Abstract
Proinsulin is expressed prior to development of the pancreas and promotes cell survival. Here we study the mechanism affecting the translation efficiency of a specific embryonic proinsulin mRNA. This transcript shares the coding region with the pancreatic form, but presents a 32 nt extended leader region. Translation of proinsulin is markedly reduced by the presence of two upstream AUGs within the 5′ extension of the embryonic mRNA. This attenuation is lost when the two upstream AUGs are mutated to AAG, leading to translational efficiency similar to that of the pancreatic mRNA. The upstream AUGs are recognized as initiator codons, because expression of upstream ORF is detectable from the embryonic transcript, but not from the mutated or the pancreatic mRNAs. Strict regulation of proinsulin biosynthesis appears to be necessary, since exogenous proinsulin added to embryos in ovo decreased apoptosis and generated abnormal developmental traits. A novel mechanism for low level proinsulin expression thus relies on upstream AUGs within a specific form of embryonic proinsulin mRNA, emphasizing its importance as a tightly regulated developmental signal.
Keywords: developmental cell death/proinsulin translation/upAUGs
Introduction
Insulin is considered an essential anabolic hormone, produced and secreted into the blood by the adult pancreas. In chickens and in humans, proinsulin is encoded by a single gene (Perler et al., 1980). The presence of proinsulin mRNA variants differing in their 5′ untranslated regions (5′ UTRs) was recently demonstrated. In chick embryos, we found a novel proinsulin transcript generated by alternative transcription start sites whose synthesis is developmentally regulated (Hernández-Sánchez et al., 2002). In humans, an intron 1 splice variant is metabolically regulated and increases translation efficiency (Shalev et al., 2002).
In adult organisms, insulin biosynthesis by pancreatic β-cells is a paradigm of tissue-specific expression. The mature β-cells synthesize the ∼9 kDa proinsulin precursor, which is processed to insulin (∼6 kDa) by the endopeptidases PC2 and PC3 in the secretory granules. Circulating insulin is responsible for maintaining glucose homeostasis, and plasma hormone levels are largely dependent on food intake. Glucose is, in turn, the central regulator of pancreatic insulin biosynthesis at multiple steps (Robbins et al., 1984; Rhodes, 2000). Plasma glucose concentration controls insulin mRNA levels by stimulating insulin gene transcription, as well as insulin mRNA processing and stability (Brunstedt and Chan, 1982; Nielsen et al., 1985; Welsh et al., 1985; Leibiger et al., 1998); in addition, glucose modulates insulin protein synthesis initiation and elongation rates (Permutt, 1974; Itoh and Okamoto, 1980; Welsh et al., 1986).
In the past decade, several observations have suggested that low levels of prepancreatic and extrapancreatic proinsulin expression displayed physiological functions (reviewed in De Pablo and Roth, 1990; De Pablo and de la Rosa, 1995; Varela-Nieto et al., 2003). Proinsulin expression during early development occurs prior to formation of the pancreatic primordium in several vertebrates (Serrano et al., 1989; Shuldiner et al., 1991; Deltour et al., 1993). Indeed, insulin-related molecules are expressed and control vital functions in organisms without a pancreas, such as nematodes (Pierce et al., 2001) and flies (Brogiolo et al., 2001). In the neurulating chick embryo and retina (a part of the central nervous system), proinsulin mRNA is expressed and translated to proinsulin. In contrast to the pancreatic protein, this proinsulin remains unprocessed, due to the absence PC2 convertase, and is rapidly secreted (Hernández-Sánchez et al., 1995, 2002; Morales et al., 1997; Alarcón et al., 1998; Díaz et al., 1999). In the chick, embryonic proinsulin mRNA levels are ∼1000-fold lower than those of the postnatal pancreas (Serrano et al., 1989), but are nevertheless essential for correct development. Proinsulin, present at embryonic stages when insulin-like growth factor I (IGF-I) is very low, promotes cell survival during neurulation and retinal neurogenesis. Interference with proinsulin and insulin-receptor biosynthesis by antisense oligonucleotides, as well as blockade of proinsulin signalling by anti-insulin and anti-insulin receptor antibodies, induces apoptosis in early chick embryos (De Pablo et al., 1985a; Morales et al., 1997; Díaz et al., 2000; Hernández-Sánchez et al., 2002).
Proinsulin gene products appear to have a dual biological function. First, as a tissue factor in early development, low local proinsulin levels are essential to regulate programmed cell death, a highly-regulated physiological process required for correct body pattern formation and development (reviewed by Jacobson et al., 1997; Vaux and Korsmeyer, 1999; de la Rosa and De Pablo, 2000). In postnatal organisms, mature insulin assumes its much better-characterized hormonal role, acting on newly acquired target tissues such as adipose tissue. At that stage, the pancreas functions as an insulin ‘factory’ (Orci et al., 1988), able to respond to rapidly changing metabolic requirements. In parallel with these very distinct functions, we found different embryonic and pancreatic proinsulin transcripts (Hernández-Sánchez et al., 2002). Embryonic proinsulin mRNA shares its coding region with the pancreatic form, but has an extended 5′ UTR. In addition, embryonic proinsulin mRNA levels are developmentally regulated, but are not glucose dependent, a difference from pancreatic mRNA (Pérez-Villamil et al., 1994), suggesting distinct regulation of expression. Pancreatic proinsulin mRNA shows the typical 5′ UTR of higher eukaryotic genes, as it is short, unstructured and without additional AUGs. In contrast, genes that display critical proliferation or survival functions, such as oncogenes, cytokines, signal transduction proteins and transcription factors, harbour upstream AUGs (upAUGs) and associated open reading frames (upORF) (Sachs and Geballe, 2000; Morris and Geballe, 2000; Kozak 2002a).
Here we characterize a novel form of control of the proinsulin gene expression by generating alternative transcript structure. In the specific embryonic form, proinsulin translation initiation is stringently repressed by the presence of two upAUGs within the extended 5′ UTR. In ovo, proinsulin addition to chick embryos modulates apoptosis and morphogenesis, suggesting that fine control of the specific embryonic proinsulin transcript structure is part of the regulated signalling required for normal embryo development in vertebrates.
Results
Embryonic and pancreatic proinsulin transcription initiation sites are adjacent in the genome
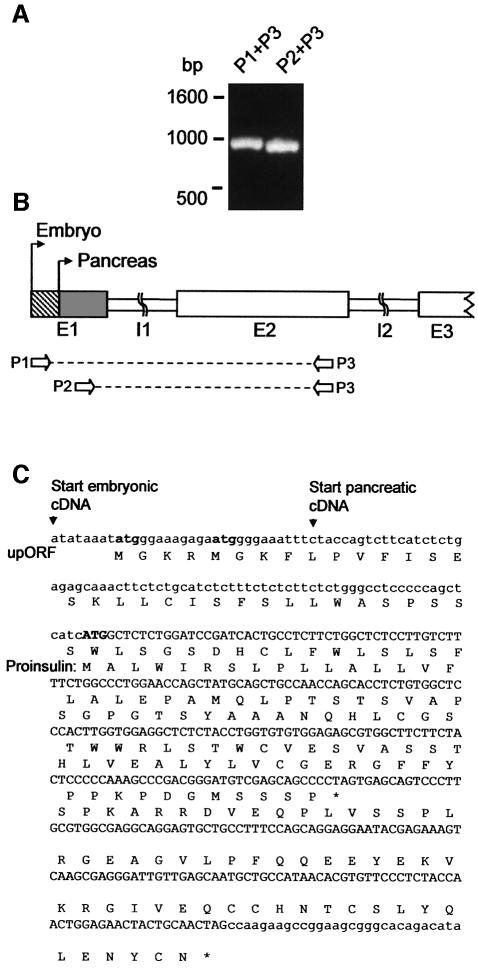
We recently reported that embryonic proinsulin mRNA shows a 32-nt extension in its 5′ UTR, whereas the coding region is shared with the pancreatic mRNA (Hernández-Sánchez et al., 2002). These alternative transcription start sites mapped in adjacent positions in the genome, as demonstrated by sequencing of genomic PCR products (Figure 1A and B). Sequence analysis of the embryonic proinsulin 5′ UTR (Figure 1C) revealed an upORF with two potential start codons (upAUG1/2) within the 32-nt extension of this mRNA. The first upAUG is located 12 nt upstream of the second, and the upORF overlaps 183 nt with the proinsulin ORF, which is shifted by –1 nt in its phase. Notably, upATG1/2 as well as the proinsulin initiator codon are located in good Kozak consensus sequence (Kozak, 1986).
Fig. 1. Characterization of pancreatic and embryonic proinsulin leader regions. PCR of the 5′ region of the chicken proinsulin genomic locus (A). Genomic organization of the amplified products (B) according to sequence analysis of the chicken proinsulin gene 5′ region (C). Arrows indicate embryonic and pancreatic transcription start sites; exon (E), intron (I) and primer (P) locations are shown. The proinsulin cDNA and predicted amino acid sequence (C) contain the proinsulin ATG (bold capitals), as well as the two upATGs (bold lowercase) and predicted upORF. Note that upORF is out of the proinsulin reading frame.
Embryonic and pancreatic proinsulin mRNAs show distinct translational activity
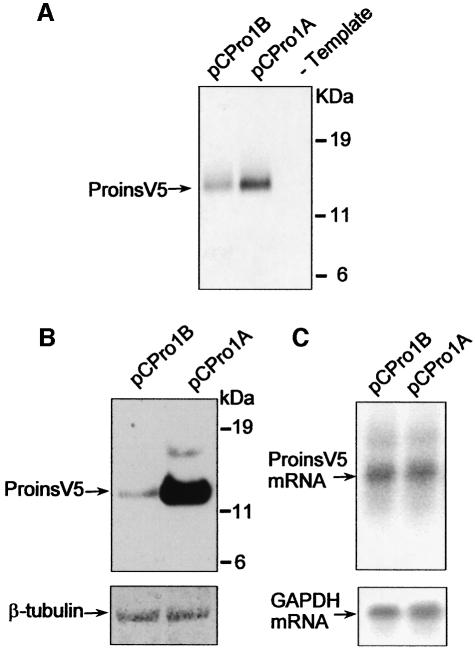
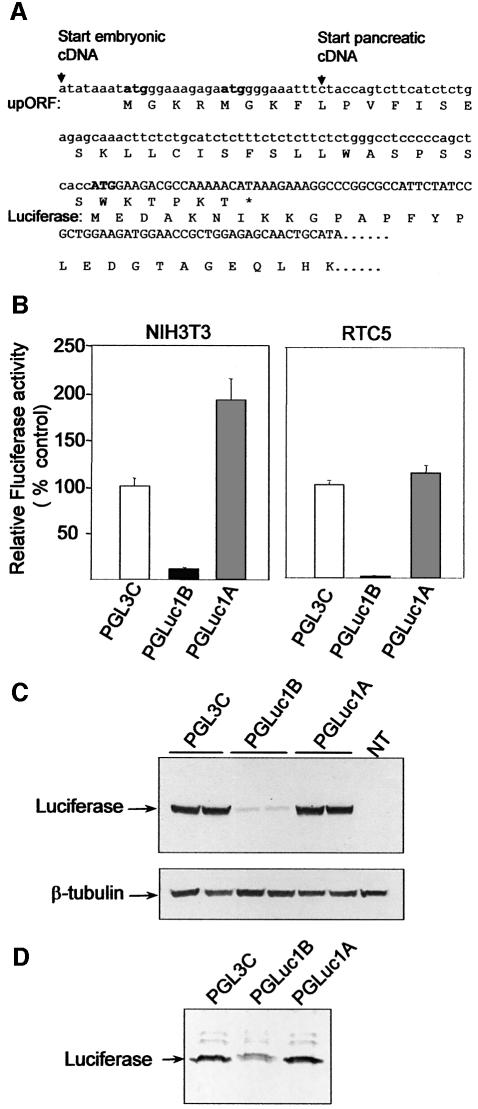
The presence of upAUGs and upORF in embryonic proinsulin mRNA suggests possible differential translational efficiency of this transcript compared with the pancreatic mRNA. We therefore studied the translational activity of embryonic and pancreatic proinsulin mRNAs in vitro and in vivo. To facilitate detection of proinsulin, the embryonic or the pancreatic proinsulin cDNAs were fused to the V5 epitope tag in constructs termed pCPro1B (embryonic) and pCPro1A (pancreatic). Transcription–translation assay of these constructs in a reticulocyte lysate system rendered a 13 kDa protein common to the embryonic and pancreatic constructs that corresponded to the proinsulin V5-tagged protein, as verified by anti-V5 immunoprecipitation (Figure 2A). The amount of proinsulin generated by the embryonic construct (pCPro1B) was nonetheless lower than that generated by the pancreatic construct (pCPro1A). Once it was confirmed that both transcripts were able to generate proinsulin in vitro (also in an un-coupled transcription and translation system, not shown), the differences observed in translation levels were analysed in vivo. NIH3T3 cells were transfected with each of the constructs and proinsulin-V5 was detected by western blot. Untransfected cells or cells transfected with an unrelated construct showed no V5 immunoreactivity (not shown). The amount of proinsulin generated with the embryonic construct was much lower than that in cells transfected with the pancreatic construct (Figure 2B). To assess whether the dramatic difference in proinsulin expression was mediated by translational efficiency or by differences in mRNA levels, the integrity and abundance of the transcripts were analysed by northern blot. Both embryonic and pancreatic proinsulin constructs were expressed at comparable levels relative to endogenous GAPDH mRNA (Figure 2C), suggesting that the differential embryonic and pancreatic proinsulin expression is regulated at a postrancriptional level.
Fig. 2. Translation of embryonic and pancreatic proinsulin. In vitro-coupled transcription–translation and immunoprecipitation with anti-V5 epitope antibody of embryonic (pCPro1B) and pancreatic (pCPro1A) proinsulin constructs (A). The same constructs were transfected into NIH3T3 cells. Transfected cells were analysed for proinsulin expression by western blotting using anti-V5 epitope antibody (B). β-tubulin was used as a loading control (bottom panel). Northern blot of NIH3T3 transfected cells (C). Membranes were probed for proinsulin (top panel) and GAPDH (bottom panel) mRNA. Proinsulin and GAPDH mRNA levels were quantified by a Fuji image analyzer and proinsulin mRNA levels corrected by that of endogenous GAPDH mRNA levels (proinsulin mRNA/GAPDH mRNA, 1.01 for embryonic and 1 for pancreatic constructs).
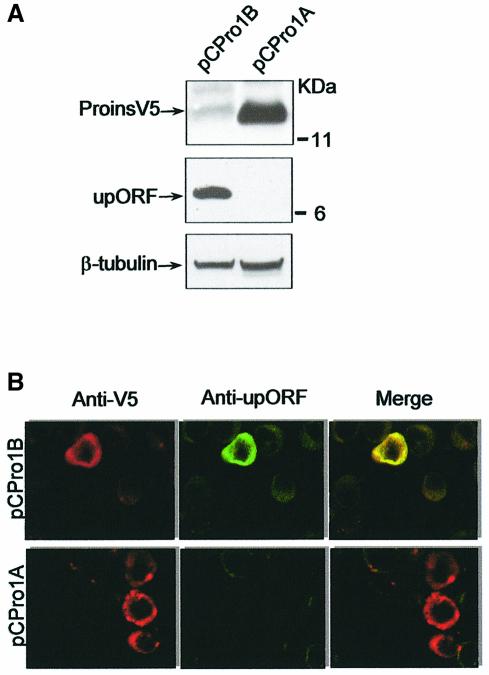
The good Kozak consensus of the upAUGs suggests that they mediate translation initiation. To examine upORF expression, we generated an antibody against a C-terminal peptide of the upORF. An ∼10 kDa protein was detected by western blot of NIH3T3 cells transfected with the embryonic, but not with the pancreatic, construct (Figure 3A). A single band was observed in standard gels whereas in long gels from experiments in vitro a doublet could be resolved (not shown). Immunostaining with anti-V5 and anti-upORF demostrated cytoplasmic expression of both proinsulin and upORF in the same cells transfected with the embryonic construct whereas only proinsulin was detected in cells transfected with the pancreatic construct (Figure 3B).
Fig. 3. Expression of upORF in transfected cells. Western blot of NIH3T3 transfected cells with the embryonic (pCPro1B) and pancreatic (pCPro1A) proinsulin constructs using anti-V5 epitope or anti-upORF antibodies for proinsulin and upORF expression, respectively (upper and middle panels) (A). β-tubulin was used as a loading control (bottom panel). Immunostaining of NIH3T3 transfected cells with pCPro1B and pCPro1A constructs using anti-V5 epitope antibody in red and anti-upORF antibody in green (B). Images were captured by a confocal microscope with a 40× objective.
Proinsulin is expressed in immortalized embryonic neuroepithelial cells
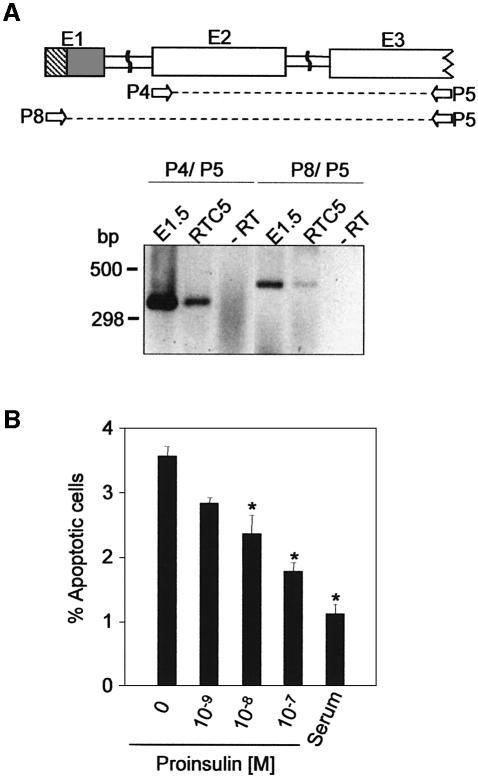
The embryonic proinsulin transcript is translated to a lesser extent than the pancreatic form in the reticulocyte lysate system and in the NIH3T3 cell line. To confirm these observations and relate them to physiological function, we used a cellular system that reflects embryonic proinsulin expression and cell response. Quail RTC5 is a retinal cell line originated from quail embryonic retina undergoing neurogenesis, immortalized by retroviral transduction with the myc oncogene. Under controlled conditions, proliferating cells can be induced to differentiate into cells with mature neuronal characteristics (Pollerberg and Eickholt, 1995; Pollerberg et al., 1995). Embryonic proinsulin mRNA expression was compared in RTC5 cells with incubation day 1.5 (E1.5) whole chick embryos undergoing neurulation. Proinsulin mRNA was characterized by RT–PCR of total RNA using primers directed to the proinsulin coding region (P4, P5) or primers that specifically amplified the embryonic proinsulin mRNA variant (P8, P5). Gel analysis and sequencing (not shown) of the PCR products demonstrated the presence of the embryonic proinsulin transcript in RTC5 cells (Figure 4A).
Fig. 4. Expression and effect of proinsulin in RTC5 cells. Proinsulin amplification products obtained by RT–PCR of total RNA from E1.5 embryos and RTC5 cells (A). –RT, control without reverse transcriptase. Location of primers (P) used is shown in the upper panel. Apoptosis of RTC5 cells (B). Apoptotic cells were scored by DAPI staining of pyknotic nuclei after 20 h growing in complete medium (serum) or serum-free medium supplemented with the indicated proinsulin concentrations. A minimum of 1000 cells was counted per experimental point. Results represent the mean ± SEM of three replicates. *P ≤ 0.001 with respect to non-proinsulin treatment.
Proinsulin attenuates programmed cell death in the neurulating embryo and developing retina (Díaz et al., 2000; Hernández-Sánchez et al., 2002); we therefore analysed the anti-apoptotic effect of proinsulin on RTC5 cells cultured in serum-free medium. As predicted, proinsulin rescued RTC5 cells from growth factor deprivation-induced apoptotic cell death in a dose-dependent manner. In the presence of 10–7 M proinsulin, the percentage of apoptotic cells decreased from 3.6 to 1.8%, close to the level observed in the presence of serum (1.1%; Figure 4B). In contrast to the antiapoptotic effect, proinsulin addition produced only a slight, non-significant increase in total cell number, even at the highest concentration. Serum starvation for long periods (up to 32 h) did not induce apoptosis in NIH3T3 cells (not shown). All together these data validate the use of RTC5 cells as a model to study regulation of embryonic proinsulin.
The embryonic proinsulin mRNA leader region determines translation efficiency
Translation of the embryonic proinsulin mRNA yielded a reduced amount of proinsulin concomitant with the synthesis of a polypeptide generated from the upORF. It is therefore possible that translation attenuation was the consequence of the primary 5′ UTR sequence or the presence of the polypeptides originated at upAUG1/2. To determine whether the 5′ leader region was responsible for this effect, we analysed chimeric constructs containing either the embryonic (pGLuc1B) or pancreatic (pGLuc1A) proinsulin 5′ UTRs fused to the luciferase ORF. In this scheme, the upORF encoded in the embryonic chimeric construct differs at the C-terminal end in sequence and length from the upORF encoded in the proinsulin cDNA (compare Figures 5A and 1C). A strong reduction in luciferase activity was observed in both NIH3T3 and RTC5 cells transfected with the embryonic chimeric construct, but not with the pancreatic chimera (Figure 5B). The pancreatic chimera construct was translated efficiently, showing even higher luciferase activity than the luciferase control vector lacking proinsulin 5′ UTR sequences in the NIH3T3 cells, an effect that was not seen in RTC5 cells (Figure 5B). Similar results were obtained when luciferase protein was detected by western blot (Figure 5C). Additionally, pulse labelling of pGLuc1B-transfected NIH3T3 cells consistently showed a lower luciferase translation rate than the pancreatic or control constructs (Figure 5D).
Fig. 5. Influence of embryonic and pancreatic proinsulin leader region on luciferase chimeric construct translation. Nucleotide and amino acid sequences of the luciferase chimeric constructs (A). The luciferase ATG (bold uppercase), as well as the two upATG (bold lowercase) are labelled. Note that upORF is out of the luciferase reading frame. NIH3T3 and RTC5 cells were transfected with control (pGL3C), embryonic (pGLuc1B) or pancreatic (pGLuc1A) firefly luciferase expression constructs (B). Firefly luciferase (Fluciferase) activity was corrected by Renilla luciferase activity, cotransfected to normalize transfection efficiency. Results represent the mean ± SEM of six samples. Luciferase western blot of NIH3T3 cells transfected with pGL3C, pGLuc1B and pGLuc1A constructs (C). β-tubulin was used as a loading control (bottom panel). [35S]Met/Cys pulse labelling of NIH3T3 cells transfected with the indicated constructs. Cell lysates were immunoprecipitated with anti-luciferase antibody, separated in a polyacrylamide gel and visualized using a Fuji image analyser (D).
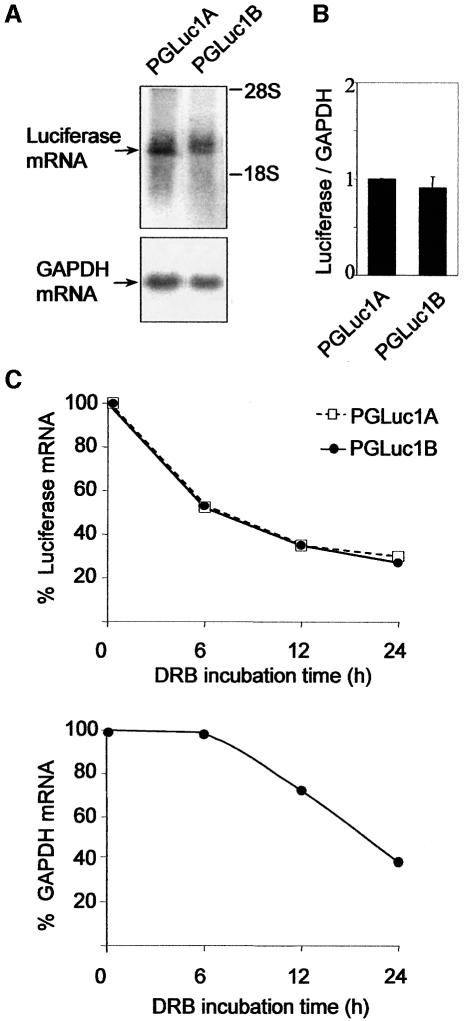
To study whether the dramatic variance in luciferase activity was mediated by differences in transcription levels and/or mRNA stability, the abundance of the transcripts were analysed by northern blot in transfected NIH3T3 cells. Both embryonic and pancreatic luciferase chimeric constructs were expressed as a single band of about 2 kb, at comparable levels relative to endogenous GAPDH mRNA (Figure 6A and B). To examine the mRNA degradation rate, we treated transfected NIH3T3 cells with the RNA polymerase II inhibitor 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) for different periods of time. The decay curves for both constructs were identical (Figure 6C), indicating that the embryonic leader region does not influence mRNA degradation levels. Similar results were obtained by inhibiting transcription with actinomycin D (not shown). The GAPDH mRNA degradation rate showed no general transcript degradation in the presence of DRB, albeit the luciferase mRNA was less stable than the GAPDH mRNA. All together, these results point to translational inhibition mediated by the embryonic proinsulin mRNA leader region.
Fig. 6. mRNA levels and stability of luciferase chimeric constructs with embryonic or pancreatic proinsulin leader regions. Northern blot of NIH3T3 cells transfected with the embryonic (pGLuc1B) or pancreatic (pGluc1A) firefly luciferase expression constructs (A). Membranes were probed for luciferase (top panel) and GAPDH (bottom panel) mRNA. Luciferase and GAPDH mRNA levels were quantified by a Fuji image analyser and luciferase mRNA levels corrected by that of endogenous GAPDH mRNA levels (B). Results represent the mean ± range of two experiments. The RNA polymerase II inhibitor DRB (100 µM) was added 48 h after transfection. RNA was extracted at the times indicated and hybridized in a northern blot with a luciferase probe (C). The luciferase mRNA decay curves are shown after Fuji image analyser quantitation (top panel). The filter was rehybridized with a GAPDH probe (bottom panel). An identical GAPDH mRNA decay curve was obtained for cells transfected with the pGluc1A construct.
A low level of embryonic proinsulin mRNA translation depends on the presence of upAUGs
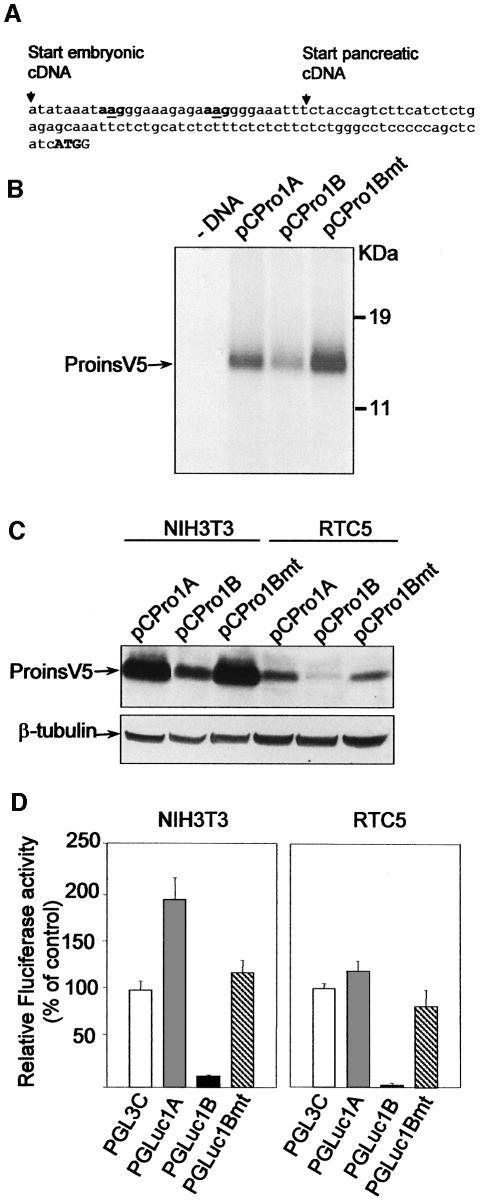
Increasing evidence suggests that upAUGs and upORFs significantly influence mRNA translational efficiency (for review see Geballe and Sachs, 2000; Morris and Geballe, 2000; Dever, 2002). The two upAUGs in the 32-nt extension of the embryonic proinsulin mRNA were thus candidates to mediate the low level of translation characterized for the embryonic proinsulin transcript. As hypothesized above, mutation of the two upAUG initiation codons to AAG (Figure 7A) abolished the inhibitory effect caused by the embryonic proinsulin mRNA leader region. The mutated embryonic construct (pCPro1Bmt) produced proinsulin levels comparable to those yielded by the pancreatic construct in the three experimental assays, the reticulocyte lysate system, as well as the transfected NIH3T3 and RTC5 cells (Figure 7B and C). Identical results were obtained with the luciferase constructs in transfected cells. The mutated embryonic chimeric construct (pGLuc1Bmt) rendered luciferase activity comparable to that of the control vector and much higher than the embryonic chimeric construct (Figure 7D). Moreover, expression of the upORF in the embryonic construct was abolished when the two upAUG were mutated to AAG as observed by western blotting (Figure 8) and immuno cytochemistry staining of transfected cells (not shown).
Fig. 7. Translational effect of upAUG mutation in the embryonic proinsulin leader sequence. Nucleotide sequence of the mutated embryonic proinsulin leader region (A). The proinsulin ATG (bold uppercase) and the two mutated AAG (bold lowercase) are labelled. The mutated nucleotides are underlined. In vitro translation and immunoprecipitation with anti-V5 epitope antibody of pancreatic (pCPro1A), embryonic (pCPro1B) and mutated embryonic (pCPro1Bmt) proinsulin expression constructs (B). The same constructs were transfected into NIH3T3 and RTC5 cells, and proinsulin expression analysed by western blotting. β-tubulin was used as a loading control (bottom panel) (C). NIH3T3 and RTC5 cells were transfected with control (pGL3C), pancreatic (pGLuc1A), embryonic (pGLuc1B) or mutated embryonic (pGLuc1Bmt) firefly luciferase expression constructs (D). Firefly luciferase (Fluciferase) activity was corrected by that of Renilla, cotransfected to normalize transfection efficiency. Results represent the mean ± SEM of four replicates.
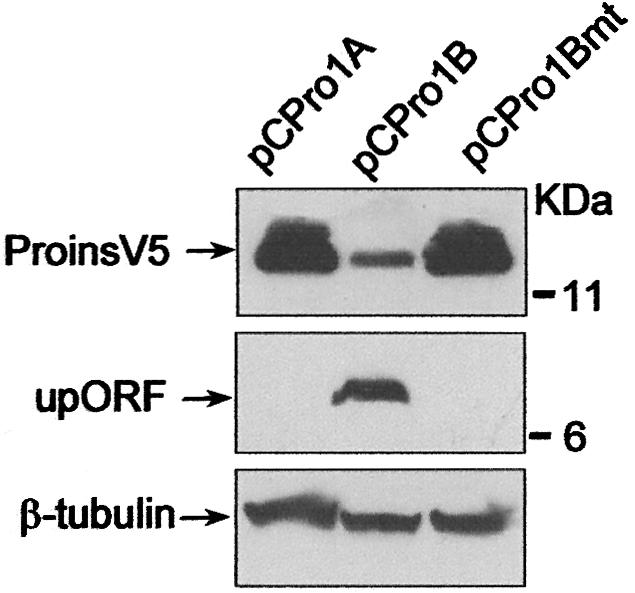
Fig. 8. Effect of upAUG mutation in upORF expression. NIH3T3 cells were transfected with pancreatic (pCPro1A), embryonic (pCPro1B) and mutated embryonic (pCPro1Bmt) proinsulin expression constructs. Cell extracts were analysed by western blotting using anti-V5 epitope or anti-upORF antibodies for proinsulin and upORF expression, respectively (upper and middle panels). β-tubulin was used as a loading control (bottom panel).
Proinsulin excess decreases naturally occurring apoptosis and causes embryonic malformations
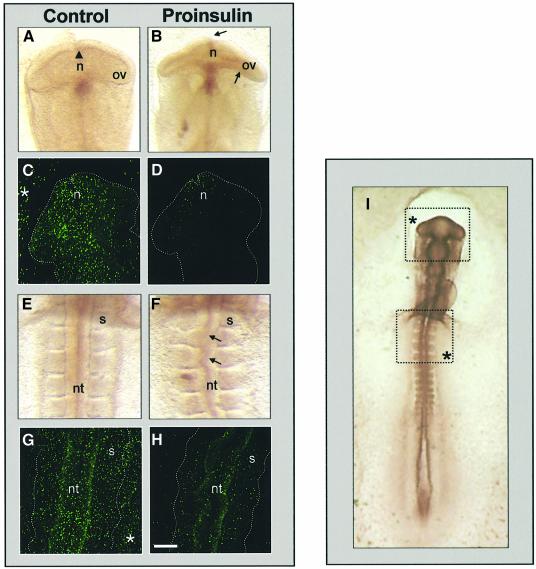
Thus far, the results demonstrate that the embryonic proinsulin transcript presents low translational activity due to two upAUGs not found in pancreatic proinsulin mRNA. To support the physiological relevance of these observations, we tried to confirm in vivo that an excess of proinsulin is as deleterious as its deficit, shown in the past in neurulating embryos (Morales et al., 1997). The effect of moderate addition of exogenous proinsulin was evaluated in neurulating chick embryos. Proinsulin at 10–7 M signals through hybrid insulin/IGF-I receptors and homodimeric insulin receptors, but not through IGF-I receptors (García-de Lacoba et al., 1999; see Discussion for details). Strikingly, gross developmental abnormalities were observed after 8 h treatment in most (12 of 15) proinsulin-treated embryos, but none of the control embryos (Table I). Developmental progression, as measured by number of new somites formed during treatment, was nonetheless identical in control and proinsulin-treated embryos (six to seven somites). Frequent abnormalities included neural defects such as asymmetric anterior neuropore closure, collapsed optic vesicles and flexures in the neural tube (Figure 9B and F). Proinsulin-treated embryos showed a strong reduction in TdT-mediated dUTP nick end labelling (TUNEL) staining in the regions in which malformations had been observed, compared with control embryos (Figure 9C, D, G and H). This concurs with the effect described for proinsulin in attenuation of cell death at this stage (Morales et al., 1997; Hernández-Sánchez et al., 2002).
Table I. Developmental abnormalities induced by proinsulin excess in E1.5 chick embryos.
| No. of embryos | |
|---|---|
| Control embryos abnormal | 0/13 |
| Proinsulin treated embryos abnormal | 12/15 |
| Abnormalities | |
| Asymmetric anterior neuropore closure | 5/15 |
| Flattened optic vesicles | 8/15 |
| Asymmetric rhombomers | 6/15 |
| Neural tube flexures | 9/15 |
| Reduction of apoptosis in proinsulin treated embryos | 5/5 |
Fig. 9. In ovo treatment with exogenous proinsulin. Embryos were treated in ovo with 50 µl of vehicle (A, C, E and G) or with 50 µl 10–7 M proinsulin (B, D, F and H). After 8 h incubation, the morphological aspect of the embryos was evaluated by counterstaining with neutral red (A, B, E and F). Apoptotic cells were visualized by TUNEL staining of whole embryos (C, D, G and H). Serial images were captured in a confocal microscope every 10 µm with a 10× objective and compiled. The bright dots represent TUNEL-stained pyknotic bodies. Different regions shown are boxed in the whole embryo picture (I); the upper box corresponds to the prosencephalon region (A–D) and the lower box to neural tube and rostral somites (E–H). A few dots around the prosencephalon (C) and the somites (G) indicated by asterisks in the boxes (I) are apoptotic cells in the extraembryonic membranes. The embryo is delimited by dotted lines in (C), (D), (G) and (H). The main morphological features are labelled: n, anterior neuropore (arrowhead in A); ov, optic vesicle; nt, neural tube; s, somite; arrows in (B) indicate the main morphological abnormalities. Bar, ∼120 µm (A–H) and ∼250 µm in (I).
Discussion
We have characterized a novel regulatory mechanism of proinsulin expression mediated by variations in transcript structure. The 32-nt extension of the embryonic proinsulin leader region limits proinsulin mRNA translation. This effect, which resides in two upAUGs absent in the pancreatic transcript, contributes to the maintenance of low embryonic proinsulin levels compatible with the need for programmed cell death for normal development, since moderate proinsulin excess induces embryonic malformations.
Prior to the onset of its endocrine function in maintaining glucose homeostasis, proinsulin attenuates cell death during embryonic and retinal development (Hernández-Sánchez et al., 1995, 2002; Morales et al., 1997; Díaz et al., 1999, 2000). Consistent with its dual biological role, regulation of proinsulin transcription, translation and posttranslational modifications appears to depend on the stage of development. Transcription of the chicken proinsulin gene generates embryonic and pancreatic mRNAs that differ in their 5′ UTR sequences. We show that translation of embryonic proinsulin is attenuated by the presence of upAUGs in the leader region of the embryonic transcript in whole cells and in a cell-free system. Concordant with this inhibitory role, mutation of the upAUG triplets to AAG abrogated the translation attenuation. These effects were observed in two unrelated cell types, NIH3T3 cells, which do not express endogenous proinsulin (not shown), and the embryonic quail retina RTC5 cell line, which expresses embryonic proinsulin mRNA constitutively.
The effect of upAUGs and upORFs on downstream translation initiation is not fully understood, although recent evidence reveals complex and diverse regulatory mechanisms (Gaba et al., 2001; Dever, 2002). Translation of the embryonic upORF, located out-of-frame relative to the proinsulin or the luciferase coding region, generated two peptides that share 34 amino acids in the N-terminal domain, but differ in 60 amino acids in their C-terminal regions. The fact that the inhibitory effect of the embryonic proinsulin leader region was independent of the sequence fused to its 3′ end suggests that the translational attenuation is due to efficient recognition of the upAUGs. Nonetheless, we cannot exclude the possibility that the common N-terminal fragment of the nascent peptide may mediate the regulatory effect. A protein homology computer search did not reveal conservation of the nascent upORF with proteins known to be involved in RNA biology. No evidence is yet available for other mechanisms by which ribosomes can reach the proinsulin downstream translation start codon, such as internal initiation (Hellen and Sarnow, 2001; Martínez-Salas et al., 2001). Moreover, reinitiation (Morris and Geballe, 2000) seems very unlikely, as there is an extensive overlapping sequence (183 nt) between the upORF and proinsulin ORF. Leaky scanning usually operates in mRNAs with poor Kozak consensus in the upAUGs, allowing relatively efficient recognition of the downstream start codon. In contrast, the consensus sequence around the upAUGs in the embryonic proinsulin matches that operating in the scanning mechanism (Kozak, 2002b). Accordingly, only a very small percentage of ribosomal subunits entering the 5′ end of the mRNAs is predicted to ignore them, gaining access to the proinsulin start codon.
It has been reported that AUGs closer than 8 nt to the 5′ end may not be recognized by scanning ribosomes (Pestova and Kolupaeva, 2002), although leakiness was suppressed when the leader sequence was lengthened to 20 nt (Kozak, 1991). The proinsulin upAUG1/2 are located at 8 and 20 nt from the 5′ end, respectively. The ribosome subunits that bypassed the first upAUG would thus not skip the second upAUG. It is nonetheless possible that the strong effect of the upAUGs studied here has been somewhat magnified by the increased length of the 5′ UTR due to vector polylinker sequences.
The situation described for chicken embryos also appears to apply to mammalian proinsulin. In preliminary experiments, a proinsulin transcript with an extended leader containing upAUG in good translational context was localized by conventional RT–PCR in 8-week fetal human head and 7.5-day mouse embryo total RNA (not shown). Furthermore, attenuation of translational efficiency by the presence of upORF in leader regions, a novel feature for the proinsulin gene, is well known for genes encoding proteins controlling cell growth and differentiation, whose translation efficiency is tightly regulated (Morris and Geballe, 2000; Geballe and Sachs, 2000; Kozak, 2002a). The physiology underlying the constrained translation imposed by the upORFs is thought to be selected to prevent inappropriate expression of these proteins, as appears to be the case of embryonic proinsulin. We previously showed that reduction with proinsulin levels or signalling induces cell death in the neurulating chick embryo and retina. Here we show that an increase in proinsulin availability causes malformations in vivo, correlating with an observed reduction in cell death. Even larger amounts of proinsulin and insulin were shown to be teratogenic in embryos in early organogenesis (De Pablo et al., 1985b).
Programmed cell death is a natural process required for correct morphogenesis during embryonic development (Jacobson et al., 1997; Vaux and Korsmeyer, 1999; de la Rosa and De Pablo, 2000). Cell death levels must be finely regulated, since either a reduction in naturally occurring cell death during development or its increase results in dramatic phenotypes (Kuida et al., 1996, 1998; Barnes et al., 1998; Gao et al., 1998; Roth and D’Sa, 2001). This fine regulation is also maintained at the cellular level, where a balance among pro- and anti-apoptotic signalling elements is established through several mechanisms, including regulation of pro- and anti-apoptotic growth factors, pro- and anti-apoptotic members of the Bcl-2 family, caspases, caspase inhibitors (IAP) and inhibitors of caspase inhibitors (Diablo) (Strasser et al., 2000; Joza et al., 2002). In our view, embryonic proinsulin expression is subject to fine, stringent regulation to ensure sufficient protein levels for its survival role, but not in excess, which would prevent necessary developmental cell death. The amounts and regulation of insulin required for its endocrine role are completely different, as the pancreas must rapidly secrete relatively large amounts into the bloodstream in response to food-derived glucose and other metabolites. Our results suggest that the very distinct required proinsulin levels are achieved by regulation of the transcript structure. First, the extended upAUG-containing embryonic transcript ensures very low proinsulin expression levels. Secondly, the onset of an alternative initiation of transcription in pancreas (Hernández-Sánchez et al., 2002), leading to a transcript that avoids the upAUGs, allows for high production of proinsulin and insulin. Furthermore, the signalling receptor used in embryogenesis is also regulated posttranscriptionally. In the embryonic retina, we previously characterized a hybrid heterodimeric insulin-IGF receptor able to bind proinsulin at stages when the mature dimeric insulin receptor is low or absent. The hybrid receptor decreases as development proceeds, after which homodimeric insulin receptors appear (Garcia-de Lacoba et al., 1999).
All together, these data show constrained translational activity of proinsulin prior to the onset of its pancreatic production operating on the embryonic specific transcript. This translational restriction of embryonic proinsulin, expressed at very low levels as many other growth factors, further reflects the need for fine regulation of the cell death process during early development.
Materials and methods
Chick embryos
Fertilized White Leghorn eggs (Granja Rodríguez-Serrano, Salamanca, Spain) were incubated at 38.4°C and 60–90% relative humidity for the periods indicated. Embryos were staged according to Hamburger and Hamilton (1951).
Cell lines, transfections and pulse labelling
NIH3T3 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine and antibiotics (all from Gibco-BRL, Gaithersburg, MD), at 37°C and 5% CO2. RTC5 cells (Pollerberg et al., 1995) were cultured in DMEM:F12 (Sigma, St Louis, MO) supplemented with 8% FBS, 2% chicken serum (Sigma), 2 mM glutamine, antibiotics, 2 µM β-estradiol (Sigma) and 0.5% Biomatrix EHS solution (Serva, Heidelberg, Germany), at 39°C and 6% CO2. For proinsulin treatment, RTC5 cells were cultured in serum-free medium (DMEM:F12, 0.1% bovine serum albumin, 20 mM HEPES, pH 7.5, 2 mM glutamine and antibiotics). NIH3T3 and RTC5 cells were transfected using Lipofectamine Plus or Lipofectamine 2000, respectively (Invitrogen, Carlsbad, CA), and analysed 48 h after transfection. Where indicated, cells were co-transfected with the pCMVRenilla luciferase vector (Promega, Madison WI) to correct for transfection efficiency variation. Firefly and Renilla luciferase activities were measured with the dual luciferase system (Promega) according to manufacturer’s instructions. When required, NIH3T3 cells were pulse-labelled 20 h after transfection with 100 µCi [35S]Met/Cys ProMix (Amersham) in 500 µl of medium per 35 mm dish. Following incubation for 3 h at 37°C cells were lysed in 50 mM Tris–HCl pH 7.5, 120 mM NaCl, 0.5% NP-40 and protease inhibitor cocktail.
Genomic PCR
High molecular weight DNA was isolated from E4 chicken embryos by proteinase K digestion and phenol extraction. PCR was performed using sense primers 5′-ATATGGGAAAGAGAATGGGG-3′ (P1) or 5′-CCAGTCTTCATCTCTGAGAG-3′ (P2), the antisense primer 5′-GCTCTCCACACACCAGGTAG-3′ (P3) and the High Fidelity system (Roche Diagnostics, Mannheim, Germany). The DNA sequence of the PCR products was determined following cloning into the pCRII-TOPO vector (Invitrogen).
Plasmids
Embryonic and pancreatic chicken proinsulin cDNAs were generated by standard procedures. The proinsulin coding region was amplified by reverse transcriptase reaction of E1.5 embryo total RNA using the Invitrogen Superscript II kit and the oligo-dT primer, followed by PCR with the 5′-ATGGCTCTCTGGATCCGATCACTG-3′ (sense, P4) and 5′-GCTAGTTGCAGTAGTTCTCCAGTT-3′ (antisense, P5) primers and cloning of the PCR product into the pCRII TOPO vector. The embryonic and pancreatic proinsulin 5′ UTR constructs (e1B and e1A, respectively) were described previously (Hernández-Sánchez et al., 2002). The BamHI fragment corresponding to 321 nt of the proinsulin coding region was then cloned in BamHI-digested pCRII TOPO vector, downstream of the embryonic or pancreatic 5′ UTRs.
To generate the embryonic and pancreatic proinsulin expression constructs (pCPro1B and pCPro1A, respectively), cDNAs were amplified using sense primers 5′-ATATAAATATGGGAAAGAGAATG-3′ (P6) and 5′-CTACCAGTCTTCATCTCTGA-3′ (P7) for embryonic and pancreatic sequences, respectively, and a common antisense primer 5′-GTTGCAGTAGTTCTCCAGTT-3′.
The PCR products were then cloned in-frame 5′ to the V5 epitope into pcDNA3.1/V5-His TOPO vector (Invitrogen). The transcripts produced from T7 and CMV promoters contain 71 or 124 nt, respectively, upstream of the proinsulin cDNA sequence, corresponding to the vector polylinker. These sequences were verified for the absence of upAUG codons or the formation of stable hairpin structures. Translation from the proinsulin AUG predicts a 122 amino acid protein fused to the V5 epitope, whereas the upstream AUGs predict two peptides of 94 and 90 amino acids.
The pGLuc1B and PGLuc1A luciferase constructs containing the embryonic and pancreatic 5′ UTRs, respectively, upstream of the firefly luciferase cDNA, were based on the pGL3 vector (Promega). Both 5′ UTRs were amplified using the P6 and P7 sense primers for the embryonic and the pancreatic forms respectively, and a common antisense primer 5′-AGAGAGCCATGGTGAGCTGG-3′. The antisense primer introduced a single substitution (underlined) to create a NcoI site (bold) that allowed subcloning of this fragment immediately upstream of the luciferase ATG. The PCR products were cloned into the pCRII TOPO shuttle vector, subsequently excised with HindIII and NcoI and cloned in similarly digested pGL3. The point mutations of the upstream ATGs were generated by the ExSite PCR-based site-directed mutagenesis method (Stratagene, La Jolla, CA). The identity and orientation of all constructs was verified by sequencing.
In vitro transcription/translation and immunoprecipitation
Constructs (2 µg each) were transcribed and translated with the TNT T7-coupled Reticulocyte Lysate System (Promega) and [35S]Cys (Amersham Pharmacia Biotech, Essex, UK), following the manufacturer’s protocol. The translation reaction was immunoprecipitated with anti-V5 epitope antibody (4°C, 16 h), precipitated with protein A–Sepharose (Amersham) and resolved in a 10% polyacrylamide NuPAGE gel (Invitrogen).
Antibody generation and immunoblotting
Anti-upORF antiserum was raised in rabbits using a synthetic peptide extending from amino acids 80 to 94 of the upORF (CASSTPPKPDGMSSSP). For proinsulin and upORF expression analysis, 48 h post-transfection NIH3T3 and RTC5 cells were lysed in 50 mM Tris–HCl (pH 7.5), 300 mM NaCl, 10 mM EDTA, 1% Triton X-100 and protease inhibitor cocktail (Roche). For luciferase analysis, 24 h post-transfection NIH3T3 cells were lysed in 50 mM Tris–HCl (pH 7.5), 120 mM NaCl, 0.5% NP-40 and protease inhibitor cocktail. The homogenates were clarified by centrifugation, equal amounts of proteins fractionated in a 10% NuPAGE gel and transferred to nitrocellulose or immobilon-P membranes. For proinsulin detection, we used a mouse anti-V5 epitope antibody (1/5000; Invitrogen) followed by anti-mouse Ig-HRP antibody (1/10 000; Jackson Immunoresearch Laboratories, West Grove, PA) and visualized by chemiluminiscence (Pierce, Rockford, IL). For upORF, the anti-upORF antibody (1/1000) followed by anti-rabbit Ig-HRP antibody (1/20 000; Jackson) was used. For luciferase, we used a rabbit anti-luciferase antibody (1/2000; Cortex Biochem, San Leandro, CA). After stripping, membranes were analysed with mouse anti-β-tubulin antibody (1/10 000; Sigma) as protein loading control.
Immunocytochemistry
NIH3T3 transfected cells were detached from the plate by trypsin incubation. An aliquot of fixed cells was deposited on a slide using a cytospin (66 g, 7 min). Cells were incubated with mouse anti-V5 epitope (1/150) and rabbit anti-upORF (1/0) antibodies followed by Texas red-conjugated anti-mouse (1/100; Molecular Probes, Eugene, OR) and Alexa 488-conjugated anti-rabbit (1/100; Molecular Probes) antibodies.
RT–PCR analysis
Total RNA was isolated using Trizol reagent (Invitrogen) and RNA integrity was verified by electrophoresis. Typically, the reverse transcriptase reaction was performed with 5 µg total RNA, the Invitrogen Superscript II kit and the oligo-dT primer. The pro insulin coding region was amplified using P4 and P5 primers. The embryonic transcript was amplified using the 5′-GAATGG GGAAATTTCTACCAGT-3′ (sense, P8) and the antisense P5 primers.
Northern blot
Equal amounts of RNA samples were fractionated by electrophoresis through formaldehyde–agarose gels and transferred to nylon membranes. For proinsulin, membranes were hybridized with a 32P-labelled chicken embryonic cDNA probe (449 bp). For luciferase, we used a 32P-labelled firefly luciferase cDNA probe (1656 bp). After stripping, membranes were hybridized with a 32P-labelled mouse GAPDH probe (330 bp). The GAPDH sequence was generated by RT–PCR of mouse pancreas RNA using the Invitrogen Superscript II kit and the oligo-dT primer, followed by amplification with the 5′-GCAATGCATCCTGCACCACC-3′ (sense) and 5′-TGTCATCATACTTGGCAGGT-3′ (antisense) primers. The PCR product was subsequently cloned into a pCRII TOPO vector.
Apoptosis detection
RTC5 cells were cultured for 20 h without Biomatrix in the presence or absence of serum, supplemented with the indicated concentrations of human recombinant proinsulin (a kind gift of Eli Lilly, Indianapolis, IN). Cells were detached from the plate by trypsin incubation. An aliquot of fixed cells was deposited on a slide using a cytospin (66 g, 7 min). Cells were stained and mounted with Vectashield reagent containing DAPI (Vector Laboratories Inc., Burlingame, CA) and apoptotic cell death was determined by counting pyknotic nuclei.
Apoptosis in embryos treated in ovo was assessed by TUNEL (Promega) of the fragmented DNA, performed in whole-mount embryos as described (Rubio et al., 2002).
Proinsulin application in ovo and embryo evaluation
A window was cut in the eggshell after 1.5 days of incubation. To assess the embryonic developmental stage, either 20 µl of neutral red (1% of a saturated solution in ethanol diluted in PBS) was applied to cover the embryo, or 100 µl of diluted India ink (1:1 in PBS) was injected beneath the blastoderm. Basal medium (50 µl; DMEM/F12 supplemented with 100 µg/ml transferrin, 16 µg/ml putrescin, 6 ng/ml progesterone, 5.2 ng/ml sodium selenite, 2 mM glutamine, 50 µg/ml gentamicin; all from Sigma) containing 0.7% (w/v) methylcellulose (Serva, Heidelberg, Germany), alone or supplemented with 10–7 M recombinant human proinsulin were applied over the embryo. Note that the proinsulin bolus diffuses into the extraembryonic fluid and the egg albumin (around 25 ml), thus decreasing the concentration acting on the embryo. Eggs were sealed with cellophane tape and incubated for a further 8 h, after which embryos were evaluated for macroscopic development traits under a Leica Wild MZ8 binocular microscope or processed to quantify apoptotic cell death by TUNEL, as above.
Acknowledgments
Acknowledgements
We thank Ms I.Alvarez for technical assistance and Ms M.T.Chavarría for helping with RTC5 cell culture. We also thank the UK HGMP Resource Center (Babraham, Cambridge) for providing the human fetus RNA and Eli Lilly for providing recombinant human proinsulin. This study was funded by grants BMC 2001-2132 from the Dirección General de Investigación y Desarrollo (MCYT, Spain), FIS 01/0952 and Red de Grupos RGDM G03/212 from the Instituto de Salud Carlos III (Spain) to F.P. and PM99-0094 to E.J.R. and BMC-2002-00983 to E.M.S. This work was also supported in part by the Deutsche Forschungsgemeinschaft, Collaborative Research Center 488: Molecular and Cellular Bases of Neural Development, Heidelberg. The fellowship to A.M. was awarded by the Ministerio de Ciencia y Tecnología. C.H.-S. is a Ramón y Cajal Investigator of the Ministerio de Ciencia y Tecnología (Spain).
References
- Alarcon C., Serna,J., Perez-Villamil,B. and De Pablo,F. (1998) Synthesis and differentially regulated processing of proinsulin in developing chick pancreas, liver and neuroretina. FEBS Lett., 436, 361–366. [DOI] [PubMed] [Google Scholar]
- Barnes D.E., Stamp,G., Rosewell,I., Denzel,A. and Lindahl,T. (1998) Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol., 8, 1395–1398. [DOI] [PubMed] [Google Scholar]
- Brogiolo W., Stocker,H., Ikeya,T., Rintelen,F., Fernandez,R. and Hafen,E. (2001) An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol., 20, 213–221. [DOI] [PubMed] [Google Scholar]
- Brunstedt J. and Chan,S.J. (1982) Direct effect of glucose on the preproinsulin mRNA level in isolated pancreatic islets. Biochem. Biophys. Res. Commun., 106, 1383–1389. [DOI] [PubMed] [Google Scholar]
- de la Rosa E.J. and De Pablo,F. (2000) Cell death in early neural development: beyond the neurotrophic theory. Trends Neurosci., 23, 454–458. [DOI] [PubMed] [Google Scholar]
- De Pablo F., Girbau,M., Gomez,J.A., Hernandez,E. and Roth,J. (1985a) Insulin antibodies retard and insulin accelerates growth and differentiation in early embryos. Diabetes, 34, 1063–1067. [DOI] [PubMed] [Google Scholar]
- De Pablo F., Hernandez,E., Collia,F. and Gomez,J.A. (1985b) Untoward effects of pharmacological doses of insulin in early chick embryos: through which receptor are they mediated? Diabetologia, 28, 308–313. [DOI] [PubMed] [Google Scholar]
- De Pablo F. and Roth,J. (1990) Endocrinization of the early embryo: an emerging role for hormones and hormone-like factors. Trends Biochem. Sci., 15, 339–342. [DOI] [PubMed] [Google Scholar]
- De Pablo F. and de la Rosa,E.J. (1995) The developing CNS: a scenario for the action of proinsulin, insulin and insulin-like growth factors. Trends Neurosci., 18, 143–150. [DOI] [PubMed] [Google Scholar]
- Deltour L., Leduque,P., Blume,N., Madsen,O., Dubois,P., Jami,J. and Bucchini,D. (1993) Differential expression of the two nonallelic proinsulin genes in the developing mouse embryo. Proc. Natl Acad. Sci. USA, 90, 527–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E. (2002) Gene-specific regulation by general translation factors. Cell, 108, 545–556. [DOI] [PubMed] [Google Scholar]
- Diaz B., Pimentel,B., De Pablo,F. and de la Rosa,E.J. (1999) Apoptotic cell death of proliferating neuroepithelial cells in the embryonic retina is prevented by insulin. Eur. J. Neurosci., 11, 1624–1632. [DOI] [PubMed] [Google Scholar]
- Diaz B., Serna,J., De Pablo,F. and de la Rosa,E.J. (2000) In vivo regulation of cell death by embryonic (pro)insulin and the insulin receptor during early retinal neurogenesis. Development, 127, 1641–1649. [DOI] [PubMed] [Google Scholar]
- Gaba A., Wang,Z., Krishnamoorthy,T., Hinnebusch,A.G. and Sachs,M.S. (2001) Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J., 20, 6453–6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y. et al. (1998) A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell, 95, 891–902. [DOI] [PubMed] [Google Scholar]
- Garcia-de Lacoba M., Alarcon,C., de la Rosa,E.J. and De Pablo,F. (1999) Insulin/insulin-like growth factor-I hybrid receptors with high affinity for insulin are developmentally regulated during neurogenesis. Endocrinology, 140, 233–243. [DOI] [PubMed] [Google Scholar]
- Geballe A.P. and Sachs,M.S. (2000) Translational control by upstream open reading frames. In Sonenberg,N., Hershey,J.W. and Mathews,M.B. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, New York, NY, pp. 595–614. [Google Scholar]
- Hamburger V. and Hamilton,H.L. (1951) A series of normal stages in the development of the chick embryo. J. Morphol., 88, 49–92. [PubMed] [Google Scholar]
- Hellen C.U. and Sarnow,P. (2001) Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev., 15, 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hernandez-Sanchez C., Lopez-Carranza,A., Alarcon,C., de la Rosa,E.J. and De Pablo,F. (1995) Autocrine/paracrine role of insulin-related growth factors in neurogenesis: local expression and effects on cell proliferation and differentiation in retina. Proc. Natl Acad. Sci. USA, 92, 9834–9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Sanchez C., Rubio,E., Serna,J., de la Rosa,E.J. and De Pablo,F. (2002) Unprocessed proinsulin promotes cell survival during neurulation in the chick embryo. Diabetes, 51, 770–777. [DOI] [PubMed] [Google Scholar]
- Itoh N. and Okamoto,H. (1980) Translational control of proinsulin synthesis by glucose. Nature, 283, 100–102. [DOI] [PubMed] [Google Scholar]
- Jacobson M.D., Weil,M. and Raff,M.C. (1997) Programmed cell death in animal development. Cell, 88, 347–354. [DOI] [PubMed] [Google Scholar]
- Joza N., Kroemer,G. and Penninger,J.M. (2002) Genetic analysis of the mammalian cell death machinery. Trends Genet., 18, 142–149. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1986) Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell, 44, 283–292. [DOI] [PubMed] [Google Scholar]
- Kozak M. (1991) A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr., 1, 111–115. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. (2002a) Emerging links between initiation of translation and human diseases. Mamm. Genome, 13, 401–410. [DOI] [PubMed] [Google Scholar]
- Kozak M. (2002b) Pushing the limits of the scanning mechanism for initiation of translation. Gene, 299, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuida K., Zheng,T.S., Na,S., Kuan,C., Yang,D., Karasuyama,H., Rakic,P. and Flavell,R.A. (1996) Decreased apoptosis in the brain and premature lethality in CPP32-deficient mice. Nature, 384, 368–372. [DOI] [PubMed] [Google Scholar]
- Kuida K., Haydar,T.F., Kuan,C.Y., Gu,Y., Taya,C., Karasuyama,H., Su,M.S., Rakic,P. and Flavell,R.A. (1998) Reduced apoptosis and cytochrome c-mediated caspase activation in mice lacking caspase 9. Cell, 94, 325–337. [DOI] [PubMed] [Google Scholar]
- Leibiger B., Moede,T., Schwarz,T., Brown,G.R., Kohler,M., Leibiger,I.B. and Berggren,P.O. (1998) Short-term regulation of insulin gene transcription by glucose. Proc. Natl Acad. Sci. USA, 95, 9307–9312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas E., Ramos,R., Lafuente,E. and Lopez,D.Q. (2001) Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol., 82, 973–984. [DOI] [PubMed] [Google Scholar]
- Morales A.V., Serna,J., Alarcon,C., de la Rosa,E.J. and De Pablo,F. (1997) Role of prepancreatic (pro)insulin and the insulin receptor in prevention of embryonic apoptosis. Endocrinology, 138, 3967–3975. [DOI] [PubMed] [Google Scholar]
- Morris D.R. and Geballe,A.P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol., 20, 8635–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen D.A., Welsh,M., Casadaban,M.J. and Steiner,D.F. (1985) Control of insulin gene expression in pancreatic beta-cells and in an insulin-producing cell line, RIN-5F cells. I. Effects of glucose and cyclic AMP on the transcription of insulin mRNA. J. Biol. Chem., 260, 13585–13589. [PubMed] [Google Scholar]
- Orci L., Vassalli,J.D. and Perrelet,A. (1988) The insulin factory. Sci. Am., 259, 85–94. [DOI] [PubMed] [Google Scholar]
- Perez-Villamil B., de la Rosa,E.J., Morales,A.V. and De Pablo,F. (1994) Developmentally regulated expression of the preproinsulin gene in the chicken embryo during gastrulation and neurulation. Endocrinology, 135, 2342–2350. [DOI] [PubMed] [Google Scholar]
- Perler F., Efstratiadis,A., Lomedico,P., Gilbert,W., Kolodner,R. and Dodgson,J. (1980) The evolution of genes: the chicken preproinsulin gene. Cell, 20, 555–566. [DOI] [PubMed] [Google Scholar]
- Permutt M.A. (1974) Effect of glucose on initiation and elongation rates in isolated rat pancreatic islets. J. Biol. Chem., 249, 2738–2742. [PubMed] [Google Scholar]
- Pestova T.V. and Kolupaeva,V.G. (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev., 16, 2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce S.B. et al. (2001) Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev., 15, 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollerberg G.E. and Eickholt,B.J. (1995) Target preference of embryonic retina cells and retinal cell lines is cell-autonomous, position-specific, early determined and heritable. Eur. J. Neurosci., 7, 1431–1441. [DOI] [PubMed] [Google Scholar]
- Pollerberg G.E., Kuschel,C. and Zenke,M. (1995) Generation of cell lines from embryonic quail retina capable of mature neuronal differentiation. J. Neurosci. Res., 41, 427–442. [DOI] [PubMed] [Google Scholar]
- Rhodes C.J. (2000) Processing of the insulin molecule. In Le Roith,D., Taylor,S.I. and Olefsky,J.M. (eds), Diabetes Mellitus: A Fundamental and Clinical Text. Lippincott Williams & Wilkins, Philadelphia, PA, pp. 20–38. [Google Scholar]
- Robbins D.C., Tager,H.S. and Rubenstein,A.H. (1984) Biologic and clinical importance of proinsulin. N. Engl. J. Med., 310, 1165–1175. [DOI] [PubMed] [Google Scholar]
- Roth K.A. and D’Sa,C. (2001) Apoptosis and brain development. Ment. Retard. Dev. Disabil. Res. Rev., 7, 261–266. [DOI] [PubMed] [Google Scholar]
- Rubio E., Valenciano,A.I., Segundo,C., Sanchez,N., De Pablo,F. and de la Rosa,E.J. (2002) Programmed cell death in the neurulating embryo is prevented by the chaperone heat shock cognate 70. Eur. J. Neurosci., 15, 1646–1654. [DOI] [PubMed] [Google Scholar]
- Serrano J., Bevins,C.L., Young,S.W. and De Pablo,F. (1989) Insulin gene expression in chicken ontogeny: pancreatic, extrapancreatic, and prepancreatic. Dev. Biol., 132, 410–418. [DOI] [PubMed] [Google Scholar]
- Shalev A., Blair,P.J., Hoffmann,S.C., Hirshberg,B., Peculis,B.A. and Harlan,D.M. (2002) A proinsulin gene splice variant with increased translation efficiency is expressed in human pancreatic islets. Endocrinology, 143, 2541–2547. [DOI] [PubMed] [Google Scholar]
- Shuldiner A.R., De Pablo,F., Moore,C.A. and Roth,J. (1991) Two nonallelic insulin genes in Xenopus laevis are expressed differentially during neurulation in prepancreatic embryos. Proc. Natl Acad. Sci. USA, 88, 7679–7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A., O’Connor,L. and Dixit,V.M. (2000) Apoptosis signaling. Annu. Rev. Biochem., 69, 217–245. [DOI] [PubMed] [Google Scholar]
- Varela-Nieto I., de la Rosa,E.J., Valenciano,A.I. and León,Y. (2003) Cell death in the nervous system. Lessons from insulin and insulin-like growth factors. Mol. Neurobiol., 28, 23–50. [DOI] [PubMed] [Google Scholar]
- Vaux D.L. and Korsmeyer,S.J. (1999) Cell death in development. Cell, 96, 245–254. [DOI] [PubMed] [Google Scholar]
- Welsh M., Nielsen,D.A., MacKrell,A.J. and Steiner,D.F. (1985) Control of insulin gene expression in pancreatic beta-cells and in an insulin-producing cell line, RIN-5F cells. II. Regulation of insulin mRNA stability. J. Biol. Chem., 260, 13590–13594. [PubMed] [Google Scholar]
- Welsh M., Scherberg,N., Gilmore,R. and Steiner,D.F. (1986) Translational control of insulin biosynthesis. Evidence for regulation of elongation, initiation and signal-recognition-particle-mediated translational arrest by glucose. Biochem. J., 235, 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]