Abstract
High Mobility Group 1 protein (HMGB1) is a chromatin component that, when leaked out by necrotic cells, triggers inflammation. HMGB1 can also be secreted by activated monocytes and macrophages, and functions as a late mediator of inflammation. Secretion of a nuclear protein requires a tightly controlled relocation program. We show here that in all cells HMGB1 shuttles actively between the nucleus and cytoplasm. Monocytes and macrophages acetylate HMGB1 extensively upon activation with lipopolysaccharide; moreover, forced hyperacetylation of HMGB1 in resting macrophages causes its relocalization to the cytosol. Cytosolic HMGB1 is then concentrated by default into secretory lysosomes, and secreted when monocytic cells receive an appropriate second signal.
Keywords: histones/HMGB/inflammation/NLS/NES/sepsis
Introduction
High Mobility Group protein 1 (HMGB1) is a highly conserved component of eukaryotic nuclei (reviewed by Bustin, 1999; Thomas and Travers, 2001; Agresti and Bianchi, 2003). HMGB1 is almost ubiquitous and only 10 times less abundant than core histones, at 106 molecules per typical mammalian cell. It has a tripartite structure, composed of two homologous DNA-binding domains, the HMG boxes, and a C-terminal domain of aspartic and glutamic acids.
In most cells HMGB1 is located in the nucleus, where it acts as an architectural protein that can bend DNA. DNA bending promotes the formation of complexes comprising several transcription factors, and the sliding of nucleosomes (Bonaldi et al., 2002). The phenotype of Hmgb1 knockout mice confirmed the functional importance of HMGB1 as a regulator of transcription: they die shortly after birth and show a defect in the transcriptional control exerted by the glucocorticoid receptor (Calogero et al., 1999).
We have recently shown that HMGB1 is passively leaked out from cells when the integrity of membranes is lost during necrosis (Scaffidi et al., 2002). The presence of a nuclear protein in the extracellular space is a good indication for cellular death; moreover, apoptotic cells retain HMGB1 firmly bound to their chromatin even when they lose the integrity of their membranes (late apoptosis or secondary necrosis), and do not release it. In this way, extracellular HMGB1 is a reliable indicator of necrosis, and an immediate trigger for inflammation (Scaffidi et al., 2002) and a panoply of responses in cells that have receptors to it (Andersson et al., 2000; Degryse et al., 2001; Fiuza et al., 2002; Sappington et al., 2002).
Remarkably, HMGB1 can also be secreted by monocytes, developing neurons and a few other cell types (reviewed by Müller et al., 2001b). Macrophages and monocytes secrete HMGB1 as a delayed response to activation by lipopolysaccharide (LPS), IL-1 or TNF (Wang et al., 1999): they therefore use HMGB1 as a proinflammatory cytokine.
The secretion of a nuclear protein poses formidable challenges. HMGB1 lacks a secretory signal peptide, and does not traverse the ER–Golgi system. We recently showed that activation of monocytes results in the accumulation of HMGB1 into cytoplasmic vesicles that display the features of secretory lysosomes (Gardella et al., 2002). Secretory lysosomes are Ca2+-regulated secretory organelles, particularly abundant in hematopoietic cells where they participate in inflammatory and immune responses by mobilizing their content into the external milieu in response to triggering signals (Blott and Griffiths, 2002). Exocytosis of HMGB1-containing secretory lysosomes is triggered by lysophosphatidylcholine (LPC), a lipid generated by phospholipase A2 at inflammation sites (Gardella et al., 2002).
The present work provides a molecular characterization of the mechanism of HMGB1 transfer from the nucleus to secretory lysosomes. We found that in activated monocytes HMGB1 is extensively modified by acetylation, and that the two major clusters of lysines are important both for acetylation and nuclear localization. Since HMGB1 also has two non-classical nuclear export signals (NESs), in most cells HMGB1 shuttles continually from the nucleus to the cytoplasm, but the equilibrium is almost completely shifted towards a nuclear accumulation. Fibroblasts forced to hyperacetylate HMGB1 with deacetylase inhibitors reduce its nuclear import: HMGB1 is then relocated to the cytoplasm. In monocytic cells, HMGB1 is moved further, and is concentrated into secretory lysosomes. Thus, monocytes have recruited a nuclear reshuttling mechanism and (de)acetylating activities to regulate the cellular localization of HMGB1, switching a chromatin protein into a cytokine in response to inflammatory stimuli.
Results
Bi-dimensional (2D) electrophoresis resolves several spots corresponding to multiply acetylated HMGB1 protein
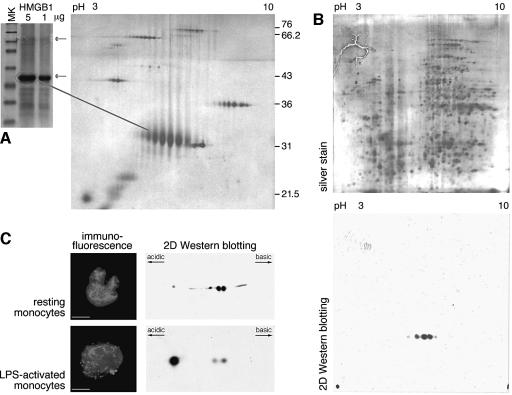
We reasoned that the alternative localization of a protein in two different cellular compartments must be subject to precise control, and must correspond to alternative states of the protein. We therefore investigated possible post-translational modifications of HMGB1 using 2D electrophoresis. HMGB1 can be easily purified in large amounts from calf thymus; purified HMGB1 separates into two bands in monodimensional SDS–PAGE, a major band corresponding to an apparent molecular weight of 29 kDa, and a minor band containing HMGB1 modified with ADP-ribosyl moieties (Figure 1A, left gel, and results not shown). In 2D electrophoresis, the faster migrating band resolves into 8 to 10 different spots (Figure 1A, right) with similar molecular weights and regular spacing along the pH gradient. A total extract of thymus from a 17-day-old mouse embryo was loaded onto twin 2D gels (Figure 1B), one of which was silver stained while the other was blotted and probed with anti-HMGB1 antibodies. The pattern of HMGB1 spots detected on the blot was similar to that obtained with purified HMGB1, taking into consideration that the most acidic and most basic spots were below the detection limit of a 2D blot from whole tissue.
Fig. 1. HMGB1 is multiply acetylated in thymus and activated monocytes. (A) HMGB1 purified from calf thymus gives rise on a monodimensional gel (SDS–PAGE, Coomassie stain, right panel) to two bands (arrows). The minor band contains ADP-ribosylated HMGB1. Fifty micrograms of the same sample of HMGB1 was subjected to 2D gel electrophoresis (silver stain, left panel); 2D Protein Marker from Bio-Rad was loaded together with HMGB1, generating a matrix of spots at the molecular weights listed on the right. (B) Total mouse thymus contains many isoforms of HMGB1. About 300 µg of protein from a thymus total extract (from a 17-day-old mouse embryo) were loaded on twin 2D gels; the gel shown at the top was silver stained, while the one on the bottom was blotted and assayed with anti-HMGB1 antibody. (C) LPS-activated human monocytes hyperacetylate HMGB1 and accumulate it in cytoplasmic vesicles. Monocytes purified from peripheral blood were cultured overnight, with or without LPS. Aliquots of activated and control monocytes were then fixed and immunostained with anti-HMGB1 antibody (red). HMGB1 is nuclear in unstimulated monocytes, as opposed to nuclear plus vesicular in LPS-activated monocytes. Bar represents 7 µm. Aliquots of untreated and LPS-activated monocytes were freeze-thawed, and about 400 µg of total protein extract was loaded onto 2D gels, blotted and immunodetected with anti-HMGB1. Note the major additional HMGB1 spot in activated monocytes.
We then verified that the alternative modification states of HMGB1 correlated with the activation state of freshly isolated monocytes (Figure 1C). Remarkably, while resting monocytes show only two isoforms of HMGB1, LPS-activated monocytes exhibit a major additional HMGB1 spot but at lower pI.
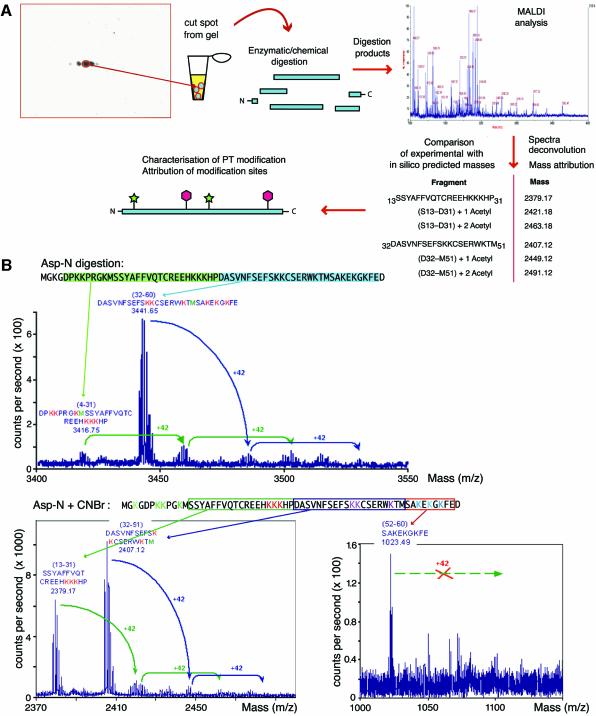
HMGB1 has long been known to be acetylated on lysines 2 and 11 (Sterner et al., 1979). In fact, in three different cell lines (3T3, HeLa and HEK) and in resting monocytes we found only two isoforms of HMGB1, consistent with previous knowledge; however, histone deacetylase (HDAC) inhibitors trichostatin A (TSA), sodium butyrate and HC-toxin generated multiple HMGB1 spots, similar to the ones observed in thymus extracts (results not shown). To localize the sites of post-translational modification, we started from the most abundantly available material, HMGB1 purified from thymus. We excised the four most abundant spots from Coomassie-stained 2D gels, and subjected them to trypsin digestion and mass spectrometry (MS). The mass difference among peptides was frequently 42 Da, or a multiple of this value, which corresponds to acetyl moieties. We could not find evidence of methylation, phosphorylation or glycosylation. The existence of up to 10 HMGB1 isoforms suggested that at least as many lysines would be acetylated. HMGB1 contains 43 lysines over a length of just 214 amino acids: to identify the acetylated lysines, we used different site-specific proteolytic agents [Asp-N, cyanogen bromide (CNBr), trypsin, Glu-C, Asp-C], both singly and in combination, and analyzed the peptides by MALDI (Figure 2A). One or more acetylations were assigned to large fragments of the protein, and their positions were then progressively restricted inside smaller fragments. In the example provided in Figure 2B, HMGB1 was first digested with Asp-N, and the non-acetylated fragments expected from the digestion were identified in the complex pattern of MS data. Peaks corresponding to the masses of the non-acetylated peptides plus 42 or n-multiples of 42 were presumptively assigned as 1-to-n acetylations of that specific peptide. Then, aliquots of the same peptides were further cleaved with CNBr, and smaller fragments were obtained. The assignment procedure was iterated, until the acetylations could be attributed to specific lysines.
Fig. 2. Strategy for 2D/MALDI-MS analysis of multiply modified HMGB1. (A) Spots were excised from 2D gels, proteolysed and analysed by MALDI-TOF. A mass was attributed to each peptide in the mixture; the masses corresponding to peptides predicted in silico were selected as ‘anchors’, and we searched the complex spectra for further peaks corresponding to anchor masses plus multiples of 42 (the molecular weight of an acetyl group). The analysis of two peptides is shown; the procedure was iterated for all peptides. We also mined the spectra for evidence of phosphorylation, methylation and glycosylation, without finding any. (B) An example of the multi-step digestion strategy developed to assign acetylation sites in HMGB1. Spots from 2D gels were digested in gel with protease Asp-N. One aliquot was analysed by MALDI-TOF: the arrows identify the peaks corresponding to unmodified fragment. We then looked for peaks with the molecular weight of unmodified fragments plus multiples of 42. The maximum number of acetyl moieties on each fragment was thus determined. Another aliquot of Asp-N digested HMGB1 was further digested with CNBr, and the products were analysed similarly. We proceeded with further cleavages until we could obtain identifiable fragments where all lysines were acetylated, or none was.
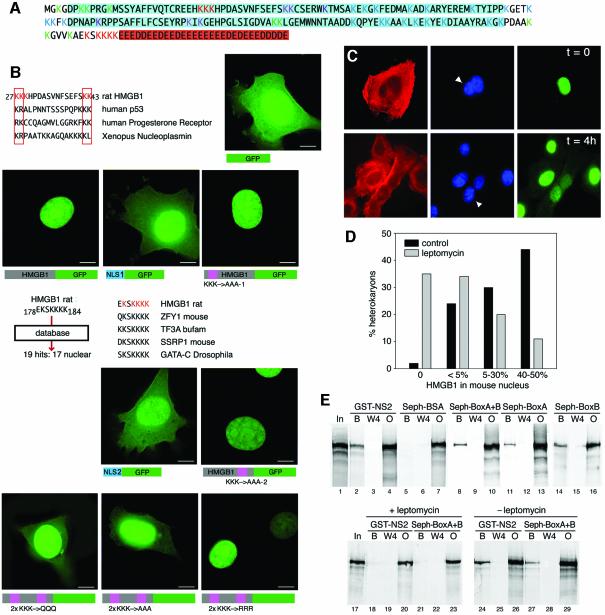
After analysing a large set of digestion products (not shown), we identified 17 acetylated lysines and excluded modification of 20 lysines; only 6 lysines remained uncharacterized (Figure 3A). If each of 17 lysines can be acetylated independently, there can exist 217 (>100 000) molecular species of HMGB1; however, two sets of lysines (red in Figure 3A) appeared to be acetylated concomitantly.
Fig. 3. HMGB1 has two segments with NLS activity and two NESs. (A) Final attribution of acetylation sites on the HMGB1 sequence. Lysines marked in red (8 out of 43) are frequently modified; lysines in blue are never modified (20/43); lysines in green (9/43) are modified with a low frequency; lysines in violet (6/43) are uncharacterized, because the peptides that contained them were not found in spectra. HMG boxes are highlighted in blue and the acidic tail in red. (B) Identification of peptides with NLS activity. Amino acids 27–43 of HMGB1 match perfectly with bipartite NLSs. This presumptive bipartite NLS was fused to GFP and expressed in mouse fibroblasts: while the NLS1–GFP fusion is predominantly nuclear, GFP alone is broadly diffuse. Mutation of lysines (K) 27, 28 and 29 of HMGB1–GFP into alanines (KKK→AAA-1) does not alter the nuclear localization. Sequence segments of HMGB1 were then launched into the PredictNLS database: the 178–184 segment matched with 19 proteins, 17 of them nuclear. This constitutes a good indication for a potential NLS, which we named NLS2. Its fusion to GFP (NLS2–GFP) showed a predominantly nuclear distribution. Mutation of lysines 181, 182 and 183 of HMGB1–GFP into alanines (KKK→AAA-2) does not alter the nuclear localization of the fluorescent protein. Mutation of all six lysines in the two clusters into alanines (2×KKK→AAA) or glutamines as a mimic of acetyl-lysine (2×KKK→QQQ) caused a partial cytosolic localization. Mutation of the six lysines to arginines (2×KKK→RRR) did not alter the nuclear localization. Bar represents 10 µm. (C) HMGB1 diffuses rapidly from the nucleus to the cytoplasm. Heterokaryons were formed by fusing HMGB1-expressing HeLa cells (human) and Hmgb1–/– mouse embryonic fibroblasts. Human cytokeratin and HMGB1 were stained red and green, respectively; nuclei were stained blue with Hoechst 33342. Cells with human cytokeratin staining and two nuclei, one of which with bright Hoechst-positive heterochromatic spots characteristic of mouse cells, were considered heterokaryons. Initially, only the human nucleus in the heterokaryons contained HMGB1 (t = 0). After incubation at 37°C for 4 h (t = 4 h), HMGB1 re-equilibrated from human to mouse nuclei. (D) HMGB1 exits the nucleus both by passive and active transport. Leptomycin B substantially reduces, but does not abolish, HMGB1 transfer between human and mouse nuclei in heterokaryons. For each treatment (leptomycin and control) 150 heterokaryons were evaluated; the 50% level is equivalent to complete equilibration. (E) Both HMG boxes interact directly with CRM1 exportin. Labelled CRM1 protein was mixed with beads bearing GST–NS2 immobilized onto glutathione–Sepharose or BSA, tail-less HMGB1 (BoxA+B) or individual boxes covalently linked to Sepharose. Aliquots representing the input material (In), the fourth wash (W4), the material remaining bound to beads after five washes (bound, B), and the unbound material (output, O) were electrophoresed and autoradiographed. CRM1 binds to all beads save the negative control BSA. Inclusion of 0.4 µM leptomicin B in the binding buffer prevents binding of CRM1 to both GST–NS2 and the NESs contained in the HMG boxes (lanes 18–23).
HMGB1 has a complex set of localization signals
Examination of the first acetylated cluster suggested that the lysines between residues 27 and 43 (Figure 3B) might represent a classical bipartite nuclear localization signal (NLS) (Cokol et al., 2001). We fused amino acids 27–43 of HMGB1 to the N-terminus of green fluorescent protein (GFP): unfused GFP was distributed throughout the cell, whereas ∼90% of fluorescence from the fusion protein (NLS1–GFP) was concentrated in the nucleus. Likewise, the peptide drove β-galactosidase to the nucleus (data not shown). Thus, amino acids 27–43 represent a functional NLS. We then mutated lysines 27–29 of the complete HMGB1–GFP fusion into alanines: surprisingly, nuclear localization was unaffected (KKK→AAA-1; Figure 3B). Visual inspection of the HMGB1 sequence did not reveal any other canonical NLS, but the program PredictNLS, which offers a collection of experimentally verified plus in silico-generated NLSs (Cokol et al., 2001), matched amino acids 178–184 with several nuclear proteins (Figure 3B). When fused to the N-terminus of GFP (NLS2–GFP), amino acids 178–184 showed clear NLS activity. Again, when lysines 181, 182, 183 of the HMGB1–GFP fusion were changed into alanines (KKK→AAA-2; Figure 3B), the fusion protein remained nuclear. Double mutants of HMGB1–GFP where both lysine clusters were changed to alanines or glutamines were clearly present in the cytoplasm (Figure 3B, 2×KKK→AAA and 2×KKK→QQQ), whereas the change of lysines into arginines did not affect the nuclear localization of the protein (Figure 3B, 2×KKK→RRR). Remarkably, however, even the double HMGB1 mutants were present in higher amount in the nucleus than in the cytoplasm. Apparently, HMGB1 is endowed with several regions that can contribute to nuclear localization, albeit not as isolated peptides but only in the context of the whole protein, as was previously shown for HMGB2 (Shirakawa et al., 1997).
HMGB1 migrates to the cytoplasm both by passive and active transport
We noticed the coincidence of acetylation clusters in HMGB1 purified from calf thymus and segments that, by themselves, had NLS activity on GFP fusions. The partial cytoplasmic localization of lysine-to-glutamine double mutants suggested that cells might exclude HMGB1 from the nucleus by acetylating those lysines, thereby neutralizing their basic charge and rendering them unable to function as NLSs. However, such a mechanism might only prevent the movement of HMGB1 from the cytoplasm to the nucleus, but could not promote the opposite movement, from nucleus to cytoplasm. To verify that HMGB1 could also traverse the nuclear membrane in the export direction, we fused Hmgb1–/– mouse embryonic fibroblasts (Calogero et al., 1999) with HMGB1-positive human HeLa cells (Figure 3C). Cells with human cytoplasm (staining red with anti-human cytokeratin antibodies) and a mouse nucleus (identified by bright Hoechst-positive heterochromatic spots) were heterokaryons. Shortly after fusion, mouse nuclei in heterokaryons had very little HMGB1, but 4 h at 37°C (in the presence of cycloheximide to inhibit protein synthesis) was generally sufficient to equilibrate HMGB1 (green) between the human and the mouse nucleus. We then treated heterokaryons with leptomycin B, an inhibitor of active export that involves the CRM1 exportin: re-equilibration of HMGB1 between human and mouse nuclei was strongly reduced (Figure 3D). To prove that HMGB1 has a CRM1-dependent nuclear export signal (NES), we synthesized labelled CRM1 in a reticulocyte extract, and passed it over columns that contained NS2 (a strong NES as positive control; Askjaer et al., 1999), BSA (a negative control), tail-less HMGB1 (BoxA+B), BoxA or BoxB (Figure 3E). CRM1 bound to both HMG boxes, indicating that each contains an NES. When 400 nM leptomycin was added to the reticulocyte extract containing CRM1, the labelled protein did not bind to either NS2 or BoxA+B, which contains both NESs. HMGB1 is a small protein (mol. wt, 25 kDa) and potentially it could migrate to and from the nucleus by simple diffusion across the nuclear pores: to test for passive diffusion, we incubated HeLa cells at 4°C for 4 h, a condition that impedes active, GTP-driven nuclear import/export. After cold incubation, a significant fraction of HMGB1 was localized in the cytoplasm (not shown).
Acetylation of HMGB1 determines its relocation to the cytoplasm in fibroblasts and to vesicles in monocytic cells
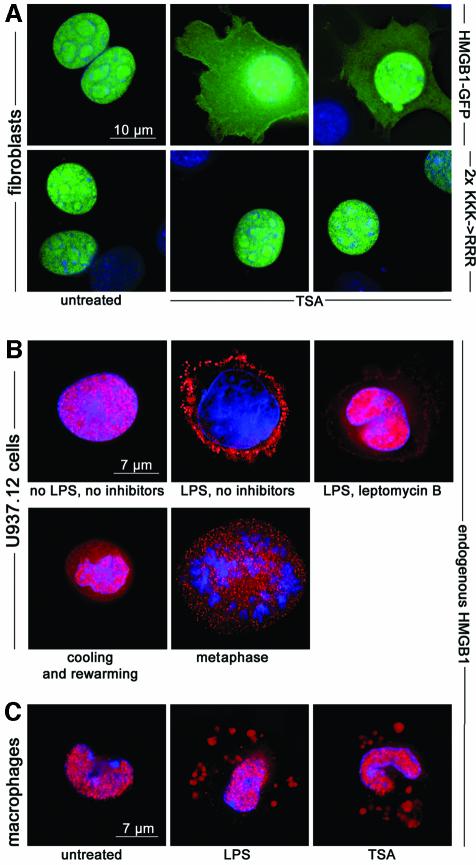
These results indicated that HMGB1 can traverse the nuclear membrane in both directions, by both passive and active mechanisms. However, all cultured cells and resting monocytes contain the vast majority of HMGB1 in the nucleus, indicating that in baseline conditions import is much more effective than export. To confirm that acetylation can tilt the balance by inhibiting nuclear import, we treated mouse fibroblasts expressing HMGB1–GFP with TSA for 1 h: a significant amount of fluorescent protein was relocated to the cytoplasm (Figure 4A). The 2×KKK→RRR mutant, which cannot be acetylated on six lysines, did not respond to TSA treatment and remained nuclear.
Fig. 4. HMGB1 moves to the cytoplasm following TSA treatment. (A) Exposure of mouse fibroblasts to 10 ng/ml TSA for 1 h causes a significant relocation of HMGB1–GFP to the cytoplasm; no vesicles are recognizable. The mutation of six lysines to arginines (2×KKK→RRR) prevents the cytoplasmic accumulation of HMGB1–GFP, even after TSA treatment. (B) U937.12 cells were cultured for 3 h with or without LPS or leptomycin B. Cells were then fixed and immunostained with anti-HMGB1 antibody (red). HMGB1 is exclusively nuclear in resting U937.12 cells, whereas it is predominantly vesicular in LPS-activated cells. Leptomycin blocks nuclear export, and consequently vesicular accumulation. A significant fraction of HMGB1 is contained in the cytoplasm and in vesicles in resting U937.12 cells that have been incubated 3 h at 4°C to promote passive diffusion of hypoacetylated HMGB1 to the cytoplasm and rewarmed to 37°C for 5 min to resume active transport (‘cooling and rewarming’). Likewise, a significant fraction of HMGB1 is contained in the cytoplasm and in vesicles when U937.12 cells enter M phase and free hypoacetylated HMGB1 into the cytoplasm after nuclear membrane breakdown (‘metaphase’). (C) Mouse peritoneal macrophages contain HMGB1 in the nucleus, but after stimulation with LPS, a fraction of HMGB1 is transferred to cytoplasmic vesicles. Incubation with TSA, but in the absence of LPS, also causes the transfer of HMGB1 to vesicles.
To assess the role of acetylation in monocytic cells, we used the U937 cell line and freshly isolated mouse peritoneal macrophages. Different subclones of U937 cells perform differently in cytokine secretion; for this reason, we chose a defined subclone, 12[-], with a well established profile of plasma membrane molecular markers and functional responses (Bovolenta et al., 1999). Resting U937.12 cells contained HMGB1 in the nucleus, whereas LPS-activated cells redistributed a significant fraction of the protein into cytoplasmic vesicles within 1 h, sometimes emptying their nucleus completely (Figure 4B). HMGB1 relocation in U937.12 cells is not blocked by cycloheximide (results not shown), but is sensitive to leptomycin B.
HMGB1 was also relocated in LPS-stimulated mouse macrophages (Figure 4C). The distinct morphology of HMGB1-containing vesicles in macrophages (fewer and much larger than in U937.12 cells) allowed us to test whether HMGB1 hyperacetylation per se can redirect HMGB1 from the nucleus towards secretion. Resting macrophages treated with 10 ng/ml TSA for 1 h appeared indistinguishable from macrophages activated with LPS.
We then tested whether hyperacetylation is necessary for accumulation into secretory lysosomes (Figure 4B). We incubated U937.12 cells for several hours at 4°C, causing the passive diffusion of a significant fraction of HMGB1 to the cytoplasm, and then raised the temperature back to 37°C. Within 5 min, a constellation of small vesicles became positive for HMGB1. Likewise, HMGB1 liberated into the cytoplasm by the breakdown of the nuclear membrane during mitosis (Falciola et al., 1997) was also associated with vesicles. We conclude that secretory lysosomes are equipped to accumulate HMGB1 as a default activity, and do so whenever HMGB1 is accessible to them in the cytosol.
HMGB1 is acetylated by PCAF, CBP and p300
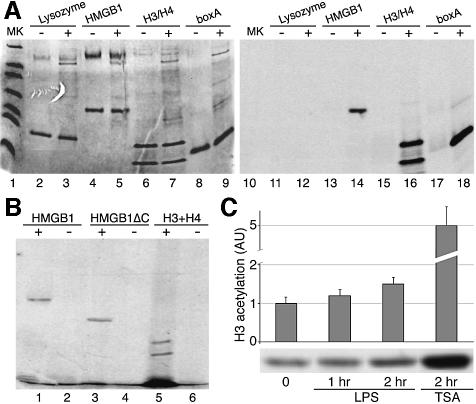
We also investigated whether HMGB1 can be acetylated by the most abundant histone acetylases. In vitro, a glutathione S-transferase (GST)–PCAF fusion acetylated bacterially produced HMGB1 about 50% as efficiently as recombinant histones H3 and H4, used as positive controls (Figure 5A). The single boxes, boxA and boxB, were both acetylated as efficiently. Lysozyme, a negative control, was not acetylated. Similar results were obtained with recombinant CBP and p300 HAT domains (data not shown). Immunoprecipitated PCAF, CBP and p300 also acetylate HMGB1 (Figure 5B and results not shown). Interestingly, U937 cells stimulated with LPS showed a marked increase in bulk H3 acetylation (Figure 5C) and a smaller but significant increase in bulk H4 acetylation (∼20% after 2 h; results not shown). These results indicate that monocytic cells respond to activation by tilting the acetylation/deacetylation balance in favour of acetylation, and suggest that HMGB1 acetylation might be part of a more general acetylation wave.
Fig. 5. HMGB1 is acetylated by histone acetyltransferases. (A) The indicated purified proteins were incubated with gluthatione–Sepharose-bound GST–PCAF (+), or control gluthatione–Sepharose (–), and [14C]acetyl-CoA. After incubation, the reaction products in the supernatant were separated from the beads and electrophoresed. The gel was Coomassie stained (left), dried and autoradiographed (right). (B) Native PCAF was immunoprecipitated from mouse 3T3 fibroblasts and incubated with recombinant histones H3 and H4, recombinant full-length HMGB1 and its tail-less derivative HMGB1ΔC. [14C]acetyl-CoA was added to the reaction mix in lanes 2, 4 and 6. (C) U937 cells were incubated with LPS for the indicated times, and the cell extracts assayed by western blotting with anti-acetyl H3 antibody. For comparison, the extent of H3 acetylation after incubation with TSA is also shown. The histogram shows the average and standard error of three independent experiments (the difference after LPS treatment is significant, P < 0.05).
Discussion
HMGB1 is unique among nuclear proteins in that it can be secreted, both by cells of myeloid origin (which use it as a proinflammatory cytokine) and by developing neural cells (where its role is much less well understood) (Müller et al., 2001b). The present work establishes that the alternative subcellular locations of HMGB1 depend on acetylation. This is based on the following evidence. (i) HMGB1 is post-translationally modified in monocytic cells. The pattern of migration of modified HMGB1 from monocytic cells is compatible with multiple lysine acetylation. HMGB1 purified from calf thymus is indeed multiply acetylated. (ii) Deacetylase inhibitors cause the relocalization of a fraction of HMGB1 from the nucleus to the cytoplasm. Mutation of six lysines to glutamine, which mimics acetylated lysine in that it has no positive charge, also causes the relocalization of a fraction of HMGB1 from the nucleus to the cytoplasm. Mutation of the same six lysines to arginine (which cannot be acetylated) makes HMGB1 localization unresponsive to deacetylase inhibitors. (iii) Once in the cytoplasm of monocytic cells, HMGB1 is accumulated by default in secretory lysosomes that undergo regulated exocytosis (Gardella et al., 2002). Thus, secretion depends indirectly on protein acetylation.
Our data show that the predominant nuclear localization of HMGB1 in the majority of cell types is the result of a steady state where HMGB1 molecules are continually shuttled between cytosol and the nucleus by energy-driven transport processes. Very early reports indicated that a variable proportion of HMGB1 is located in the cytoplasm (Bustin and Neihart, 1979): this observation might be due to the cooling of cells, a condition that blocks energy-driven transport.
The regulation of nuclear versus cytoplasmic protein localization by acetylation has been described before for transcription factors HNF4 and CTIIA (Soutoglou et al., 2000; Spilianakis et al., 2000); however, in these cases lysine acetylation promotes nuclear accumulation, in a process that appears to be related to NES function and protein export mechanisms. Very recently, acetylation of the NLS of viral protein E1A has been shown to cause its cytoplasmic accumulation (Madison et al., 2002). The process we describe is similar, save for the scale: a small fraction of E1A is acetylated on a single lysine, whereas around one million HMGB1 molecules per cell are multiply acetylated in monocytic cells.
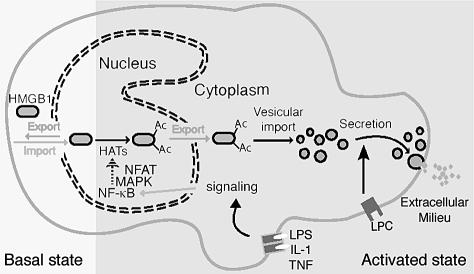
Monocytic cells must have developed a specific ability to acetylate massively a chromatin component such as HMGB1 in response to inflammatory stimuli. Figure 6 summarizes the information gathered so far. In monocytic cells, binding of proinflammatory signals to surface receptors activates a number of signaling pathways, including calcium signalling through calmodulin and NFAT/calcineurin, NF-κB and all MAP kinases. These signalling pathways must impinge directly on the enzymes responsible for acetylation/deacetylation of nuclear proteins, rather than indirectly through the expression of specific genes, since cycloheximide treatment of monocytic cells does not prevent histone acetylation or HMGB1 relocation. When the balance between HATs and HDACs is tilted in favour of acetylation, nuclear import is reduced because many positive charges are lost; as a consequence, a significant fraction of HMGB1 cannot re-enter the nucleus once it has been exported through the action of its NESs and the CRM1 exportin. Once HMGB1 is cytosolic, its accumulation into vesicles might simply require the presence of specific organelles, the secretory lysosomes. These are abundant in hematopoietic cells and melanocytes, but not in other cell types (Blott and Griffiths, 2002). Developing neurons can secrete HMGB1, but no accumulation in vesicles is apparent: how they secrete HMGB1 is still unknown.
Fig. 6. The control of HMGB1 secretion in professional inflammatory cells. In all cells, including resting inflammatory cells, HMGB1 shuttles between nucleus and cytoplasm; nuclear import is active, and the protein migrates back to the cytoplasm via passive diffusion and CRM1-mediated active export. When HMGB1 is underacetylated, the rate of nuclear import exceeds that of rediffusion plus export, and the protein appears predominantly or solely nuclear. Upon activation of inflammatory cells through binding of IL-1β, TNF-α, LPS or HMGB1 itself to their own receptors, the NF-κB and MAP kinase (MAPK) pathways are activated. Phosphorylated MAPKs migrate to the nucleus, where directly or via adaptor proteins they activate histone acetylases or inhibit deacetylases. This in turn promotes acetylation of HMGB1. Exported acetyl-HMGB1 cannot return to the nucleus. Myeloid cells are equipped with secretory lysosomes, a variety of lysosomes that can be secreted upon appropriate stimulation and that can accumulate IL-1β or HMGB1, presumably through specific transporters embedded in the lysosomal membrane. Upon binding of LPC (an inflammatory lipid) to its own receptor, the secretory lysosomes carrying HMGB1 fuse with the plasma membrane and secrete their cargo.
In our interpretation, professional inflammatory cells have redeveloped an ancient, non-specific necrosis signal into a real cytokine. This did not require extensive re-engineering, since HMGB1 shuttles between the nucleus and cytoplasm in all cells, can be acetylated by nuclear acetyltransferases, and secretory lysosomes are present in hematopoietic cells and serve for the secretion of other leaderless cytokines such as IL-1β and IL-18 (Andrei et al., 1999; Gardella et al., 2000). The only likely innovation was the grand scale of the chromatin acetylation response. The molecular mimicry between passive HMGB1 release and active HMGB1 secretion is not perfect, however: HMGB1 is highly acetylated for secretion, whereas passively released HMGB1 is not. The repeated reinvention of the chromatin protein HMGB1, as a necrosis signal first and as an inflammatory cytokine later, is remarkable in itself; potentially, it has also involved enough post-translational modification for therapeutic intervention. It might be possible to target the acetylation process itself to block harmful secretion of HMGB1 from monocytes/macrophages, for example during sepsis (Wang et al., 1999) or arthritis (Pullerits et al., 2003; Taniguchi et al., 2003).
Materials and methods
Protein expression and purification
Bacterially produced full-length HMGB1 and fragments thereof were purified as described (Müller et al., 2001a). HMGB1ΔC is a truncation that lacks the acidic tail (Bonaldi et al., 2002). Calf thymus HMGB1 was purified according to the protocol kindly provided by J.Bernués (CSIC, Barcelona). Briefly, thymus was minced in Buffer 1 (0.14 M NaCl, 0.5 mM EDTA, 0.1 mM PMSF), after removing the fat and the connective tissue. The homogenate was subjected to three 5% PCA extractions; supernatants collected after centrifuging were pooled and clarified with TCA (18% final concentration). Fractionated precipitations with acetone–HCl (400:1 v/v) and acetone alone eliminated histone H1. The sediment dissolved in borate buffer pH 9.0 was then passed once or twice (according to the purity of the sample) on a CM-Sephadex C25 column, and eluted with a NaCl gradient from 0.11 to 0.2 M.
Throughout this paper, residues in HMGB1 have been numbered according to Allfrey and coworkers (Sterner et al., 1979): the first amino acid of mature HMGB1 is a glycine, since the first methionine encoded by the gene is cleaved off after synthesis.
Plasmids
Plasmid pEGFP-HMGB1 expressing HMGB1-GFP (Scaffidi et al., 2002) was used as template to generate the mutants in NLS1 and NLS2 in multi-step PCR mutagenesis, using HMG-GFPdir 5′-ATCCTCgagACAT gggCAAAggAgATCC-3′ and HMG-GFPrev 5′- ggCCCgcGGTTCAT CATCATCATCTTCTTCTTC-3′ as external primers and six pairs of internal mutagenic primers (substitutions in bold): NLS1KQdir 5′-gAg gAgCACCAgCAgCAgCACCCggATg-3′ and NLS1KQrev 5′-CATCC gggTgCTgCTgCTggTggTCCTC-3′; NLS1KRdir 5′-CACAggAggAgg CACCCggATgCTTCTgTC-3′ and NLS1KRrev 5′-gTgCCTCCTCCT gtgCTCCTCCCgGCAg-3′; NLS1KAdir 5′-gAggAgCACgCggCggCg CACCCggATgC-3′ and NLS1KArev 5′-gCATCCgggTgCgCCgCCgC gTgCTCCTC-3′; NLS2KQdir 5′-AgCCAgCAACAgAAggAAgAggAA gACgACgAg-3′ and NLS2KQrev 5′-CTTCTgTTgCTggCTCTTCTC AgCCTTgAC-3′; NLS2KRdir 5′-AgCAggAgAAggAAggAAgAggAAg ACgACgAg-3′and NLS2KRrev 5′-CTTCCTTCTCCTgCTCTTCTCA gCCTTgAC-3′; NLS2KAdir 5′-AgCgCggCAgCgAAggAAgAggAAg ACgAC-3′ and NLS2KArev 5′-CgCTgCCgCgCTCTTCTCAgCCTTg AC-3′. The final PCR products were then cloned into pEGFP-HMGB1 cut with XhoI and SacII to obtain the mutant plasmids.
We generated the constructs for NLS1–GFP and NLS2–GFP fusions by cloning between the XhoI/SacII sites of the pEGFP-N1 vector two cassettes, produced by annealing the oligonucleotide pairs NLS1dir 5′-TCTACTCgAgACATGAAgAAgAAgCACCCggATgCTTCTgTCAAC TTCTCAgAgTTCTCCAAgAAgCCgCggCTAA-3′ and NLS1rev 5′-TT AgCCgCggCTTCTTggAgAACTCTgAgAAgTTgACAgAAgCATCCgg gTgCTTCTTCT-TCATgTCTCgAgTAgA-3′; NLS2dir 5′-TCTACTCgAg ACATGAAgAgCAAgAAAAAg-AAggAACCgCggCTCA-3′ and NLS2rev 5′-TgAgCCgCggTTCCTTCTTTTTCTTgCTCTTCTAgTCTCgAgTAgA-3′.
All constructs were verified by sequencing.
2D gel electrophoresis
About 50 µg of purified HMGB1 or 250 µg of total cellular protein were added to 350 µl of rehydration buffer containing 8 M urea, 2% CHAPS, 20 mM dithioerythritol, 0.8% IPG buffer [carrier ampholytes, pH 3–10 non-linear (NL) or pH 4–7 linear (L)]. Samples were applied on 18 cm polyacrylamide (PA) gel strips (pH range 3–10 NL, or pH 4–7 L). Isoelectrophocusing (IEF) in IPGphor (Pharmacia Biotech) was stopped at 75 000–90 000 VoltHours. Second dimension runs were performed using a Protean II apparatus (Bio-Rad). After IEF, strips were soaked first in equilibration buffer (EB; 6 M urea, 3% SDS, 375 mM Tris pH 8.6, 30% glycerol, 2% DTE), then in EB containing 3% iodoacetamide and traces of bromophenol blue. Strips were then applied onto 10–12% PA gels. Gels were run at 90 V for ∼16 h, and were then either silver stained or transferred onto nitrocellulose membranes (ECL, Amersham) in 25 mM Tris pH 7.5, 0.192 M glycine, 20% methanol. We used the software ImageMaster 2D to calculate the pI of a spot from its position in a gel and to estimate the number of specific modifications corresponding to a defined ΔpI.
Mass spectrometry
Isolated protein spots were excised from 2D gels stained with colloidal Coomassie, and reduced and alkylated as described (Shevchenko et al., 1996). We then performed sequential proteolytic or chemical cleavages. In particular, trypsin, Asp-N, Glu-C digestions were performed in 50 mM NH4HCO3 buffer pH 8.0 at 37°C, with shaking, for 3 h to overnight according to the efficiency of cleavage. Chemical cleavage (Asp-C) was achieved by incubating spots overnight at 56°C in 2% formic acid. Spots blotted on PVDF membranes were incubated in the presence of CNBr in 70% trifluoroacetic acid for 1 h at room temperature in the dark. One microlitre of digestion products was loaded onto the MALDI target using the dried droplet technique and α-cyano-4-hydroxycinnamic acid or sinapinic acid as matrix.
MALDI-TOF mass measurements were performed on a Voyager-DE STR time-of-flight (TOF) mass spectrometer (Applied Biosystems, Framingham, MA) operated in the delayed extraction and reflector mode. Internally calibrated spectra were processed via the Data Explorer software.
Cell cultures and transfections
Mouse fibroblasts and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (Gibco), 100 IU/ml of penicillin and 100 µg/ml streptomycin, in 5% CO2 humidified atmosphere. HeLa cells and mouse fibroblasts were transfected with FuGene (Roche). To express fluorescent proteins, 105 cells were plated on coverslips in 35 mm dishes and were transiently transfected with 1 µg of the appropriate plasmid. Cells were observed 24 h after transfection. About 24 h after transfection, fibroblasts were treated for 1 h with 10 ng/ml trichostatin A (TSA; Sigma) before fixing and imaging. Subclone 12[-] of U937 promoyelocitic cells, kindly provided by M.Alfano and G.Poli (HSR, Milano), was grown in RPMI medium (Gibco) supplemented with 10% FCS, l-glutamine, 100 U/ml penicillin 100 µg/ml streptomycin, exposed for 3 days to 0.1 µM vitamin D3 (Roche) to promote CD14 expression, and stimulated with 100 ng/ml LPS (Sigma L4391) or with 10 ng/ml TSA for 1 h. Mouse macrophages, recovered by peritoneal lavage, and human monocytes, purified from peripheral blood, were maintained in RPMI medium supplemented as described above, and were activated with 100 ng/ml LPS.
Heterokaryon assays
HeLa cells were plated on glass coverslips in a six-well dish (100 000 cells/dish). After 16 h, 200 000 Hmgb1–/– mouse fibroblasts were plated on the same coverslips. Following a 3 h incubation in the presence of 100 µg/ml cycloheximide, the cells were washed with PBS and treated with 100 µl of prewarmed 50% PEG-6000 in PBS for 1 min. After three washes with PBS, the cells were incubated with DMEM containing 100 µg/ml cycloheximide for 4 h, and then fixed with 4% paraformaldehyde (PFA). Immunofluorescence was performed using anti-human cytokeratin (Santa Cruz) and anti-HMGB1 antibodies, and chromatin was visualized by Hoechst staining. When indicated, 150 nM leptomycin B (a kind gift of Barbara Wolff, Novartis, Vienna) was added to the medium together with cycloheximide.
For testing passive diffusion of HMGB1, HeLa cells were pretreated with 100 µg/ml cycloheximide at 37°C for 30 min, incubated at 4°C for 4 h, fixed with 4% PFA, and stained with anti-HMGB1 antibodies.
Pull-down assays with CRM1
CRM1 protein was in vitro transcribed–translated with the TnT Coupled Reticulocyte Lysate System (Promega) following the manufacturer’s protocol, using pSGCRM1 plasmid as template (Askjaer et al., 1999). Freshly made [35S]Met labeled CRM1 (7 µl) was incubated in 15 µl RAN Buffer (50 mM Tris–HCl pH 7.5, 200 mM NaCl, 2 mM MgCl2, 10% glycerol), 5 µl of 6× CRM1 buffer [20 mM HEPES–KOH pH 7.5, 80 mM CH3COOK, 4 mM Mg (CH3COO)2, 250 mM sucrose, 2.5 mM DTT], 1 mg/ml BSA, 400 nM leptomycin B where indicated, and ∼10 µl of beads bearing immobilized GST-NS2 (Askjaer et al., 1999), BSA, recombinant tail-less HMGB1 (identified as HMGB1ΔC in Bonaldi et al., 2002), BoxA, or BoxB. GST–NS2 was coupled to gluthatione–Sepharose (Amersham), the other proteins were covalently cross-linked to activated Sepharose–CH (Amersham). The incubation was for 1 h at 4°C on a rotating wheel. The beads were centrifuged, supernatants were dried in Savant. Beads were then washed five times at 4°C with 50 vol of PBS containing 9% glycerol, 5 mM MgCl2 and 1% NP-40. Beads were then boiled in 10 µl SDS–PAGE loading buffer, and loaded on a 8% SDS–PA gel together with dried supernatants (output), the fourth wash (W4) and equivalent amount of input as a reference. The gel was then blotted on nylon filter and exposed to X-ray film to detect labeled CRM1.
Immunofluorescence and GFP imaging
Cell cultured in LabTek II chambers (Nalgene) were directly fixed in 3.7% PFA in PHEM buffer (36.8 g/l PIPES, 13 g/l HEPES, 7.6 g/l EGTA, 1.99 g/l MgSO4, buffered to pH 7.0 with KOH) for 10 min at room temperature. After fixation, cells were washed with PBS and incubated for 3 min at 4°C with HEPES-based permeabilization buffer containing 300 mM sucrose and 0.2% Triton X-100. 15 min of incubation in blocking solution (BlS; 0.2% BSA in PBS) followed. Primary antibodies were then diluted in BlS to the final concentration and incubation was prolonged for 1 h at room temperature. After three rinses with BlS, cells were incubated with secondary antibodies in BlS for 1 h, washed three times with BlS, and then incubated with PBS containing 0.5 µg/ml Hoechst. The polyclonal rabbit anti-HMGB1 was purchased from BD PharMingen (Torrey Pines, CA), and used at 1:1600 dilution. Goat polyclonal antibodies against rabbit IgG (H+L) conjugated to Alexa Fluor 488 or 594 (working dilution 1:1000) were purchased from Molecular Probes (Eugene, OR).
Cells expressing HMGB1–GFP, its derivatives and the NLSs–GFP fusions were PFA-fixed as described above, then incubated in PBS containing Hoechst 33342 to stain the nuclei. Cells were imaged using Olympus 60× or 100× 1.4NA Plan Apo oil immersion objective lenses on a DeltaVision Restoration Microscopy System (Applied Precision, Issaquah, WA) built around an Olympus IX70 microscope equipped with mercury-arc illumination. Filters were from Chroma Technology Corp. (Brattleboro, VT) Hoechst 33342 excitation 360/40, emission 457/50; Alexa 488 or GFP excitation 490/20, emission 528/38; AlexaFluor 594, excitation 555/30, emission 617/73. Forty optical sections spaced by 0.4 µm were collected with a Coolsnap_Hq/ICX285 CCD camera (Photometrix, Tucson, AZ) and deconvolved by the constrained iterative algorithm available in the SoftWoRx 2.50 package (Applied Precision) using 10 iterations and standard parameters. Each image measured 512 × 512 pixels, and effective pixel size was 106 nm (60×) or 63 nm (100×).
In vitro and in vivo acetylation assays
Assays were performed in HAT buffer (50 mM Tris–HCl pH 8.0, 10% glycerol v/v, 1 mM DTT, 0.1 mM EDTA, 0.1 mM PMSF, 10 mM sodium butyrate), with 0.1–0.2 mg/ml substrate proteins, 18 mM [14C]acetyl-CoA (55 mCi/mmol; Amersham Pharmacia Biotech), and ∼20 ng/ml GST–PCAF and GST–CBP fusion proteins. Reactions were performed at 37°C for 20 min followed by 10 min at 4°C. Beads were separated from the substrate by centrifugation at 1000 g for 5 min. Supernatants were directly mixed with SDS–PAGE loading buffer, boiled and loaded on 10% tricine–PAGE. Gels were stained with Coomassie, then dried and autoradiographed. Films were scanned with Personal Densitometer IS (Molecular Dynamics) and bands were quantified with ImageQuant software.
Rabbit antibodies against PCAF, CBP and p300 (Santa Cruz Biotechnology) were used to specifically immunoprecipitate HATs. About 200 µg of total protein from 3T3 extracts were first precleared by 3 h of incubation with protein A–Sepharose, then were incubated overnight at 4°C with antibodies (1:100 dilution, corresponding to ∼1.5 µg Ig) in 300 µl RIPA buffer (10 mM Tris–HCl pH 8.0, 1% Triton, 0.1% SDS, 0.1% sodium deoxycholate, 140 mM NaCl, 1 mM DTT, 1 mM PMSF, 1 mM EDTA, 60 µg/ml protease inhibitors cocktail). 20 µl protein A–Sepharose slurry (50% beads in RIPA buffer) was added; the mixtures were incubated at 4°C for 3 h on a rotating wheel; the beads were then centrifuged and washed five times in RIPA buffer while the supernatants were precipitated with acetone. Aliquots of the beads (IP samples) were loaded on an 8% SDS–PAGE gel, together with supernatants (Non-IP samples) and input, to check the efficiency of immunoprecipitation. Aliquots of the immunoprecipitated HATs, linked to the protein A–Sepharose, were incubated with about 0.5 µg of protein substrate and 80 mM [14C]acetyl-CoA in a total volume of 40 µl buffer HAT. Reactions were carried out at 37°C for 30 min, plus 10 min at 4°C. Reaction products were electrophoresed and quantified as indicated above.
In vivo acetylation of HMGB1 in cultured cells (HeLa, 3T3, HEK Phoenix) was performed incubating 70% confluent cells in DMEM to which various HDAC inhibitors were added: 5 ng/ml TSA, 10 ng/ml HC toxin, 5 mM sodium butyrate; cells were harvested at different time points by trypsinization and freeze–thawed three times in lysis buffer (50 mM PIPES pH 7.0, 50 mM KCl, 5 mM EGTA, 2 mM MgCl2, 1 mM DTT, 60 µg/ml protease inhibitor cocktail).
To monitor chromatin acetylation in LPS-treated U937 cells, 10 µg of cells was boiled in SDS–PAGE buffer and electrophoresed as indicated above. As a control, a sample of cells was incubated with 10 ng/ml TSA. After transfer to a filter, the upper part of the filter was stained for loading control, and the lower part was subjected to western blotting using rabbit anti-acetyl-H4 antibody kindly provided by Professor B.Turner (Birmingham) and rabbit anti-acetyl-H3 from Santa Cruz.
Bioinformatics
HMGB1 was inspected for potential NLSs using PredictNLS at http://cubic.bioc.columbia.edu/predictNLS, an automated tool for the analysis and determination of NLSs (Cokol et al., 2001).
Acknowledgments
Acknowledgements
We thank B.M.Turner and L.Vandel for helpful suggestions and reagents, A.J.Bannister, J.Bernues, C.Kilstrup-Nielsen and M.Vujanac for reagents and protocols, M.Alfano and G.Poli for U937.12 cells, L.Ronfani and I.Benzoni for mouse peritoneal macrophages and N.Collu for technical assistance. L.Falciola contributed unpublished results. This work was supported by the Italian Association for Cancer Research (AIRC) and by the Ministries for Health and for Education, University and Research.
References
- Agresti A. and Bianchi,M.E. (2003) HMGB proteins and gene expression. Curr. Opin. Genet. Dev., 13, 170–178. [DOI] [PubMed] [Google Scholar]
- Andersson U. et al. (2000) High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J. Exp. Med., 192, 565–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei C., Dazzi,C., Lotti,L., Torrisi,M.R., Chimini,G. and Rubartelli,A. (1999) The secretory route of the leaderless protein interleukin 1β involves exocytosis of endolysosome-related vesicles. Mol. Biol. Cell, 10, 1463–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askjaer P. et al. (1999) RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol., 19, 6276–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blott E.J. and Griffiths,G.M. (2002) Secretory lysosomes. Nat. Rev. Mol. Cell Biol., 3, 122–131. [DOI] [PubMed] [Google Scholar]
- Bonaldi T., Längst,G., Strohner,R., Becker,P.B. and Bianchi,M.E. (2002) The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J., 21, 6865–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovolenta C., Lorini,A.L., Mantelli,B., Camorali,L., Novelli,F., Biswas,P. and Poli,G. (1999) A selective defect of IFN-γ- but not of IFN-α-induced JAK/STAT pathway in a subset of U937 clones prevents the antiretroviral effect of IFN-γ against HIV-1. J. Immunol., 162, 323–330. [PubMed] [Google Scholar]
- Bustin M. (1999) Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol., 19, 5237–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin M. and Neihart,N.K. (1979) Antibodies against chromosomal HMG proteins stain the cytoplasm of mammalian cells. Cell, 16, 181–189. [DOI] [PubMed] [Google Scholar]
- Calogero S., Grassi,F., Aguzzi,A., Voigtländer,T., Ferrier,P., Ferrari,S. and Bianchi,M.E. (1999) The lack of chromosomal protein HMG1 does not disrupt cell growth, but causes lethal hypoglycaemia in newborn mice. Nat. Genet., 22, 276–280. [DOI] [PubMed] [Google Scholar]
- Cokol M., Nair,R. and Rost,B. (2001) Finding nuclear localization signals. EMBO Rep., 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degryse B., Bonaldi,T., Scaffidi,P., Müller,S., Resnati,M., Sanvito,F., Arrigoni,G. and Bianchi,M.E. (2001) The high mobility group (HMG) boxes of the nuclear protein HMG1 induce chemotaxis and cytoskeleton reorganization in rat smooth muscle cells. J. Cell Biol., 152, 1197–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciola L., Spada,F., Calogero,S., Längst,G., Voit,R., Grummt,I. and Bianchi,M.E. (1997) High mobility group 1 (HMG1) protein is not stably associated with the chromosomes of somatic cells. J. Cell Biol., 137, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiuza C., Bustin,M., Talwar,S., Tropea,M., Gerstenberger,E., Shelhamer,J.H. and Suffredini,A.F. (2002) Inflammatory promoting activity of HMGB1 on human microvascular endothelial cells. Blood, 27, 2652–2660. [DOI] [PubMed] [Google Scholar]
- Gardella S., Andrei,C., Poggi,A., Zocchi,M.R. and Rubartelli,A. (2000) Control of interleukin-18 secretion by dendritic cells: role of calcium influxes. FEBS Lett., 481, 245–248. [DOI] [PubMed] [Google Scholar]
- Gardella S., Andrei,C., Ferrera,D., Lotti,L.V., Torrisi,M.R., Bianchi,M.E. and Rubartelli,A. (2002) The nuclear protein HMGB1 is secreted by monocytes via a non-classical, vesicle-mediated secretory pathway. EMBO Rep., 3, 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison D.L., Yaciuk,P., Kwok,R.P. and Lundblad,J.R. (2002) Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J. Biol. Chem., 277, 38755–38763. [DOI] [PubMed] [Google Scholar]
- Müller S., Bianchi,M.E. and Knapp,S. (2001a) Thermodynamics of HMG1 interaction with duplex DNA. Biochemistry, 40, 10254–10261. [DOI] [PubMed] [Google Scholar]
- Müller S., Scaffidi,P., Degryse,B., Bonaldi,T., Ronfani,L., Agresti,A., Beltrame,M. and Bianchi,M.E. (2001b) The double life of HMGB1 chromatin protein architectural factor and extracellular signal. EMBO J., 20, 4337–4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullerits R., Jonsson,I.M., Verdrengh,M., Bokarewa,M., Andersson,U., Erlandsson-Harris,H. and Tarkowski,A. (2003) High mobility group box chromosomal protein 1, a DNA binding cytokine, induces arthritis. Arthritis Rheum., 48, 1693–1700. [DOI] [PubMed] [Google Scholar]
- Sappington P.L., Yang,R., Yang,H., Tracey,K.J., Delude,R.L. and Fink,M.P. (2002) HMGB1 B box increases the permeability of Caco-2 enterocytic monolayers and impairs intestinal barrier function in mice. Gastroenterology, 123, 790–802. [DOI] [PubMed] [Google Scholar]
- Scaffidi P., Misteli,T. and Bianchi,M.E. (2002) Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature, 418, 191–195. [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm,M., Vorm,O. and Mann,M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem., 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Shirakawa H., Tanigawa,T., Sugiyama,S., Kobayashi,M., Terashima,T., Yoshida,K., Arai,T. and Yoshida,M. (1997) Nuclear accumulation of HMG2 protein is mediated by basic regions interspaced with a long DNA-binding sequence and retention within the nucleus requires the acidic carboxyl terminus. Biochemistry, 36, 5992–5999. [DOI] [PubMed] [Google Scholar]
- Soutoglou E., Katrakili,N. and Talianidis,I. (2000) Acetylation regulates transcription factor activity at multiple levels. Mol. Cell, 5, 745–751. [DOI] [PubMed] [Google Scholar]
- Spilianakis C., Papamatheakis,J. and Kretsovali,A. (2000) Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol., 20, 8489–8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner R., Vidali,G. and Allfrey,V.G. (1979) Studies of acetylation and deacetylation in high mobility group proteins. J. Biol. Chem., 254, 11577–11583. [PubMed] [Google Scholar]
- Taniguchi N. et al. (2003) High mobility group box chromosomal protein 1 plays a role in the pathogenesis of rheumatoid arthritis as a novel cytokine. Arthritis Rheum., 48, 971–981. [DOI] [PubMed] [Google Scholar]
- Thomas J.O. and Travers,A.A. (2001) HMG1 and 2 and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci., 26, 167–174. [DOI] [PubMed] [Google Scholar]
- Wang H. et al. (1999) HMG-1 as a late mediator of endotoxin lethality in mice. Science, 285, 248–251. [DOI] [PubMed] [Google Scholar]