854a031. [DOI] [PubMed] [Google Scholar]
- Sahl H. G., Trüper H. G. Malic enzyme of chromatium vinosum. Arch Microbiol. 1980 Aug;127(1):17–24. doi: 10.1007/BF00414350. [DOI] [PubMed] [Google Scholar]
- Scawen M. D., Darbyshire J., Harvey M. J., Atkinson T. The rapid purification of 3-hydroxybutyrate dehydrogenase and malate dehydrogenase on triazine dye affinity matrices. Biochem J. 1982 Jun 1;203(3):699–705. doi: 10.1042/bj2030699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K., Sundaram T. K., Kernick M. Malate dehydrogenases from actinomycetes: structural comparison of Thermoactinomyces enzyme with other actinomycete and Bacillus enzymes. J Bacteriol. 1984 Feb;157(2):684–687. doi: 10.1128/jb.157.2.684-687.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprott G. D., McKellar R. C., Shaw K. M., Giroux J., Martin W. G. Properties of malate dehydrogenase isolated from Methanospirillum hungatii. Can J Microbiol. 1979 Feb;25(2):192–200. doi: 10.1139/m79-030. [DOI] [PubMed] [Google Scholar]
- TUBOI S., KIKUCHI G. ENZYMIC CLEAVAGE OF MALYTL-COENZYME A INTO ACETYL-COENZYME A AND GLYOXYLIC ACID. Biochim Biophys Acta. 1965 Jan;96:148–153. doi: 10.1016/0005-2787(65)90618-0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Weisburg W. G., Paster B. J., Madigan M. T., Fowler V. J., Hahn C. M., Blanz P., Gupta R., Nealson K. H. The phylogeny of purple bacteria: the alpha subdivision. Syst Appl Microbiol. 1984;5:315–326. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- Wright I. P., Sundaram T. K. Simple efficient methods for the isolation of malate dehydrogenase from thermophilic and mesophilic bacteria. Biochem J. 1979 Feb 1;177(2):441–448. doi: 10.1042/bj1770441. [DOI] [PMC free article] [PubMed] [Google Scholar]