Abstract
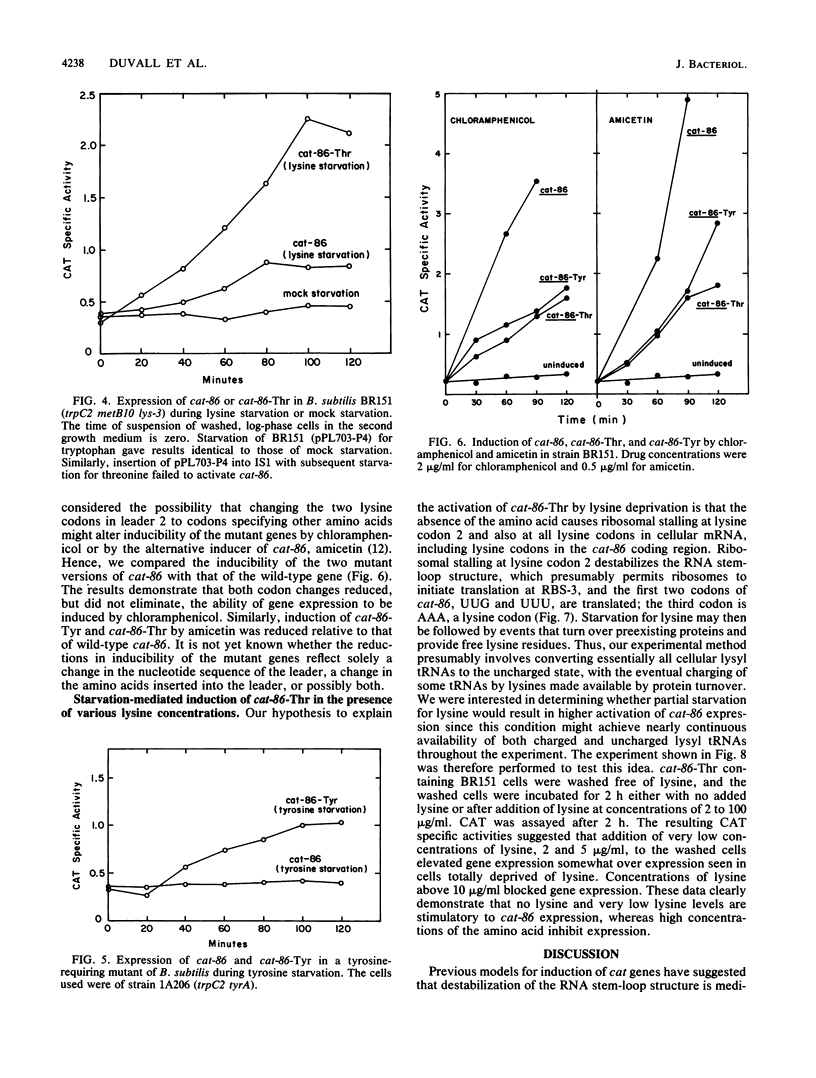
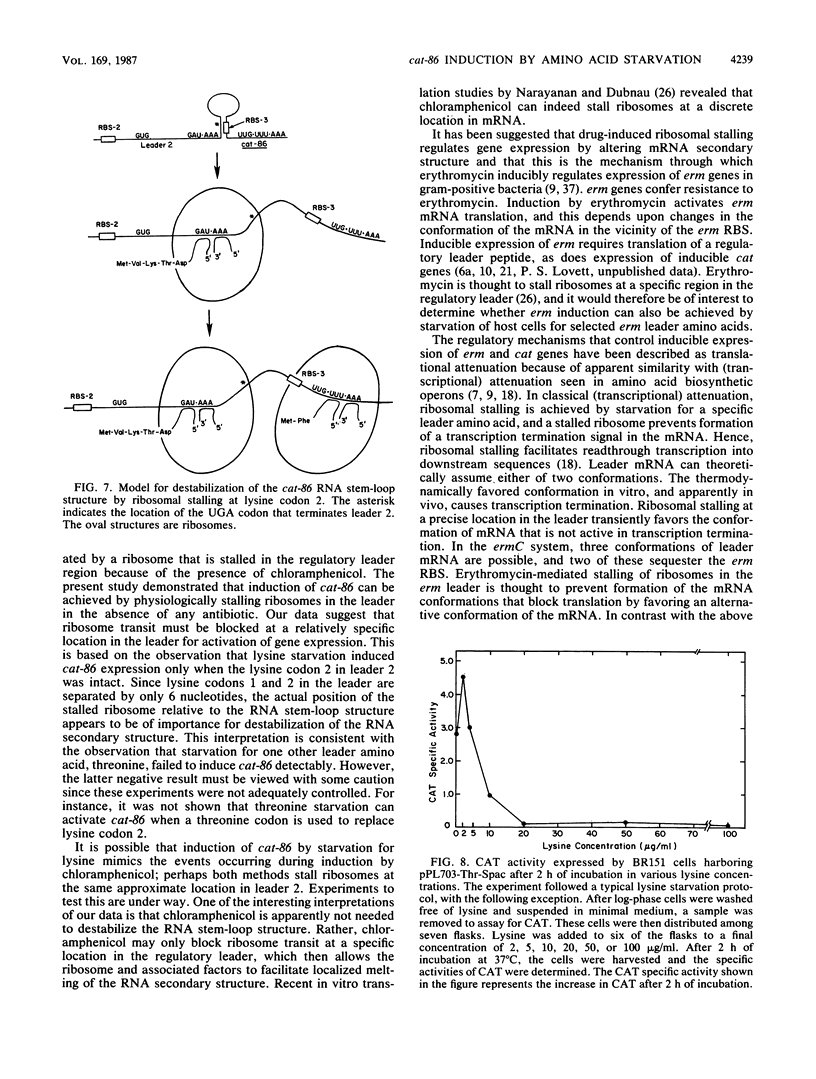
The plasmid gene cat-86 is induced by chloramphenicol in Bacillus subtilis, resulting in the synthesis of the gene product chloramphenicol acetyltransferase. Induction is due to a posttranscriptional regulatory mechanism in which the inducer, chloramphenicol, activates translation of cat-86 mRNA. We have suggested that chloramphenicol allows ribosomes to destabilize a stem-loop structure in cat-86 mRNA that sequesters the ribosome-binding site for the coding sequence. In the present report we show that cat-86 expression can be activated by stalling ribosomes in the act of translating a regulatory leader peptide. Stalling was brought about by starving host cells for specific leader amino acids. Ribosomal stalling, which led to cat-86 expression, occurred upon starvation for the amino acid specified by the leader codon located immediately 5' to the RNA stem-loop structure and was independent of whether that codon specified lysine or tyrosine. These observations support a model for chloramphenicol induction of cat-86 in which the antibiotic stalls ribosome transit in the regulatory leader. Stalling of ribosomes in the leader can therefore lead to destabilization of the RNA stem-loop structure.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos N. P., Jr, Chow J. H., Mongkolsuk S., Preis L. H., Vollmar W. R., 2nd, Lovett P. S. Constitutive variants of the pC194 cat gene exhibit DNA alterations in the vicinity of the ribosome binding site sequence. Gene. 1984 May;28(2):171–176. doi: 10.1016/0378-1119(84)90254-3. [DOI] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Duvall E. J., Lovett P. S. Analysis of the regulatory sequences needed for induction of the chloramphenicol acetyltransferase gene cat-86 by chloramphenicol and amicetin. J Bacteriol. 1986 Sep;167(3):842–849. doi: 10.1128/jb.167.3.842-849.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Duvall E. J., Lovett P. S. The mRNA for an inducible chloramphenicol acetyltransferase gene is cleaved into discrete fragments in Bacillus subtilis. J Bacteriol. 1987 Mar;169(3):967–972. doi: 10.1128/jb.169.3.967-972.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Mongkolsuk S., Kaufman J. D., Lovett P. S. Chloramphenicol-induced translation of cat-86 mRNA requires two cis-acting regulatory regions. J Bacteriol. 1985 Nov;164(2):696–703. doi: 10.1128/jb.164.2.696-703.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambulos N. P., Jr, Mongkolsuk S., Lovett P. S. A transcription termination signal immediately precedes the coding sequence for the chloramphenicol-inducible plasmid gene cat-86. Mol Gen Genet. 1985;199(1):70–75. doi: 10.1007/BF00327512. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brückner R., Dick T., Matzura H. Dependence of expression of an inducible Staphylococcus aureus cat gene on the translation of its leader sequence. Mol Gen Genet. 1987 May;207(2-3):486–491. doi: 10.1007/BF00331619. [DOI] [PubMed] [Google Scholar]
- Brückner R., Matzura H. Regulation of the inducible chloramphenicol acetyltransferase gene of the Staphylococcus aureus plasmid pUB112. EMBO J. 1985 Sep;4(9):2295–2300. doi: 10.1002/j.1460-2075.1985.tb03929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byeon W. H., Weisblum B. Post-transcriptional regulation of chloramphenicol acetyl transferase. J Bacteriol. 1984 May;158(2):543–550. doi: 10.1128/jb.158.2.543-550.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Induction of ermC requires translation of the leader peptide. EMBO J. 1985 Feb;4(2):533–537. doi: 10.1002/j.1460-2075.1985.tb03661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Lovett P. S. Chloramphenicol induces translation of the mRNA for a chloramphenicol-resistance gene in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3939–3943. doi: 10.1073/pnas.83.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Mongkolsuk S., Kim U. J., Lovett P. S., Henkin T. M., Chambliss G. H. Induction of the chloramphenicol acetyltransferase gene cat-86 through the action of the ribosomal antibiotic amicetin: involvement of a Bacillus subtilis ribosomal component in cat induction. J Bacteriol. 1985 Feb;161(2):665–672. doi: 10.1128/jb.161.2.665-672.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Lovett P. S., Rudolph C., Vasantha N., Guyer M. Chloramphenicol-inducible gene expression in Bacillus subtilis. Gene. 1983 Oct;24(2-3):171–177. doi: 10.1016/0378-1119(83)90077-x. [DOI] [PubMed] [Google Scholar]
- Duvall E. J., Williams D. M., Mongkolsuk S., Lovett P. S. Regulatory regions that control expression of two chloramphenicol-inducible cat genes cloned in Bacillus subtilis. J Bacteriol. 1984 Jun;158(3):784–790. doi: 10.1128/jb.158.3.784-790.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keggins K. M., Lovett P. S., Duvall E. J. Molecular cloning of genetically active fragments of Bacillus DNA in Bacillus subtilis and properties of the vector plasmid pUB110. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1423–1427. doi: 10.1073/pnas.75.3.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolter R., Yanofsky C. Attenuation in amino acid biosynthetic operons. Annu Rev Genet. 1982;16:113–134. doi: 10.1146/annurev.ge.16.120182.000553. [DOI] [PubMed] [Google Scholar]
- Lovett P. S., Keggins K. M. Bacillus subtilis as a host for molecular cloning. Methods Enzymol. 1979;68:342–357. doi: 10.1016/0076-6879(79)68025-4. [DOI] [PubMed] [Google Scholar]
- Mayford M., Weisblum B. Messenger RNA from Staphylococcus aureus that specifies macrolide-lincosamide-streptogramin resistance. Demonstration of its conformations and of the leader peptide it encodes. J Mol Biol. 1985 Oct 20;185(4):769–780. doi: 10.1016/0022-2836(85)90061-0. [DOI] [PubMed] [Google Scholar]
- Mongkolsuk S., Ambulos N. P., Jr, Lovett P. S. Chloramphenicol-inducible gene expression in Bacillus subtilis is independent of the chloramphenicol acetyltransferase structural gene and its promoter. J Bacteriol. 1984 Oct;160(1):1–8. doi: 10.1128/jb.160.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Chiang Y. W., Reynolds R. B., Lovett P. S. Restriction fragments that exert promoter activity during postexponential growth of Bacillus subtilis. J Bacteriol. 1983 Sep;155(3):1399–1406. doi: 10.1128/jb.155.3.1399-1406.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongkolsuk S., Duvall E. J., Lovett P. S. Transcription termination signal for the cat-86 indicator gene in a Bacillus subtilis promoter-cloning plasmid. Gene. 1985;37(1-3):83–90. doi: 10.1016/0378-1119(85)90260-4. [DOI] [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Demonstration of erythromycin-dependent stalling of ribosomes on the ermC leader transcript. J Biol Chem. 1987 Feb 5;262(4):1766–1771. [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Evidence for the translational attenuation model: ribosome-binding studies and structural analysis with an in vitro run-off transcript of ermC. Nucleic Acids Res. 1985 Oct 25;13(20):7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W. L., Chambliss G. H., Buckbinder L., Ambulos N. P., Jr, Lovett P. S. Isolation and expression of a constitutive variant of the chloramphenicol-inducible plasmid gene cat-86 under control of the Bacillus subtilis 168 amylase promoter. Gene. 1985;35(1-2):113–120. doi: 10.1016/0378-1119(85)90163-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V., Brenner D. G., LeGrice S. F., Skinner S. E., Hawkins A. R. Chloramphenicol acetyltransferase gene of staphylococcal plasmid pC221. Nucleotide sequence analysis and expression studies. FEBS Lett. 1985 Jan 1;179(1):101–106. doi: 10.1016/0014-5793(85)80200-3. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase: enzymology and molecular biology. CRC Crit Rev Biochem. 1983;14(1):1–46. doi: 10.3109/10409238309102789. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. The enzymatic acetylation of chloramphenicol by extracts of R factor-resistant Escherichia coli. J Biol Chem. 1967 Feb 25;242(4):687–693. [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. W., Ott J., Eckstein F. The rapid generation of oligonucleotide-directed mutations at high frequency using phosphorothioate-modified DNA. Nucleic Acids Res. 1985 Dec 20;13(24):8765–8785. doi: 10.1093/nar/13.24.8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. M., Duvall E. J., Lovett P. S. Cloning restriction fragments that promote expression of a gene in Bacillus subtilis. J Bacteriol. 1981 Jun;146(3):1162–1165. doi: 10.1128/jb.146.3.1162-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yansura D. G., Henner D. J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Jan;81(2):439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis of DNA fragments cloned into M13 vectors. Methods Enzymol. 1983;100:468–500. doi: 10.1016/0076-6879(83)00074-9. [DOI] [PubMed] [Google Scholar]



