Abstract
Flagella and cilia are elaborate cytoskeletal structures conserved from protists to mammals, where they fulfil functions related to motility or sensitivity. Here we demonstrate novel roles for the flagellum in the control of cell size, shape, polarity and division of the protozoan Trypanosoma brucei. To investigate the function of the flagellum, its formation was perturbed by inducible RNA interference silencing of com ponents required for intraflagellar transport, a dynamic process necessary for flagellum assembly. First, we show that down-regulation of intraflagellar transport leads to assembly of a shorter flagellum. Strikingly, cells with a shorter flagellum are smaller, with a direct correlation between flagellum length and cell size. Detailed morphogenetic analysis reveals that the tip of the new flagellum defines the point where cytokinesis is initiated. Secondly, when new flagellum formation is completely blocked, non-flagellated cells are very short, lose their normal shape and polarity, and fail to undergo cytokinesis. We show that flagellum elongation controls formation of cytoskeletal structures (present in the cell body) that act as molecular organizers of the cell.
Keywords: cytokinesis/flagellum/morphogenesis/polarity/trypanosome
Introduction
Flagella and cilia are eukaryotic organelles whose core structure (a set of microtubules and associated proteins called the axoneme) shows remarkable conservation from protists to mammals (Silflow and Lefebvre, 2001). Despite their highly conserved structural architecture, flagella and cilia are involved in widely different functions: motility (of the cell or of material in its vicinity), reproduction and sensitivity (Bastin et al., 2000b; Silflow and Lefebvre, 2001). Recently, an essential role for a combination of motile and sensory cilia has been demonstrated in asymmetric development of vertebrate (McGrath et al., 2003).
Trypanosomes are flagellated protozoa belonging to the order of Kinetoplastida, and are responsible for tropical diseases such as sleeping sickness. They possess a single flagellum, with a classic axoneme and an additional lattice-like structure called the paraflagellar rod (PFR) (Gull, 1999). In recent years, trypanosomes have turned out to be a useful model to study flagellum assembly and targeting (Bastin et al., 1999a; Godsel and Engman, 1999; Ersfeld and Gull, 2001). With the exception of its distal tip, the flagellum is attached along the length of the cell body. Within the cytoplasm adjacent to the flagellum, a defined set of cytoskeletal structures (called flagellum attachment zone or FAZ) lies underneath the plasma membrane and follows the path of the flagellum. The FAZ is composed of two individual structures: (i) a set of four microtubules, of distinct biochemical characteristics, connected to the smooth endoplasmic reticulum; and (ii) an electron-dense filament (Sherwin and Gull, 1989a). Both structures are initiated from the basal body area and run towards the anterior of the cell right up to its end. FAZ function is unknown, but its positioning suggests a potential role in cellular organization (Robinson et al., 1995). During cell replication, the old flagellum is maintained, and a new one is assembled from a new basal body complex, always localized at the posterior end of the cell. FAZ structures are also replicated and associated to the new flagellum (Kohl et al., 1999). As the new flagellum elongates, its distal tip remains in constant contact with the old flagellum. In Trypanosoma brucei, a discrete cytoskeletal structure, termed the flagellum connector (FC), has been identified at this point, and is only present during cell duplication (Moreira-Leite et al., 2001). It has been postulated that the FC would anchor the tip of the new flagellum to the old flagellum. As a result, new flagellum assembly would take place following the path of the existing one and act as a guide to cell formation, hence replicating the existing organization. Cytokinesis is initiated at the anterior end of the cell and proceeds in a helical manner, cutting the cell in two following the new flagellum/FAZ as axis (Vaughan and Gull, 2003).
The trypanosome flagellum is highly motile, and the involvement of both the paraflagellar rod and the axoneme has been demonstrated previously (Bastin et al., 1998, 1999b, 2000a; Kohl et al., 2001; McKean et al., 2003). However, trypanosomes with reduced motility remain viable in culture. Judging from the omnipresence of the flagellum during the trypanosome life and cell cycle, we postulated that this structure could fulfil other functions than motility and could be involved in the trypanosome cell cycle. Therefore, we considered the possibility of perturbing flagellum construction by interfering with intraflagellar transport (IFT). IFT is defined as the transport of protein particles (‘rafts’) between the outer doublet microtubules and the membrane of flagella or cilia (Rosenbaum and Witman, 2002). Transport towards the distal tip of the flagellum is powered by heterotrimeric kinesin II, whereas particles are returned to the basal body by a dynein motor. Inhibition of IFT prevents flagella or cilia assembly in most eukaryotes examined to date. It has been suggested that IFT is used to transport axoneme precursors to the distal tip of the flagellum, which is the site of assembly of the organelle (Rosenbaum and Witman, 2002).
Here we report the identification of T.brucei IFT proteins and the consequences of knocking down their expression by inducible RNA interference (RNAi). First, trypanosomes with shorter flagella were obtained, demonstrating the importance of IFT for flagellum length control. Cells bearing shorter flagella were smaller, with a linear relationship between flagellum length and cell size. These short cells retain normal organization and could divide, with the anterior tip of their flagellum apparently defining the point of initiation of cytokinesis. Secondly, cells without a flagellum were even shorter, lost both their shape and polarity, and could not divide. These results demonstrate novel roles for the flagellum in cell morphogenesis and division.
Results
IFT is required for flagellum formation in trypanosomes
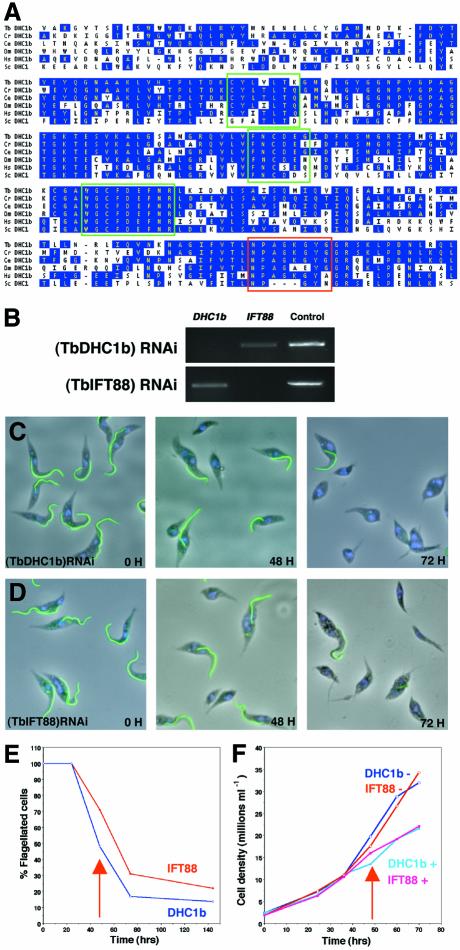
By mining the T.brucei genome databases, we identified several genes coding for homologues of proteins involved in IFT: either motor proteins or proteins of unknown function present in cargoes. All the corresponding trypanosome proteins show a high degree of conservation, suggesting that IFT could be functional in trypanosomes. For example, TbDHC1b exhibits all specific signatures of dynein heavy chains involved in IFT (Figure 1A). One gene from each category was selected for this work: TbDHC1b (motor) and TbIFT88 (cargo).
Fig. 1. TbDHC1b and TbIFT88 are required for flagellum assembly. (A) Alignment of protein sequences of the DHC1b central domain from T.brucei (Tb), Chlamydomonas reinhardtii (Cr), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (Hs) and the cytoplasmic dynein from Saccharomyces cerevisae (Sc). Conserved residues are shown in blue, and green and red blocks indicate dynein and DHC1b signatures, respectively. (B) RT–PCR analysis performed on total RNA extracted from non-induced and 48 h-induced (TbDHC1b) RNAi or (TbIFT88) RNAi mutants using specific primers for TbDHC1b, TbIFT88 or an unrelated RNA as a control. Knockdown of TbDHC1b (C) or of TbIFT88 (D) lead to production of non-flagellated cells over time. Cells were induced for the indicated periods and stained with the anti-PFRA antibody L8C4 as a marker of the flagellum (green), and with DAPI (blue) showing nucleus and kinetoplast. (E) RNAi was induced in (TbDHC1b) RNAi or (TbIFT88) RNAi cell lines, and cells were fixed at the indicated times and processed for immunofluorescence with anti-PFRA. Cells possessing at least one flagellum were scored as positive. Arrows indicate when non-flagellated cells were first detected. (F) In the same experiment, cell growth was monitored in the presence (+) or absence (–) of tetracycline.
To address the question of a potential involvement of the flagellum in the trypanosome cell cycle, we decided to perturb flagellum formation by silencing TbDHC1b or TbIFT88 via RNAi (Fire et al., 1998; Ngô et al., 1998; Bastin et al., 2000a; Wang and Englund, 2001; Morris et al., 2002). Trypanosomes were transformed with plasmids able to express TbDHC1b or TbIFT88 double-stranded (ds) RNA under the control of an inducible promoter. After transfection and selection, recombinant cell lines were induced to express TbDHC1b or TbIFT88 dsRNA by growth in the presence of tetracycline for 48 h. In both cases, RT–PCR demonstrated a specific reduction in the amount of targeted RNA (Figure 1B). Cells from non-induced (TbDHC1b) RNAi or (TbIFT88) RNAi mutants behaved as normal trypanosomes. During the course of induction, cells from (TbDHC1b) RNAi and (TbIFT88) RNAi mutants were examined by light microscopy and the flagellum was identified by immunostaining with an anti-paraflagellar rod antibody. In both cases, non-flagellated cells were detected 48 h after induction of RNAi (Figure 1C and D). Non-flagellated cells possessed an apparently normal basal body but no flagellum (see Supplementary material, available at The EMBO Journal Online) (Figures 3C and 5H). Phenotypes looked very similar for both mutants, except that kinetics appeared faster for (TbDHC1b)RNAi (Figure 1E). The proportion of non-flagellated cells increased rapidly to reach a maximum of 80% of the population after 3 days (Figure 1E). This demonstrates the essential role of TbDHC1b (a motor protein) and of TbIFT88 (an unrelated protein present in IFT particles) in flagellum formation. A progressive reduction in both (TbDHC1b) and (TbIFT88) RNAi mutant growth rates was noticed during the course of induction of RNAi (Figure 1F), suggesting an important role for flagellum formation in trypanosome development.
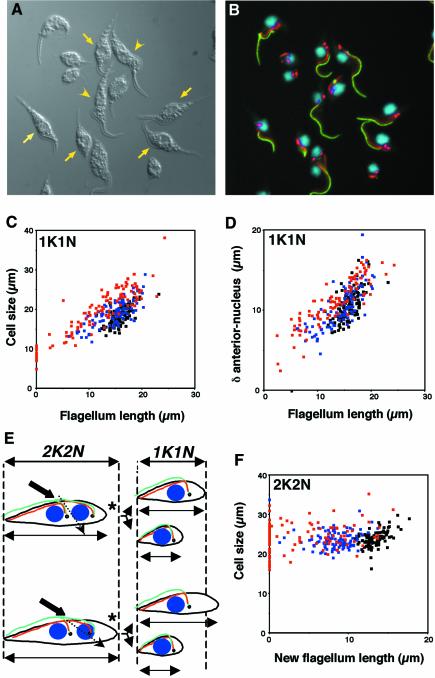
Fig. 3. Flagellum length controls cell size. (A and B) Field of (TbDHC1b) RNAi trypanosomes induced for 48 h. Flagellum was labelled with L8C4 (green), FAZ filament with L6B3 (red lines), basal body with BBA4 (red spots) and DNA was stained with DAPI (blue). (A) DIC image; (B) merged fluorescence. Cells with a shorter flagellum appeared smaller (arrows) than cells with a normal flagellum (arrowheads). (C, D and F) (TbDHC1b) RNAi trypanosomes were non-induced (black squares), or induced for 24 h (blue) or 48 h (red). (C) Mononucleated cells. Cell body size was measured and plotted versus the measured flagellum length. Each point represents an individual cell. (D) In the same uniflagellated trypanosomes, the distance between the anterior tip of the cell and the kinetoplast was measured and plotted versus flagellum length. Non-flagellated cells were not included, as recognition of the posterior/anterior end is virtually impossible. (E) Two models for the generation of small cells with short flagella. The nucleus is shown in blue, the flagellum in green, the FAZ in red and the basal body/kinetoplast (shown as a single circle for simplicity) in black. The asterisk indicates the posterior end. Normal cell size of a binucleated (‘2K2N’) or of a uninucleated (‘1K1N’) trypanosome is delimited by the dashed lines. Point of cleavage initiation is indicated by the thick arrow and probable cleavage progression by the dashed arrow (see text). (F) Binucleated cells. Cell size was measured and plotted versus new flagellum length.
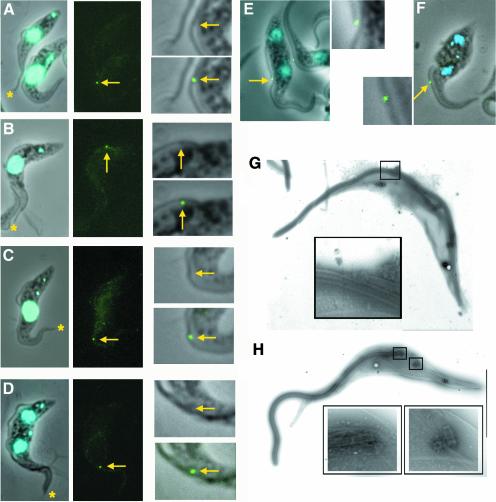
Fig. 5. The FC in wild-type and in induced (TbIFT88) RNAi cells. Normal wild-type (A–D) and (TbIFT88) RNAi trypanosomes induced for 36 h (E–F) were stained with an anti-FC antiserum. Left panels, phase contrast image merged with DAPI staining (blue). Central panels, FC staining only; right panels, magnification of the area surrounding the FC. Top, phase contrast image; bottom, phase contrast image merged with FC staining (green). Stars, tip of the old flagellum; arrows, tip of the new flagellum. (A) The left cell has only one flagellum, and its tip is not stained. The right cell has an elongating new flagellum and its distal tip shows defined staining. In contrast, the distal tip of the old flagellum is not stained. Similar observations were made as the new flagellum elongated (B–D) and stained with the anti-FC antiserum (green). (E) Binucleated cell whose new flagellum is too short (compare with D). (F) Binucleated cell without a new flagellum. FC is still present on the old flagellum of ∼30% of these cells. (G) Electron micrograph of a negatively stained cytoskeleton of (TbDHC1b) RNAi non-induced trypanosome. The discrete pyramidal structure of the FC is shown in the enlarged box. (H) Electron micrograph of a negatively stained cytoskeleton of (TbDHC1b) RNAi trypanosome induced for 48 h. The old basal body lies adjacent to the flagellum (enlarged left box), whereas no flagellum at all is visible on the new basal body (enlarged right box). In these conditions, the FC is not detected.
The flagellum is required for cell shape and polarity
Normal trypanosomes are long and slender cells (Figure 2A). They swim with the distal end of the flagellum leading, defining a narrow anterior end and a wider posterior end. The nucleus is in a central position and the mitochondrial genome is localized at the posterior end. Trypanosomes possess a single mitochondrion with a concatenated DNA network called the kinetoplast that is tightly linked to the basal body of the flagellum (Robinson and Gull, 1991). This cellular organization was severely perturbed in non-flagellated cells: the distinct shape was lost and the kinetoplast appeared extremely close to the nucleus (Figure 2B). No posterior/anterior end could be recognized, neither for the whole cell nor for its microtubular cytoskeleton (Figure 2B and C), demonstrating dramatically that the flagellum is required for definition of cell shape.
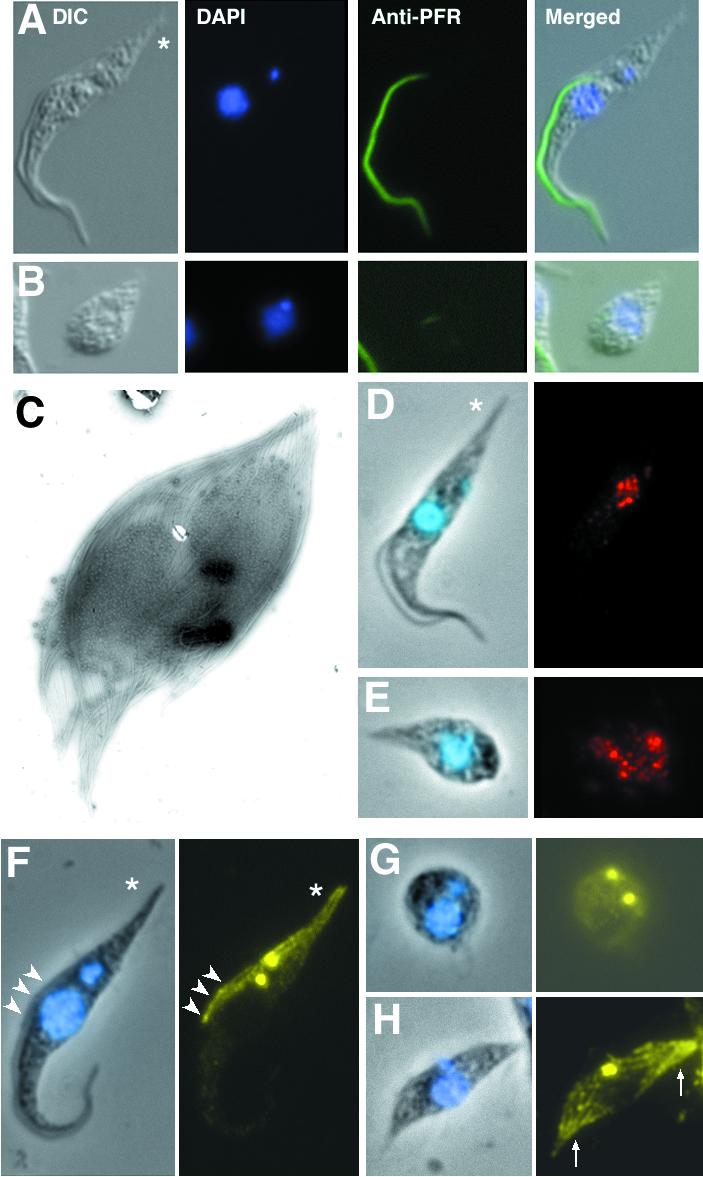
Fig. 2. Non-flagellated trypanosomes lose cell shape and polarity. (A and B) Cells were stained with the L8C4 antibody (green) and with DAPI (blue). (A) A non-induced (TbDHC1b) RNAi cell showing the typical trypanosome polarity. The asterisk indicates the posterior end of the cell. (B) (TbDHC1b) RNAi trypanosome induced for 48 h without a flagellum did not exhibit normal cell shape. (C) Electron micrograph of a (TbDHC1b) RNAi non-flagellated cell (48 h induction), revealing the absence of polarity and partial microtubule disorganization. (D and E) Phase contrast images of trypanosomes stained with DAPI (blue, left panels) and with anti-clathrin antibodies (red, right panels). Non- induced (TbIFT88) RNAi cells (D) showed a defined signal at the posterior end, whereas non-flagellated (TbIFT88) RNAi cells induced for 72 h (E) showed dispersion of clathrin vesicles throughout the cytoplasm. (F–H) Non-flagellated trypanosomes exhibit abnormal cell growth. Phase contrast images of (TbIFT88) RNAi trypanosomes stained with DAPI (blue, left panels) and with the anti-tyrosinated tubulin antibody YL1/2 (yellow, right panels). (F) Non-induced trypanosome, showing normal polarized cell growth that takes place by microtubule elongation at the posterior end (arrows). The arrowheads indicate the position of the new flagellum. (G and H) (TbIFT88) RNAi cells induced for 72 h: incorporation of tyrosinated tubulin was seen throughout the cytoskeleton (G) or at opposite ends (H).
We then asked whether such cells were still polarized. As a polarity marker, we used an anti-clathrin antibody (Morgan et al., 2001). In trypanosomes, endocytosis takes place via a restricted area of the cell surface situated at the base of the flagellum. In non-induced trypanosomes (Figure 2D), clathrin localized almost exclusively at the posterior end of the cell, between the kinetoplast and the nucleus. In non-flagellated cells from (TbDHC1b) RNAi mutants (data not shown) and (TbIFT88) RNAi mutants (Figure 2E) induced for 72 h, clathrin was no longer localized at its defined position. Instead, it was found dispersed throughout the cytoplasm, suggesting severe defects in cellular organization and polarity.
Polarized growth is another feature of the trypanosome cell, proceeding via extension and insertion of microtubules at the posterior end of the cell. This is illustrated by means of a specific monoclonal antibody that recognizes only tyrosinated α-tubulin, a marker of tubulin recently incorporated in microtubules (Sherwin and Gull, 1989b). In replicating normal cells (identified by the presence of two basal body complexes), this antibody stained the basal bodies, the posterior end of the cell and the distal tip of the new flagellum (Figure 2F). In contrast, in (TbIFT88) RNAi mutants induced for 72 h, 60.5% (n = 162) of the non-flagellated cells having duplicated their basal bodies did not show the typical polar incorporation of tyrosinated tubulin. Instead, relatively weak non-distinct staining was observed throughout the cytoskeleton (Figure 2G). In addition, 19.1% of replicating non-flagellated cells exhibited incorporation of tyrosinated tubulin at both ends of their cytoskeleton (Figure 2H). These trypanosomes often display a nut-like shape. These results demonstrate that non-flagellated cells have lost cues for correct replication of their cytoskeleton.
Flagellum length determines cell size and cytokinesis
During the course of RNAi knockdown targeting TbDHC1b or TbIFT88 RNA, we noticed the presence of a large number of cells with unusually short flagella (Figure 3A and B). The associated FAZ filament (red staining on figure) also looked shorter. Curiously, these cells looked smaller. To determine whether both phenomena were related, the total cell body size of 100 uniflagellated trypanosomes was measured in (TbDHC1b) and (TbIFT88) RNAi cell lines (Figure 3C and data not shown, respectively) either non-induced or induced for 24 or 48 h, and plotted versus the length of the flagellum. In both cases, there was a striking and almost linear correlation (r2 = 0.834) between the length of the flagellum and the size of the cell. In other words, as trypanosomes grew shorter and shorter flagella, their cell body size also decreased. To evaluate whether this reduction was affecting all cell parts equally, we measured distances from the anterior tip of the cell to the nucleus, from the nucleus to the kinetoplast and from the kinetoplast to the posterior end in the same cells, as above (Figure 3D). These data showed that the reduction in size was mostly due to a reduction of the anterior part of the cell, i.e. the zone along which the flagellum is attached. In contrast, the distance between the kinetoplast and the posterior end of the cell was not modified. This correlation suggests that flagellum elongation could control cell size.
In order to understand how the length of the flagellum could be involved in this process, we turned our attention to the ‘mother’ cell from which shorter trypanosomes are derived. Trypanosomes divide by binary fission after having replicated basal bodies and kinetoplasts, FAZ and flagella and nuclear mitosis (Figure 3E). One simple explanation for the smaller size observed in cells with a short flagellum would be to postulate that elongation of the new flagellum is required for normal cell growth at the posterior end (Figure 3E, top). In these conditions, the posterior end of the mother cell would be too short and the progeny inheriting the new flagellum would be too small. To evaluate this possibility, we measured cell body size and new flagellum length in trypanosomes with two nuclei from (TbDHC1b) RNAi and (TbIFT88) RNAi cell lines (Figure 3F and data not shown, respectively) either non-induced or induced for 24 or 48 h. These data showed that despite the production of shorter and shorter new flagella (and finally no new flagellum at all), cell elongation proceeded normally to reach ∼25 µm. Therefore, cell elongation at the posterior end is not modified and cannot account for the observed results.
A second possibility to explain the smaller size of the cell with the short flagellum is that the tip of the new flagellum defines the position of cell cleavage (Figure 3E, bottom). In these conditions, cell cleavage would be ‘too posterior’, producing a smaller daughter cell with the short new flagellum, and, since cell elongation proceeded normally, a larger daughter cell inheriting the old flagellum (of normal length). To evaluate this possibility we examined again measurements of cell size and flagellum length of the 100 uniflagellated cells from (TbDHC1b) and (TbIFT88) RNAi cell lines (Figure 3C and data not shown, respectively) either non-induced or induced for 24 or 48 h, as described above. Remarkably, the size of the cell inheriting the old flagellum (that of normal length) progressively increased during the course of induction (notice the shift of most red squares in Figure 3C). This strongly favours the hypothesis that the tip of the new flagellum defines the initiation of cytokinesis (Figure 3E, bottom).
As the new flagellum appears critical for cell division, it was surprising to find that non-flagellated cells could be produced. To understand how these non-flagellated cells were born, we first examined the normal trypanosome cell cycle (Figure 4A–C). Cells in the early phase of their cell cycle possess a mature basal body, with a single flagellum, and a pro-basal body, a FAZ, a kinetoplast and a nucleus (Figure 4A). Basal bodies duplicate first, followed by mitochondrial genome replication and segregation. The new basal body and the associated kinetoplast migrate towards the posterior end of the cell, which is increasing in length. A new FAZ is assembled, presumably nucleated from the basal body area, prior to the formation of the new flagellum. The mature posterior basal body lies adjacent to the new flagellum, which elongates towards the anterior end of the cell (Figure 4B). FAZ is also elongated at that stage, following the path of the new flagellum. Mitosis then occurs and the most posterior nucleus migrates between the two kinetoplasts (Figure 4C). Cytokinesis is initiated from the anterior tip and proceeds in a helical manner towards the posterior end to generate two siblings, each inheriting a single nucleus, kinetoplast/basal body and flagellum/FAZ.
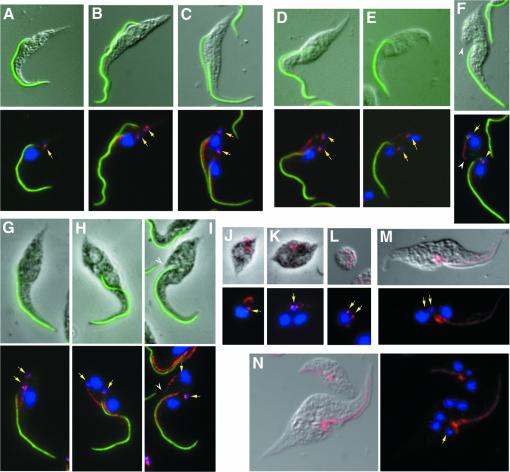
Fig. 4. New and old flagella are required for cell morphogenesis. (TbDHC1b) RNAi cells are shown by DIC and (TbIFT88) RNAi cells are shown by phase contrast. Cell cycle of normal, non-induced, (TbDHC1b) RNAi trypanosomes (A–C), of 48-h-induced (TbDHC1b) RNAi (D–F) and (TbIFT88) RNAi (G–I). Flagellum was labelled with L8C4 (green), FAZ filament with L6B3 (red lines), basal body with BBA4 (red spots) and DNA was stained with DAPI (blue). Top panels, DIC merged with flagellum staining (green); bottom image, merged fluorescence. Yellow arrows indicate kinetoplast DNA position. After 48 h of induction, trypanosomes cannot assemble a new flagellum, but nevertheless formation of a short new FAZ (tip indicated by the white arrowhead) is observed. (J and K) Non-flagellated (TbIFT88) RNAi cells (induced for 72 h) stained with the anti-basal body BBA4 and anti-FAZ L6B3 (both in red), and with DAPI (blue). Kinetoplasts segregation failed, even after completion of nuclear mitosis (K). (L) Non-flagellated (TbDHC1b) RNAi cells (induced for 72 h) stained with the anti-basal body BBA4 and anti-FAZ L6B3 (both in red), and with DAPI (blue). Although basal body duplication and segregation occurred, no new FAZ formation was detected. (M and N) After 72 h of induction, flagellated cells were immunolabelled using antibody 1B41 (red) and stained with DAPI (blue), showing lack of assembly of FAZ-associated microtubules, accumulation of patches of material around the basal bodies area and absence of cytokinesis.
After 48 h of induction, many (TbDHC1b) or (TbIFT88) RNAi mutant cells (Figure 4D–F and G–I, respectively) in duplication possessed an old flagellum, but no new flagellum was visible, either by direct Nomarski or phase contrast observation or by immunolabelling of flagellar components. Despite the absence of a new flagellum, these trypanosomes were able to assemble a short FAZ, initiated from the expected localization at the basal body area and extending towards the anterior end where its tip seemed in contact with the old FAZ. However, this FAZ extended to roughly only one-third of its normal length. Its shape looked different: instead of the harmonious, undulating aspect observed in flagellated, non-induced trypanosomes, it looked straight and almost stretched. This structure was stained by markers of both the FAZ filament (Figure 4D–I) and of the four specialized microtubules (data not shown).
These cells underwent apparently normal nuclear mitosis, but the posterior nucleus was positioned beyond the posterior kinetoplast/basal body (Figure 4E and H) instead of being between the two kinetoplasts/basal bodies (compare with Figure 4C). Cell measurements showed that nuclear positioning relative to the posterior end was correct, but kinetoplasts/basal bodies segregation was often incomplete. The interkinetoplast distance in binucleated cells without a new flagellum was 2.67 ± 0.46 µm for induced (TbDHC1b) RNAi (n = 40), or 3.60 ± 0.94 µm in the case of (TbIFT88) RNAi (n = 54), compared with 5.33 ± 0.47 µm (n = 101) in normal non-induced binucleated cells. As shown previously for cells assembling a short flagellum, cell body elongation at the posterior end proceeded normally (Figure 3F, cells with no new flagellum or flagellum length = 0). Such cells divided to generate a short, non-flagellated sibling, with a limited FAZ filament (Figure 4J), and a longer, apparently normal flagellated sibling (see below). Interestingly, the anterior tip of the new, short FAZ was always positioned at the initiation of the cleavage furrow (Figure 4F and I, arrowheads). These results show that even in the absence of a new flagellum, FAZ formation can be initiated but its elongation cannot proceed fully. However, these cells can divide, suggesting that the tip of the FAZ would be the critical determinant for initiation of cell division.
Flagellum connector in flagellated and non-flagellated trypanosomes
Recently, a particular cytoskeletal structure connecting old and new flagella during cell duplication of T.brucei has been identified (Moreira-Leite et al., 2001). This structure could play an essential role in the cell cycle by directing the replication of the existing helical pattern typical of trypanosomes. Therefore, we looked at whether the FC was present during the production of non-flagellated trypanosomes. First, we used a rabbit antiserum as a marker of the FC in an immunofluorescence assay. In wild type, as in non-induced duplicating (TbDHC1b) RNAi or (TbIFT88) RNAi cells, this antibody stained the tip of the new flagellum and did not produce any significant signal in uniflagellated cells (Figure 5A–D). The tip of the old flagellum was not stained either. The signal was detected as soon as the new flagellum exited the flagellar pocket (Figure 5B) and was present throughout flagellum elongation (Figure 5C and D). In (TbDHC1b) or (TbIFT88) RNAi mutant cells induced for 24–48 h assembling a shorter new flagellum than usual, the FC was always detected by immunofluorescence analysis with the anti-FC marker antibody (Figure 5E). Surprisingly, about one-third of cells without a new flagellum still possessed an FC signal on the old flagellum (Figure 5F), suggesting that at least some components of that structure could still be present. Finally, no FC signal at all could be visualized in non-flagellated cells.
Electron microscopy analysis of negatively stained cytoskeletons (detergent-extracted cells) of biflagellated cells from the non-induced (TbDHC1b) RNAi cell line confirmed the typical lamellar structure of the FC (Moreira-Leite et al., 2001), sitting on the axoneme (slightly bent at this point) from the old flagellum, with some fuzzy extensions towards the tip of the axoneme from the new flagellum (Figure 5G). In contrast, we could not identify an FC by electron microscopy examination of trypanosomes with an old flagellum, but without a new one (Figure 5H). This result differs from the immunofluorescence analysis with the marker antibody, and may be due to differences in the methods used for sample preparation (cells need to be detergent-extracted for electron microscopy analysis) or in the assay employed. Immunofluorescence could identify components present at the FC position, but these might not be assembled in a stable structure visible by electron microscopy. These images also revealed the total absence of axonemal microtubules from the new basal body in induced (TbDHC1b) RNAi cells (Figure 5H).
Formation of FAZ structures is required for basal body segregation and cytokinesis
We next asked whether non-flagellated trypanosomes with only a short FAZ could re-enter the cell cycle, and duplicate and divide. In agreement with the role of the flagellum in the control of cell size, non-flagellated cells were tiny, showing a 2-fold reduction of their cell body compared with normal trypanosomes [9.8 ± 2.2 µm (n = 121) compared with 19.4 ± 2.5 µm (n = 106)]. Non-flagellated cells could duplicate their basal bodies, but no new FAZ filament was assembled (Figure 4K and L). Segregation of the duplicated basal bodies and kinetoplasts was seriously reduced in binucleated, non-flagellated (TbIFT88) RNAi mutants induced for 72 h. Only 28.4% of these cells showed clear segregation of both basal bodies and kinetoplasts (Figure 4L). In contrast, 57.4% segregated their basal bodies, but these failed to migrate apart and had a single large kinetoplast (Figure 4K), and the remaining 14.2% did not segregate either their basal bodies or their kinetoplasts. These data can be correlated with the poor basal bodies/kinetoplasts segregation observed during the generation of non-flagellated cells and strongly suggest that FAZ/flagellum elongation is crucial for this segregation process. These cells could complete nuclear mitosis, but they failed to divide, their cell cycle appeared to have arrested, and they presumably died. These data demonstrate that the presence of the old flagellum is required for formation of a new FAZ [(cells with an old flagellum could produce a short FAZ (see Figure 4D–I)] and reveal that in the absence of a new FAZ, cytokinesis cannot occur.
The importance of the FAZ for cell division was demonstrated further in the flagellated siblings derived from cells like those shown in Figure 4F and I. These cells could reiterate the same cycle once or twice, but then encountered some amazing difficulties. They retained the old flagellum and could duplicate their basal bodies and kinetoplast; however, segregation and migration were highly inefficient, and no new FAZ filament or FAZ-associated microtubules were assembled. Instead, FAZ proteins accumulated in patches of material dispersed around the basal bodies area, whereas FAZ proteins associated to the filament along the old flagellum remained in place (Figure 4M). These cells did not undergo cytokinesis, yet nuclear mitosis still proceeded, producing multinucleated cells (Figure 4M and N). Taken together, these data show that formation of a new FAZ is required for cytokinesis. Mitosis without cytokinesis has been observed previously and is probably explained by the absence of that particular cell cycle checkpoint in trypanosomes (Robinson et al., 1995; Grellier et al., 1999; Ploubidou et al., 1999).
Discussion
IFT is required for flagellum formation
In this report, we have shown that separate silencing of two different proteins, presumably associated with the IFT machinery, leads to the inability of trypanosomes to form a new flagellum despite the presence of a new basal body (Figure 5H). Together with the presence of trypanosome orthologues of all IFT genes identified so far in other organisms, these data strongly suggest that IFT is functional in trypanosomes and is involved in flagellum formation. We could not directly identify movement of IFT rafts as elegantly demonstrated in Chlamydomonas (Kozminski et al., 1993), but structures morphologically resembling IFT rafts have been identified in cross-sections of wild-type trypanosome flagella (Sherwin and Gull, 1989a; Bastin et al., 2000b). As in Chlamydomonas flagella, these rafts localize between the B tubule of the peripheral doublets of the axoneme and the plasma membrane.
The requirement for the IFT machinery in cilia or flagella formation has been demonstrated in different flagellated organisms: Chlamydomonas, Tetrahymena, sea urchin, nematodes and mouse (Rosenbaum and Witman, 2002). To date, trypanosomes represent the oldest eukaryotes in which the role of IFT proteins in flagella formation has been demonstrated.
The old flagellum appears unaffected by RNAi knockdown of either TbDHC1b or TbIFT88. This may seem surprising given the knowledge that in Chlamydomonas, IFT is required for both the formation and maintenance of flagella (Kozminski et al., 1995). This result is probably explained by the fact that RNAi targets only RNA for degradation, hence proteins existing before expression of dsRNA are not affected (Bastin et al., 2000a; Moreira-Leite et al., 2001). This means that TbDHC1b and TbIFT88 proteins associated with the old flagellum should remain present and presumably functional.
IFT controls flagellum length and cell size
During the course of induction of RNAi silencing of TbDHC1b or TbIFT88, many cells assembled shorter flagella than usual. These flagella appeared functional and looked normal, except for their length. In Chlamydomonas, IFT is required to control flagellum length by continuously balancing microtubule turnover at the distal end of the axoneme. This is illustrated by the temperature-sensitive mutant FLA10, which fails to assemble or to maintain its flagella when grown at the restrictive temperature. However, incubation of these cells at intermediate temperatures between restrictive and permissive temperatures leads to flagella of intermediate lengths (Marshall and Rosenbaum, 2001). These results suggest that alterations in IFT could lead to changes in flagellum length. In T.brucei RNAi mutants, inducible expression of dsRNA leads to destruction of RNA and inhibition of new protein synthesis, but existing proteins synthesized before the onset of RNAi are not affected (Bastin et al., 2000a). As a result, when RNAi takes effect, the amount of proteins coded by the targeted RNA is progressively reduced before eventually becoming absent. In the case of our T.brucei IFT mutants, one could imagine that as the pool of TbDHC1b or TbIFT88 protein is reduced, it is still sufficient for initiating flagellum assembly but not enough to support the construction of a full-length flagellum. This result suggests that modifying the amount of available IFT proteins could act as a system to regulate flagellum length.
The link between flagellum length and cell size was even more spectacular. As the cells grew shorter flagella, cell body size appeared to be smaller. The correlation was almost linear and is explained by a shorter distance between the anterior tip and the kinetoplast, i.e. the part of the cell body flanked by the flagellum. Our measurements have shown that this was not due to a failure in cell body elongation, but rather to mispositioning of the cleavage furrow, suggesting that the tip of the flagellum (or of the associated FAZ) could define the point of initiation of division.
Trypanosomes are parasitic organisms with a complex life cycle, where they alternate between an insect vector and a mammalian host. Due to the changes in environment, trypanosomes need to adapt to each condition (Matthews, 1999). These changes are accompanied by extensive modulation of cell size and shape (Vickerman, 1985), and interestingly the flagellum follows these changes closely. The ‘procyclic’ trypanosome form used for this study colonizes the midgut of a tsetse fly, and its size varies between 20 and 25 µm. In the following part of the life cycle, trypanosomes become much longer (up to 40 µm), and this increase in cell size is correlated with an increase in flagellum length (Van Den Abbeele et al., 1999). When trypanosomes are present in the bloodstream of their mammalian host, two stages can be discriminated: the long, slender, proliferating form, and the short stumpy, non-proliferating but differentiating form (from bloodstream to insect stage). The slender to stumpy differentiation program incorporates a round of cell division. Interestingly, when such dividing cells were examined just before cytokinesis, the average length of the new flagellum was shorter than the one measured from replicating slender cells at the same stage (21 µm instead of 25 µm) (Tyler et al., 2001). It is tempting to speculate that regulating the amount of functional IFT particles could be a system to control both flagellum length and cell size.
Interestingly, the amastigote form of Trypanosoma cruzi, a related trypanosome species, possesses a very short flagellum not extending beyond the flagellar pocket, and its cell body size and shape resemble that of non-flagellated T.brucei mutants. Amastigotes show a simpler mode of cell division, not displaying the helical aspect (de Souza, 1984). Leishmania are other trypanosomatid organisms, but their flagellum is not attached along the cell body. Since flagellum length defines cell size by determining the position of initiation of cytokinesis, it seems unlikely that a similar role would exist in Leishmania, as the tip of the flagellum is separated from the cell body. However, several amazing observations have been reported. First, differentiation of Leishmania braziliensis from the long-flagellated promastigote stage to the short-flagellated amastigote stage is concomitant with drastic reduction of cell body length and alteration in cell shape (Stinson et al., 1989). Secondly, Leishmania mexicana cells expressing a modified G-protein display much shorter flagella than usual. Although cell body/flagellum length measurements have not been reported, cells with short flagella appear smaller than cells with normal flagella (see Figure 4G and H in Cuvillier et al., 2000). Thirdly, metacyclic stages possess a longer flagellum and, although their cell body length is barely modified, they display a much thinner shape (Zakai et al., 1998).
A model for the role of IFT, flagellum and FAZ in trypanosome cell cycle and division
The contribution of the flagellum and its associated structures (FAZ and FC) to cell morphogenesis and the cell cycle can be summarized in the following working model. First, basal body duplicates (Sherwin and Gull, 1989a) and a new FAZ is assembled, prior to flagellum exit from the flagellar pocket (Kohl et al., 1999). These steps are independent from the formation of the new flagellum as they still take place in (TbDHC1b) and (TbIFT88) RNAi mutant cells with an old flagellum but without a new one. Next, the flagellum elongates and somehow drives FAZ elongation. From that point in the cell cycle, FAZ elongation is controlled by flagellum growth as production of a flagellum that does not reach wild-type length also leads to incomplete FAZ. As the new FAZ elongates, it could participate in basal body segregation. In the absence of a new flagellum, the new FAZ is much shorter and basal body segregation is less efficient. During this whole process, the elongation of the cytoskeleton continues at the posterior end (Sherwin and Gull, 1989b). Once flagellum growth is terminated, FAZ elongation also finishes, the FC is disassembled and the cell initiates cleavage at the anterior end of the FAZ.
In the absence of a new flagellum (but when the old one is still present), binucleated cells are still able to divide. Hence, the presence of the new flagellum alone is not responsible for cleavage initiation. We propose that this function is actually fulfilled by the FAZ. In all cells without a new flagellum but with an old one, a new FAZ is still made. The extremity of that structure always appears to be in contact with the old FAZ. The new FAZ does not elongate properly, but cell growth continues unabated, nuclear mitosis takes place and the posterior nucleus migrates in a position that is normal with respect to its distance from the posterior end, suggesting that these processes are independent of flagellum formation. The presence of a short FAZ, whose anterior end is positioned at the point where cleavage is initiated, is sufficient to initiate division. When it cleaves, it produces a shorter, non-flagellated progeny, with its basal body/kinetoplast complex very close to the nucleus, and a longer, flagellated progeny with an excessively long posterior end. FAZ could also be involved in the establishment and maintenance of cell polarity, as non-flagellated cells show rapid loss of polarity. The non-flagellated sibling can duplicate its basal body/kinetoplast complex, but these organelles segregate either poorly or not at all and the cell fails to divide. No new FAZ is assembled in these cells, suggesting that the presence of the old flagellum is required for formation of FAZ. The flagellated progeny can reiterate once or twice the cycle described above, but then also fails to assemble a new FAZ and to undergo cytokinesis. This model is in agreement with a previous hypothesis suggesting a role for FAZ in cytokinesis (Robinson et al., 1995), as cells that are not able to produce a new FAZ fail to undergo cytokinesis.
Materials and methods
Identification of TbDHC1b and TbIFT88 genes and generation of RNAi mutants
The TIGR and Sanger Centre T.brucei databases [TIGR genome project (http://www.tigr.org/tdb/mdb/tbdb) and the Sanger genome project (http://www.sanger.ac.uk/Projects/T_brucei/)] were screened by a BLAST search using the full-length sequence of the Chlamydomonas DHC1b and IFT88 genes. Homologous sequences were identified and the genes were reconstructed and control-sequenced. Sequences have been submitted to the EMBL/DDBJ/GenBank database under accession codes BK000491 (TbDHC1b) and AF521959 (TbIFT88). For RNAi, a segment of the TbDHC1b or of the TbIFT88 gene was amplified by PCR from genomic DNA and cloned between two facing T7 promoters in the pZJM vector, allowing tetracycline-inducible expression of dsRNA (Wang et al., 2000). Primers designed to amplify an 832-bp internal region of TbDHC1b [position 399–1230 of the T.brucei nucleic acid sequence, corresponding to amino acids 1801–2076 of the Chlamydomonas DHC1b sequence; accession number CAB56748 (Porter, 1999)] were 5′-CGATGAATTCCTCGAGTCAAAATAGATCAGCTTTCAG-3′ and 5′-CGATCGAAGCTTCCCAAAAGCTGCTGTCGGTG-3′. This region is only conserved in DHC1b proteins and not in other dyneins. Highest overall identity of that region with other trypanosome dyneins reaches 53–57%, with no regions totalling >14 bp of total identity. Such an identity does not generate cross RNAi in our system (Durand-Dubief et al., 2003). For TbIFT88, a 941-bp internal region [positions 301–1241 of the T.brucei nucleic acid sequence, corresponding to amino acids 353–668 of the Chlamydomonas IFT88 sequence; accession number AF298884 (Pazour et al., 2000)] was amplified using primers 5′-CGATGAATTCCTCGAGGATTAAGGAGGAACGTACAC-3′ and 5′-CGATCGAAGCTTAGCGCCTC GTAGAGTCGCTTG-3′. No related sequence could be identified in T.brucei. Amplified fragments were ligated in the pZJM vector and control-sequenced. Plasmids were transfected in the 29-13 cell line that expresses T7 RNA polymerase and tet-repressor (Wirtz et al., 1999). For screening (Bastin et al., 1999a), resistant cells were grown with or without 2 µg tetracycline per millilitre for 48 h, fixed and processed by immunofluorescence using the anti-PFRA antibody L8C4 as a marker of the flagellum. For longer induction experiments, 1 µg of fresh tetracycline was added daily.
Immunofluorescence and phenotype analysis
Cells were spread on poly-l-lysine-coated slides and fixed in methanol at –20°C before processing for immunofluorescence as described previously (Sherwin et al., 1987). The following antibodies were used: L8C4 (Kohl et al., 1999) (marker of the flagellum), L3B2, L6B3 (Kohl et al., 1999) and DOT-1 (Woods et al., 1989) (markers of the FAZ filament), 1B41 (Gallo et al., 1988) (marker of the four specialized microtubules), BBA4 (Woods et al., 1989) (marker of the basal body), YL1/2 (Kilmartin et al., 1982) (marker of recently assembled tubulin) and anti-clathrin heavy chain (Morgan et al., 2001). Slides were viewed using a DMR Leica microscope and images were captured with a Cool Snap HQ camera (Roper Scientific). Images were analysed and cell parameters were measured using the IPLab Spectrum software (Scanalytics).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank K.Gull, M.Field and P.Grellier for providing various antibodies, P.Englund for the pZJM plasmid, G.Cross for the 29-13 cell line, C.Walsh and H.Ngô for helpful discussions about protists cell biology, and E.Charlier for technical assistance. We are grateful to M.Gèze and M.Dellinger for providing help with microscopy at the initial stages of this work. Sequencing of T.brucei was accomplished at TIGR and the Sanger Institute with support from NIAID and the Wellcome Trust, respectively. This work was funded by CNRS ATIPE grants and by ‘Aides à l’Implantation de Nouvelles Equipes’ awarded by the Fondation pour la Recherche Médicale.
References
- Bastin P., Sherwin,T. and Gull,K. (1998) Paraflagellar rod is vital for trypanosome motility. Nature, 391, 548. [DOI] [PubMed] [Google Scholar]
- Bastin P., MacRae,T.H., Francis,S.B., Matthews,K.R. and Gull,K. (1999a) Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol. Cell. Biol., 19, 8191–8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P., Pullen,T.J., Sherwin,T. and Gull,K. (1999b) Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci., 112, 3769–3777. [DOI] [PubMed] [Google Scholar]
- Bastin P., Ellis,K., Kohl,L. and Gull,K. (2000a) Flagellum ontogeny studied via an inherited and regulated RNA interference system. J. Cell Sci., 113, 3321–3328. [DOI] [PubMed] [Google Scholar]
- Bastin P., Pullen,T.J., Moreira-Leite,F.F. and Gull,K. (2000b) Inside and outside of the trypanosome flagellum: a multifunctional organelle. Microbes Infect., 2, 1865–1874. [DOI] [PubMed] [Google Scholar]
- Cuvillier A., Redon,F., Antoine,J.C., Chardin,P., DeVos,T. and Merlin,G. (2000) LdARL-3A, a Leishmania promastigote-specific ADP-ribosylation factor-like protein, is essential for flagellum integrity. J. Cell Sci., 113, 2065–2074. [DOI] [PubMed] [Google Scholar]
- de Souza W. (1984) Cell biology of Trypanosoma cruzi. Int. Rev. Cytol., 86, 197–283. [DOI] [PubMed] [Google Scholar]
- Durand-Dubief M., Kohl,L. and Bastin,P. (2003) Efficiency and specificity of RNA interference generated by intra- and intermolecular double stranded RNA in Trypanosoma brucei. Mol. Biochem. Parasitol., 129, 11–21. [DOI] [PubMed] [Google Scholar]
- Ersfeld K. and Gull,K. (2001) Targeting of cytoskeletal proteins to the flagellum of Trypanosoma brucei. J. Cell Sci., 114, 141–148. [DOI] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Gallo J.M., Precigout,E. and Schrevel,J. (1988) Subcellular sequestration of an antigenically unique beta-tubulin. Cell Motil. Cytoskeleton, 9, 175–183. [DOI] [PubMed] [Google Scholar]
- Godsel L.M. and Engman,D.M. (1999) Flagellar protein localization mediated by a calcium-myristoyl/palmitoyl switch mechanism. EMBO J., 18, 2057–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grellier P., Sinou,V., Garreau-de Loubresse,N., Bylen,E., Boulard,Y. and Schrevel,J. (1999) Selective and reversible effects of vinca alkaloids on Trypanosoma cruzi epimastigote forms: blockage of cytokinesis without inhibition of the organelle duplication. Cell Motil. Cytoskeleton, 42, 36–47. [DOI] [PubMed] [Google Scholar]
- Gull K. (1999) The cytoskeleton of trypanosomatid parasites. Annu. Rev. Microbiol., 53, 629–655. [DOI] [PubMed] [Google Scholar]
- Kilmartin J.V., Wright,B. and Milstein,C. (1982) Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol., 93, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl L., Sherwin,T. and Gull,K. (1999) Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol., 46, 105–109. [DOI] [PubMed] [Google Scholar]
- Kohl L., Durand-Dubief,M. and Bastin,P. (2001) Genetic inhibition of intraflagellar transport in trypanosomes. Mol. Biol. Cell, 12, 445a. [Google Scholar]
- Kozminski K.G., Johnson,K.A., Forscher,P. and Rosenbaum,J.L. (1993) A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl Acad. Sci. USA, 90, 5519–5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Beech,P.L. and Rosenbaum,J.L. (1995) The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol., 131, 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F. and Rosenbaum,J.L. (2001) Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol., 155, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K.R. (1999) Developments in the differentiation of Trypanosoma brucei. Parasitol. Today, 15, 76–80. [DOI] [PubMed] [Google Scholar]
- McGrath J., Somlo,S., Makova,S., Tian,X. and Brueckner,M. (2003) Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell, 114, 61–73. [DOI] [PubMed] [Google Scholar]
- McKean P.G., Baines,A., Vaughan,S. and Gull,K. (2003) γ-tubulin functions in the nucleation of a discrete subset of microtubules in the eukaryotic flagellum. Curr. Biol., 13, 598–602. [DOI] [PubMed] [Google Scholar]
- Moreira-Leite F.F., Sherwin,T., Kohl,L. and Gull,K. (2001) A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science, 294, 610–612. [DOI] [PubMed] [Google Scholar]
- Morgan G.W., Allen,C.L., Jeffries,T.R., Hollinshead,M. and Field,M.C. (2001) Developmental and morphological regulation of clathrin-mediated endocytosis in Trypanosoma brucei. J. Cell Sci., 114, 2605–2615. [DOI] [PubMed] [Google Scholar]
- Morris J.C., Wang,Z., Drew,M.E. and Englund,P.T. (2002) Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J., 21, 4429–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngô H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G.J., Dickert,B.L., Vucica,Y., Seeley,E.S., Rosenbaum,J.L., Witman,G.B. and Cole,D.G. (2000) Chlamydomonas IFT88 and its mouse homologue, Polycystic Kidney Disease Gene Tg737, are required for assembly of cilia and flagella. J. Cell Biol., 151, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploubidou A., Robinson,D.R., Docherty,R.C., Ogbadoyi,E.O. and Gull,K. (1999) Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci., 112, 4641–4650. [DOI] [PubMed] [Google Scholar]
- Porter M.E. (1999) Cytoplasmic dynein heavy chain 1b is required for flagellum assembly in Chlamydomonas. Mol. Biol. Cell, 10, 693–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.R. and Gull,K. (1991) Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature, 352, 731–733. [DOI] [PubMed] [Google Scholar]
- Robinson D.R., Sherwin,T., Ploubidou,A., Byard,E.H. and Gull,K. (1995) Microtubule polarity and dynamics in the control of organelle positioning, segregation and cytokinesis in the trypanosome cell cycle. J. Cell Biol., 128, 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum J.L. and Witman,G.B. (2002) Intraflagellar transport. Nat. Rev. Mol. Cell. Biol., 3, 813–825. [DOI] [PubMed] [Google Scholar]
- Sherwin T. and Gull,K. (1989a) The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Philos. Trans. R. Soc. Lond. B Biol. Sci., 323, 573–588. [DOI] [PubMed] [Google Scholar]
- Sherwin T. and Gull,K. (1989b) Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell, 57, 211–221. [DOI] [PubMed] [Google Scholar]
- Sherwin T., Schneider,A., Sasse,R., Seebeck,T. and Gull,K. (1987) Distinct localization and cell cycle dependence of COOH terminally tyrosinolated α-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol., 104, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow C.D. and Lefebvre,P.A. (2001) Assembly and motility of eukaryotic cilia and flagella. Lessons from Chlamydomonas reinhardtii. Plant Physiol., 127, 1500–1507. [PMC free article] [PubMed] [Google Scholar]
- Stinson S., Sommer,J.R. and Blum,J.J. (1989) Morphology of Leishmania braziliensis: changes during reversible heat-induced transformation from promastigote to an ellipsoidal form. J. Parasitol., 75, 431–440. [PubMed] [Google Scholar]
- Tyler K.M., Matthews,K.R. and Gull,K. (2001) Anisomorphic cell division by African trypanosomes. Protist, 152, 367–378. [DOI] [PubMed] [Google Scholar]
- Van Den Abbeele J., Claes,Y., van Bockstaele,D., Le Ray,D. and Coosemans,M. (1999) Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology, 118, 469–478. [DOI] [PubMed] [Google Scholar]
- Vaughan S. and Gull,K. (2003) The trypanosome flagellum. J. Cell Sci., 116, 757–759. [DOI] [PubMed] [Google Scholar]
- Vickerman K. (1985) Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull., 41, 105–114. [DOI] [PubMed] [Google Scholar]
- Wang Z. and Englund,P.T. (2001) RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J., 20, 4674–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Morris,J.C., Drew,M.E. and Englund,P.T. (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem., 275, 40174–40179. [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal,S., Ochatt,C. and Cross,G.A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol., 99, 89–101. [DOI] [PubMed] [Google Scholar]
- Woods A., Sherwin,T., Sasse,R., MacRae,T.H., Baines,A.J. and Gull,K. (1989) Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci., 93, 491–500. [DOI] [PubMed] [Google Scholar]
- Zakai H.A., Chance,M.L. and Bates,P.A. (1998) In vitro stimulation of metacyclogenesis in Leishmania braziliensis, L. donovani, L. major and L. mexicana. Parasitology, 116, 305–309. [DOI] [PubMed] [Google Scholar]