Abstract
Replication represents a key step in the infectious cycles of RNA viruses. Here we describe a regulatory RNA element, termed replication silencer, that can down-regulate complementary RNA synthesis of a positive-strand RNA virus via an RNA–RNA interaction. This interaction occurs between the 5-nucleotide-long, internally positioned replication silencer and the extreme 3′-terminus of the viral RNA comprising part of the minimal minus-strand initiation promoter. Analysis of RNA synthesis in vitro, using model defective interfering (DI) RNA templates of tomato bushy stunt virus and a partially purified, RNA-dependent RNA polymerase preparation from tombusvirus-infected plants, revealed that this interaction inhibits minus-strand synthesis 7-fold. This functional interaction was supported further by: (i) RNA structure probing; (ii) phylogenetic analysis; (iii) inhibition of activity by short complementary DNAs; and (iv) compensatory mutational analysis. The silencer was found to be essential for accumulation of DI RNAs in protoplasts, indicating that it serves an important regulatory role(s) in vivo. Because similar silencer–promoter interactions are also predicted in other virus genera, this type of RNA-based regulatory mechanism may represent a widely utilized strategy for modulating replication.
Keywords: cis-acting RNA element/RNA-dependent RNA polymerase/RNA promoter/RNA synthesis/virus replication
Introduction
Positive-strand RNA viruses are significant pathogens of humans, animals and plants. Despite their diverse host ranges, the replication strategies of this class of viruses show remarkable similarities, including the involvement of viral-coded, RNA-dependent RNA polymerases (RdRps) and the dual roles of viral genomes as mRNAs for viral protein synthesis and templates for the production of progeny genomes (Buck, 1996; Ahlquist, 2002). Following entry into a host cell, viral proteins are translated from the genome and, subsequently, the genome is used as a template for its own replication. Studies have shown that replication cannot proceed if the genome is being actively translated (Gamarnik and Andino, 1998).
Genome replication in plus-strand RNA viruses is considered to be a two-step process that leads to the preferential production of plus-strand progeny (Buck, 1996, 1999). The first step in this scheme involves the production of small amounts of minus-strand RNAs. In the second step, these intermediates are used as templates to synthesize much larger amounts of plus-stranded progeny RNAs. RNA elements that regulate the level, polarity and timing of RNA synthesis are present within both the plus- and minus-strand templates (Buck, 1996; Lai, 1998; Kao et al., 2001). The best-characterized cis-acting elements are the promoters, which serve to specify the site of initiation in addition to influencing the level of complementary RNA synthesis (Buck, 1996; Dreher, 1999; Kao et al., 2001). Promoters are generally found at 3′-terminal locations in both the plus- and minus-strand RNA templates and, because their sequences and secondary structures are different, it is believed that intrinsic promoter strength could account for the observed strand asymmetry in RNA replication. The emerging picture, however, is more complex. The discovery of novel cis-acting replication enhancer elements for turnip crinkle virus (TCV) (Nagy et al., 1999, 2001), tomato bushy stunt virus (TBSV) (Ray and White, 1999, 2003; Panavas and Nagy, 2003) and other viruses (Esteban et al., 1989; Ranjith-Kumar, 2003) suggests that basal promoter activity can be stimulated significantly. This mechanism is distinct from other types of enhancer elements such as those in brome mosaic virus, which serve to stabilize and recruit RNA templates to sites of replication (Sullivan and Ahlquist, 1999).
Tombusviruses, including TBSV, are plus-stranded RNA viruses of plants that are frequently associated with parasitic defective interfering (DI) RNAs. These DI RNAs are derived from the parental viral genome by multiple sequence deletions and do not encode any essential replication proteins (Hillman et al., 1987; White and Morris, 1999). As a result, they are replicated only in the presence of the parental genome, which provides viral RdRp. Their efficient replication and non-coding nature make them excellent surrogate templates for studying cis-acting RNA elements required for genome replication (White and Morris, 1999). Indeed, previous in vivo and in vitro studies using TBSV-associated DI RNAs revealed that short 3′-terminal promoters and enhancer-like elements are present within these molecules (Ray and White, 1999, 2003; Panavas et al., 2002a,b; Panavas and Nagy, 2003).
In this work, we describe a novel cis-acting RNA element—termed a replication silencer—that down- regulates minus-strand RNA synthesis in vitro from plus-stranded genomic or DI RNA-based templates. The inhibitory activity of the replication silencer is mediated by its base-pairing to the 3′-terminal portion of the minus-strand initiation promoter and leads to almost an order of magnitude reduction in promoter activity. The interaction is also critical for tombusvirus DI RNA replication in protoplasts, confirming an essential role of the silencer in vivo. The potential for similar types of interactions in other viruses suggests that silencer- mediated regulation of viral RNA replication may represent a widespread phenomenon.
Results
Rationale
Tombusviruses, like other plus-strand RNA viruses, produce significantly (up to ∼100-fold; J.Pogany and P.D.Nagy, unpublished data) more plus-strands than minus-strands during their replication. This asymmetrical RNA synthesis cannot be explained by the ‘strength’ of the separate promoters present on plus- or minus-stranded templates, because they supported similar levels of RNA synthesis in vitro using a partially purified tombusvirus RdRp preparation (Nagy and Pogany, 2000; Panavas et al., 2002a,b). Although the minus-stranded template contains two replication enhancers (Panavas and Nagy, 2003; Panavas et al., 2003; Ray and White, 2003), these regulatory RNA elements can only enhance the production of plus-strands by ∼10- to 20-fold, which is less than the ∼100-fold difference in plus- versus minus-strand synthesis. To explain this discrepancy, we reasoned that the plus-strand templates might contain inhibitory sequences that would decrease the use of plus-strands for minus-strand synthesis. Overall, the combined effect of inhibitory sequences reducing minus-strand synthesis and the enhancer sequences increasing plus-strand synthesis would result in less competition between these processes for the viral RdRp, and it might explain how the asymmetrical replication could be achieved in tombusviruses.
Inhibition of minus-strand synthesis in vitro by a stem–loop structure
The presence of putative inhibitory sequence in plus-strand templates is supported by previous in vitro studies using DI RNA templates and a partially purified tombusvirus RdRp preparation, which revealed that the full-length positive-strand DI RNAs are poor templates for complementary strand synthesis (Nagy and Pogany, 2000). In contrast, the 3′-terminal ‘core’ minus-strand initiation promoter (gPR, the 3′-terminal 19 nucleotide) supported high levels of complementary RNA synthesis (Nagy and Pogany, 2000; Panavas et al., 2002a,b). Based on these results, we hypothesized that a sequence in the full-length template may be responsible for inhibiting complementary RNA synthesis.
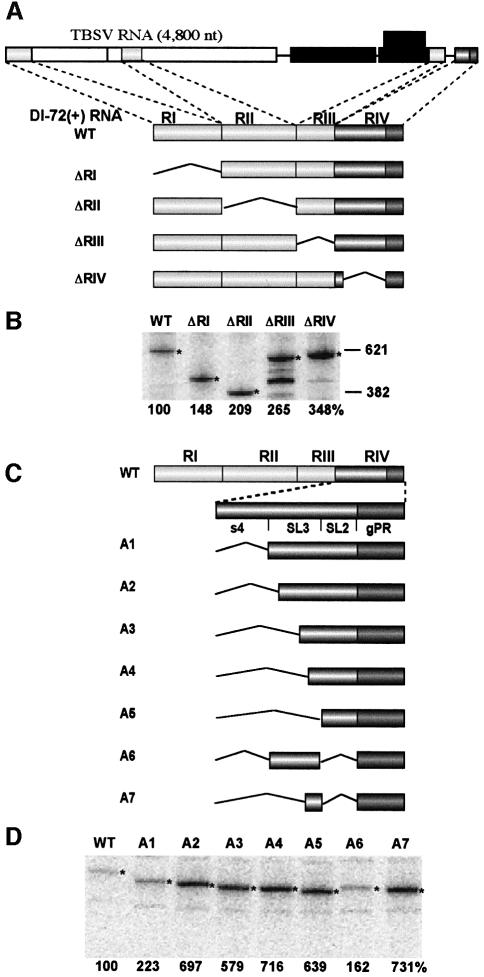
To test this concept, we made a series of DI RNA deletion constructs, each of which lacked one of the four conserved regions (RI to RIV), and tested their in vitro template activities in the tombusvirus RdRp system (Figure 1A). For the region IV deletion mutant, ΔRIV, only the 5′-proximal portion of RIV was removed so that it would maintain the minus-strand promoter, gPR (Panavas et al., 2002a, b). Analysis of the complementary RNA products generated from these RNA templates revealed the ΔRIV template was the most active, while deletions of regions I, II or III had lesser stimulatory effects (Figure 1B). These results suggest that all regions inhibit RNA synthesis to varying degrees, but that the 5′ half of RIV is most effective at down-regulating RNA synthesis.
Fig. 1. Mapping inhibitory sequences of minus-strand synthesis in tombusviruses. (A) Schematic representation of a typical TBSV genomic RNA, the prototypical DI-72 RNA and its deletion derivatives. Tombusvirus genomes contain five open reading frames (ORFs), of which two are translated directly from the genome (shown by open boxes) and three (shown by black boxes) are translated from two subgenomic RNAs (not shown). The four non-contiguous regions (indicated by roman numerals) from which DI-72 RNA is derived are depicted with gray boxes. RI (169 nucleotides) is derived from the 5′-non-translated region of the genomic TBSV RNA. RII is 239 nucleotides long and originated from the coding region (namely the p92 ORF), while RIII (82 nucleotides) represents the end of the p22 ORF plus part of the 3′-non-coding region. RIV is 131 nucleotides long and derived from the very 3′-terminal non-coding region of the TBSV RNA. Each region of DI-72(+) was deleted separately to generate the four constructs shown. (B) Relative template activities of the above RNA constructs in an in vitro tombusvirus RdRp assay. The radiolabeled RdRp products, synthesized by in vitro transcription with CNV RdRp, were analyzed on denaturing gels, quantified using a phosphoimager and normalized based on the number of templated uridylates since [32P]UTP was used for labeling in the RdRp reaction. RNA templates were used in equal molar amounts. Half of the RdRp products were treated with single-strand-specific ribonuclease to confirm the double-stranded nature of the RdRp products (data not shown; see also Nagy and Pogany, 2000). The expected template-sized products are depicted with asterisks. The relative efficiencies of template activities are shown at the bottom. Each experiment was repeated two or three times. (C) Schematic representation of the DI-72(+)-derived constructs with deletions in RIV. Portions of deleted RIV are shown with broken lines. The gPR sequence, represented by a dark gray box, was not deleted in these constructs because it is required for the initiation of minus-strand synthesis (Panavas et al., 2002a, b). Note that constructs A1 to A7 also contain RI, RII and RIII (not shown). (D) Representative denaturing gel analyses of radiolabeled RNA products synthesized on templates shown in (C) by in vitro transcription with CNV RdRp. The CNV (a tombusvirus closely related to TBSV) RdRp can efficiently and correctly recognize DI-72 RNA in vitro (Nagy and Pogany, 2000). Asterisks depict the RdRp products generated by de novo initiation from the 3′-terminus.
To map the inhibitory element in RIV more precisely, we made an additional set of deletions in this region (Figure 1C). Based on secondary structure analysis that will be presented in a separate publication (Fabian, et al., 2003), RIV of DI-72(+) can be divided into four segments (Figure 1C). In the mutants constructed, various combinations of s4, SL3 and SL2 were deleted from the full-length DI-72(+), however gPR was not altered because it is required for initiation of complementary RNA synthesis (Panavas et al., 2002a). Analysis of the various deletion mutants revealed that the most active templates were those that lacked either part of, or the entire SL3 sequence (i.e. templates A2 to A5; Figure 1C and D). In contrast, deletion of s4 or SL2/s4 changed the template activity only marginally (constructs A1 and A6; Figure 1C). The ∼7-fold increase in complementary strand synthesis observed when SL3 (templates A3, A4 and A5; Figure 1C) was deleted implicated this structure as a key determinant of minus-strand inhibition.
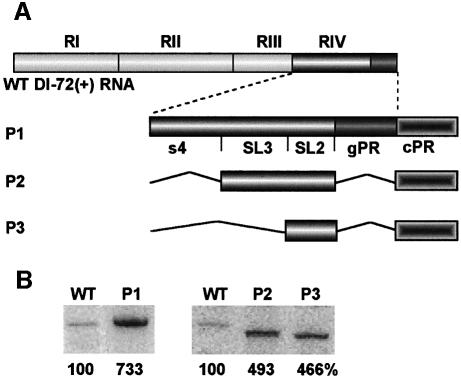
To determine if SL3 represented a non-specific inhibitor of RNA synthesis, we investigated whether it could down-regulate the activity of a different promoter. To test this, the plus-strand initiation promoter of TBSV (termed cPR, which includes the 21-nucleotide long 3′ terminal sequence of the minus strands; see construct cPR21 in Panavas et al., 2002a) was fused to the 3′ end of DI-72(+)RNA (construct P1 in Figure 2A, which also carries the gPR promoter). The resulting P1 construct, containing cPR and gPR in tandem, supported minus-strand synthesis ∼7-fold more efficiently than the wild-type DI-72(+) (Figure 2B). The increase in activity and the size of the RdRp product indicated efficient use of the cPR promoter in the presence of SL3. Similar high levels of activity were also observed with P2, in which gPR was replaced with cPR (Figure 2A). Construct P3, a derivative of P2 that lacks SL3, also supported RNA synthesis at the same level as P2 (Figure 2B). Thus, SL3 does not seem to modulate initiation from the cPR promoter. Overall, we conclude that the inhibitory activity of SL3 is specific for initiation from the gPR promoter.
Fig. 2. Initiation from the plus-strand promoter is not inhibited by the SL3 sequence. (A) Schematic representation of the constructs tested in the in vitro CNV RdRp assays. A 21-nucleotide long plus-strand initiation sequence (labeled as cPR) was fused to the 3′ end of full-length DI-72(+)RNA or its derivatives, as shown. The sequences not present in the constructs are shown with broken lines. Note that constructs P1 to P3 also contain RI, RII and RIII (not shown). (B) Representative denaturing gel analyses of radiolabeled RNA products synthesized by in vitro transcription with CNV RdRp. See the legend to Figure 1 for details.
A five-nucleotide sequence within SL3 is essential for replication silencing in vitro
The inhibition of only the gPR promoter by the SL3 suggested a specific mechanism for suppression. One means of conferring selective communication between RNA elements is via RNA–RNA interactions. For example, base pairing between these structures could mask critical elements in the gPR and/or sterically hinder accessibility to the RdRp—either of which would lead to reduced template activity.
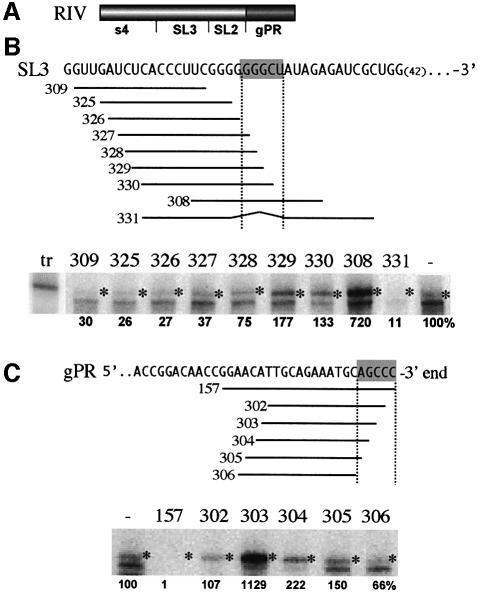
As a first step to identifying sequences in SL3, which could potentially participate in an RNA–RNA interaction, we performed oligonucleotide-based inhibition studies. In these experiments, 16-base single-stranded (ss) DNAs that were complementary to various portions of the SL3 segment were included, together with a minimal RNA template (construct RIV; Figure 3A), and tested in the in vitro RdRp system. It was anticipated that ssDNA that base-paired efficiently to the critical sequence within the SL3 region would suppress its inhibitory activity. In turn, the RNA sequence complementary to that oligonucliotide would be a good candidate for participating in an RNA–RNA interaction with the gPR promoter. The results of the assay revealed that the SL3 segment was unable to inhibit minus-strand synthesis when either ssDNAs 329, 330 or 308 were present, with the latter being the most effective (Figure 3B). Interestingly, there is a five-nucleotide stretch in SL3 (5′-GGGCU; boxed in Figure 3B), corresponding to the middle of ssDNA 308, which is complementary to the very 3′-terminal sequence of the gPR promoter (5′-AGCCC; boxed in Figure 3C). This GGGCU sequence therefore represented a potential element for inhibiting the gPR promoter by base-pairing with the 3′ end sequence. To investigate this possibility, we designed ssDNA 331 to hybridize only to the sequence adjacent to the GGGCU segment, leaving the GGGCU sequence unbound and available to participate in other interactions (Figure 3B). The RdRp assay revealed that minus-strand synthesis was very low (11% of the wild type; Figure 3B) in the presence of ssDNA 331, suggesting that the GGGCU sequence needed to be unbound by ssDNA in order to effectively inhibit RNA synthesis.
Fig. 3. Mapping the location of the replication silencer using DNA oligonuclotides in an in vitro tombusvirus RdRp assay. (A) Schematic representation of the RIV template, which includes only RIV sequences (Figure 1A). (B) The actual sequence of SL3 in construct RIV is shown on the top, and the complementary ssDNAs are shown below the target sequence. The critical nucleotides in SL3 are boxed. The position of the last nucleotide shown is indicated. A representative denaturing gel analysis of radiolabeled RNA products synthesized by in vitro transcription with CNV RdRp is also shown. Each RdRp reaction contained the same amounts of the RIV template and the ssDNA competitor indicated. The lane marked ‘–’ represents the control, with no ssDNA competitor added. Asterisks depict the RdRp products generated by de novo initiation from the 3′-terminus. The band migrating below the template-sized band (marked with asterisk) is the result of internal initiation, probably at an internal cytidylate (position 6 from the 3′ end). The molecular marker (the RIV construct) is shown as ‘tr’ on the left. See Figure 1 for further details. (C) The actual sequence of gPR is shown on the top and the complementary ssDNAs are shown below the target sequence. The critical sequence in gPR is boxed. See (A) for further details.
To evaluate the role of the 3′ end of the gPR promoter in the replication silencing phenomenon, we analyzed it using the oligonucleotide-based inhibition approach used for SL3. These experiments demonstrated that ssDNA 306, 305 and 304, which left five, four and three nucleotides at the 3′ end non-base-paired to ssDNA, respectively, either did not enhance minus-strand synthesis or did so only marginally (1.5- to 2-fold) when compared with the control ssDNA-free control (Figure 3C). Interestingly, ssDNA 303, which left only two of the 3′ terminal cytidylates unbase-paired, supported minus-strand synthesis 11-fold more efficiently (Figure 3C). This increased template activity is consistent with interference by ssDNA 303 with the SL3/gPR promoter interaction. Taken together, these data indicate that the 3′-terminal five nucleotides within the gPR promoter need to be exposed (non-base-paired to ssDNA) for inhibition via the SL3/gPR promoter interaction to occur. Consistent with this notion is the observation that in the presence of ssDNA 302 and 157, which left one or no nucleotides exposed at the 3′ end, minus-strand synthesis was inhibited at control levels or was blocked completely (Figure 3C). In these latter cases, the ssDNAs are likely acting to effectively block the 3′-terminal sequences in a manner similar to that proposed for the pentanucleotide GGGCU sequence in SL3.
Solution structure probing and sequence comparison of 3′ ends of tombusviruses support the SL3/gPR promoter base-pairing interaction
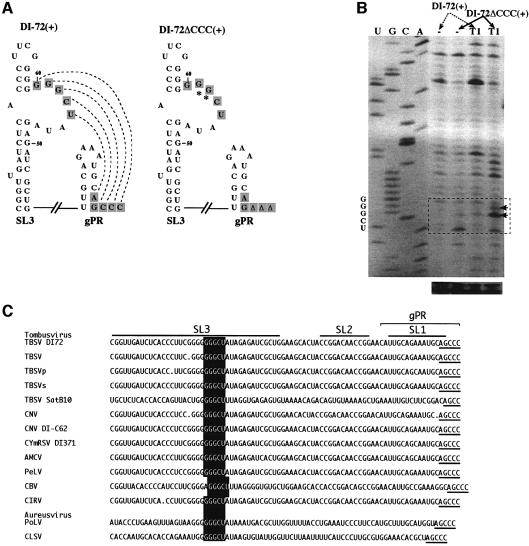
The proposed mechanism of action of complementary strand inhibition relies on a base-pairing interaction between 5′-GGGCU in SL3 and the complementary sequence 5′-AGCCCOH at the end of the template (within the gPR promoter). MFOLD-predicted secondary structures for SL3 and gPR are presented in Figure 4, and these structures are consistent with solution structure probing and compensatory mutational analysis of RIV that will be presented in a separate publication (Fabian, et al., 2003). In the present study we focused our solution structure analysis on the sequences in the proposed interaction. To provide physical evidence for this interaction, we performed RNase T1 analysis (Figure 4B) of either wild-type full-length DI-72(+) or DI-72ΔCCC(+) transcripts. The latter transcript lacks the three 3′-terminal cytidylates and therefore should not be able to form the SL3/gPR promoter interaction efficiently, if at all. The solution structure data (Figure 4B) are consistent with the SL3/gPR promoter interaction, because the G residues in the 5′-GGGCU sequence in SL3 become sensitive to cleavage by ssRNA-specific T1 ribonuclease in the DI-72ΔCCC(+)RNA.
Fig. 4. An RNA–RNA interaction between gPR and SL3 is supported by enzymatic probing of DI-72(+). (A) Schematic representation of the putative interaction, indicated with dotted lines, between the interior loop in SL3 and the 3′ end in DI-72(+), and the lack of interaction in DI-72ΔCCC(+).The predicted secondary structures of gPR and SL3 are shown with complementary sequences boxed. The RNase T1 (T1) sensitive nucleotides in the internal loop are indicated with arrows (see below). (B) Enzymatic probing of DI-72(+) and DI-72ΔCCC(+). The in vitro-transcribed RNAs were treated in vitro with T1 or not treated (control), followed by analysis by reverse transcription-based primer extension using primer PMF59S in the presence of [35S]dATP, and separated on an 8% sequencing gel. The areas boxed with dotted lines include the interior-loop region of SL3. (C) Sequence comparison of the replication silencer region and the 3′ ends in tombusviruses. Sequences are shown in the 5′-to-3′direction, with AGCCC as the 3′ end. The predicted secondary structure for DI-72 is shown schematically on the top. The conserved silencer sequence is boxed, while the complementary 3′ terminal pentanucleotide is underlined.
Comparison of 3′-end plus-strand sequences in tombusviruses and related DI or satellite RNAs revealed the conserved nature of the replication silencer (GGGCU, boxed in Figure 4C) and the 3′-end sequences (AGCCC, underlined). The conservation of these sequences and their ability to base pair is consistent with their functional importance during replication.
Compensatory mutations between gPR and SL3 demonstrate that down-regulation of replication is based on an RNA–RNA interaction
Additional sets of mutants were constructed to obtain further evidence that replication silencing functions through base pairing. Various substitutions were introduced into the SL3 5′-GGGCU sequence that was predicted to decrease the stability of its proposed interaction with the gPR promoter (SL3-U, UC and UUC; Figure 5A). In all cases there were significant increases in RNA synthesis (11- to 16-fold), indicating defective inhibitory activity (Figure 5B). The substitution of all three 3′-terminal cytidylates to adenylates in gPR-AAA resulted in no template-sized RdRp product visible in the gels, indicating that these cytidylates are necessary for de novo initiation from the 3′ end in vitro (Figure 5A and B). Disruption of the predicted interaction with a two-nucleotide substitution in gPR-GA supported a 45-fold increase in template activity (Figure 5B). Collectively, these data are consistent with the requirement of the proposed interaction for inhibition of RNA synthesis.
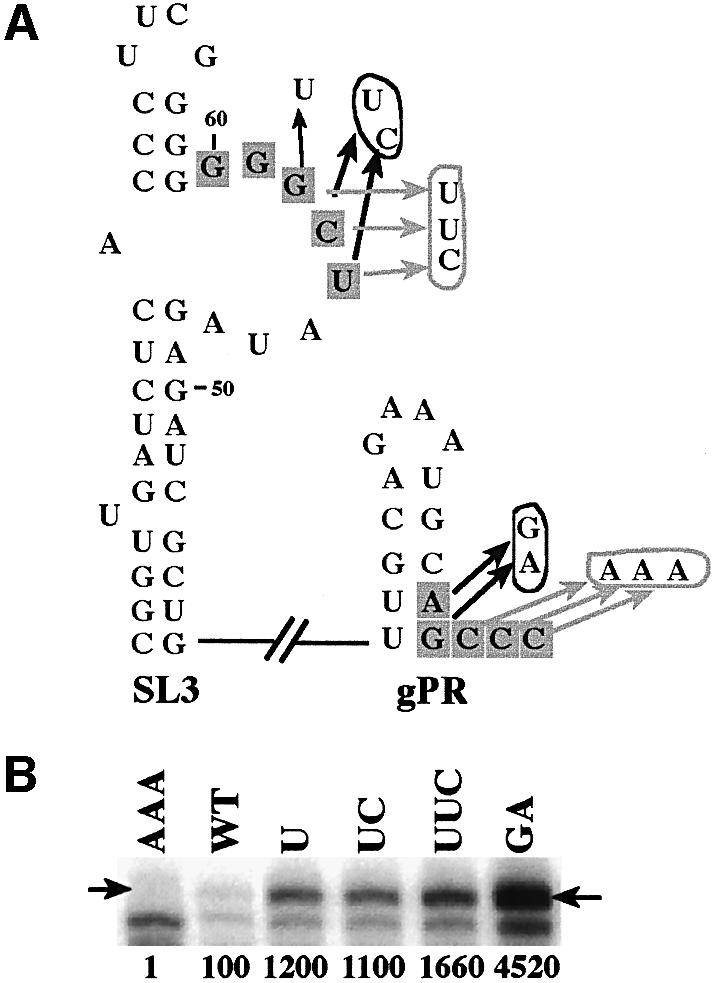
Fig. 5. Mutations in gPR and SL3 sequences affect minus-strand synthesis in the in vitro CNV RdRp assay. (A) Schematic representation of the mutation series of constructs generated from RIV (Figure 3). The mutated nucleotides are indicated with arrows, with the double and triple mutants circled (see Figure 3 for a detailed description of the individual sequence elements). The names of the constructs indicate the type of mutation. (B) Representative denaturing gel analyses of radiolabeled RNA products synthesized by in vitro transcription with CNV RdRp. Arrows point to the RdRp products generated by initiation from the 3′-terminus. The band migrating below the template-sized band is the result of internal initiation, probably at an internal cytidylate (position 6 from the 3′ end). Further details are as described in the legend to Figure 1.
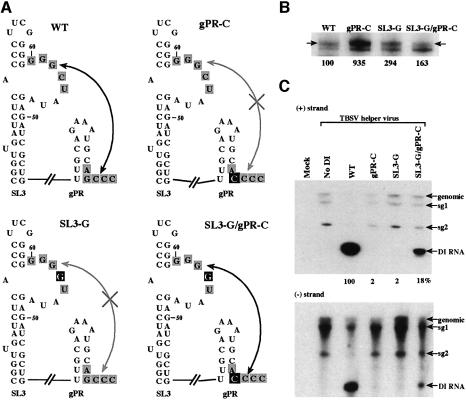
To obtain more compelling evidence for this interaction, we used the compensatory mutagenesis approach. Changing the SL3 sequence from GGGCU to GGGGU (SL3-G; Figure 6A) or the 3′-terminal sequence from AGCCC to ACCCC (gPR-C), both of which would disrupt the interaction, resulted in 3- and 9-fold increases, respectively, in RNA synthesis (Figure 6B). However, combining these independent substitutions in the same template (SL3-G/gPR-C), which would restore the interaction, reduced RNA synthesis from the gPR-C promoter by ∼6-fold, resulting in near wild-type levels of RNA synthesis (Figure 6B). This clear correlation between base-pairing potential and down-regulation of RNA synthesis supports the concept that the inhibitory activity is mediated via the proposed RNA–RNA interaction. Consequently, we have termed the critical GGGCU sequence in SL3 a ‘replication silencer’.
Fig. 6. Compensatory mutations in gPR and SL3 sequences inhibit minus-strand synthesis in vitro and essential for replication in vivo. (A) Schematic representation of the mutation series of constructs generated from RIV (Figure 3A). The mutated nucleotides are indicated with letters in black boxes (see Figure 3 for a detailed description of the individual sequence elements). The names of the constructs indicate the position and type of mutation. Black arrows indicate a possibly strong gPR–SL3 interaction, while gray arrows (crossed in the middle) indicate weak interactions. (B) Representative denaturing gel analyses of radiolabeled RNA products synthesized by in vitro transcription with CNV RdRp. Arrows point to the RdRp products generated by initiation from the 3′-terminus. The band migrating below the template-sized band is the result of internal initiation, probably at an internal cytidylate (position 6 from the 3′ end). Further details are as described in the legend to Figure 1. (C) In vivo analysis of the accumulation of DI RNAs containing single mutations in gPR or SL3, or compensatory mutations in gPR and SL3. Northern blot analysis of positive strands (top) and negative strands (bottom) is performed with oligonucleotide PR29. Total RNA was harvested from cucumber protoplasts, which had been inoculated with TBSV genomic RNA and the respective DI RNAs, as described in the Materials and methods. The positions of the genomic, the two subgenomic (sg1 and sg2) and the DI RNAs are indicated with arrows. The relative accumulation of DI RNAs is shown below the autoradiogram.
Interaction between the replication silencer and the promoter is essential for DI RNA replication in vivo
To determine if the replication silencer and gPR promoter interaction is also relevant for viral RNA replication in vivo, we generated full-length DI-72 RNA templates carrying the same set of compensatory mutations as those described in Figure 6A. DI RNA mutants were tested by co-infection with helper genome (i.e. transcripts of the TBSV genome) into cucumber protoplasts, and accumulation of DI RNA plus- and minus-strands was monitored by northern blotting (Figure 6C). These experiments revealed that constructs DI-72SL3-G and DI-72gPR-C, which are expected to have weak RNA–RNA interactions between the SL3 replication silencer and the gPR promoter, replicated extremely poorly in protoplasts (Figure 6C). In contrast, the double mutant restoring base pairing, DI-72SL3-G/gPR-C, replicated to readily detectable levels, reaching ∼20% of the wild-type level (Figure 6C). Therefore, the RNA–RNA interaction shown to influence complementary RNA synthesis in vitro is also important for in vivo-replication of a model viral RNA.
The replication silencer is also functional on the TBSV genomic RNA
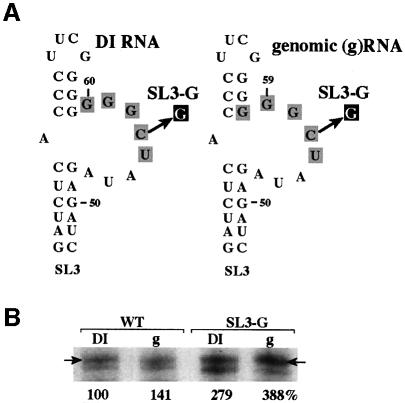
Sequences in DI RNAs are almost identical to the corresponding regions in the genomic RNA, leading to the assumption that common cis-acting elements are functional on both DI and genomic RNAs. Since the full-length, plus-stranded genomic RNA, similar to the DI-72(+)RNA, is a poor template for RNA synthesis in vitro (not shown), it is likely that the SL3 replication silencer is also functional on the genomic RNA. We tested this assumption in the in vitro system, because the genomic RNA contains one less guanylate within the replication silencer region compared with DI-72 RNA (see the run of six versus seven Gs in the genomic and DI RNA, respectively; Figure 7A). We made the corresponding minimal SL3/gPR constructs, without or with a C→G mutation (see Figure 7A, constructs SL3-G), representing either the DI RNA or the genomic RNA sequences, and tested their in vitro template activities in the standard tombusvirus RdRp assay. These experiments revealed that the genomic and DI RNA sequences supported increased levels of RNA synthesis for the mutated constructs (Figure 7B). This indicates that the replication silencer is functional in both the genomic and DI(+)RNAs.
Fig. 7. Comparison of the replication silencers in TBSV genomic and DI RNAs. (A) The sequences of the genomic (g)RNA and the DI RNA differ at one position within the replication silencer region, which is the presence of an extra guanylate within the internal loop in DI-72 as shown (two versus three Gs, boxed). Note that a letter in a black box represents a single mutation (see Figure 6). (B) Representative denaturing gel analyses of radiolabeled RNA products synthesized by in vitro transcription with CNV RdRp. Arrows point to the RdRp products generated by initiation from the 3′-terminus. Further details are as described in the legend to Figures 1 and 3.
Discussion
The replication of the genomes of positive-strand RNA viruses is regulated by cis-acting RNA elements present in both plus- and minus-stranded viral RNAs (Buck, 1996; Dreher, 1999; Kao et al., 2001). In this paper, we present evidence for the existence of a novel type of cis-acting element, which can down-regulate the level of complementary RNA synthesis from a promoter element in an in vitro RdRp system. This element functions via an RNA–RNA interaction with the 3′ portion of the gPR promoter. Based on our analyses, we have termed the SL3 segment a replication silencer since it: (i) inhibits RNA synthesis from the gPR minus-strand initiation promoter; but (ii) it is not an essential component of the 3′-terminal gPR promoter.
From a mechanistic perspective, we provided physical and functional evidence (Figures 3–6) that the replication silencer operates by base-pairing with the very 3′-end of the template RNA, which includes part of the gPR promoter. The interaction between the replication silencer (GGGCU) and the gPR promoter are consistent with a mechanism of inhibition of RNA synthesis mediated by ‘promoter masking’. Previous studies with tombusvirus RdRp in vitro have shown that minus-strand synthesis initiates at the ultimate or penultimate 3′-terminal cytidylate, and that these residues need to be present for efficient RNA synthesis (Figure 5, lane 1; Nagy and Pogany, 2000). Thus, it is not surprising that base pair-mediated masking of these 3′-terminal residues by the silencer would reduce their accessibility to the RdRp and have an inhibitory affect on initiation of complementary RNA synthesis. This mechanism is further supported by the complete inhibition of RNA synthesis obtained with ssDNA 157, which also binds to all three 3′-terminal cytidylates. Conversely, the ssDNA 303 that left two of the 3′-terminal cytidylates unbase-paired supported RNA synthesis 11-fold more efficiently than the control template. This result can be explained by ssDNA 303 efficiently blocking silencer binding while allowing RdRp access to the two unpaired 3′-terminal cytidylates in the template. These results, combined with (i) the solution structure mapping that supports a physical interaction between these regions (Figure 4), (ii) the conserved nature of the silencer region and the complementary 3′ terminal sequences among tombusviruses (Figure 4C) and (iii) compensatory mutational analysis (Figure 5) that provides correlative structure/function data, support the proposed mechanism for down-regulation of minus-strand synthesis in the in vitro tombusvirus RdRp system. Detailed analysis of additional compensatory mutations (such as UC–GA mutations within the silencer and the gPR, respectively), the role of neighboring sequences and the effect of the length of base-pairing between the silencer region and gPR will be published elsewhere. Importantly, it is unlikely that the replication silencer down-regulates complementary RNA synthesis by disrupting the essential stem–loop in gPR. This is because: (i) the two base pairs within the lower stem (i.e. U–G and U–A) in gPR are not essential for complementary RNA synthesis in vitro (J.Pogany and P.D.Nagy, unpublished); and (ii) ssDNA 303, although it disrupts the stem–loop in gPR, even enhanced RNA synthesis (Figure 3C). Based on these observations, we conclude that the replication silencer most likely functions via forming a double-stranded structure with the 3′ end, thus masking the initiation site from recognition by the tombusvirus RdRp.
Inhibition of RNA synthesis by the replication silencer appears to be promoter-specific, because it inhibited RNA synthesis from the gPR promoter, but not from the cPR plus-strand initiation promoter. Therefore, the replication silencer does not appear to represent a generic ‘road block’ for the tombusvirus RdRp or an element that reduces processivity of RNA synthesis. Instead, the replication silencer likely acts at the level of initiation of RNA synthesis by specifically masking the promoter initiation sequences from RdRp. The promoter specificity observed is also consistent with the proposed silencer/gPR base pair-mediated mechanism of inhibition because the cPR does not contain sequences that are complementary to the silencer, which would make cPR immune to the inhibitory activity.
Sequences in DI RNAs are almost identical to the corresponding regions in the genomic RNAs of tombusviruses, leading to the assumption that common cis-acting elements are functional in both DI and genomic RNAs. Accordingly, we observed that the TBSV genomic RNA sequence, similar to the DI-72(+)RNA sequence, was a poor template for RNA synthesis in the in vitro RdRp assay, while mutations within the silencer sequence increased template activities (Figure 7). Therefore, the replication silencer is likely important for the replication of both DI and genomic RNAs.
The in vivo experiments with mutated DI RNA templates in protoplasts confirmed that mutations within either the silencer or gPR could debilitate replication, while compensatory mutations could restore the replication ability of the DI RNA, albeit at a reduced level (Figure 6C). We found that accumulation of minus-strands mirrored the accumulation of plus-strands. Although this observation is different from the in vitro results, this may not be surprising, because the 3′-end, in addition to the minus-strand synthesis, could play multiple roles in many essential processes in replication (see below). Never theless, the in vivo experiments confirmed that the replication silencer–gPR promoter interaction is essential for tombusvirus replication.
The purpose of down-regulation of minus-strand synthesis by the replication silencer is currently unknown. However, the requirement for this interaction for productive DI RNA accumulation in protoplasts supports an important role(s) for this element in vivo (Figure 6C). Accordingly, we propose four different models, all of which might be important, that could explain why replication silencers may be advantageous for tombusviruses. The first possible function of the replication silencer could be to promote asymmetrical replication of the tombusviral RNA. This model predicts that, due to the presence of the replication silencer, the plus-stranded RNAs could only support the synthesis of limited amounts of minus-strand in the infected cells. Limiting the use of the abundant plus-strands for minus-strand synthesis might be important to reduce the level of competition between minus- and plus-strands for the tombusvirus RdRp, which should mainly be involved in the robust plus-strand synthesis process. Furthermore, the production of large amounts of non-infectious intermediate minus-strands would not be economical in a system where the ribonucleotide pool may be limiting. Finally, but perhaps most relevant, the production of small amounts of minus-strands would delay recognition of the viral infection by the host surveillance system, termed postranscriptional gene silencing (PTGS) in plants, which is activated by double-stranded RNAs (Vance and Vaucheret, 2001; Waterhouse et al., 2001). Production of a large amount of minus-strands during tombusvirus or DI RNA replication would certainly trigger rapid PTGS response from the host.
A second role of the conserved replication silencer–gPR interaction might be to serve as a recognition signal for viral replicase proteins or for possible host factors. This might be important for either targeting the RNA to the site of replication or assembly of the replicase complex (Ahlquist, 2002). Since these steps occur prior to the minus-strand synthesis, disruption of the replication silencer–gPR interaction could prevent replication.
A third role for the replication silencer–gPR interaction may be to protect the 3′ end of the RNA from degradation. Maintenance of the 3′-terminal cytidylates is important because they are required for de novo initiation in vitro (see Figure 5 construct gPR-AAA) and efficient replication in vivo (M.Fabian and K.A.White, unpublished data). Without base-pairing with the replication silencer, the 3′ end of gPR would be single-stranded and would likely be vulnerable to digestion by ribonuclease during various stages of the viral replication cycle. However, formation of the silencer–promoter interaction would make the 3′ end double-stranded and more resistant to ssRNA-specific ribonucleases (Mitchell and Tollervey, 2000). This may be especially important in the early stages of infection (prior to replication) when the viral RNA is not yet involved in replication and therefore not yet protected by binding to either host or viral proteins (e.g. RdRp), or localization to specialized compartments within the cell. It is interesting to note that tombus- and carmoviruses have an additional 3′-terminal protection mechanism that is based on 3′-end repair (Dalmay et al., 1993; Nagy et al., 1997; Guan and Simon, 2000). In fact the protection and repair systems in tombusviruses may be linked because the base-pairing interaction that could protect the 3′ sequences from nucleases could also act to prime and provide a template for extension and repair of the 3′ end. These redundant mechanisms for protection of the 3′-end sequences would be beneficial to these viruses in the hostile host environment.
A fourth function of the replication silencer could be in the temporal regulation of the different roles of the genome in translation and replication during the virus replication cycle. It is possible that the interaction, which inhibits replication, could conversely facilitate translation. Indeed, the 3′-half of region IV, containing both the silencer element and gPR, is required for efficient translation of TBSV mRNAs (Wu and White, 1999). The down-regulation of replication and the up-regulation of translation would act to prevent competition between these two processes and the potential for collision between the 5′-to-3′-moving ribosome and the 3′-to-5′-moving RdRp on the same template. After completion of sufficient rounds of translation, the silencer–gPR complex may be recognized (possibly by a viral or a host factor) to recruit the viral RNA to the site of replication. Temporal regulation of translation and replication is also proposed in other viral systems, including poliovirus (Gamarnik and Andino, 1998) and alfalfa mosaic virus (Olsthoorn et al., 1999), although the mechanisms of molecular switches between translation and replication are likely different in those systems.
We propose that in vivo the replication silencer may be involved in any, or all, of the four functions described above. Clearly, its function in a cellular environment will be complex, however our demonstration of the requirement of this interaction for productive DI RNA replication provides us with a system to pursue these proposed activities in future studies. Furthermore, silencer-mediated regulation of RNA synthesis may extend to other plus-strand RNA viruses since similar replication silencer–promoter interactions are predicted in the related aureusvirus, carmovirus (not shown) and luteovirus genera (Koev et al., 2002). A somewhat similar structure, involving the 3′ end and an internal region, is also proposed to exist for the Qβ bacteriophage (Klovins et al., 1998; Klovins and van Duin, 1999). Future experiments will be needed to investigate whether these sequences play similar roles in these other viruses.
Materials and methods
Tombusvirus RdRp preparation
Nicotiana benthamiana plants were inoculated with cucumber necrosis virus (CNV; a tombusvirus closely related to TBSV) genomic RNA transcripts obtained by standard T7 RNA transcription using a SmaI-linerized clone of pK2/M5p20STOP for CNV (Rochon, 1991). CNV RdRp preparations were purified from systemically infected leaves of N.benthamiana as described previously (Nagy and Pogany, 2000). The CNV RdRp can efficiently and correctly recognize DI-72(+)RNA in vitro (Nagy and Pogany, 2000).
Preparation of RNA templates
TBSV-associated DI-72(+)RNA template and its derivatives were obtained by in vitro transcription with T7 RNA polymerase using PCR-amplified DNA templates (Nagy et al., 1999; Panavas et al., 2002a). The templates and the primers for each PCR amplification are listed in Supplementary table I (available at The EMBO Journal Online), while the sequences of the primers are shown in Supplementary table II. Several constructs were obtained by using sequential PCR (Panavas et al., 2002a), where the product of the first PCR reaction was gel-purified and used for a second round of PCR as shown in Supplementary table I.
After T7 RNA transcription and phenol–chloroform extraction, unincorporated nucleotides were removed by repeated ammonium-acetate–isopropanol precipitation (Nagy et al., 1998; Nagy and Pogany, 2000). The amounts and sizes of the obtained RNA transcripts were measured by a UV spectrophotometer and 5% polyacrylamide/8 M urea gel (denaturing PAGE) analysis (Nagy et al., 1998; Nagy and Pogany, 2000).
Tombusvirus RdRp assay
CNV RdRp reactions were carried out as described previously (Nagy and Pogany, 2000). Briefly, each RdRp reaction contained 100 nM template RNA, 50 mM Tris–HCl pH 8.2, 10 mM MgCl2, 10 mM dithiothreitol, 100 mM potassium glutamate, 1.0 mM each of ATP, CTP and GTP, 0.01 mM UTP (final concentration) and 0.3 µl of [32P]UTP (specific activity 800 Ci/mmol from ICN) in a 50 µl total volume. The RdRp reaction was performed at 25°C for 120 min. After phenol–chloroform extraction and ammonium-acetate–isopropanol precipitation, half the amount of the RdRp products were analyzed on a 20 cm long denaturing 5% PAGE/8 M urea gel, followed by phosphorimager analysis as described previously (Nagy and Pogany, 2000).
Preparation of RNA for protoplast inoculation
Mutant plasmid constructs were derived from the DI RNA clone DI-72 (White and Morris, 1994), utilizing PCR- and restriction enzyme-based procedures. The following PCR products were generated using a DI-72 template with specific oligonucleotide pairs in order to construct gPR-C, SL3-G, SL3-G/gPR-C and DI-72ΔCCC: PCR1, PR21/PMF61; PCR2, PR21/PMF62; PCR3, PR21/PMF63; and PCR4, PR21/PMF34. In order to construct gPR-C, SL3-G, SL3-G/gPR-C and DI72ΔCCC, PCR products 1–4, respectively, were digested with NsiI and SphI and inserted into DI-72 vectors digested with NsiI and SphI. Viral transcripts were synthesized in vitro by transcription of linearized template DNAs using an Ampliscribe T7 RNA polymerase transcription kit (Epicentre Technologies) as described previously (Oster et al., 1998). Viral cDNA clones were either linearized with SmaI or with PvuII for DI-72ΔCCC only.
Protoplast inoculation and viral RNA analysis
Protoplasts were purified from 7-day-old cucumber cotyledons and inoculated with viral transcripts using polyethylene glycol (PEG) and CaCl2 as described previously (White and Morris, 1994). Approximately 2 µg of full-length TBSV helper virus and/or 1 µg of DI-72-derived transcripts were used for inoculations and nucleic acid was isolated from protoplasts as described (White and Morris, 1994). Aliquots of total nucleic acid preparations were separated in denaturing 4.5% polyacrylamide gels containing 8 M urea, and viral RNAs were detected by electrophoretic transfer to nylon (Hybond-N; Amersham) followed by northern blot analysis using complementary 32P-end-labelled oligonucleotides (PR29).
Structural probing of RNA
Approximately 2 µg of transcript, either DI-72 or DI-72ΔCCC, were treated with RNase T1 as described previously (Wu et al., 2001). Enzymatically treated RNAs were analyzed by primer extension using oligonucleotide PMF59S and the products were separated in denaturing 6% (w/v) acrylamide gels containing 8 M urea. A DNA sequencing ladder generated from a DI-72 template using PMF59S was separated alongside extension products.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Drs Tom Pirone and John Shaw for critical reading of the manuscript, and Dr Mark Farman for very helpful suggestions. This work was supported by the National Science Foundation (MCB0078152) and by the University of Kentucky to P.D.N., and NSERC, PREA and CRC to K.A.W. M.F. was supported by an NSERC postgraduate scholarship.
References
- Ahlquist P. (2002) RNA-dependent RNA polymerases, viruses and RNA silencing. Science, 296, 1270–1273. [DOI] [PubMed] [Google Scholar]
- Buck K.W. (1996) Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res., 47, 159–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck K.W. (1999) Replication of tobacco mosaic virus RNA. Philos. Trans. R Soc. Lond. B Biol. Sci., 354, 613–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay T., Russo,M. and Burgyan,J. (1993) Repair in vivo of altered 3′ terminus of cymbidium ringspot tombusvirus RNA. Virology, 192, 551–555. [DOI] [PubMed] [Google Scholar]
- Dreher T.W. (1999) Functions of the 3′-untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol., 37, 151–174. [DOI] [PubMed] [Google Scholar]
- Esteban R., Fujimura,T. and Wickner,R.B. (1989) Internal and terminal cis-acting sites are necessary for in vitro replication of the L-A double-stranded RNA virus of yeast. EMBO J., 8, 947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M.R., Na,H., Ray,D. and White,K.A. (2003) 3′-terminal RNA secondary structures are important for accumulation of tomato bushy stunt virus DI RNAs. Virology, 313, 567–580. [DOI] [PubMed] [Google Scholar]
- Gamarnik A.V. and Andino,R. (1998) Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev., 12, 2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan H. and Simon,A.E. (2000) Polymerization of nontemplate bases before transcription initiation at the 3′ ends of templates by an RNA-dependent RNA polymerase: an activity involved in 3′ end repair of viral RNAs. Proc. Natl Acad. Sci. USA, 97, 12451–12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman B.I., Carrington,J.C. and Morris,T.J. (1987) A defective interfering RNA that contains a mosaic of a plant virus genome. Cell, 51, 427–433. [DOI] [PubMed] [Google Scholar]
- Kao C.C., Singh,P. and Ecker,D.J. (2001) De novo initiation of viral RNA-dependent RNA synthesis. Virology, 287, 251–260. [DOI] [PubMed] [Google Scholar]
- Klovins J. and van Duin,J. (1999) A long-range pseudoknot in Qβ RNA is essential for replication. J. Mol. Biol., 294, 875–884. [DOI] [PubMed] [Google Scholar]
- Klovins J., Berzins,V. and van Duin,J. (1998) A long-range interaction in Qβ RNA that bridges the thousand nucleotides between the M-site and the 3′ end is required for replication. RNA, 4, 948–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koev G., Liu,S., Beckett,R. and Miller,W.A. (2002) The 3′-terminal structure required for replication of Barley yellow dwarf virus RNA contains an embedded 3′ end. Virology, 292, 114–126. [DOI] [PubMed] [Google Scholar]
- Lai M.M. (1998) Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology, 244, 1–12. [DOI] [PubMed] [Google Scholar]
- Mitchell P. and Tollervey,D. (2000) mRNA stability in eukaryotes. Curr. Opin. Genet. Dev., 10, 193–198. [DOI] [PubMed] [Google Scholar]
- Nagy P.D. and Pogany,J. (2000) Partial purification and characterization of Cucumber necrosis virus and Tomato bushy stunt virus RNA-dependent RNA polymerases: similarities and differences in template usage between tombusvirus and carmovirus RNA-dependent RNA polymerases. Virology, 276, 279–288. [DOI] [PubMed] [Google Scholar]
- Nagy P.D., Carpenter,C.D. and Simon,A.E. (1997) A novel 3′-end repair mechanism in an RNA virus [published erratum appears in Proc. Natl Acad. Sci. USA (1997), 94, 5980–5982]. Proc. Natl Acad. Sci. USA, 94, 1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Zhang,C. and Simon,A.E. (1998) Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J., 17, 2392–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany,J. and Simon,A.E. (1999) RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J., 18, 5653–5665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Pogany,J. and Simon,A.E. (2001) In vivo and in vitro characterization of an RNA replication enhancer in a satellite RNA associated with turnip crinkle virus. Virology, 288, 315–324. [DOI] [PubMed] [Google Scholar]
- Olsthoorn R.C., Mertens,S., Brederode,F.T. and Bol,J.F. (1999) A conformational switch at the 3′ end of a plant virus RNA regulates viral replication. EMBO J., 18, 4856–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster S.K., Wu,B. and White,K.A. (1998) Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol., 72, 5845–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T. and Nagy,P.D. (2003) The RNA replication enhancer element of tombusviruses contains two interchangeable hairpins that are functional during plus-strand synthesis. J. Virol., 77, 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T., Pogany,J. and Nagy,P.D. (2002a) Analysis of minimal promoter sequences for plus-strand synthesis by the cucumber necrosis virus RNA-dependent RNA polymerase. Virology, 296, 263–274. [DOI] [PubMed] [Google Scholar]
- Panavas T., Pogany,J. and Nagy,P.D. (2002b) Internal initiation by the cucumber necrosis virus RNA-dependent RNA polymerase is facilitated by promoter-like sequences. Virology, 296, 275–287. [DOI] [PubMed] [Google Scholar]
- Panavas T., Panaviene,Z., Pogany J. and Nagy,P.D. (2003) Generation of a novel cis-acting replication element by promoter duplication in tombusviruses. Virology, 310, 118–129. [DOI] [PubMed] [Google Scholar]
- Ranjith-Kumar C.T., Zhang,X. and Kao,C.C. (2003) Enhancer-like activity of a brome mosaic virus RNA promoter. J. Virol., 77, 1830–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D. and White,K.A. (1999) Enhancer-like properties of an RNA element that modulates Tombusvirus RNA accumulation. Virology, 256, 162–171. [DOI] [PubMed] [Google Scholar]
- Ray D. and White,K.A. (2003) An internally located RNA hairpin enhances replication of Tomato bushy stunt virus RNAs. J. Virol., 77, 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon D.M. (1991) Rapid de novo generation of defective interfering RNA by cucumber necrosis virus mutants that do not express the 20-kDa nonstructural protein. Proc. Natl Acad. Sci. USA, 88, 11153–11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M.L. and Ahlquist,P. (1999) A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol., 73, 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance V. and Vaucheret,H. (2001) RNA silencing in plants—defense and counterdefense. Science, 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Waterhouse P.M., Wang,M.B. and Lough,T. (2001) Gene silencing as an adaptive defence against viruses. Nature, 411, 834–842. [DOI] [PubMed] [Google Scholar]
- White K.A. and Morris,T.J. (1994) Recombination between defective tombusvirus RNAs generates functional hybrid genomes. Proc. Natl Acad. Sci. USA, 91, 3642–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K.A. and Morris,T.J. (1999) Defective and defective interfering RNAs of monopartite plus-strand RNA plant viruses. Curr. Top. Microbiol. Immunol., 239, 1–17. [DOI] [PubMed] [Google Scholar]
- Wu B. and White,K.A. (1999) A primary determinant of cap-independent translation is located in the 3′-proximal region of the tomato bushy stunt virus genome. J. Virol., 73, 8982–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Vanti,W.B. and White,K.A. (2001) An RNA domain within the 5′ untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication. J. Mol. Biol., 305, 741–756. [DOI] [PubMed] [Google Scholar]