Abstract
Down-regulation of activated signaling receptors in response to their ligands plays a key role in restricting the extent and duration of the signaling. Mechanisms underlying down-regulation of the type I interferon receptor consisting of IFNAR1 and IFNAR2 subunits remain largely unknown. Here we show that IFNAR1 interacts with the Homolog of Slimb (HOS) F-box protein in a phosphorylation-dependent manner, and that this interaction is promoted by interferon α (IFNα). IFNAR1 is ubiquitinated by the Skp1-Cullin1-HOS-Roc1 (SCFHOS) ubiquitin ligase in vitro. HOS expression and activities are required for IFNα-stimulated ubiquitination of IFNAR1, endocytosis of the type I interferon receptor, down-regulation of IFNAR1 levels, and IFNAR1 proteolysis via the lysosomal pathway. Furthermore, modulations of HOS activities affect the extent of Stat1 phosphorylation and Stat-mediated transcriptional activities as well as the extent of antiproliferative effects of type I interferons. These findings characterize SCFHOS as an E3 ubiquitin ligase that is essential for ubiquitination, proteolysis and down-regulation of IFNAR1, and implicate HOS in the regulation of cellular responses to IFNα.
Keywords: F-box/interferon/receptor/Slimb/ubiquitin ligase
Introduction
A key issue in the regulation of signal transduction has to do with how cells limit the duration and magnitude of signaling. Modification of signaling proteins via covalent attachment of ubiquitin polypeptide (ubiquitination) is an important mechanism that underlies the rapid changes in protein stability, localization and activity. Substrate specificity and timing of ubiquitin conjugation are determined by E3 ubiquitin protein ligases, which are the rate-limiting factors in the enzymatic cascade that also requires ubiquitin-activating (E1) and ubiquitin-conjugating (E2) enzymes. Proteins modified by lysine 48 polyubiquitin chains are efficiently degraded via a 26S proteasome-mediated pathway (reviewed in Weissman, 2001). Among E3 species that ubiquitinate proteasomal substrates, the Skp1-Cullin1-F-box-Roc1 (SCF) family members represent the ligases, which specifically recognize and ubiquitinate phosphorylated substrates. Within SCF complexes, the ligase activity is mediated by the Cullin1 and Roc1 (also termed Rbx1 or Hrt1) complex, which is tethered via Skp1 to a specificity-conferring F-box protein. F-box proteins bind to Skp1 via the F-box motif and they interact with phosphorylated substrates through other protein–protein binding domains, including WD40 repeats (reviewed in Deshaies, 1999).
In addition to proteasomal degradation, ubiquitination plays a role in endocytosis of plasma membrane proteins followed by their sorting and lysosomal degradation. A few known ligases that mediate this ubiquitination (including c-Cbl/Hakai and Rsp5/NEDD4 family members) are important in regulating the magnitude and duration of various signal transduction pathways (Hicke, 1999; Katzmann et al., 2001). However, the ubiquitin ligases that mediate degradation of many receptors, including receptors for interferons, remain to be identified.
Type I interferons (IFN) have become widely used therapeutic agents due to their potent anti-tumor, anti-viral and immunomodulatory activities (Biron, 2001; Kirkwood, 2002). These cytokines trigger their signaling by binding type I IFN receptor at the cell surface and inducing assembly of a receptor complex consisting of IFNAR1 and IFNAR2 subunits (Uze et al., 1990; Novick et al., 1994). Down-regulation and degradation of these subunits in response to the ligand are expected to play a key role in restricting the extent and duration of IFN signaling (Constantinescu et al., 1994; Stark et al., 1998), However, the mechanisms of ligand-induced down-regulation and degradation of type I IFN receptor have not been determined.
IFNAR1 is rapidly degraded upon treatment of cells with IFNα (Constantinescu et al., 1994). Cellular responses to IFNα require an adequate expression of the IFNAR1 subunit and its genetic disruption in mice compromises innate immunity (Muller et al., 1994; Hwang et al., 1995). Conversely, IFNAR1 mutants that bear deletions in their C-terminus inhibit internalization of the type I IFN receptor, and these mutants mediate enhanced responses to IFNα (Gibbs et al., 1996; Basu et al., 1998). Among putative negative regulatory domains located within the distal cytoplasmic tail of IFNAR1 is a motif that may be recognized by the Homologue of Slimb (HOS) F-box-containing protein, which serves as a receptor for SCFHOS E3 ligase in ubiquitination of phosphorylated IκB and β-catenin (Fuchs et al., 1999; Tan et al., 1999). HOS (also termed β-TrCP2 or Fbw1b) recruits E3 activity via the F-box-dependent binding to Skp1 and recognizes phospho-motifs (DpSGXXpS) in its substrates via WD40 domains (Fuchs et al., 1999; Suzuki et al., 1999). Here we show that IFNAR1 undergoes ubiquitination and degradation in a manner dependent on HOS and SCFHOS ligase activity, and that HOS contributes to the endocytosis of the type I IFN receptor and down-regulation of IFNAR1, as well as to regulation of cellular responses to IFNα.
Results
HOS interacts with IFNAR1 in a phosphorylation-dependent manner
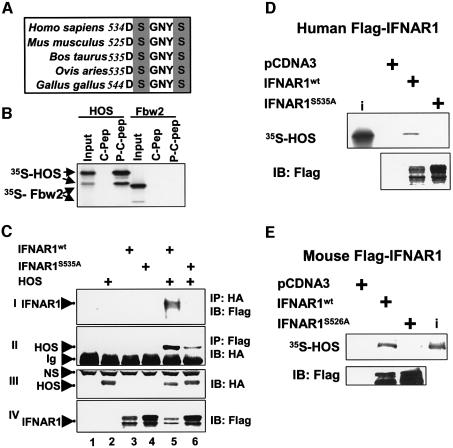
A putative HOS recognition motif within the C-terminus of IFNAR1 is conserved among different vertebrate species (Figure 1A). HOS interacted with synthetic doubly phosphorylated peptide encompassing this motif (P-C-Pep), but not with the non-phosphorylated peptide (C-Pep) in vitro (Figure 1B). Another F-box/WD40 repeat-containing protein Fbw2 (Cenciarelli et al., 1999; Winston et al., 1999a) did not associate with phospho-peptide, suggesting that the interaction of HOS with P-C-Pep was highly specific. Recombinant HA-tagged HOS and Flag-tagged IFNAR1 expressed in 293T cells were co-immunoprecipitated with respective antibodies (Figure 1C). Mutation of potentially phosphorylated serine 535 within HOS recognition motif rendered IFNAR1S535A resistant to down-regulation in response to HOS co-expression (Figure 1C, panel IV, lane 3 compared with lane 5, and lane 4 compared with lane 6) and noticeably inhibited IFNAR1 binding to HOS (panels I and II, lane 5 compared with lane 6). A residual binding of HOS to IFNAR1S535A mutant (lane 6) may be attributed to recruitment of HOS by endogenous proteins that partake in plasma membrane complexing with IFNAR1S535A. Furthermore, human and mouse Flag-IFNAR1wt interacted with HOS in vitro, and this interaction was not observed with either human IFNAR1S535A or murine IFNAR1S526A mutant (Figure 1D and E). These data indicate that IFNAR1 interacts with HOS in vivo and in vitro depending on the integrity of HOS recognition motif, which likely requires phosphorylation of IFNAR1 within this motif.
Fig. 1. Interaction between HOS and IFNAR1. (A) Multiple alignment of a putative HOS recognition motif in IFNAR1 of different species. Potentially phosphorylated serines are shaded. (B) Binding of an in vitro-translated (IVT) and 35S-labeled HOS and Fbw2 to the beads coated with phosphorylated (P-C-Pep) or non-phosphorylated (C-Pep) IFNAR1-derived peptides. (C) Co-immunoprecipitation of HA-tagged HOS and human Flag-tagged IFNAR1 (wild type or S535A mutant) expressed in 293T cells as indicated. Ig, immunoglobulins; NS, non-specific band. (D–E) Binding of an IVT HOS to human (D) or mouse (E) Flag-IFNAR1 proteins expressed in 293T cells and immunopurified with Flag antibody. Aliquots of reactions were analyzed by autoradiography (upper panel) or immunoblotting with Flag antibody (lower panel). Input of radioactive HOS (10% of the amount added to the reactions) is also shown (i).
Phosphorylation of IFNAR1 and its ability to interact with HOS is promoted by IFNα
We further tested a role of IFNAR1 phosphorylation in binding to HOS. Phosphatase treatment of Flag-IFNAR1 decreased its ability to bind HOS in vitro (Figure 2A), indicating that specific phosphorylation of IFNAR1 was essential for its affinity to HOS. This result also indicates that recombinant Flag-IFNAR1wt overexpressed in 293T cells is already phosphorylated and capable of binding to HOS in the absence of added ligand. This does not rule out a role for IFNα in HOS–IFNAR1 interaction since mammalian cells often secret type I IFN (Pestka, 2000) and overexpression of cytokine receptors alone (e.g. tumor necrosis factor α receptor; Heller et al., 1992) is known to mediate signaling events. Indeed, treatment of cells with IFNα promoted the interaction of endogenous IFNAR1 and HOS in 293T (Figure 2B) or HeLa (data not shown) cells. Furthermore, endogenous IFNAR1 purified from the cells treated with the ligand exhibited a higher capacity for binding HOS in vitro compared with IFNAR1 from untreated cells. This interaction was abolished by pre-treatment of IFNAR1 with phosphatase λ (Figure 2C), indicating that phosphorylation of IFNAR1 is required for its association with HOS.
Fig. 2. Treatment with IFNα promotes phosphorylation-dependent binding of HOS to IFNAR1. (A) Binding of an IVT HOS to human Flag-IFNAR1 proteins expressed in 293T cells and immunopurified with Flag antibody before or after treatment with protein phosphatase λ. Aliquots of reactions were analyzed by autoradiography (upper panel) or immunoblotting with Flag antibody (lower panel). (B) Co-immunoprecipitation of endogenous HOS and IFNAR1 from 293T cells (8 mg of lysates) either treated or not with IFNα (4000 IU/ml for 30 min) using naïve rabbit serum (NRS) or antibodies against HOS or IFNAR1 (4B1). Immunoblotting analysis with antibodies against HOS or IFNAR1 (SC) is shown. (C) Binding of an IVT HOS to endogenous IFNAR1 immunopurified from 293T cells (treated or not with IFNα, 1000 IU for 30 min) with 4B1 antibody before or after treatment with protein phosphatase λ. Aliquots of reactions were analyzed by autoradiography (upper panel) or immunoblotting with IFNAR1 antibody (SC, lower panel). Control immunoprecipitation with Flag antibody is also shown. (D) Binding of GST–IFNAR1 proteins phosphorylated in vitro in the presence of [32P]γ-ATP by extracts from 293T cells (pre-treated with IFNα as indicated) with SCFHOS complexes immobilized on HA-agarose. Input of GST–IFNAR1 proteins (100%, ‘i’) and their amount bound to SCFHOS complexes and retained on the beads after washing was analyzed by autoradiography. (E) Binding of an IVT HOS to the beads coated with C-Pep IFNAR1-derived peptide pre-phosphorylated by the extracts from 293T cells (either pre-treated or not with IFNα, as indicated). Reactions were analyzed by autoradiography. Input of radioactive HOS (10% of the amount added to the reactions) is also shown (‘i’). (F) Phosphorylation of IFNAR1-derived peptides by the extracts from 293T cells, which were either pre-treated (black bars) or not (white bars) with IFNα in the presence of [32P]γ-ATP, was analyzed by scintillation counter. A representative of two independent experiments (each in triplicate) is shown. *P < 0.05 (Student’s t-test) (compared with extracts from non-treated cells).
These results also imply that IFNα induces phosphorylation of IFNAR1 within the HOS recognition motif. Thus, we tested the ability of the glutathione S-transferase (GST)–IFNAR1 (cytoplasmic tail) fusion protein, phosphorylated in vitro by extracts from 293T or HeLa cells, to bind to an immobilized SCFHOS complex. Consistent with previous findings (Colamonici et al., 1994), GST–IFNAR1 proteins were efficiently phosphorylated by cell extracts in an IFNα-independent manner (Figure 2D, odd lanes). However, binding of SCFHOS to GST–IFNAR1 phosphorylated by extracts from the cells pre-treated with the ligand was increased compared with the extracts from untreated cells (lane 4 versus 2). Omitting HOS from the SCFHOS complex or mutating Ser535 to alanine within GST–IFNAR1 abrogated this binding (Figure 2D, lanes 6 and 8). Furthermore, whereas IFNAR1-derived non-phosphorylated C-Pep peptide failed to interact with HOS in vitro (Figure 1B), phosphorylation of C-Pep by cell extracts enabled this binding, and its efficiency was increased when the extracts were from the ligand-treated cells (Figure 2E). Additionally, a kinase activity assay (with cell extracts as a source of a kinase and C-Pep as a substrate) showed that treating cells with IFNα induced the ability of cell extracts to phosphorylate C-Pep (Figure 2F). This induction was less evident when using P-C-Pep (in which Ser535 and Ser539 are pre-phosphorylated and, thus, protected from incorporation of 32P) as a substrate, indicating that IFNα induced phosphorylation of one or both of these serines (Figure 2F). These results collectively suggest that IFNα induces a kinase activity that phosphorylates IFNAR1 within the HOS recognition motif and enables HOS recruitment.
Ubiquitination of IFNAR1 by SCFHOS E3 ubiquitin protein ligase
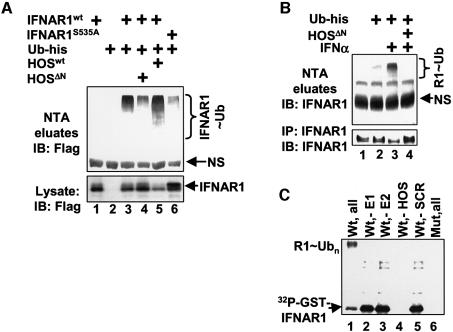
Co-transfection of Flag-tagged IFNAR1 with His-tagged ubiquitin in 293T cells, followed by purification of ubiquitinated proteins on nickel resins under stringent denaturing conditions, allowed detection of ubiquitinated Flag-IFNAR1 in vivo (Figure 3A). Whereas the overall levels of Flag-IFNAR1 were reduced by expression of HOS (most likely due to IFNAR1 degradation), the extent of ubiquitin-conjugated Flag-IFNAR1 was noticeably increased (Figure 3A, lane 3 versus lane 5). Expression of the dominant-negative HOSΔN mutant, which is known to compete with endogenous HOS for substrates and to inhibit the activities of SCFHOS ubiquitin ligase (Fuchs et al., 1999; Suzuki et al., 1999), elevated Flag-IFNAR1 levels and decreased the extent of its ubiquitination (Figure 3A, lanes 3 and 4). Compared with Flag-IFNAR1wt, mutant IFNAR1S535A protein exhibited less ubiquitination (Figure 3A, lane 6 versus lane 3) and its extent was not affected by expression of HOSΔN (Supplementary figure 1, available at The EMBO Journal Online).
Fig. 3. IFNAR1 is ubiquitinated by SCFHOS E3 ubiquitin protein ligase. (A) In vivo ubiquitination of Flag-IFNAR1 in 293T cells co-transfected with His-tagged ubiquitin and HOS constructs as indicated. Cell lysates (lower panel) and ubiquitinated proteins purified on Ni-NTA agarose under denaturing conditions (upper panel) were analyzed by immunoblotting with Flag antibody. Ubiquitinated Flag-IFNAR1 species (IFNAR1∼Ub) are indicated. (B) In vivo ubiquitination of endogenous IFNAR1 in 293T cells transfected with His-tagged ubiquitin and HOSΔN constructs and treated with IFNα as indicated. Endogenous IFNAR1 immunoprecipitated with 4B1 antibody (from ∼10% of harvested cells, lower panel) and ubiquitinated proteins purified on Ni-NTA agarose under denaturing conditions (from ∼90% of harvested cells, upper panel) were analyzed by immunoblotting with IFNAR1 antibody (SC). Ubiquitinated endogenous IFNAR1 species (R1∼Ub) are indicated. (C) In vitro ubiquitination of GST–IFNAR1 proteins (wild type or S535A, ‘mut’) phosphorylated in the presence of [32P]γ-ATP by the extracts from 293T cells (pre-treated with IFNα) and pre-bound to SCFHOS complexes or HOS only (‘-SCR’) immobilized on HA-agarose. After washing away the non-bound proteins, the ubiquitination reaction was carried out in the presence of ubiquitin, ATP and E1/E2 (as indicated), and analyzed by autoradiography. Ubiquitinated GST–IFNAR1 species (R1∼Ub) are indicated.
Ubiquitination of endogenous IFNAR1 in 293T cells transfected with His-ubiquitin was promoted by the treatment of cells with IFNα, concurrent with a decrease in the levels of IFNAR1 (Figure 3B). Expression of HOSΔN inhibited ubiquitination and prevented down-regulation of endogenous IFNAR1. These results indicate that ubiquitination of IFNAR1 in vivo is promoted by the ligand and mediated by HOS, although the putative contribution of other ubiquitin ligase(s) to this process cannot be ruled out. These data also suggest that HOS activities are required for down-regulation of IFNAR1 in response to IFNα.
The extracts from 293T cells pre-treated with IFNα were used to phosphorylate GST–IFNAR1 fusion protein captured on immobilized recombinant SCFHOS ubiquitin ligase (Tan et al., 1999) for recapitulation of IFNAR1 ubiquitination in vitro. This reaction yielded a high molecular weight smear characteristic of ubiquitinated proteins (Figure 3C, lane 1). Omission of either E1 or E2 severely reduced the intensity of this smear (lanes 2 and 3), indicating that it represented in vitro ubiquitinated phospho-GST–IFNAR1. The extent of ubiquitination was decreased when Skp1, Cullin1 and Roc1 were omitted from the immobilized material (Figure 3C, lane 5). Consistent with the data shown in Figure 2D, omitting HOS from immobilized complexes or the use of the mutant GST–IFNAR1S535A prevented capturing phospho-GST–IFNAR1 (lanes 4 and 6). These observations provide biochemical evidence suggesting that SCFHOS is capable of serving as an E3 protein ubiquitin ligase for phosphorylated IFNAR1.
HOS is required for IFNAR1 degradation
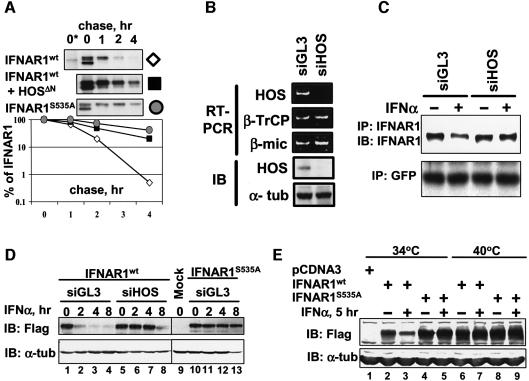
We next investigated whether HOS regulates proteolysis of IFNAR1. As analysis of the half-life of endogenous IFNAR1 is technically challenging (Gauzzi et al., 1997; data not shown), we determined the half-life of Flag-IFNAR1 expressed in 293T cells, which interacted with HOS (Figure 1C and D) and underwent ubiquitination (Figure 3A) in the absence of exogenously added IFNα. Overexpressed IFNAR1 appeared in the form of a doublet (Figures 1C–E and 3A), which was consistent with observations of another group who clearly demonstrated that a slower migrating form of IFNAR1 represents the mature IFNAR1 species (Ragimbeau et al., 2003). As evident from the pulse–chase experiments (Figure 4A), Flag-IFNAR1S535A mutant exhibited a substantially longer half-life than Flag-IFNAR1wt, which might reflect poor ability of this mutant to interact with HOS (Figure 1C and D) and undergo ubiquitination (Figure 3A). Moreover, degradation of Flag-IFNAR1wt was delayed by co-expression of HOSΔN (Figure 4A). These results suggest that HOS plays a major role in regulating IFNAR1 proteolysis.
Fig. 4. SCFHOS regulates IFNAR1 stability and levels. (A) Pulse–chase analysis of human Flag-IFNAR1 proteins expressed in 293T cells (metabolically labeled with [35S]methionine/[35S]cysteine) and immunoprecipitated with Flag antibody, and analyzed by autoradiography. Asterisk denotes the mock-transfected cells. A representative of four independent experiments is shown. (B) Inhibition of HOS expression by siRNA against HOS (siHOS) or luciferase (siGL3) transfected into HeLa cells. Depicted are the levels of HOS, and β-TrCP1 and β-microglobulin mRNA (measured by RT–PCR, three upper panels), and HOS and α-tubulin proteins (measured by immunoblotting, two lower panels). (C) Effect of IFNα on the levels of endogenous IFNAR1 in 293T cells co-transfected with the indicated siRNA oligonucleotides and GFP plasmid. IFNAR1 was immunoprecipitated and analyzed by immunoblotting (with 4B1 antibody and SC antibody, respectively). Levels of GFP analyzed by immunoblotting are also shown. (D) Effect of IFNα (2000 IU/ml) on the levels of human Flag-IFNAR1 proteins co-transfected with the indicated siRNA oligonucleotides and β-gal construct in HeLa cells. Loading was normalized per β-gal activity and the levels of proteins were analyzed by immunoblotting with Flag antibody (upper panel) or α-tubulin antibody (lower panel). (E) Effect of IFNα (1000 IU/ml for 5 h) on the levels of human Flag-IFNAR1 proteins expressed in hamster ts41 cells grown at the indicated temperatures. Immunoblotting with Flag antibody (upper panel) or α-tubulin antibody (lower panel) is depicted.
HOS levels and Cullin1 activity are essential for down-regulation of IFNAR1 in response to IFNα
We further examined the role of HOS in down-regulation of IFNAR1 in response to its ligand using short inhibitory RNA (siRNA) technology. Transfection of HeLa (Figure 4B) or 293T cells (data not shown) with siRNA against HOS (siHOS) decreased the steady-state levels of HOS mRNA and protein (without affecting the expression of closely related β-TrCP1 gene) compared with the control siGL3. Treatment of cells with IFNα decreased the levels of endogenous IFNAR1, and this decrease was abrogated by siHOS in 293T cells (Figure 4C) and in HeLa (not shown) cells. These data, together with our findings that the HOSΔN mutant inhibits IFNα-induced down-regulation of endogenous IFNAR1 (Figure 3B), suggest that HOS expression and activities are essential for this down-regulation.
Treatment with IFNα decreased the levels of IFNAR1wt (but not of IFNAR1S535A mutant) co-transfected with siGL3, whereas co-transfection with siHOS (Figure 4D), but not with siβTrCP (directed against β-TrCP1; Supplementary figure 2), substantially delayed IFNAR1wt down-regulation in HeLa cells. These results together with the data shown in Figure 4A indicate that down-regulation of overall levels of IFNAR1 in response to IFNα parallels degradation of IFNAR1. These observations provide genetic evidence suggesting that HOS is essential for degradation of IFNAR1 in response to its ligand.
Stability of SCFHOS substrates depends on activity of Cullin1 (Fuchs et al., 1999; Tan et al., 1999), which requires its modification by ubiquitin-like protein NEDD8 (Read et al., 2000; Wu et al., 2000). The recent finding that the NEDD8 conjugating system is impaired in hamster ts41 cells at a non-permissive temperature (40°C) (Chen et al., 2000) makes these cells a convenient model for studying Cullin-dependent protein degradation (Ohh et al., 2002). Levels of Flag-IFNAR1wt were down-regulated by IFNα in ts41 cells at a permissive temperature (34°C). However, shifting the temperature to 40°C led to the accumulation of IFNAR1wt and rendered it insensitive to the ligand (Figure 4E), although IFNα activity was not impaired in ts41 cells [as assessed by the induction of interferon-stimulated response element (ISRE)-luciferase reporter; data not shown]. Neither temperature shift nor IFNα treatment affected the levels of IFNAR1S535A mutant, indicating that alterations in the levels of IFNAR1wt most likely resulted from changes in the rate of proteolysis. These data indirectly point to a role for Cullin proteins in IFNAR1 down-regulation and (together with other findings) implicate SCFHOS in regulating the stability of IFNAR1.
HOS regulates internalization of the type I IFN receptor and cell surface levels of IFNAR1
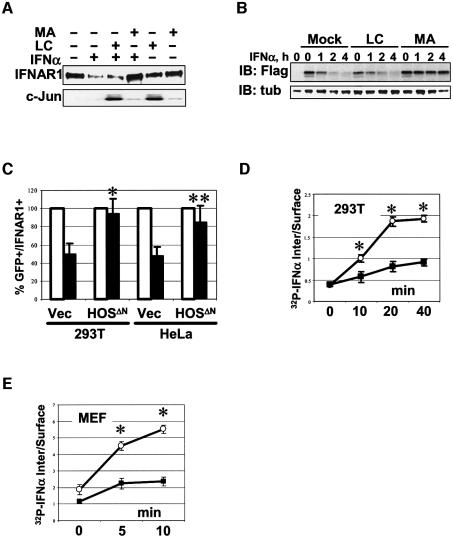
Ubiquitination of cell surface receptors is implicated in their endocytosis and degradation via the lysosomal pathway (Hicke, 1999; Katzmann et al., 2001). Treatment of cells with methylamine HCl, which inhibits the lysosomal pathway (Reijngoud et al., 1976), delayed the degradation of endogenous IFNAR1 (Figure 5A) and transiently expressed Flag-IFNAR1wt (Figure 5B) in cells treated with IFNα. Methylamine HCl (as well as with chloroquine) also prolonged the half-life of Flag-IFNAR1wt expressed in 293T cells (Supplementary figure 3). Inhibition of proteasomes by lactacystin led to the accumulation of c-Jun (Figure 5A), but did not affect degradation of IFNAR1 (Figure 5A and B). These results indicate that IFNAR1 is mainly degraded via the lysosomal pathway, although the role of proteasomes cannot be entirely ruled out.

Fig. 5. Down-regulation of IFNAR1 cell surface levels and endocytosis of type I IFN receptor in response to IFNα depends on HOS. (A) Inhibition of lysosomal pathway prevents down-regulation of endogenous IFNAR1 by IFNα (1000 IU/ml for 2 h) in 293T cells. Cells were treated with IFNα with or without adding either proteasome inhibitor lactacystin (LC, 50 µM) or lysosomal pathway inhibitor methylamine HCl (MA, 20 mM), as indicated. Immunoblotting analyses of IFNAR1 (immunoprecipitated with 4B1 antibody), as well as c-Jun are shown. (B) Inhibition of lysosomal pathway prevents down-regulation of human Flag-IFNAR1 expressed in HeLa cells together with β-gal construct. Cells were treated with IFNα (1000 IU/ml) and either lactacystin or methylamine HCl (as in Figure 5A), as indicated. Lane 1 represents mock-transfected cells. Loading was normalized per β-gal activity and the levels of proteins were analyzed by immunoblotting with Flag antibody (upper panel) or α-tubulin antibody (lower panel). (C) The levels of endogenous IFNAR1 on the surface of HeLa and 293T cells transfected with GFP-expressing plasmid and either HOSΔN or empty vector (‘Vec’), and treated with IFNα (1000 IU/ml for 2 h) for the times indicated. Average data of five experiments are shown. Depicted are the percentages of GFP-positive and IFNAR1-positive cells (analyzed by FACS with AA3 monoclonal antibody against the extracellular domain of IFNAR1) in the absence (white bars) or presence (black bars) of IFNα. *P < 0.01 and **P < 0.05 compared with empty vector (Student’s t-test). (D) Rate of the type I IFN receptor endocytosis as measured by internalization of 32P-labeled IFNα in 293T cells transfected with either HOSΔN (closed squares) or empty vector (open circles). Ratio of acid-resistant internal radioactivity to cell surface radioactivity was assessed in two independent experiments (each in triplicate). *P < 0.01 (Student’s t-test). (E) Rate of type I IFN receptor endocytosis (measured as in Figure 5D) in IFNAR1-null MEF transfected with mouse IFNAR1 constructs (wild type, open circles; S526A mutant, closed squares). *P < 0.01 (Student’s t-test). (F) The levels of flag-tagged murine IFNAR1wt and IFNAR1S526A proteins expressed in NIH 3T3 cells treated with IFNα (1000 IU/ml) and cycloheximide (50 µg/ml) for the indicated times, fixed, permeabilized and analyzed by immunocytochemistry with M2 Flag antibody (red) and counterstaining with DAPI (blue) for detection of nuclei. Results representative of two independent experiments are shown.
Inhibiting HOS activities by expression of HOSΔN prevented IFNα-mediated down-regulation of endogenous cell surface IFNAR1 (Figure 5C). We next assessed a role of HOS in the endocytosis of type I IFN receptor by determining the initial rates of its internalization. Expression of HOSΔN significantly inhibited internalization of endogenous receptor in 293T cells (Figure 5D). Moreover, a slower rate of type I IFN receptor internalization was detected in mouse embryo fibroblasts (MEF) from IFNAR1-null cells transfected with mouse IFNAR1S526A mutant (which did not interact with HOS; Figure 1E) compared with MEF transfected with IFNAR1wt (Figure 5E). This result is in line with the failure of IFNα to down-regulate the levels of surface receptor in cells expressing IFNAR1Δ525–557 mutant (Basu et al., 1998). These data collectively indicate that HOS is required for down-regulation of cell surface IFNAR1 and internalization of the entire type I IFN receptor in response to IFNα.
Mouse Flag-tagged IFNAR1 proteins were detected at the plasma membrane and were distributed throughout the cell (Figure 5F). Treatment of cells with IFNα and cycloheximide (to inhibit de novo protein synthesis) led to a noticeable re-distribution of the wild-type Flag-IFNAR1 from the cell surface to the perinuclear compartments within 30–60 min, followed by a drastic decrease in overall IFNAR1 levels. In contrast, mouse Flag-IFNAR1S526A protein exhibited a prominent decoration of the cell surface up to 60 min after treatment, and a higher level of expression compared with the wild-type protein (Figure 5F). These data indicate that down-regulation of IFNAR1 is likely to depend on the ability of IFNAR1 to interact with HOS.
HOS is involved in the regulation of cellular responses to IFNα
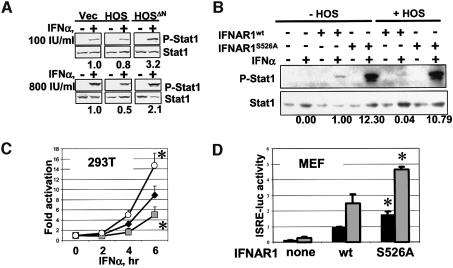
The extent of cellular responses to IFNα depends on the dose of IFNAR1 gene (Hwang et al., 1995), suggesting that HOS-mediated alteration of IFNAR1 levels via changes in its stability is expected to influence the outcome of IFNα signaling. Indeed, phosphorylation of Stat1 at Tyr701 in IFNα-treated cells was moderately decreased by HOS overexpression (at higher doses of IFNα), but augmented by the expression of HOSΔN (especially at lower doses of IFNα; Figure 6A). In IFNAR1-null MEF, re-expression of mouse Flag-IFNAR1 proteins (although undetectable by Flag immunoblotting) allowed analysis of Stat1 phosphorylation in response to IFNα, and the IFNAR1S526A mutant was more efficient in this assay than IFNAR1wt (Figure 6B). Moreover, whereas co-expression of HOS noticeably decreased the ability of IFNAR1wt to transduce signals, resulting in Stat1 phosphorylation, the effects of the IFNAR1S526A mutant were hardly affected. These data suggest that HOS contributes to the regulation of the extent of IFNα signaling, most likely by controlling IFNAR1 stability and abundance.
Fig. 6. HOS regulates the IFNα signaling pathway. (A) Immunoblotting analysis of phospho-Stat1/Stat1 in 293T cells transfected and treated with IFNα (100 IU/ml, upper panel or 800 IU/ml, lower panel) for 30 min as indicated. Relative ratio between phospho-Stat1 and Stat1 densitometric values is depicted in numbers below. Vec, empty vector. (B) Immunoblotting analysis of phospho-Stat1/Stat1 in IFNAR1-null MEF transfected with either mIFNAR1wt or mIFNAR1S526A mutant (with or without HOS) as indicated. Cells were treated with IFNα (1000 IU/ml for 30 min) as indicated. Relative ratio between phospho-Stat1 and Stat1 densitometric values is depicted in numbers below. (C) IFNα-dependent activation of 5× ISRE-luc reporter in 293T cells transfected with either vector (black diamonds) or HOS (gray squares), or HOSΔN (white circles) from three independent experiments (each in triplicate). Depicted is the fold activation of normalized luciferase activity values (over values in cells not treated with IFNα). *P < 0.05 compared with cells transfected with vector (Student’s t-test). (D) 5× ISRE-luc reporter activity in IFNAR1-null MEF transfected with either mIFNAR1wt or mIFNAR1S526A mutant as indicated. Depicted (in arbitrary units) are the normalized luciferase values from the cells treated with IFNα (500 IU for 6 h, gray bars) and from untreated cells (black bars). *P < 0.05 compared with mIFNAR1wt (Student’s t-test).
We next determined the effects of HOS on transcriptional activity of the Stat1–Stat2–p48IRF9 complex (ISGF3) measured by ISRE-driven luciferase reporter activity. Expression of HOS significantly inhibited the induction of this activity by IFNα in 293T cells, whereas HOSΔN mutant produced an opposite result (Figure 6C). Furthermore, the experiments with re-expression of mouse IFNAR1 in IFNAR1-null MEF showed that mouse Flag-IFNAR1S526A mutant was more efficient in activation of ISGF3 than the wild-type mouse IFNAR1 (Figure 6D). These data collectively suggest that regulation of IFNAR1 stability by HOS contributes to the outcome of cellular responses to IFNα.
In the absence of the ligand, expression of IFNAR1 inhibited the proliferation of K562 human leukemia cells (Colamonici et al., 1994), and the growth rate of K562 cells stably transfected with IFNAR1S535A underwent more pronounced inhibition compared with IFNAR1wt (Figure 7A). Since IFNAR1S535A mutant is insensitive to HOS-mediated ubiquitination and degradation (Figures 3 and 4), this finding indicates that the stability of IFNAR1 contributes to the extent of its antiproliferative potential. Furthermore, expression of HOSΔN sensitized WM115 human melanoma cells to growth inhibition by IFNα (Figure 7B) or IFNβ (Figure 7C), presumably via the combination of augmenting ISGF3 transcriptional activity (Figure 6C and D) and inhibiting NF-κB transcriptional activity (Fuchs et al., 1999), which is otherwise increased by IFNα (Yang et al., 2000).
Fig. 7. HOS regulates cellular responses to IFNα. (A) Proliferation of K562 cells stably transfected with either vector (‘vec’) or IFNAR1 (wild type or S535A mutant), as assessed 3 days after plating with the aid of the WST-1 Cell Proliferation kit. Depicted is the percentage of growth inhibition (compared with the cells transfected with vector) as average data from five experiments. *P <0.05 compared with vector, and **P <0.05 compared with IFNAR1wt (Student’s t-test). (B–C) Colony-forming efficiency of WM115 human melanoma cells co- transfected with pBABE-puro and either vector (white circles) or HOSΔN (black squares). Cells were grown in the presence of puromycin and either IFNα (B) or IFNβ (C), and the number of colonies was scored. Average data (in percentage of cells grown in the absence of IFN) obtained from three independent experiments (in triplicates) are depicted. *P <0.05 compared with cells transfected with vector (Student’s t-test).
Discussion
The data presented here collectively delineate the mechanisms underlying the ligand-induced down-regulation of IFNAR1 that include phosphorylation-dependent binding (Figures 1 and 2) and ubiquitination (Figure 3) of IFNAR1 by SCFHOS E3 ubiquitin protein ligase, followed by internalization of the cell surface receptor and degradation of IFNAR1 via the lysosomal pathway (Figures 4 and 5). Our findings also implicate SCFHOS ubiquitin ligase in the regulation of the extent of IFNα signaling (Figure 6) and cellular responses to type I interferons (Figure 7). SCFHOS-dependent regulation of IFNAR1 is likely to represent an important mechanism that contributes to negative regulation of cytokine signaling among other such mechanisms, including suppressor of cytokine signaling- and phosphatase-mediated inhibition of Janus kinase levels and activity, and the effects of protein inhibitors of activated STAT (reviewed in Greenhalgh and Hilton, 2001). As a conserved HOS recognition motif is also found within the intracellular domains of some other cytokine receptor subunits that are related to IFNAR1 (e.g. interleukin-10 receptor α chain), future studies will show whether SCFHOS-dependent ubiquitination may represent a common mechanism in the regulation of stability of these receptors. Further detailed studies are also required to identify and characterize an IFNα-inducible kinase(s) that phosphorylates IFNAR1 at the HOS recognition site.
SCFHOS E3 ubiquitin ligase mediates ubiquitination and degradation of IκB and β-catenin (Fuchs et al., 1999; Suzuki et al., 1999). Here we identify IFNAR1 as a novel substrate for this ligase. Another F-box/WD40 domain-containing protein β-TrCP1 (Margottin et al., 1998), which is closely related to HOS, also recognizes phosphorylated DpSGXXpS motifs in IκB and β-catenin (Yaron et al., 1998; Spencer et al., 1999; Winston et al., 1999b), and is capable of binding to the IFNAR1-derived phospho-peptide (data not shown). Evidence that specific inhibition of HOS (but not β-TrCP1) expression by siRNA suffices to block IFNAR1 down-regulation (Figure 4 and Supplementary figure 2), together with the findings of other groups that β-TrCP1 is primarily localized to the nucleus (Sadot et al., 2000; Lassot et al., 2001; Davis et al., 2002) and no apparent alterations of IFNα signaling are observed in β-TrCP1-knockout mice (Guardavaccaro et al., 2003; Nakayama et al., 2003), suggest that HOS plays a predominant role in regulating IFNAR1 stability. Nevertheless, a potential contribution of SCFβ-TrCP1 to IFNAR1 ubiquitination and degradation cannot be ruled out, especially under conditions that robustly increase β-TrCP1 levels (e.g. activation of Wnt signaling; Spiegelman et al., 2000). Ubiquitin ligases of the SCF family are involved in control of stability of many substrates, including cell cycle regulators, transcription factors and signaling proteins. Whereas the role of these E3s in proteasomal degradation of these substrates has been well established (Deshaies, 1999), this report implicates SCF ligases in regulating endocytosis and lysosomal proteolysis of a cytokine receptor.
Our data show that HOS regulates an initial rate of internalization of the type I IFN receptor (Figure 5). It remains to be determined how SCFHOS-mediated ubiquitination of IFNAR1 targets this receptor for its degradation via the lysosomal pathway. It is conceivable that, similar to the effect of ubiquitination on the yeast α-factor receptor Ste2 (Terrell et al., 1998) or mammalian growth factor receptors (Haglund et al., 2003), conjugation of ubiquitin within the intracellular tail of IFNAR1 creates an internalization signal, which is recognized by the endocytosis machinery. IFNAR2 is likely to co-internalize within the context of the entire receptor, and our data (Figure 5D and E) indirectly support this possibility. In addition, ubiquitination of IFNAR1 may also affect post-internalization sorting/recycling of IFNAR1 in a manner similar to the mechanism that contributes to the degradation of the epidermal growth factor receptor ubiquitinated by c-Cbl (Levkowitz et al., 1998).
Until recently, little was known about the mechanisms of type I IFN receptor down-regulation. It has been reported that IFNAR1 protein levels are decreased in human (Gauzzi et al., 1997) but not murine (Karaghiosoff et al., 2000; Shimoda et al., 2000) cells, which lack Tyk2 kinase expression. Recent evidence indicates that restoration of Tyk2 expression maintains IFNAR1 cell surface levels and inhibits basal internalization and degradation of IFNAR1 proteins, including an IFNAR1 deletion mutant that lacks an HOS recognition motif (Ragimbeau et al., 2003). Our data point to the role of the SCFHOS ubiquitin ligase in the ligand-dependent degradation of IFNAR1 in human and murine cells with no apparent defects in Tyk2. Future studies will determine physiological or pathological conditions under which the Tyk2-dependent and HOS-dependent mechanisms of IFNAR1 down-regulation and degradation may overlap.
Our data show that inhibition of HOS sensitizes human melanoma cells to the anti-proliferative effects of type I IFN. Considering a wide use of these cytokines in the therapy of human tumors (Kirkwood, 2002), chronic viral infections (Davis, 2001) and multiple sclerosis (Khan et al., 2002), our findings imply that targeting HOS-mediated IFNAR1 proteolysis may provide novel means for optimization of these therapeutic efforts.
Materials and methods
A detailed description of the materials and methods is provided as Supplementary data (available at The EMBO Journal Online).
Materials
Recombinant IFNα A/D that is active in human and rodent cells was purchased from Sigma. Recombinant IFNα A/D P1 containing a phosphorylation site is described elsewhere (Wang et al., 1994). Double phosphorylated [P-C-Pep; EDHKKYSSQTSQDS-(PO3)-GNYS-(PO3)-NEDESES] or non-phosphorylated (C-Pep) IFNAR1-derived synthetic peptides were immobilized on agarose beads.
Plasmids
Human or mouse IFNAR1 cDNA (kind gifts from J.Krolewski and G.Uze), with the addition of a 3× Flag tag encoding sequence at the C-terminus, were subcloned into pCDNA3 vector (Invitrogen) via PCR, and mutations were introduced by site-directed mutagenesis. Constructs for the expression of HA-HOS and HOSΔN (Fuchs et al., 1999), His-ubiquitin (Treier et al., 1994), Fbw2 (Cenciarelli et al., 1999), Cul1 and Skp1 (Ohta et al., 1999), Roc1 (Tan et al., 1999) and 5× ISRE-luciferase reporter (Parisien et al., 2002) were described previously. Chemically synthesized siRNA duplexes against luciferase (siGL3), HOS (siHOS) and β-TrCP1 (siβTrCP) were purchased from Dharmacon.
Tissue culture and transfections
293T, HeLa, NIH 3T3 and WM115 cells were kindly provided by Z.Ronai. Hamster ts41 cells (Chen et al., 2000) and mouse embryo fibroblasts (MEF) from IFNAR1-knockout mice (Muller et al., 1994) were generous gifts from R.Neve and S.Hemmi. Cells were grown in DMEM or DMEM/F-12 (1:1) in the presence of 10% fetal bovine serum (FBS) and antibiotics at 37°C and 5% CO2. Hamster ts41 cells were grown in DMEM supplemented with 10% calf serum, antibiotics and fungizone at 34°C and 5% CO2. These cells were shifted to 40°C 24 h before harvesting, where indicated. Transfections were performed using calcium phosphate procedure or lipofection (with Lipofectamine Plus or Lipofectamine 2000; Invitrogen) 24–48 h before harvesting.
Antibodies and immunotechniques
4B1 (Constantinescu et al., 1994) and AA3 (Goldman et al., 1999) (a kind gift of Dr M.Zafari) monoclonal antibodies against the extracellular domain of IFNAR1 have been described previously. Antibodies against HA (Roche), Flag (M2, Sigma), GFP (Clontech), c-Jun and IFNAR1 (against the intracellular domain; Santa Cruz), as well as against phospho-Stat1 and Stat1 (Cell Signaling Technology) were purchased. Antibody against HOS, as well as immunoprecipitation, immunocytochemistry and immunoblotting procedures, have been described previously (Spiegelman et al., 2002).
In vitro binding assays
Recombinant IFNAR1 expressed in 293T cells or endogenous IFNAR1 proteins were immunopurified with Flag or 4B1 antibodies, respectively, and protein A beads, and stringently washed. IFNAR1 proteins or IFNAR1-derived synthetic peptides immobilized on the beads were incubated with in vitro-translated and 35S-labeled HOS, followed by washing and analysis by SDS–PAGE and autoradiography.
Ubiquitination and degradation assays
In vivo ubiquitination assays were carried out as described previously (Treier et al., 1994). For in vitro ubiquitination assay, GST–IFNAR1 proteins were expressed in bacteria and phosphorylated by the extracts from IFNα-treated cells in the presence of [32P]γ-ATP. Proteins were captured on immobilized SCFHOS (Tan et al., 1999), washed and incubated with ubiquitin, E1, Cdc34 and ATP at 37°C for 60 min followed by SDS–PAGE and autoradiography. Pulse–chase analysis was carried out on 293T cells transfected with indicated constructs as described elsewhere (Fuchs et al., 1999). Briefly, cells were grown in 100 mm dishes and transfected with indicated plasmids. After metabolic labeling with 35S[methionine]/35[S-cysteine] mixture, cells from one-quarter of the sector of each plate were harvested at each time point of the chase. Harvested cells were lysed and IFNAR1 proteins were immunoprecipitated with Flag antibody, separated on SDS–PAGE and analyzed by autoradiography.
Endocytosis assays
Bacterially expressed IFNα A/D P1 was phosphorylated by cAMP-dependent kinase in the presence of [32P]γ-ATP as described elsewhere (Wang et al., 1994). 293T cells or IFNAR1-null MEF (transfected with indicated plasmids) were incubated with labelled IFNα A/D P1 on ice, washed and incubated at 37°C for different time points to allow ligand internalization. Cells were washed, the cell surface-bound IFNα was removed by acid wash, and the cells were lysed to release the internalized IFNα. Amounts of radioactivity in both fractions were determined using a scintillation counter.
Luciferase reporter assays
293T cells were transfected with 5× ISRE-luc and pBABE-puro plasmids, and selected in media supplemented with puromycin (1 µg/ml) for 5 days. Then the cells were co-transfected with renilla luciferase vector and HOS plasmids or empty vector and treated with IFNα (500 IU/ml) for the indicated periods of time before harvesting. IFNAR1-null MEF were co-transfected with 5× ISRE-luc and renilla luciferase vector and treated with IFNα (1000 IU/ml for 6 h). Luciferase activities were determined using the Dual Luciferase Assay Kit (Promega).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are indebted to D.Bohmann, S.Hemmi, C.Horvath, J.Krolewski, R.Neve, M.Pagano, Z.Ronai, G.Uze and M.Zafari for providing reagents. We thank M.Hochstrasser, C.Horvath, Z.Ronai and V.Spiegelman for comments on the manuscript and L.Izotova for technical assistance. The authors declare that they have no competing financial interests. This work was supported by grants from the National Cancer Institute (CA 92900) and The University of Pennsylvania Research Foundation to S.Y.F.
References
- Basu L., Yang,C.H., Murti,A., Garcia,J.V., Croze,E., Constantinescu,S.N., Mullersman,J.E. and Pfeffer,L.M. (1998) The antiviral action of interferon is potentiated by removal of the conserved IRTAM domain of the IFNAR1 chain of the interferon α/β receptor: effects on JAK-STAT activation and receptor down-regulation. Virology, 242, 14–21. [DOI] [PubMed] [Google Scholar]
- Biron C.A. (2001) Interferons alpha and beta as immune regulators—a new look. Immunity, 14, 661–664. [DOI] [PubMed] [Google Scholar]
- Cenciarelli C., Chiaur,D.S., Guardavaccaro,D., Parks,W., Vidal,M. and Pagano,M. (1999) Identification of a family of human F-box proteins. Curr. Biol., 9, 1177–1179. [DOI] [PubMed] [Google Scholar]
- Chen Y., McPhie,D.L., Hirschberg,J. and Neve,R.L. (2000) The amyloid precursor protein-binding protein APP-BP1 drives the cell cycle through the S–M checkpoint and causes apoptosis in neurons. J. Biol. Chem., 275, 8929–8935. [DOI] [PubMed] [Google Scholar]
- Colamonici O.R., Porterfield,B., Domanski,P., Handa,R.K., Flex,S., Samuel,C.E., Pine,R. and Diaz,M.O. (1994) Ligand-independent anti-oncogenic activity of the α subunit of the type I interferon receptor. J. Biol. Chem., 269, 27275–27279. [PubMed] [Google Scholar]
- Constantinescu S.N., Croze,E., Wang,C., Murti,A., Basu,L., Mullersman,J.E. and Pfeffer,L.M. (1994) Role of interferon α/β receptor chain 1 in the structure and transmembrane signaling of the interferon α/β receptor complex. Proc. Natl Acad. Sci. USA, 91, 9602–9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis G.L. (2001) Current treatment for chronic hepatitis C. Rev. Gastroenterol. Disord., 1, 59–72. [PubMed] [Google Scholar]
- Davis M. et al. (2002) Pseudosubstrate regulation of the SCF(β-TrCP) ubiquitin ligase by hnRNP-U. Genes Dev., 16, 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell. Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Fuchs S.Y., Chen,A., Xiong,Y., Pan,Z.Q. and Ronai,Z. (1999) HOS, a human homolog of Slimb, forms an SCF complex with Skp1 and Cullin1 and targets the phosphorylation-dependent degradation of IκB and β-catenin. Oncogene, 18, 2039–2046. [DOI] [PubMed] [Google Scholar]
- Gauzzi M.C., Barbieri,G., Richter,M.F., Uze,G., Ling,L., Fellous,M. and Pellegrini,S. (1997) The amino-terminal region of Tyk2 sustains the level of interferon α receptor 1, a component of the interferon α/β receptor. Proc. Natl Acad. Sci. USA, 94, 11839–11844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs V.C., Takahashi,M., Aguet,M. and Chuntharapai,A. (1996) A negative regulatory region in the intracellular domain of the human interferon-α receptor. J. Biol. Chem., 271, 28710–28716. [DOI] [PubMed] [Google Scholar]
- Goldman L.A., Zafari,M., Cutrone,E.C., Dang,A., Brickelmeier,M., Runkel,L., Benjamin,C.D., Ling,L.E. and Langer,J.A. (1999) Characterization of antihuman IFNAR-1 monoclonal antibodies: epitope localization and functional analysis. J. Interferon Cytokine Res., 19, 15–26. [DOI] [PubMed] [Google Scholar]
- Greenhalgh C.J. and Hilton,D.J. (2001) Negative regulation of cytokine signaling. J. Leukoc. Biol., 70, 348–356. [PubMed] [Google Scholar]
- Guardavaccaro D. et al. (2003) Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev. Cell, 4, 799–812. [DOI] [PubMed] [Google Scholar]
- Haglund K., Sigismund,S., Polo,S., Szymkiewicz,I., Di Fiore,P.P. and Dikic,I. (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol., 5, 461–466. [DOI] [PubMed] [Google Scholar]
- Heller R.A., Song,K., Fan,N. and Chang,D.J. (1992) The p70 tumor necrosis factor receptor mediates cytotoxicity. Cell, 70, 47–56. [DOI] [PubMed] [Google Scholar]
- Hicke L. (1999) Gettin’ down with ubiquitin: turning off cell-surface receptors, transporters and channels. Trends Cell Biol., 9, 107–112. [DOI] [PubMed] [Google Scholar]
- Hwang S.Y., Hertzog,P.J., Holland,K.A., Sumarsono,S.H., Tymms,M.J., Hamilton,J.A., Whitty,G., Bertoncello,I. and Kola,I. (1995) A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferons α and β and alters macrophage responses. Proc. Natl Acad. Sci. USA, 92, 11284–11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaghiosoff M. et al. (2000) Partial impairment of cytokine responses in Tyk2-deficient mice. Immunity, 13, 549–560. [DOI] [PubMed] [Google Scholar]
- Katzmann D.J., Babst,M. and Emr,S.D. (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell, 106, 145–155. [DOI] [PubMed] [Google Scholar]
- Khan O., Zabad,R., Caon,C., Zvartau-Hind,M., Tselis,A. and Lisak,R. (2002) Comparative assessment of immunomodulating therapies for relapsing-remitting multiple sclerosis. CNS Drugs, 16, 563–578. [DOI] [PubMed] [Google Scholar]
- Kirkwood J. (2002) Cancer immunotherapy: the interferon-α experience. Semin. Oncol., 29, 18–26. [DOI] [PubMed] [Google Scholar]
- Lassot I., Segeral,E., Berlioz-Torrent,C., Durand,H., Groussin,L., Hai,T., Benarous,R. and Margottin-Goguet,F. (2001) ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(βTrCP) ubiquitin ligase. Mol. Cell. Biol., 21, 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G., Waterman,H., Zamir,E., Kam,Z., Oved,S., Langdon,W.Y., Beguinot,L., Geiger,B. and Yarden,Y. (1998) c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev., 12, 3663–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F., Bour,S.P., Durand,H., Selig,L., Benichou,S., Richard,V., Thomas,D., Strebel,K. and Benarous,R. (1998) A novel human WD protein, h-βTrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell, 1, 565–574. [DOI] [PubMed] [Google Scholar]
- Muller U., Steinhoff,U., Reis,L.F., Hemmi,S., Pavlovic,J., Zinkernagel,R.M. and Aguet,M. (1994) Functional role of type I and type II interferons in antiviral defense. Science, 264, 1918–1921. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Hatakeyama,S., Maruyama,S.I., Kikuchi,A., Onoe,K., Good,R.A. and Nakayama,K.I. (2003) Impaired degradation of inhibitory subunit of NF-κB (IκB) and β-catenin as a result of targeted disruption of the β-TrCP1 gene. Proc. Natl Acad. Sci. USA, 100, 8752–8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick D., Cohen,B. and Rubinstein,M. (1994) The human interferon α/β receptor: characterization and molecular cloning. Cell, 77, 391–400. [DOI] [PubMed] [Google Scholar]
- Ohh M., Kim,W.Y., Moslehi,J.J., Chen,Y., Chau,V., Read,M.A. and Kaelin,W.G.,Jr (2002) An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep., 3, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Michel,J.J., Schottelius,A.J. and Xiong,Y. (1999) ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell, 3, 535–541. [DOI] [PubMed] [Google Scholar]
- Parisien J.P., Lau,J.F., Rodriguez,J.J., Ulane,C.M. and Horvath,C.M. (2002) Selective STAT protein degradation induced by paramyxoviruses requires both STAT1 and STAT2 but is independent of α/β interferon signal transduction. J. Virol., 76, 4190–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. (2000) The human interferon α species and receptors. Biopolymers, 55, 254–287. [DOI] [PubMed] [Google Scholar]
- Ragimbeau J., Dondi,E., Alcover,A., Eid,P., Uze,G. and Pellegrini,S. (2003) The tyrosine kinase Tyk2 controls IFNAR1 cell surface expression. EMBO J., 22, 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read M.A. et al. (2000) Nedd8 modification of cul-1 activates SCF(βTrCP)-dependent ubiquitination of IκBα. Mol. Cell. Biol., 20, 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijngoud D.J., Oud,P.S., Kas,J. and Tager,J.M. (1976) Relationship between medium pH and that of the lysosomal matrix as studied by two independent methods. Biochim. Biophys Acta, 448, 290–302. [DOI] [PubMed] [Google Scholar]
- Sadot E., Simcha,I., Iwai,K., Ciechanover,A., Geiger,B. and Ben-Ze’ev,A. (2000) Differential interaction of plakoglobin and β-catenin with the ubiquitin-proteasome system. Oncogene, 19, 1992–2001. [DOI] [PubMed] [Google Scholar]
- Shimoda K. et al. (2000) Tyk2 plays a restricted role in IFN α signaling, although it is required for IL-12-mediated T cell function. Immunity, 13, 561–571. [DOI] [PubMed] [Google Scholar]
- Spencer E., Jiang,J. and Chen,Z.J. (1999) Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev., 13, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman V.S., Slaga,T.J., Pagano,M., Minamoto,T., Ronai,Z. and Fuchs,S.Y. (2000) Wnt/β-catenin signaling induces the expression and activity of βTrCP ubiquitin ligase receptor. Mol. Cell, 5, 877–882. [DOI] [PubMed] [Google Scholar]
- Spiegelman V.S., Tang,W., Chan,A.M., Igarashi,M., Aaronson,S.A., Sassoon,D.A., Katoh,M., Slaga,T.J. and Fuchs,S.Y. (2002) Induction of homologue of Slimb ubiquitin ligase receptor by mitogen signaling. J. Biol. Chem., 277, 36624–36630. [DOI] [PubMed] [Google Scholar]
- Stark G.R., Kerr,I.M., Williams,B.R., Silverman,R.H. and Schreiber,R.D. (1998) How cells respond to interferons. Annu. Rev. Biochem., 67, 227–264. [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. (1999) IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1 and two F-box/WD40-repeat proteins, βTrCP1 and βTrCP2. Biochem. Biophys. Res. Commun., 256, 127–132. [DOI] [PubMed] [Google Scholar]
- Tan P., Fuchs,S.Y., Chen,A., Wu,K., Gomez,C., Ronai,Z. and Pan,Z.Q. (1999) Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell, 3, 527–533. [DOI] [PubMed] [Google Scholar]
- Terrell J., Shih,S., Dunn,R. and Hicke,L. (1998) A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell, 1, 193–202. [DOI] [PubMed] [Google Scholar]
- Treier M., Staszewski,L.M. and Bohmann,D. (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the δ domain. Cell, 78, 787–798. [DOI] [PubMed] [Google Scholar]
- Uze G., Lutfalla,G. and Gresser,I. (1990) Genetic transfer of a functional human interferon α receptor into mouse cells: cloning and expression of its cDNA. Cell, 60, 225–234. [DOI] [PubMed] [Google Scholar]
- Wang P., Izotova,L., Mariano,T.M., Donnelly,R.J. and Pestka,S. (1994) Construction and activity of phosphorylatable human interferon-α B2 and interferon-α A/D. J. Interferon Res., 14, 41–46. [DOI] [PubMed] [Google Scholar]
- Weissman A.M. (2001) Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell. Biol., 2, 169–178. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Koepp,D.M., Zhu,C., Elledge,S.J. and Harper,J.W. (1999a) A family of mammalian F-box proteins. Curr. Biol., 9, 1180–1182. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Strack,P., Beer-Romero,P., Chu,C.Y., Elledge,S.J. and Harper,J.W. (1999b) The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev., 13, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Chen,A. and Pan,Z.Q. (2000) Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem., 275, 32317–32324. [DOI] [PubMed] [Google Scholar]
- Yang C.H., Murti,A., Pfeffer,S.R., Basu,L., Kim,J.G. and Pfeffer,L.M. (2000) IFNα/β promotes cell survival by activating NF-κB. Proc. Natl Acad. Sci. USA, 97, 13631–13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A. et al. (1998) Identification of the receptor component of the IκBα-ubiquitin ligase. Nature, 396, 590–594. [DOI] [PubMed] [Google Scholar]