Abstract
The role of IF2 from Escherichia coli was studied in vitro using a system for protein synthesis with purified components. Stopped flow experiments with light scattering show that IF2 in complex with guanosine triphosphate (GTP) or a non-cleavable GTP analogue (GDPNP), but not with guanosine diphosphate (GDP), promotes fast association of ribosomal subunits during initiation. Biochemical experiments show that IF2 promotes fast formation of the first peptide bond in the presence of GTP, but not GDPNP or GDP, and that IF2–GDPNP binds strongly to post-initiation ribosomes. We conclude that the GTP form of IF2 accelerates formation of the 70S ribosome from subunits and that GTP hydrolysis accelerates release of IF2 from the 70S ribosome. The results of a recent report, suggesting that GTP and GDP promote initiation equally fast, have been addressed. Our data, indicating that eIF5B and IF2 have similar functions, are used to rationalize the phenotypes of GTPase-deficient mutants of eIF5B and IF2.
Keywords: G protein/IF2/initiation of translation/protein synthesis/ribosome
Introduction
Initiation of protein synthesis is more complex in eukaryotes than in eubacteria (Roll-Mecak et al., 2001). In eubacteria, initiation of mRNA translation requires only the three initiation factors, IF1, IF2 and IF3 (Gualerzi and Pon, 1990), while there are no less than 12 initiation factors in eukaryotes (reviewed by Pestova et al., 2000; Roll-Mecak et al., 2001).
Sequence data show that IF1 and IF2 are homologues of the eukaryotic factors eIF1A (Kyrpides and Woese, 1998) and eIF5B, respectively (Roll-Mecak et al., 2001), but there is no eukaryotic homologue of IF3.
After termination of eubacterial protein synthesis by either of the class-1 peptide release factors RF1 or RF2 and its rapid dissociation by the guanosine triphosphate (GTP)-dependent action of the class-2 release factor RF3 (Freistroffer et al., 1997; Zavialov et al., 2001, 2002), the ribosome is split into subunits by ribosome recycling factor (RRF) and EF-G in a GTP-dependent manner (Karimi et al., 1999). The first step in re-initiation of the ribosome is probably association of IF3 to the 30S subunit, an event that rapidly removes the deacylated tRNA from its partial P site on the small subunit and allows the mRNA to dissociate (Karimi et al., 1999). Binding of initiator tRNA to the 30S subunit under physiological conditions at low Mg2+ concentration is strongly favoured by the presence of mRNA (K.Andersson and A.Antoun, unpublished data) and is, in addition, greatly accelerated by the presence of IF3 (Wintermeyer and Gualerzi, 1983). In the absence of IF2 and initiator tRNA, the presence of IF3 on the 30S subunit effectively prevents formation of a 70S ribosomal complex (Grunberg-Manago et al., 1975).The IF3 block against subunit association and 70S ribosome formation is overcome when the small subunit contains mRNA, initiator tRNA and IF2. Association of the 50S subunit to such a pre-initiation 30S complex leads to 70S formation and subsequent formation of the first peptide bond in the nascent protein. The role of IF1 in this process has remained obscure (Dahlquist and Puglisi, 2000), in spite of extensive research on the factor over decades (Benne et al., 1973; Hartz et al., 1989).
It was suggested from early experiments that 70S formation from the ribosomal subunits during initiation of mRNA translation requires IF2 in the GTP form, and that GTP hydrolysis is essential for release of IF2 to allow for the subsequent peptide bond formation (Dubnoff et al., 1972; Benne et al., 1973). More recently, Tomsic et al. (2000) used quench–flow techniques to follow the extent of peptide bond formation, after mixing pre-initiated 30S subunits with 50S ribosomal subunits and ternary complexes containing EF-Tu, GTP and aminoacyl-tRNA. Surprisingly, they found that the time for subunit association and formation of a post-initiation ribosomal complex ready for rapid peptidyl transfer was ∼1 s and virtually the same in the presence of either GTP or guanosine diphosphate (GDP). This experiment showed, in other words, that IF2 in the GDP form can catalyse subunit association and make the ribosome ready for the first peptide bond with the same efficiency as IF2 in the GTP form on 30S followed by GTP hydrolysis after 70S formation. This is in contrast to the corresponding eukaryotic case, where GTP and GTP hydrolysis are essential for the eIF5B-dependent subunit association and subsequent peptide bond formation (Pestova et al., 2000; Lee et al., 2002). One interpretation of these data is that eIF5B and IF2 operate according to different principles in their respective kingdoms (Roll-Mecak et al., 2001; Ramakrishnan, 2002). However, the experimental data presented in this work show that IF2 and eIF5B use GTP in a similar manner during initiation of protein synthesis in both kingdoms.
We have found that GTP and its non-cleavable analogue GDPNP promote IF2-dependent subunit association much more efficiently than GDP, and that GTP hydrolysis is necessary for the subsequent peptide bond formation on the 70S ribosome. These results are in line with what has been observed for eIF5B, but are very different from those obtained for IF2 by Tomsic et al. (2000). We use our data to discuss how IF2 and GTP overcome the IF3-dependent block against ribosome formation, to compare initiation of protein synthesis in eubacteria and eukaryotes, and to clarify the phenotypes of GTPase-deficient mutants of IF2 (Luchin et al., 1999) and eIF5B (Shin et al., 2002).
Results
fMet-tRNAfMet stabilizes the binding of GTP to IF2 in the 30S pre-initiation complex
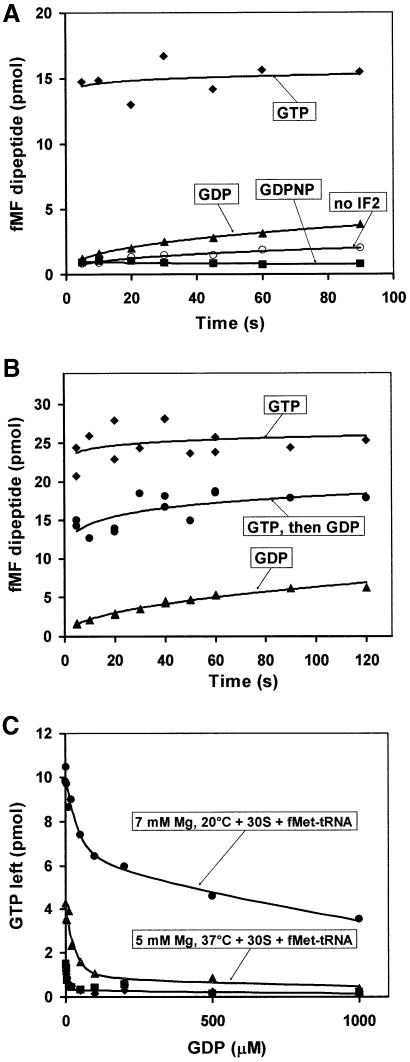
To study the roles of IF2 and GTP in initiation of protein synthesis, we used an in vitro system for mRNA translation with purified components from Escherichia coli (Pavlov et al., 1997; Zavialov et al., 2001). Initiation of protein synthesis in eubacteria requires the formation of a pre-initiation complex containing the 30S ribosomal subunit, mRNA, initiator tRNA (fMet-tRNAfMet) and the initiation factors (IFs) IF1, IF2 with GTP and IF3 (Gualerzi and Pon, 1990). We first studied the effects of the G-protein IF2 and different guanine nucleotides (GTP, GDP and the non-cleavable GTP analogue GDPNP) on the extent of fMet-tRNAfMet binding to the 30S subunit. The experiment was carried out in the presence of IF1, IF3 and an mMFTI mRNA containing a strong Shine–Dalgarno sequence, an AUG initiation codon and a small open reading frame encoding the tetrapeptide fMet-Phe-Thr-Ile-stop (Pavlov et al., 1997). In the absence of IF2, the extent of binding of [3H]fMet-tRNAfMet to 30S did not depend on the choice of guanine nucleotide. In the presence of IF2 and GTP or GDPNP, the extent of binding was equal to the amount of active 30S subunits and slightly lower (75%) in the presence GDP. In all cases, the binding of fMet-tRNAfMet to the 30S subunits reached equilibrium within 5 s of the start of incubation (experiments not shown).
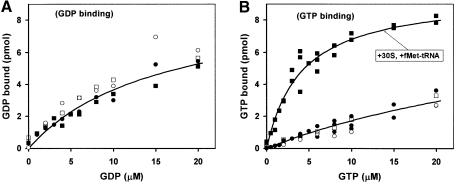
Next, we studied how fMet-tRNAfMet and/or 30S subunits, supplemented with the mMFTI mRNA, IF1 and IF3, affected the binding of either GDP or GTP to IF2. It can be seen in Figure 1A that the binding of GDP to IF2 was independent of the presence of initiator tRNA and/or 30S subunits. The binding of GTP to IF2 (Figure 1B) was weak in the absence of fMet-tRNAfMet and/or 30S subunits, and strong in the presence of both fMet-tRNAfMet and 30S subunits (Kd = 2 µM). This implies that IF2 in the GTP form stabilizes the binding of initiator tRNA to the 30S subunit much more than IF2 in the GDP or guanine nucleotide-free conformation. This follows from the principle of detailed balance (Fersht, 1999), which in this particular case states that if fMet-tRNAfMet stabilizes GTP binding to IF2 on the small subunit, then the GTP-form of 30S-bound IF2 must stabilize the binding of initiator tRNA to the 30S subunit.
Fig. 1. The effects of fMet-tRNAfMet and 30S subunits on the affinity of GDP or GTP to IF2. The extent of GDP (A) or GTP (B) binding to IF2 was measured by nitrocellulose filtration as a function of G-nucleotide concentration in the presence of combinations of fMet-tRNAfMet and 30S subunits containing mMFTI mRNA and IF1. Open circles: with neither fMet-tRNAfMet nor 30S; closed circles: with 30S; open squares: with fMet-tRNAfMet; closed squares: with fMet-tRNAfMet and 30S.
Rate of formation of 70S ribosomes from ribosomal subunits
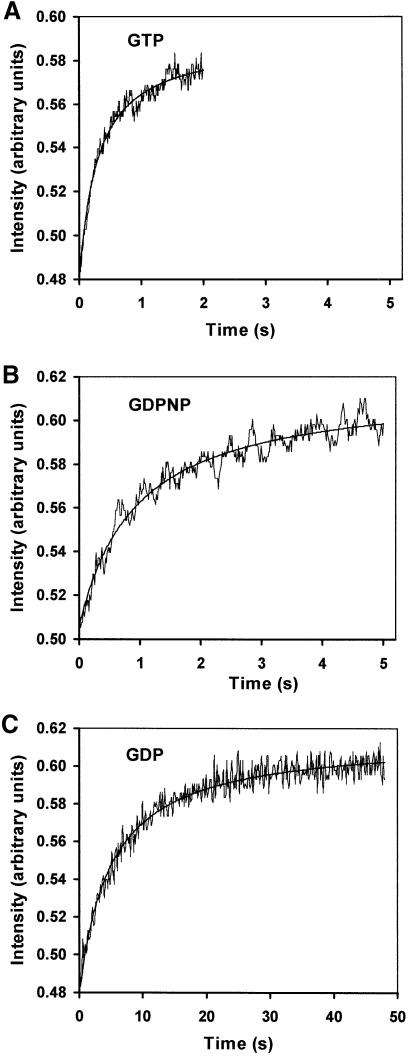
After formation of the pre-initiation 30S complex, the next step in initiation of eubacterial protein synthesis is the association of the ribosomal subunits and formation of the 70S ribosome (Grunberg-Manago et al., 1975; Blumberg et al., 1979). Pre-initiated 30S complexes with all three IFs, mMFTI mRNA and initiator tRNA in the presence of either GTP, GDPNP or GDP were rapidly mixed with 50S subunits in a stopped-flow instrument, and the intensity of the scattered light was recorded. This is a direct way to assess ribosome formation from ribosomal subunits (Grunberg-Manago et al., 1975), since the scattered intensity from each type of macromolecular complex is proportional to its concentration and to the square of its molecular weight. Subunit association took about half a second in the presence of GTP (Figure 2A), corresponding to an association rate constant of ka = 1.1 µM–1s–1, 1 s in the presence of GDPNP (ka = 0.4 µM–1s–1; Figure 2B) and 10 s in the presence of GDP (ka = 0.05 µM–1s–1; Figure 2C).
Fig. 2. The effects of G-nucleotides on the rate of association of 30S pre-initiation complexes with 50S subunits. The extent of 70S complex formation was monitored as a function of time by light scattering after rapid mixing of pre-initiation 30S complexes with 50S subunits in a stopped-flow instrument. Time curves were obtained with GTP (A), GDPNP (B) or GDP (C).
Rate of initiation of protein synthesis monitored by formation of the first peptide bond
Once 70S ribosomes have been formed (see previous section), the IFs must dissociate to allow for rapid binding of a ternary complex (consisting of an aminoacyl-tRNA, an EF-Tu and a GTP molecule) to the ribosomal A site followed by release of EF-Tu·GDP and peptidyl transfer. Peptide bond formation, which is very fast on post-initiation ribosomes (Tomsic et al., 2000), was used to monitor the rate of initiation of protein synthesis in the presence of GTP, GDP or GDPNP.
To study the rate of initiation with IF2 in recycling mode, pre-initiated 30S complexes were first prepared in the presence of the following components: either GTP or GDP, initiator tRNA, IF1 and IF3 in excess over 30S subunits and IF2 in a small amount (one IF2 to ten 30S subunits). 50S subunits were then added to start the initiation reaction and, subsequently, ternary complexes (containing EF-Tu, aminoacyl-tRNA and GTP) were added at different incubation times. In each case, peptide bond formation was stopped with formic acid 5 s after ternary complex addition. This time was enough to allow complete peptidyl transfer in already initiated 70S ribosomes (not shown).
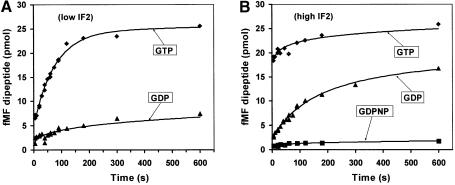
Figure 3A shows that peptide bonds were made much more rapidly in the presence of GTP than GDP, implying that the rate of recycling of IF2 was much higher with GTP than GDP. To monitor the rate of initiation with IF2 in single cycle mode, 30S complexes were first prepared in the presence of GTP, GDPNP or GDP, and with all factors, including IF2, in excess over 30S subunits. 50S subunits were then added and the formation of translation- competent 70S ribosomes was monitored by peptidyl transfer to aminoacyl-tRNA, as described above. The results (Figure 3B) show that peptide bond formation was very fast in the presence of GTP, intermediate in the presence of GDP and very slow in the presence of GDPNP (Figure 3B).
Fig. 3. The effects of G-nucleotides and IF2 concentration on the rate of peptidyl transfer, starting from pre-initiation 30S complexes and 50S subunits. Subunits were mixed in the presence of IFC and either GTP (diamonds), GDP (triangles) or GDPNP (squares). The reaction mixture contained either 2.5 pmol IF2 (A) or 120 pmol IF2 (B) per 50 pmol of ribosomes. The extent of ribosome initiation was monitored by dipeptide formation. Ternary complexes (EF-Tu–GTP–Phe-tRNAPhe) were added at different time points after subunit mixing, and the peptidyl-transfer reaction was quenched with 50% formic acid after 5 s.
In the presence of GDP, the time for 70S ribosome formation and peptidyl transfer (Figure 3B) was significantly longer than the time for subunit association (Figure 2C). This difference is accounted for by the higher concentrations of subunits used in the light-scattering experiment. This was demonstrated by using the association rate constant (ka = 0.05 µM–1s–1) for 70S ribosome formation obtained from the data in Figure 2C to accurately predict the time dependence of peptide bond formation in Figure 3B.
Initiation of protein synthesis with a non-cleavable analogue of GTP
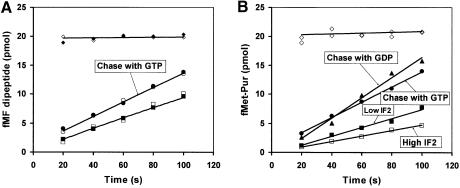
The rates of subunit association in the presence of GDPNP (Figure 2B) and GTP (Figure 2A) were similar, but the rate of peptide bond formation was almost zero in the presence of GDPNP and very fast in the presence of GTP (Figure 3B). This indicates that IF2 was able to promote rapid subunit association in the presence of GDPNP, but remained bound to the 70S ribosome, thereby blocking subsequent association of ternary complex and peptide bond formation. If true, this would suggest that addition of IF2 and GDPNP to already initiated 70S ribosomes would result in a high affinity complex between IF2–GDPNP and ribosomes. This, in turn, would strongly inhibit ternary complex binding and subsequent peptidyl transfer as in Figure 3B. To test this hypothesis, we first assembled 70S complexes (0.4 µM) from pre-initiation 30S complexes, and 50S subunits in the presence of small amounts of GTP and IF2. Then, IF2 was added to a final concentration of either 0.5 or 1.5 µM together with GDPNP to a concentration of 0.2 mM. After 1 min, peptide bond formation was initiated by the addition of ternary complex containing Phe-tRNAPhe or puromycin, and the extent of peptidyl transfer was monitored as a function of time (Figure 4). The transfer of [3H]fMet to Phe-tRNAPhe was fast in the absence of extra IF2 and GDPNP, and very slow in their presence (Figure 4A). The absence of any rapid formation of peptide bonds in the presence of IF2 and GDPNP shows that all active ribosomes contained IF2, i.e. that the rate of forming a complex between IF2–GDPNP and the ribosome was much larger than the rate of dissociation of this complex. The rate of peptide bond formation per ribosome (0.004 s–1) was the same for the two IF2 concentrations (lowest curves), showing that in the presence of ternary complex there was no significant rebinding of IF2 to the 70S ribosome after the first dissociation event. Accordingly, the overall rate constant for the dissociation of IF2–GDPNP from the ribosome must under those conditions have been 0.004 s–1. When, instead, puromycin was used as fMet acceptor in the peptidyl-transfer reaction, peptide bond formation was very fast in the absence of extra IF2 and GDPNP (Figure 4B, upper curve) and much slower in their presence (0.003 s–1) (Figure 4A). In this case, the rate of peptide bond formation decreased even further with an increasing concentration of IF2 at a fixed concentration of GDPNP, suggesting that re-binding of IF2 to ribosomes after the first dissociation event was significant. The reason why rebinding of IF2 occurred only in the presence of puromycin is, we propose, that the rate of association of the drug to a free ribosome was much smaller than the rate of association of the ternary complex, giving time for re-association of IF2 in the former but not in the latter case.
Fig. 4. Peptide bond formation in the presence of IF2 and GDPNP. The 70S complexes were assembled by mixing 0.4 µM 50S subunits and 0.4 µM 30S pre-initiation complexes in the presence of 0.04 µM IF2 and 10 µM GTP. After 10 min of incubation at 37C, either 0.5 µM IF2 and 0.2 mM GDPNP (closed squares, ‘low IF2-GDPNP’ mix), or 1.5 µM IF2 and 0.2 mM GDPNP (open squares, ‘high IF2-GDPNP’ mix), or an equivalent volume of polymix buffer (open diamonds) were added to these 70S complexes. The incubation then continued for 1 min more, after which EF-Tu–GTP–Phe-tRNA ternary complexes (A) or puromycin (B) were added to all mixes and the [3H]fMet-Phe or [3H]fMet-Pur formation was followed as a function of time. Ternary complexes (A) were also added to the ‘high’ (open circles) and ‘low’ (closed circles) IF2-GDPNP mixes, together with a large amount of GTP (1.4 mM final concentration). Puromycin (B) was also added to the ‘low IF2-GDPNP’ mix together with a large amount of GTP (closed circles) or GDP (closed triangles). As a control, 70S complexes were also assembled from 0.4 µM 50S subunits and 0.4 µM 30S pre- initiation complexes in the presence of 0.5 µM IF2 and 0.2 mM GTP. After incubation for 11 min, the extent of complex formation was checked by the addition of ternary complexes (A, closed diamonds). Note that open diamonds in (B) come from two independent experiments.
When either GTP or GDP was present in excess over GDPNP in these assays, the rate of peptide bond formation was faster than in their absence (Figure 4). This, we suggest, is because GDPNP can dissociate from a ribosome-bound IF2 and that binding of either GTP or GDP to IF2 removes it rapidly from the ribosome, while re-binding of GDPNP in the absence of GTP and GDP delays dissociation of the factor and therefore peptide bond formation.
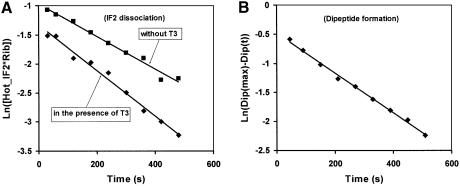
It is a common prejudice in the literature that binding of ternary complex to the ribosome and subsequent peptide bond formation cannot occur in the presence of ribosome-bound IF2 in the GTP form (La Teana et al., 2001), but direct experimental evidence for this assertion is scarce. This question is important, since interpretation of the phenotype of GTPase-deficient mutants of IF2 (Luchin et al., 1999; Gualerzi et al., 2001) and eIF5B (Shin et al., 2002) depends on the answer. To clarify the situation, we labelled IF2 with 35S and prepared ribosome complexes as in the experiments shown in Figure 4 containing [35S]IF2–GDPNP. In experiments shown in Figure 5, the radiolabelled IF2 was chased either by only a large excess of unlabelled IF2 or by an excess of both IF2 and ternary complex, and the rate constants (0.0028 s–1 and 0.0037 s–1, respectively) for the dissociation of IF2 were obtained by separating ribosome-bound from free [35S]IF2 by ultracentrifugation (Figure 5A). We also monitored the rate of peptidyl transfer in the experiment with ternary complex, and obtained a rate constant of 0.0036 s–1(Figure 5B). These experiments show that the rates of peptide bond formation and release of IF2–GDPNP in the presence of ternary complex were identical. There was a small difference between the rates of dissociation of labelled IF2–GDPNP in the presence of excess unlabelled IF2 and ternary complex, showing that the ternary complex could accelerate the dissociation of IF2–GDPNP from the ribosome by ∼50% compared with a chase with only initiation factor. This acceleration did not increase with a further increase of the concentration of GTP-containing ternary complex, nor when GTP in the ternary complex was replaced by GDPNP (not shown). This suggests that the ternary complexes could bind to ribosomes that contained IF2, possibly in a site corresponding to the first step of codon scanning by ternary complexes (Rodnina and Wintermeyer, 2001).
Fig. 5. IF2 dissociation from 70S initiation complex. The 70S initiation complexes were assembled by mixing 0.4 µM 50S subunits and 0.4 µM 30S pre-initiation complexes in the presence of 0.02 µM His-tagged cold IF2 and 10 µM GTP. After 15 min of incubation at 37°C, GDPNP was added to 0.5 mM final concentration. Then [35S]IF2 (His tagged) was added to the 0.4 µM final concentration. After 2 min of incubation, an excess of cold His-tagged IF2 (1.2 µM final concentration) was added to the reaction mix to start the exchange between hot and cold IF2 (closed squares). The exchange was also studied in the presence of 1 µM of ternary complexes (closed diamonds). Aliquots of the reaction were taken at the indicated times and the amount of ribosome-bound hot IF2 was determined as described in Materials and methods. The slope of the plot of the logarithm of this amount versus time in (A) gives the rate of IF2 dissociation. The extend of dipeptide formation in the reaction mix containing ternary complex was also measured. The slope of Log[Dip(max)-Dip(t)] (closed diamonds) versus time gives the rate of dipeptide formation in the reaction mix (B).
Factors affecting GTP to GDP exchange on IF2 in complex with 30S
Recently, Tomsic et al. (2000) used quench–flow techniques to estimate the rate of IF2-dependent association of pre-initiated 30S complexes to 50S subunits by monitoring the extent of peptide bond formation in the presence of ternary complex. They found a slightly higher rate of peptidyl transfer in the presence of GDP than GTP. This is in sharp contrast to the results in Figure 3B, showing a much lower peptidyl-transfer rate in the GDP than in the GTP case, and also to the results in Figure 2, showing a much slower subunit association in the presence of GDP (Figure 2C) than GTP (Figure 2A). Tomsic et al. performed their experiments at 20°C in Tris buffer with 7 mM Mg2+ concentration, while our experiments were performed at 37°C in polymix buffer with 5 mM Mg2+ concentration. To examine whether these different experimental conditions could explain the discrepancy, we performed experiments as in Figure 3 at 20°C with 7 mM Mg2+. The data in Figure 6A show that peptidyl transfer was much faster in the presence of GTP than GDP also under those conditions. This leaves the difference between the results unexplained, unless substituting polymix for Tris buffer drastically changed the outcome of our experiments. We noted that Tomsic et al. had used a protocol, stating that 30S pre-incubation complexes were prepared in the presence of [32P]GTP, and that, subsequently, either GTP or GDP was added in 30-fold excess over the [32P]GTP initially present, before loading the mixture to one of the syringes of the quench–flow instrument. This experimental design requires that GDP can effectively chase the GTP initially present on IF2. We found that fMet-tRNAfMet and 30S together stabilized the binding of GTP to IF2 in relation to GDP (Figure 1A and B). We also found that lowering the temperature from 37 to 20°C and increasing the Mg2+ concentration from 5 to 7 mM increased the relative affinity of GTP to IF2 even further, and dramatically slowed down the exchange of GTP for GDP (Figure 6C). This suggested that the GDP result obtained by Tomsic et al. could have been due to incomplete exchange of GTP for GDP. To test this, we performed an experiment at 20°C with 7 mM Mg2+, where the pre-initiation 30S complexes were pre-incubated with GTP (70 µM), which subsequently was chased with a 30-fold excess of GDP. Under those conditions, peptidyl transfer occurred very rapidly (as with GTP) on 60% and very slowly (as with GDP) on 40% of the active ribosomes (Figure 6B). This result agrees qualitatively with the results obtained by Tomsic et al., including their finding that the extent of rapid peptidyl transfer after the chase with GDP reached a lower plateau value (80%) than in the presence of GTP.
Fig. 6. The effects of temperature and [Mg2+] on the rate of peptidyl transfer, starting from pre-initiation 30S complexes and 50S subunits, and on the exchange of GTP for GDP on IF2 in the pre-initiation 30S complex. (A) The extent of ribosome initiation was monitored as in Figure 3 in the presence of IF2 and GTP (closed diamonds), IF2 and GDP (closed triangles), IF2 and GDPNP (closed squares), or in the absence of IF2 (open circles). The reaction mixture at 20°C contained 120 pmol IF2 per 50 pmol ribosome and 7 mM Mg(OAc)2. (B) The extent of ribosome initiation was monitored as in Figure 3 in the presence of IF2 and GTP (closed diamonds), IF2 and GDP (closed triangles), and IF2 and GDP but with the 30S pre-initiation complex first pre-incubated (10 min) with 70 µM of GTP and then (1.5 min) with 2 mM GDP (closed circles). (C) IF2, alone or with 30S in complex with mRNA, fMet-tRNAfMet, IF1 and IF3, was pre-incubated (10 min) with [3H]GTP (10 µM), and the amount of [3H]GTP remaining on IF2 after 1 min incubation following the addition of varying concentrations of unlabelled GDP was monitored by nitrocellulose filtration: closed diamonds, 5 mM Mg2+ at 37°C; closed squares, 7 mM Mg2+ at 20°C; closed triangles, 5 mM Mg2+ at 37°C with 30S complex; closed circles 7 mM Mg2+ at 20°C with 30S complex.
Discussion
We have observed that fMet-tRNAfMet enhances the affinity of GTP to IF2 on the 30S subunit (Figure 1). Using light-scattering detection in combination with stopped-flow analysis we have also shown that IF2 in complex with either GTP (Figure 2A) or the non-cleavable analogue GDPNP (Figure 2B) promotes fast association of the ribosomal subunits, while their association in the presence of GDP is slow (Figure 2C). We have demonstrated that the time for subunit association and formation of the first peptide bond is very short in the presence of GTP and considerably longer in the presence of GDP (Figure 3A and B). We have found that although IF2–GDPNP promotes fast association of subunits (Figure 2B), the subsequent rate of peptide bond formation is near zero (Figure 3B). The results of further experiments (Figure 5) show that the rate of release of IF2–GDPNP from the ribosome (Figure 5A) is identical to the rate of peptidyl transfer (Figure 5B). From these data we conclude, first, that IF2 in complex with GDPNP binds strongly to initiated ribosomes, which effectively prevents peptidyl transfer to aminoacyl-tRNA (Figure 4A) and puromycin (Figure 4B). Secondly, we conclude that GTP hydrolysis is essential for rapid removal of IF2 from initiated 70S ribosomes.
Our results are in line with observations from classical experiments with non-cleavable analogues of GTP (Dubnoff et al., 1972; Benne et al., 1973), but are in sharp contrast to recent data from Tomsic et al. (2000).
Tomsic et al. used quench–flow techniques for rapid mixing of pre-initiation 30S complexes and 50S subunits, and monitored initiation of protein synthesis by the extent of peptidyl transfer to aminoacyl-tRNA. They found the IF2-dependent rate of initiation to be equally rapid in the presence of GTP and GDP, in contrast to our observations (Figure 3). One reason why two similarly designed experiments yielded such different results is that the protocol published by Tomsic et al. to measure the rate of initiation in the presence of GDP required that the GTP, originally present on IF2 in complex with the 30S subunit, was completely exchanged with GDP. As shown in Figure 6B and C, the addition of GDP in 30-fold excess over GTP under conditions close to those employed by Tomsic et al. allows most of the GTP to stay bound to IF2. This means that the rate of initiation, nominally measured in the presence of GDP, could have been strongly affected by a residual amount of GTP present in their experiment.
We have now been informed (M.Rodnina, personal communication) that the quench–flow protocol published by Tomsic et al. is, in fact, different from the protocol actually used in their experiment. Therefore, other explanations for the discrepancy between the present results and those obtained by Tomsic et al. cannot be excluded at the present time. In any case, the present data and analysis strongly suggest that GTP is, indeed, essential for rapid initiation of eubacterial protein synthesis. How, then, does GTP accomplish this task?
To avoid sequestering of ribosomes in an improper state during initiation of protein synthesis, docking of subunits should only occur when initiator tRNA is bound to mRNA on the 30S subunit. In principle, this problem could be solved if fMet-tRNAfMet and 50S could form a very stable complex that provided the necessary force to rapidly bring the subunits together and, at the same time, eject IF3 from the 30S subunit (Pon and Gualerzi, 1986). However, this type of design is expected to sequester the ribosome in a post-initiation complex with fMet-tRNAfMet so strongly anchored that the first steps in protein elongation are slowed down, thereby creating a ‘cul-de-sac’.
The conclusion from the data in Figure 1, that the GTP form of IF2 is strongly stabilized by fMet-tRNAfMet on 30S, suggests how rapid subunit association may be made to occur only in the presence of initiator tRNA and mRNA, so that the formation of abortive 70S complexes lacking fMet-tRNAfMet is prevented. The argument can be based on a two-conformation model for IF2; one conformation with high affinity to GTP and the 50S subunit, and the other—the GDP conformation—with low affinity to GTP and the 50S subunit. The GDP conformation is favoured when IF2 is in the free state, or in complex with the 30S subunit without initiator tRNA. When fMet-tRNAfMet binds to the 30S subunit, the GTP conformation is favoured and this attracts the 50S subunit, which leads to rapid 70S ribosome formation and ejection of IF3. In accordance with this, IF2 in the GTP conformation has very high affinity to the post-initiation ribosome, as shown by the very slow dissociation of IF2–GDPNP from the 70S ribosome (Figures 4 and 5). The binding of IF2 to the ribosome is, we suggest, stabilized by favourable interactions with both initiator tRNA and the 50S subunit. This tight complex is subsequently resolved by GTP hydrolysis and rapid dissociation of IF2 in the GDP form from the ribosome. In this way, the cul-de-sac scenario is avoided.
The present findings lend strong support to the notion that the two sequence homologues, IF2 in eubacteria and eIF5B in eukaryotes, employ similar mechanisms to promote initiation of mRNA translation. For instance, Pestova et al. (2000) demonstrated that eIF5B is required for the formation of 80S ribosomes from pre-initiated 48S subunits and 60S subunits. From ultracentrifugation data they showed that eIF5B can promote subunit association catalytically in the presence of GTP, but only stoichiometrically in the presence of the non-cleavable GTP analogue GDPNP. They also demonstrated that Met-tRNAMet in the 80S initiation complex can act as a donor in the peptidyl-transfer reaction with puromycin as acceptor, provided that the initiation complex had been formed with eIF5B and GTP rather than GDPNP. Lee et al. (2002) suggested that eIF5B, in complex with either GTP or GDPNP, stabilizes Met-tRNAMet in the 48S pre-initiation complex, and promotes its docking to the 60S subunit. They also proposed that GTP hydrolysis is necessary for the removal of eIF5B from the 80S ribosome. This would explain why eIF5B can recycle in the presence GTP, but not GDPNP, and why Met-tRNAMet can react with puromycin on ribosomes that have been assembled in the presence of GTP but not GDPNP (Pestova et al., 2000). All these results are qualitatively similar to what we have found for IF2 and its GTP dependence.
Further support for a functional homology between eIF5B and IF2 comes from reports on GTPase-deficient mutants of both IF2 from E.coli (Luchin et al., 1999) and eIF5B from yeast (Shin et al., 2002). These mutants can bind GTP and promote subunit association, but dissociate very slowly from the post-initiation ribosome, which effectively inhibits the peptidyl-transfer reaction which requires that IF2 has left the ribosome (Figures 4 and 5). Another set of IF2 mutants, with impaired GTPase activity and cold-sensitive phenotype (Laursen et al., 2003), can be explained along similar lines. A deviating interpretation of the GTPase-deficient mutants of IF2 in E.coli has been suggested, but little experimental support for this alternative view has so far been presented (Gualerzi et al., 2001). The phenotype of these mutants is strikingly similar to what we observe here, when GTP is replaced by GDPNP to drive IF2 in the initiation process (Figures 2–4). Interestingly, in their search for intragenic suppressors of the original GTPase deficiency of eIF5B, Shin et al. (2002) selected a double mutant that retains its GTPase deficiency, but promotes near wild-type growth. The GTP form of this eIF5B variant has lower affinity to the 80S ribosome than the primary mutant, allowing for faster dissociation after initiation in spite of the GTPase deficiency. The data in Figure 3B can be used to interpret their results. The figure shows that the rate of subunit association and peptide bond formation is very fast in the presence of GTP, very slow in the presence of GDPNP due to the slow dissociation of IF2 (Figure 4), and intermediate in the presence of GDP due to slow association of subunits (Figure 2C). These results can be used as a metaphor for the eIF5B mutants: the GTP case corresponds to wild-type eIF5B, the GDPNP case corresponds to the primary GTPase-deficient mutant and the GDP case corresponds to the double mutant. The wild-type factor can promote both fast subunit association and be rapidly released, while the GTPase-deficient mutants can only perform either one of these tasks rapidly. Now, imagine that the GDP form of IF2 becomes more and more like the GTP form. This would lead to faster and faster subunit association in the presence of GDP, moving the middle curve in Figure 3B towards the upper GTP curve. When subunit association and factor release occur at the same rate, the middle curve will display its maximal rate, corresponding to the optimal choice for rapid initiation among the GTPase-deficient mutants. Although this rate will be considerably faster than the rates displayed by the lower curves in Figure 3B, it will still be slower than the rate in the presence of GTP. Further evolution of the GDP structure towards the GTP form will start reducing the rate of initiation, now because slow dissociation of the factor will impair the process more and more severely. In line with this argument is that although the double mutant of eIF5B selected by Shin et al. promotes initiation much more rapidly than the primary mutant, initiation is still significantly slower than for the wild type, as judged from the polysome profiles shown in their paper.
Materials and methods
Chemicals and buffers
Nucleoside triphosphates (ATP, UTP and GTP), radioactive amino acids and unlabelled nucleotides were from Amersham. GDPNP, phosphoenolpyruvate (PEP), myokinase (MK), pyruvate kinase (PK), putrescine, spermidine, puromycin dihydrochloride and non-radioactive amino acids were from Sigma. All other chemicals were of analytical grade from Merck. Guanine nucleotides GTP and GDP, when used for binding and exchange, were further purified as described previously (Zavialov et al., 2001). All experiments were carried out in polymix buffer (Jelenc and Kurland, 1979).
Components of the translation system
Synthetic mMFTI mRNA, encoding the tetrapeptide Met-Phe-Thr-Ile, was prepared according to Pavlov et al. (1997). 70S ribosomes, 50S and 30S subunits were prepared from the E.coli strain MRE 600, using sucrose gradient zonal ultracentrifugation according to Rodnina and Wintermeyer (1995). Initiation factors were purified from overproducing strains according to Soffientini et al. (1994). [3H]fMet tRNAfMet, [35S]fMet-tRNAfMet and Phe-tRNA synthetase (PheRS) were prepared according to Freistroffer et al. (1997). Elongation factors EF-Tu, EF-Ts and tRNAPhe were purified according to Ehrenberg et al. (1990).
Purification of 35S-labelled IF2
The BL21(DE3) strain with a pAF2H plasmid containing His-tagged IF2 sequence was kindly provided by Dr Anthony Forster (Forster et al., 2001). Overproducing cells were grown at 37°C in 1 l M9 minimal medium supplemented with 0.05% casamino acids (Difco), 0.08 mg/ml each of leucine, proline and tryptophan, 2 mM MgSO4 and 40 mg/l of kanamycin (Petrelli et al., 2001). At an A600 of 0.6, the overexpression of His-tagged IF2 was induced by adding 100 mg/l IPTG. At the same time, 0.3 ml of [35S]Met/[35S]Cys Promix (14.3 mCi/ml; Amersham) was added to the cell culture. The incubation then continued for 2 h, after which the cells were collected by centrifugation at 5000 r.p.m. for 30 min, washed with 40 mM Tris pH 7.5 and stored at –80°C until use. Labelled IF2 was first purified with a Talon His-tag affinity resin (Clontech), and then to apparent homogeneity by gel filtration through an S200 Sephacryl column (Pharmacia). IF2 was precipitated with ammonium sulfate, centrifuged, dialysed against polymix buffer and stored in small aliquots at –80°C.
fMet-tRNAfMet binding to 30S
Two mixes (A and B) were prepared (concentrations refer to values in the final incubation mix). Mix A: 0.2 µM [3H]fMet-tRNAfMet, 1 mM ATP, 10 mM PEP, and either 0.5 mM GTP or GDP, or 0.2 mM GDPNP. Mix B: 0.1 µM 30S subunits, 0.2 µM mMFTI mRNA, 0.1 µM IF1, 0.2 µM IF3, and either 0.005 µM, 0.2 µM IF2 or no IF2. The mixes were pre-incubated separately at 37°C for 10 min. The reaction at 37°C was started by adding 475 µl mix A to 25 µl mix B. After varying incubation times at 37°C, the reaction was stopped by adding 3 ml of ice-cold polymix buffer, immediately followed by nitrocellulose filtration (BA-85 filters pre-soaked in polymix buffer; Schleicher and Schuell). After washing twice with 3 ml ice-cold polymix, the filters were put into vials with 5 ml Filter Safe (Zinsser Analytic) scintillate cocktail and counted in an LC6500 scintillation counter (Beckman).
G-nucleotide binding to IF2
The reaction mix contained 2 µM IF2, 3H-labelled G-nucleotides at indicated concentrations and combinations of [35S]fMet-tRNA (2 µM) and 30S subunits (1 µM), together with mMFTI mRNA (2 µM) and IF1 (1 µM). After 3 min incubation at 37°C, the samples were directly passed through nitrocellulose filters, washed with 0.6 ml ice-cold polymix buffer and counted as described above.
Exchange of GTP with GDP on IF2
A pre-incubation mix was prepared containing 10 µM [3H]GTP, 2 µM IF2 and, when indicated, 2 µM [35S]fMet-tRNAfMet with 1 µM 30S subunits, supplemented with 2 µM mMFTI mRNA, 2 µM IF3 and 1 µM IF1. After 10 min pre-incubation, the [3H]GTP on IF2 was chased by addition of non-labelled GDP at varying concentrations. After 60 s incubation, the samples were directly passed through nitrocellulose filters, washed and counted, as in the previous section. Experiments were carried in polymix buffer at 37°C with 5 mM Mg (OAc)2 or at 20°C with 7 mM Mg(OAc)2.
70S complex formation in the presence of different G-nucleotides
Mixes A, B and C were prepared (concentrations refer to values in the final incubation mix). Mix A: 1 mM ATP, 10 mM PEP, 0.5 mM GTP or 0.5 mM GDP or 0.2 mM GDPNP, 0.2 µM [3H]fMet-tRNAfMet, 0.1 µM 30S subunits, 0.2 µM mMFTI mRNA, 0.1 µM IF1, 0.2 µM IF3 and IF2 as specified. Mix B: 0.1 µM 50S subunits or as indicated. Mix C: 1 µM EF-Tu, 1.5 µM tRNAPhe, 30 µM Phe, 1 U/ml PheRS, 1 µg/ml PK and 0.1 µg/ml MK. Mixes A, B and C were pre-incubated at 37°C for 10 min, then mix A (0.45 ml) was added to mix B (0.025 ml) to allow for 70S complex formation At the indicated times, mix C (0.025 ml) was added to allow for peptidyl transfer and the reaction was stopped after 5 s by the addition of 0.25 ml 50% formic acid. The samples were centrifuged and the amount of fMet-Phe-tRNAPhe in the pellet was determined by high-pressure liquid chromatography (HPLC) as described previously (Pavlov et al., 1997).
70S complex formation with GTP to GDP exchange
Mixes A, B and C were prepared as above except that they contained 7 mM Mg(OAc)2 and mix A contained 70 µM GTP. These three mixes were first pre-incubated at 37°C for 10 min, then 2 mM GDP was added to mix A to allow for GTP to GDP exchange on IF2. After 90 s incubation at 20°C, mix B was added to mix A, and then mix C was added at the indicated times to allow for peptidyl transfer during 5 s before the reaction was quenched with formic acid and the extent of dipeptide formation measured with HPLC as described above. In the control experiments without GTP/GDP exchange, mix A contained either 2 mM GTP or 2 mM GDP.
IF2 release from 70S initiation complexes
Mixes A, B and C were prepared (concentrations refer to values in the final incubation mix). Mix A: 1 mM ATP, 10 mM PEP, 0.4 µM [3H]fMet-tRNAfMet, 0.4 µM 30S subunits, 0.6 µM mMFTI mRNA, 0.4 µM IF1, 0.5 µM IF3, and IF2, GTP or GMPPNP as specified. Mix B: 0.4 µM 50S subunits. Mix C: 3 µM EF-Tu, 4 µM tRNAPhe, 30 µM Phe, 1 U/ml PheRS, 1 µg/ml PK and 0.1 µg/ml MK. Mixes A and B were first pre-incubated at 37°C for 4 min, and then mix A (0.4 ml) was added to mix B (0.05 ml). Formation of 70S proceeded for 11 min, followed by the addition of mix C or puromycin (final concentration 0.1 M). Subsequently, 0.08 ml aliquots were removed at different time points, quenched with 0.04 ml 50% formic acid and analysed for dipeptide content with HPLC. Samples with fMet-Pur were spun for 15 min at 14 000 r.p.m. in an Eppendorf centrifuge. The amount of fMet plus fMet-Pur in the supernatant was determined by scintillation counting, and the fraction of fMet-Pur in the supernatant by HPLC.
IF2 release from pre-formed 70S initiation complexes
Mixes A, B and C were prepared as above except that mix A contained only 0.04 µM IF2 and 10 µM GTP. Mixes A and B were pre-incubated for 4 min at 37°C. Then mix A (0.4 ml) was added into mix B (0.05 ml) and the reaction of 70S complex formation was allowed to proceed for 10 min. Subsequently, either 0.5 µM IF2 and 0.2 mM GDPNP, or 1.5 µM IF2 and 0.2 mM GDPNP, or an equivalent volume of polymix buffer was added to the reaction mixture. After 1 min, mix C containing ternary complexes (pre-incubated separately for 15 min at 37°C) or puromycin (up to 0.1 M final concentration) was added to reaction mixtures and the formation of fMet-Phe dipeptide or fMet-Pur was monitored as function of time as described above. In GTP or GDP chase experiments, a large amount of GTP–Mg or GDP–Mg was added together with either mix C or with puromycin so that the final concentration of GTP–Mg or GDP–Mg in the reaction mix was 1.4 mM.
Direct measurement of IF2 dissociation rate
Ribosomal initiation complexes competent in dipeptide formation were prepared similarly to that described above by adding the pre-warmed mix B to pre-warmed mix A containing 0.02 µM IF2 and 10 µM GTP. The formation of 70S initiation complexes was allowed to proceed for 15 min, after which GDPNP was added to the 0.5 mM final concentration followed by the addition of [35S]IF2 to the 0.4 µM final concentration equal to that of the ribosomes. The IF2–GDPNP exchange reaction was started by adding cold IF2 to 1.2 µM final concentration. Immediately after cold IF2 addition, the mix C containing ternary complexes in 1 µM final concentration was also added in cases when the effect of ternary complexes on IF2 dissociation was studied. The time course of dipeptide formation was monitored by taking 0.04 ml aliquots from the incubation mixture into Eppendorfs containing 0.02 ml of 50% formic acids. The amount of formed dipeptide was determined as described above. The time course of IF2 exchange was monitored by placing 0.1 ml aliquots of the reaction mix into Eppendorfs on ice containing 0.05 ml ice-cold TMK buffer [20 mM Tris pH 7.5, 60 mM NH4Cl, 60 mM KCl, 10 mM Mg(OAc)2]. The amount of ribosome-bound [35S]IF2 was determined by pelleting ribosomes in S100-AT3 rotor for 8 min at 90 000 r.p.m. using an RC 150 GX centrifuge (Sorvall). After centrifugation, the supernatants were withdrawn and spotted on GF-C filters (Whatmann). The pellets were dissolved in 0.5 M KOH and also spotted on GF-C filters. Filters was dried, put in vials with ReadyProtein scintillation cocktail (Beckman-Coulter) and counted in a Beckman LC6500 scintillation counter.
Subunit association measured by Rayleigh light scattering
The association of ribosomal subunits was detected with Rayleigh light scattering after rapid mixing in a stopped-flow instrument (Bio-sequential SX-18MV; Applied Photophysics). Two mixes were prepared: mix A contained 1 mM ATP, 10 mM PEP, 0.5 mM of either GTP, GDP or GDPNP, 6 µM [3H]fMet-tRNAfMet, 3 µM 30S, 6 µM mMFTI mRNA, 3 µM IF1, 6 µM IF2 and 6 µM IF3. Mix B contained 2 µM 50S. To remove dust particles, the mixes were centrifuged for 30 min at 14 000 r.p.m. before they were loaded into the stopped-flow instrument. The two mixes were pre-incubated at 37°C for 10 min, rapidly mixed in the stopped-flow instrument and the intensity of scattered light (436 nm, 90° angle) recorded as function of time.
Curve fitting
The dissociation constants (KD) for GTP and GDP binding to IF2 (Figure 1) were estimated by non-linear regression (Marquardt, 1963). The concentration of IF2-bound nucleotide ([IF2·G]) was assumed to depend on the concentrations of total IF2 ([IF2]0) and free guanine nucleotide ([G]) according to:
The association rate constants (ka) for subunit association in the stopped-flow light scattering experiments (Figure 2) were also estimated by non-linear regression. The concentration of ribosomes at time t ([70S]) was assumed to depend on the final ribosome concentration ([70S]0) and the initial and equal concentrations of subunits ([30S]0 = [50S]0) according to:
This equation is valid when the association of subunits is near irreversible, which was the case under our experimental conditions.
Acknowledgments
Acknowledgements
We thank Gun Stenberg and Gunnar Johansson for helping us with the light scattering experiments, and Otto Berg for valuable suggestions on the manuscript. This work was supported by the Swedish Research Council, Estonian Science Foundation and Wenner-Grenska Samfundet Foundation.
References
- Benne R., Naaktgeboren,N., Gubbens,J. and Voorma,H.O. (1973) Recycling of initiation factors IF-1, IF-2 and IF-3. Eur. J. Biochem., 32, 372–380. [DOI] [PubMed] [Google Scholar]
- Blumberg B.M., Nakamoto,T. and Kezdy,F.J. (1979) Kinetics of initiation of bacterial protein synthesis. Proc. Natl Acad. Sci. USA, 76, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist K.D. and Puglisi,J.D. (2000) Interaction of translation initiation factor IF1 with the E. coli ribosomal A site. J. Mol. Biol., 299, 1–15. [DOI] [PubMed] [Google Scholar]
- Dubnoff J.S., Lockwood,A.H. and Maitra,U. (1972) Studies on the role of guanosine triphosphate in polypeptide chain initiation in Escherichia coli. J. Biol. Chem., 247, 2884–2894. [PubMed] [Google Scholar]
- Ehrenberg M., Bilgin,N. and Kurland,C.G. (1990) Design and use of a fast and accurate in vitro translation system. In Spedding,G. (ed.), Ribosomes and Protein Synthesis: A Practical Approach. Oxford University Press, New York, NY, pp. 101–129. [Google Scholar]
- Fersht A. (1999) Structure and Mechanism in Protein Science. W.H.Freeman and Company, pp. 125–131. [Google Scholar]
- Forster A.C., Weissbach,H. and Blacklow,S.C. (2001) A simplified reconstitution of mRNA-directed peptide synthesis: activity of the epsilon enhancer and an unnatural amino acid. Anal. Biochem., 297, 60–70. [DOI] [PubMed] [Google Scholar]
- Freistroffer D.V., Pavlov,M.Y., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J., 16, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunberg-Manago M., Dessen,P., Pantaloni,D., Godefroy-Colburn,T., Wolfe,A.D. and Dondon,J. (1975) Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J. Mol. Biol., 94, 461–478. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. and Pon,C.L. (1990) Initiation of mRNA translation in prokaryotes. Biochemistry, 29, 5881–5889. [DOI] [PubMed] [Google Scholar]
- Gualerzi C.O. et al. (2001) Initiation factors in the early events of mRNA translation in bacteria. Cold Spring Harb. Symp. Quant. Biol., 66, 363–376. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters,D.S. and Gold,L. (1989) Selection of the initiator tRNA by Escherichia coli initiation factors. Genes Dev., 3, 1899–1912. [DOI] [PubMed] [Google Scholar]
- Jelenc P.C. and Kurland,C.G. (1979) Nucleoside triphosphate regeneration decreases the frequency of translation errors. Proc. Natl Acad. Sci. USA, 76, 3174–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi R., Pavlov,M.Y., Buckingham,R.H. and Ehrenberg,M. (1999) Novel roles for classical factors at the interface between translation termination and initiation. Mol. Cell, 3, 601–609. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C. and Woese,C.R. (1998) Universally conserved translation initiation factors. Proc. Natl Acad. Sci. USA, 95, 224–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Gualerzi,C.O. and Dahlberg,A.E. (2001) Initiation factor IF 2 binds to the α-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA, 7, 1173–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen B.S., Siwanowicz,I., Larigauderie,G., Hedegaard,J., Ito,K., Nakamura,Y., Kenney,J.M., Mortensen,K.K. and Sperling-Petersen,H.U. (2003) Characterization of mutations in the GTP-binding domain of IF2 resulting in cold-sensitive growth of Escherichia coli. J. Mol. Biol., 326, 543–551. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Pestova,T.V., Shin,B.S., Cao,C., Choi,S.K. and Dever,T.E. (2002) Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl Acad. Sci. USA, 99, 16689–16694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchin S., Putzer,H., Hershey,J.W., Cenatiempo,Y., Grunberg-Manago,M. and Laalami,S. (1999) In vitro study of two dominant inhibitory GTPase mutants of Escherichia coli translation initiation factor IF2. Direct evidence that GTP hydrolysis is necessary for factor recycling. J. Biol. Chem., 274, 6074–6079. [DOI] [PubMed] [Google Scholar]
- Marquardt D.W. (1963) An algorithm for least squares estimation of nonlinear parameters. J. Soc. Ind. Appl. Math., 11, 431–441. [Google Scholar]
- Pavlov M.Y., Freistroffer,D.V., MacDougall,J., Buckingham,R.H. and Ehrenberg,M. (1997) Fast recycling of Escherichia coli ribosomes requires both ribosome recycling factor (RRF) and release factor RF3. EMBO J., 16, 4134–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin,I.B., Lee,J.H., Choi,S.K., Dever,T.E. and Hellen,C.U. (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature, 403, 332–335. [DOI] [PubMed] [Google Scholar]
- Petrelli D., LaTeana,A., Garofalo,C., Spurio,R., Pon,C.L. and Gualerzi,C.O. (2001) Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J., 20, 4560–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pon C.L. and Gualerzi,C.O. (1986) Mechanism of translational initiation in prokaryotes. IF3 is released from ribosomes during and not before 70 S initiation complex formation. FEBS Lett., 195, 215–219. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V. (2002) Ribosome structure and the mechanism of translation. Cell, 108, 557–572. [DOI] [PubMed] [Google Scholar]
- Rodnina M.V. and Wintermeyer,W. (1995) GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl Acad. Sci. USA, 92, 1945–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V. and Wintermeyer,W. (2001) Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu. Rev. Biochem., 70, 415–435. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., Shin,B.S., Dever,T.E. and Burley,S.K. (2001) Engaging the ribosome: universal IFs of translation. Trends Biochem. Sci., 26, 705–709. [DOI] [PubMed] [Google Scholar]
- Shin B.S., Maag,D., Roll-Mecak,A., Arefin,M.S., Burley,S.K., Lorsch,J.R. and Dever,T.E. (2002) Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell, 111, 1015–1025. [DOI] [PubMed] [Google Scholar]
- Soffientini A., Lorenzetti,R., Gastaldo,L., Parlett,J.H., Spurio,R., La Teana,A. and Islam,K. (1994) Purification procedure for bacterial translational initiation factors IF2 and IF3. Protein Expr. Purif., 5, 118–124. [DOI] [PubMed] [Google Scholar]
- Tomsic J., Vitali,L.A., Daviter,T., Savelsbergh,A., Spurio,R., Striebeck,P., Wintermeyer,W., Rodnina,M.V. and Gualerzi,C.O. (2000) Late events of translation initiation in bacteria: a kinetic analysis. EMBO J., 19, 2127–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermeyer W. and Gualerzi,C. (1983) Effect of Escherichia coli initiation factors on the kinetics of N-Acphe-tRNAPhe binding to 30S ribosomal subunits. A fluorescence stopped-flow study. Biochemistry, 22, 690–694. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Buckingham,R.H. and Ehrenberg,M. (2001) A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell, 107, 115–124. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Mora,L., Buckingham,R.H. and Ehrenberg,M. (2002) Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol. Cell, 10, 789–798. [DOI] [PubMed] [Google Scholar]