Abstract
Translational dynamics of chromatin in interphase nuclei of living Swiss 3T3 and HeLa cells was studied using fluorescence microscopy and fluorescence recovery after photobleaching. Chromatin was fluorescently labeled using dihydroethidium, a membrane-permeant derivative of ethidium bromide. After labeling, a laser was used to bleach small (∼0.4 μm radius) spots in the heterochromatin and euchromatin of cells of both types. These spots were observed to persist for >1 h, implying that interphase chromatin is immobile over distance scales ⩾0.4 μm. Over very short times (<1 s), a partial fluorescence recovery within the spots was observed. This partial recovery is attributed to independent dye motion, based on comparison with results obtained using ethidium homodimer-1, which binds essentially irreversibly to nucleic acids. The immobility observed here is consistent with chromosome confinement to domains in interphase nuclei. This immobility may reflect motion-impeding steric interactions that arise in the highly concentrated nuclear milieu or outright attachment of the chromatin to underlying nuclear substructures, such as nucleoli, the nuclear lamina, or the nuclear matrix.
In recent years, models of the structure and organization of the interphase nucleus have changed dramatically. The interphase nucleus was once believed to be a largely homogeneous organelle, containing little structural specialization beyond the nucleoli (Manuelidis, 1990; Berezney et al., 1995). In contrast, the interphase nucleus is now widely believed to be inhomogeneous, with many nuclear substructures and functions localized to specific nuclear subdomains (for review see van Driel et al., 1995). In particular, a considerable body of evidence suggests that the most prominent nuclear constituent, chromatin, is organized so that each chromosome occupies its own discrete domain (Cremer et al., 1982; Lichter et al., 1988; Leitch et al., 1990; for review see van Driel et al., 1995; Eils et al., 1996).
Considerably less is known about the mobility of chromatin and other constituents of interphase nuclei than is known about their organization. Despite being poorly understood, mobility has been the subject of interest for at least two reasons. First, the mobility of nuclear constituents should reflect and influence important aspects of their organization. Indeed, in the case of interphase chromatin, mobility has been most commonly inferred indirectly from observed changes in organization and distribution (Manuelidis, 1985; Bartholdi, 1991; Ferguson and Ward, 1992; Janevski et al., 1995). Second, the mobility of nuclear constituents should reflect their function. In the case of interphase chromatin, a functional significance for mobility is strongly suggested by recent observations of changes in centromere and chromosome distribution in response to cell differentiation, transcription signals, and stage of the cell cycle (Manuelidis, 1984; Bartholdi, 1991; Ferguson and Ward, 1992; Funabiki et al., 1993; Janevski et al., 1995; Pluta et al., 1995; LaSalle and Lalande, 1996).
To date, data bearing on the mobility of chromatin in interphase nuclei are not only limited, they are also somewhat contradictory. For example, some studies suggest that chromatin is relatively immobile during interphase. Notable examples include observations of nonrandom organization of chromosomal substructures and of chromosome confinement to domains in fixed interphase nuclei (Moroi et al., 1981; Lichter et al., 1988; Manuelidis and Borden, 1988; Dyer et al., 1989; Leitch et al., 1990; Popp et al., 1990; for review see van Driel et al., 1995). Moreover, in living HeLa cells, centromeres are generally motionless during interphase (Shelby et al., 1996), and in living Drosophila embryos, chromosomes undergo late decondensation and subsequent condensation at the same sites on the nuclear envelope (Hiraoka et al., 1989). Finally, photobleaching studies of chromatin in isolated interphase nuclei indicate that chromatin reorientational mobility is highly restricted (Selvin et al., 1990). In contrast, some studies have shown that chromatin can reposition during interphase; notable examples include the three-dimensional movement of heterochromatin in nuclei of living interphase neurons (De Boni and Mintz, 1986), the coalescing and dispersing of centromeres in cultured cells in late G2 and early G1, respectively (Manuelidis, 1985), and the occasional slow movement of a centromere in living HeLa cells (Shelby et al., 1996).
Data bearing on the mobility of nonchromosomal molecules and other objects in interphase nuclei are also somewhat limited and contradictory. In situ hybridization studies have shown that RNA molecules may move nonrandomly along “tracks” in the nucleus, perhaps because their motion is constrained by an underlying nuclear matrix (Lawrence et al., 1989). Studies of the trajectories of large, naturally occurring cytoplasmic inclusions have shown that the mobility of inclusion-sized objects in the nuclei of newt pneumocytes is several hundredfold slower than in dilute solution (Alexander and Rieder, 1991). In contrast, translational FRAP experiments on fluorescently labeled dextrans (3–150 kD) have shown that translation of dextrans in the nuclei of Hepatoma cells is only about sevenfold slower than in dilute solution (Lang et al., 1986). These rather disparate results leave considerable uncertainty surrounding the rates and determinants of motion in the nucleus.
In this study, we have directly monitored the translational motion of fluorescently labeled chromatin in the nuclei of living Swiss 3T3 and HeLa cells using FRAP. These cell lines were chosen because microscopy studies of nuclear organization have frequently employed cultured cells, including Swiss 3T3 and HeLa, generating a considerable body of germane literature (e.g., Fey et al., 1986; He et al., 1990; Berezney et al., 1995; Shelby et al., 1996). Our results show that interphase euchromatin and heterochromatin are substantially immobile in these cell lines. This immobility is consistent with chromatin attachment to nuclear substructures and with chromosome confinement to discrete domains during interphase. Moreover, this immobility provides a dynamics-based foundation for the many observations of nonrandom organization of chromosomal substructures in fixed interphase cells.
Materials and Methods
Cell Culture and Chromatin Labeling
Swiss 3T3 fibroblasts and HeLa cells were grown on 25-mm-diam round glass No. 1 coverslips (Fisher Scientific Co., Pittsburgh, PA) in Dulbecco's modified Eagle medium (low glucose) (GIBCO BRL, Gaithersburg, MD) supplemented with 10% fetal calf serum. Cells were maintained at 37°C in a 10% CO2/90% air incubator and used before cells had reached confluence. Chromatin in living cells was labeled with ethidium by a 10–45 min incubation at 37°C with 2.5–10 μg/ml dihydroethidium (Molecular Probes, Inc., Eugene, OR). (The longest labeling times and highest dihydroethidium concentrations were used to prepare cells for photography.) Dihydroethidium is a membrane permeant, chemically reduced derivative of ethidium bromide that is dehydrogenated to ethidium bromide. After labeling, cells were washed three times and then maintained in a physiological buffer (Hanks' or Dulbecco's phosphate buffered saline; both from GIBCO BRL). In some cases, cells were fixed in buffer containing 4% paraformaldehyde for 10 min before labeling and then labeled with 0.5 μg/ml ethidium homodimer-1 (Molecular Probes, Inc.) or ethidium bromide. For FRAP experiments and imaging, cells were mounted in buffer in Sykes-Moore Chambers (Bellco Glass, Inc., Vineland, NJ). Most experiments were conducted at 23°C, but some were also conducted at 37°C to verify that the results remain valid at physiological temperatures.
Cell viability during and after photobleaching experiments was assayed in two ways. First, short-term viability during photobleaching experiments was demonstrated by simultaneously labeling cells for 10 min at 37°C with dihydroethidium and with 10 μM calcein acetoxymethyl (AM) ester (Molecular Probes, Inc.), a “premier indicator of cell viability” (Haugland, 1996). The latter molecule is nonfluorescent and membrane permeant but is converted into the green fluorescent molecule calcein by intracellular esterases in viable cells. After conversion, calcein is retained by cells only so long as their plasma membranes are not compromised. Thus, green cells are viable cells, and cell death can be assayed by monitoring for a loss of cytoplasmic green fluorescence. This was done visually and electronically to demonstrate short-term viability during photobleaching experiments. In addition, to demonstrate that calcein is not retained after cells are compromised, calcein-labeled cells were placed in buffer containing 0.1% Triton X-100 (Bio Rad Laboratories, Hercules, CA) and loss of cytoplasmic green fluorescence was then monitored.
Second, long-term viability and absence of DNA damage were demonstrated by verifying that cells were able to divide and proliferate after they were photobleached. To facilitate identification of cells over long periods of time, cells were grown on coverslips that had been scratched with a diamond pen. Cells were labeled with dihydroethidium and calcein AM well before they had reached confluence and were then transferred to a sterile Sykes-Moore chamber for photobleaching experiments. Cells in an easily identified region of the scratch (such as the end) were bleached and returned to the incubator after ∼1 h. Cells were then examined frequently under a dissecting microscope; individual bleached cells could thus be tracked at low cell density even if they moved slowly. Division and proliferation were monitored for ∼72 h after bleaching.
Antibody Samples
As a FRAP control, samples containing a fluorescently labeled macromolecule undergoing Brownian diffusion were prepared by diluting a rhodamine-labeled goat anti–rabbit antibody into Hanks' buffer.
FRAP Experiments
Chromatin dynamics was monitored using FRAP (Axelrod et al., 1976). In a FRAP experiment, a brief, intense pulse of laser light is used to bleach (render nonfluorescent) many of the fluorescent molecules in a small subregion of a fluorescent sample. The return of fluorescence to this subregion is then monitored by shining much attenuated laser light onto the bleached spot and monitoring the temporal dependence of the post-bleach (probe) fluorescence. The time-scale over which the fluorescence recovers is determined by the rate at which the fluorescently labeled molecules diffuse and can be used in simple systems to determine a diffusion coefficient. Failure of the fluorescence to recover indicates that the fluorescently labeled molecules are immobile.
The FRAP apparatus used in these experiments has been described previously (Yuan and Axelrod, 1995). Briefly, its major components are (a) an argon-ion laser, which is used as a high-intensity source of light; (b) an acousto–optic modulator, which is used to modulate the intensity of the light incident on the sample; (c) an inverted microscope, which is used to focus the light onto the sample and to collect the fluorescence emitted by the sample; and (d) an avalanche photodiode (“single photon counting module” [SPCM-100]; EG&G Optoelectronics, Vaudrevil, Quebec, Canada), which is used to detect the fluorescence. An aperture diaphragm placed in the image plane of the microscope minimizes detection of background and out-of-focus light. Data were collected on a 486-based PC using custom FORTRAN software and a custom-programmed counter/timer board (CTM-05; Keithley Metrabyte Instruments Inc., Cleveland, OH).
Light was focused onto the sample using a 63× (NA = 1.4) Zeiss objective (Carl Zeiss Inc., Thornwood, NY), producing an in-focus spot of radius ∼0.4 μm in the sample plane. The 514-nm (green) line of the argon-ion laser was used to excite fluorescence; excitation of ethidium with visible light, as contrasted with ultraviolet radiation, reduces the probability of radiation-induced damage to the cells. The bleaching power at the sample was typically in the range ∼4–60 mW. The ratio of the intensities of the bleach and probe beams was ∼10,000:1, ensuring insignificant bleaching during the probe phase of the experiment.
Photography
Photographs of cells were taken at defined times before and after photobleaching using Kodak TMX 400 film. Cells were illuminated for photography by inserting a defocusing lens into the optical path of the photobleaching apparatus, thereby expanding the laser beam to cover the entire field of view.
Data Analysis
Diffusion coefficients and mobile fractions were obtained by fitting photobleaching data to an equation describing photobleaching recovery in a system containing one immobile fraction and one mobile fraction diffusing in two dimensions (Axelrod et al., 1976). Complex generalizations of this equation describing diffusion in three dimensions have recently been derived (Blonk et al., 1993). These generalized equations show that the recovery time along each dimension is proportional to the square of the characteristic distance along that dimension. This implies that the fastest recovery time will be dominated by diffusion along the smallest dimension. For a focused spot, the smallest dimension is the characteristic e−2 bleached radius of the spot (0.4 μm here), rather than the corresponding e−2 axial distance over which substantial bleaching occurs (2.5 μm here). Thus, for the focused spot used in these experiments, the simpler photobleaching recovery equations describing diffusion in two dimensions can be used. In addition, our conclusion that a large fraction of interphase chromatin is immobile (diffusion coefficient [D]1 <10−13 cm2/s) is a model-independent result, which is also demonstrated visually (see Results).
Results
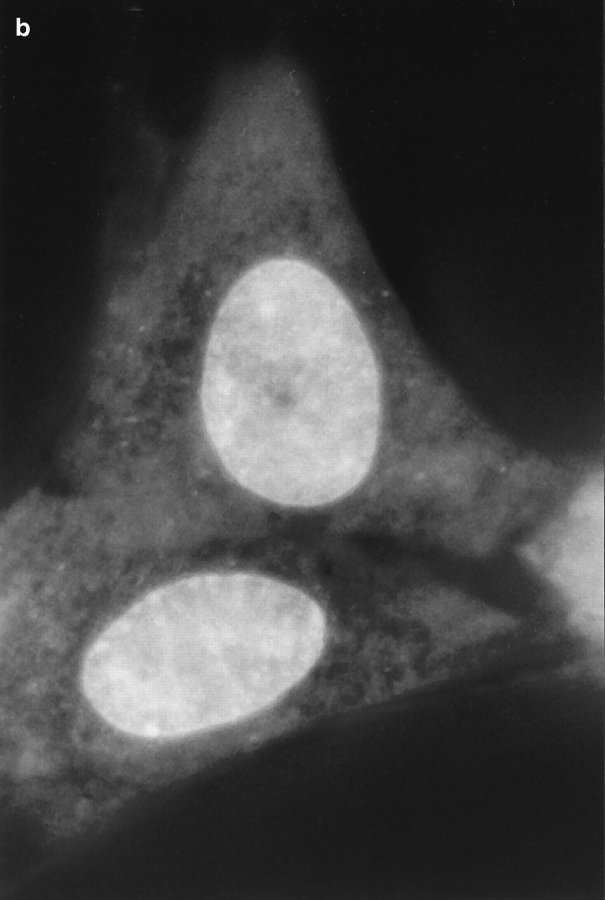
Fig. 1 shows a series of photographs of two dihydroethidium-labeled Swiss 3T3 cells taken (a) before; (b) a few min after; and (c) ∼60 min after the nucleus in the upper cell was exposed to a brief pulse of intense laser light. A several hundred millisecond bleaching pulse was sufficient to bleach the ethidium molecules in the illuminated region and create a dark spot in the euchromatin (Fig. 1, b and c). The radius of the spot was calculated to be ∼0.6 μm, using known magnification parameters and the size of the photograph. This spot persisted for at least 1 h (Fig. 1 c), indicating that a significant fraction of the interphase euchromatin in Swiss 3T3 cells is translationally immobile on the 0.6 μm distance scale. (This distance scale can be lowered to ∼0.4 μm; see below.) Moreover, the spot did not move within the nucleus, providing further evidence for chromatin immobilization during interphase. Similar results were observed after photobleaching HeLa cells and cells maintained at 37°C (data not shown).
Figure 1.
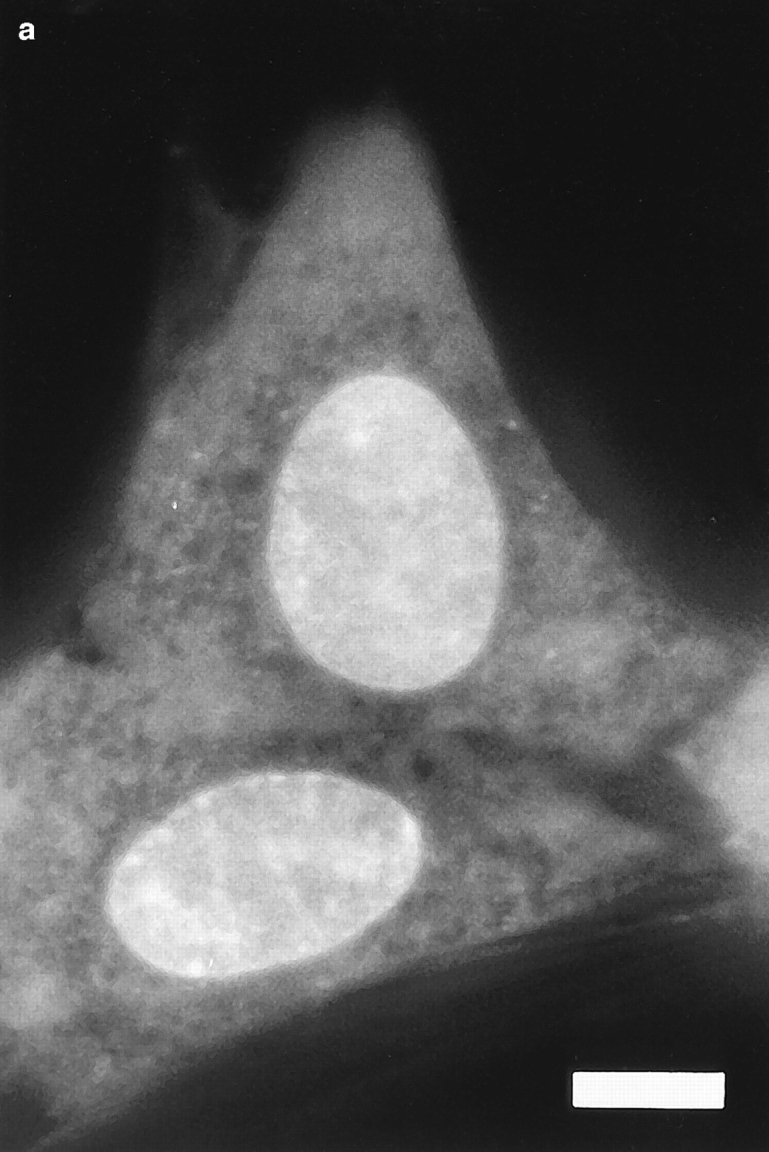
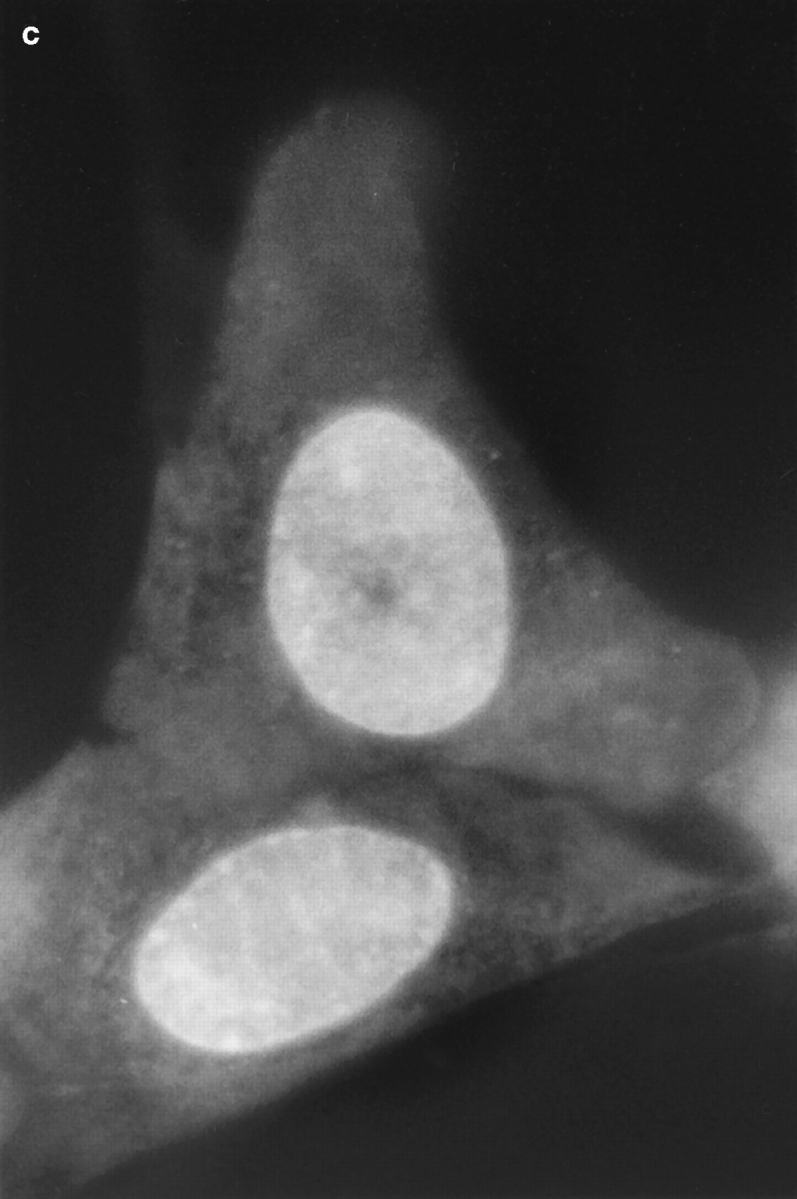
Pre- (a) and post-bleach (b and c) photographs of two dihydroethidium-labeled Swiss 3T3 cells in interphase. The ethidium stains nucleic acid in both the nucleus and cytoplasm, and reveals the distribution of euchromatin and heterochromatin within the nucleus. a shows the cells immediately before bleaching; b and c show the cells a few minutes and 1 h after bleaching the upper cell, respectively. The long-lived dark spot in the upper cell in b and c was created in an initially fluorescent region of euchromatin by illuminating the region with a 300 ms pulse of 514 nm (green) light from an argon-ion laser (Power ≈ 50 mW at the sample). The failure of this spot to fill back up with fluorescently labeled chromatin demonstrates that a large fraction of the chromatin is immobile. The focus may have shifted slightly or the cell may have moved slightly in c after the sample spent 60 min on the microscope stage. Some nonuniformity in fluorescence in the photographs may arise from interference in the illuminating light. Bar, 10 μm.
More quantitative translational mobility data were obtained by monitoring the temporal dependence of the post-bleach fluorescence using FRAP curves. Fig. 2, a and b, shows representative curves for Swiss 3T3 and HeLa cells, respectively. Both euchromatic and heterochromatic regions of the nucleus were studied. When the light used in the FRAP experiments was focused down to a small ∼0.4 μm radius spot, the recovery curves were biphasic, reflecting the presence of two fractions. One fraction was “mobile,” leading to partial recovery on a time-scale of several hundred milliseconds; the other fraction was immobile, leading to an incomplete recovery in the FRAP curves.
Figure 2.
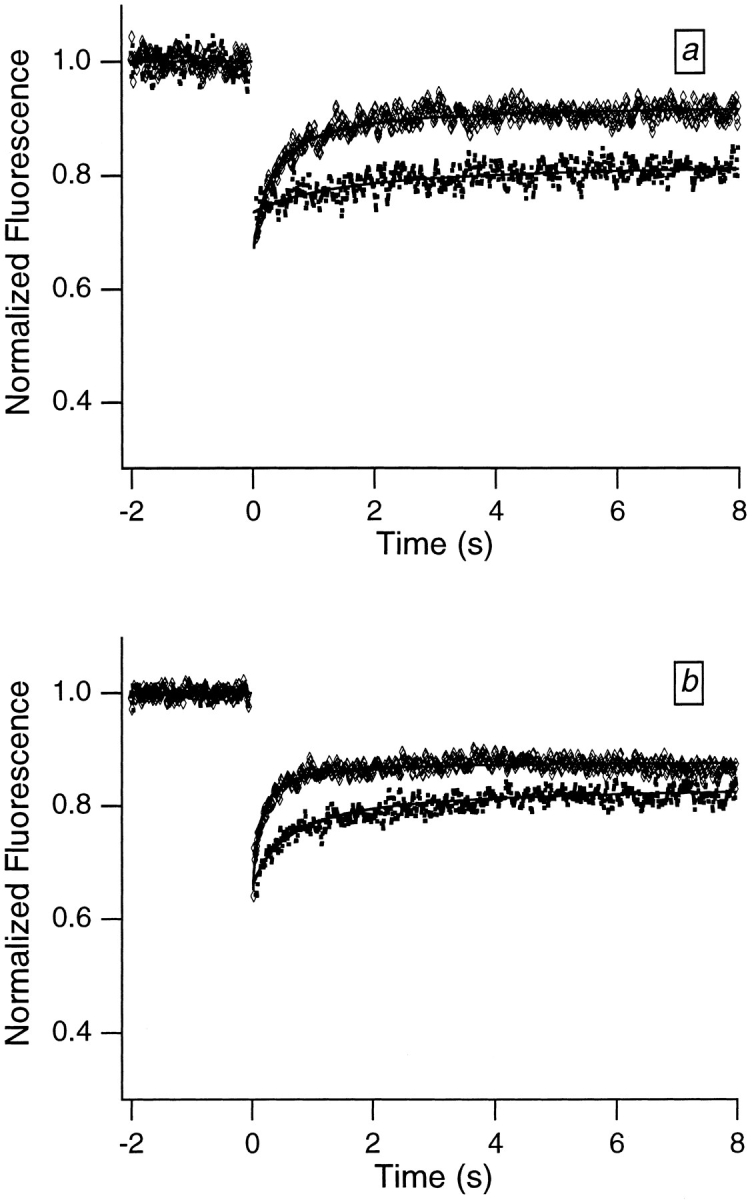
Typical FRAP curves and associated theoretical recovery curves obtained from dihydroethidium-labeled Swiss 3T3 cells (a) and HeLa cells (b). The FRAP curves were smoothed using an eleven-point fit to a fourth-order polynomial (Savitzky and Golay, 1964) to facilitate distinguishing the different recovery curves. The bleach pulse was 10 ms in duration, and data were collected in 10-ms increments. The upper (diamonds) and lower (squares) curves in each graph were obtained from relatively smaller (focused) and larger (defocused) spots, respectively. The defocused spot was obtained by translating a lens along the optical path so that the light was not focused exactly on the sample plane. Appropriate neutral density filters were inserted into the path of the bleach beam so that the bleach depths for the two curves in each graph were comparable; results did not depend on bleach depth (data not shown), because relatively shallow bleach depths were employed. Two important features are qualitatively apparent from the FRAP curves. First, a large fraction of the fluorescence fails to recover, indicating that a large fraction of the chromatin is immobile. Second, the recovery time associated with the fraction of the fluorescence that does recover increases with increasing spot size, indicating that molecular motion is the source of the initial recovery.
FRAP curves were fitted to the theoretical expression for photobleaching recovery in two dimensions, assuming one mobile and one immobile fraction. The results of the fits for dihydroethidium-labeled Swiss 3T3 cells are 27 ± 4% immobile, 73 ± 4% mobile, and D = 1.1 (± 0.5) × 10−9 cm2/s, where the quoted errors are standard deviations and typically data from ⩾20 different nuclei were averaged. The analogous results for dihydroethidium- labeled HeLa cells are 36 ± 6% immobile, 64 ± 6% mobile, and D = 2.9 (± 0.2) × 10−9 cm2/s. These results were independent of the depth of bleach. The mobile fractions obtained from the fits are fairly large but will be shown to represent a population of less tightly bound ethidium molecules, and not mobile chromatin. No more than a few percent of interphase chromatin is mobile on the ⩾0.4 μm distance scale (see Discussion).
There are two important differences between the results shown in the photographs and in the photobleaching recovery curves. First, fluorescence from out-of-focus planes inevitably appears in the photographs (Scalettar et al., 1996) but is preferentially excluded from the photobleaching recovery curves by the image–plane diaphragm. Because laser beam divergence causes the spot radius to increase in out-of-focus planes, out-of-focus pick-up may cause the spots shown in the photographs to appear larger than their in-focus size. Nevertheless, the spot radius obtained by direct measurement from the photographs closely agrees with the in-focus spot size produced by the 63× Zeiss objective.
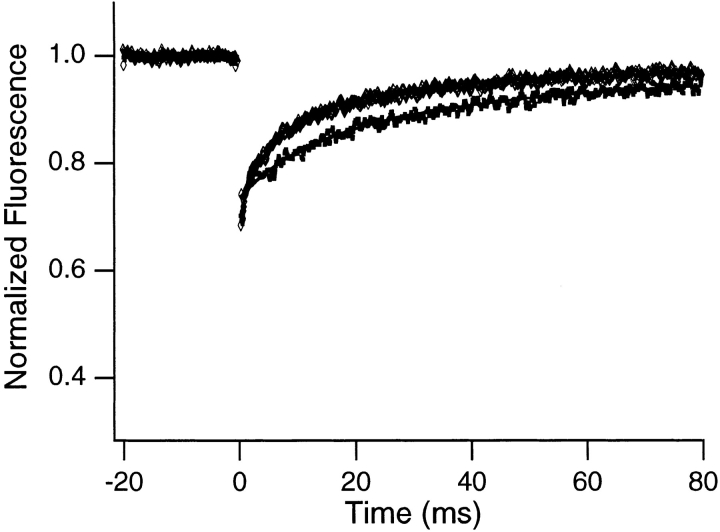
Second, the FRAP curves show an initial partial recovery of the fluorescence in the spots (mobile fractions ∼64– 73%) that is not evident from the photographs. The recovery time associated with this component was studied as a function of the size of the illuminated spot to identify its origin. If molecular motion is the source of the initial partial recovery, the recovery time should increase systematically as the size of the illuminated spot is increased. For example, for a recovery driven by Brownian diffusion, the recovery half-time is expected to increase as the square of the spot size (Axelrod et al., 1976). Fig. 3 illustrates this effect in data obtained from a fluorescently labeled antibody solution. Fig. 2, a and b, shows FRAP curves and associated fitted theoretical recovery curves obtained from two different nuclei samples for two different spot sizes. As with the antibody samples, the initial recovery time for the nuclei samples increases as spot size is increased, implying that molecular motion is the source of the initial recovery.
Figure 3.
Typical FRAP curves and associated theoretical recovery curves obtained from a rhodamine-labeled antibody undergoing Brownian diffusion in dilute solution (∼0.5 mg/ml antibody) for smaller (diamonds) and larger (squares) spots. The bleach pulse was 200 μs in duration, and data were collected in 100-μs increments. The upper curve represents an average of 1,000 experiments and the lower an average of 3,000 experiments, and each was smoothed using an 11-point fit to a fourth-order polynomial. The fluorescence recovers almost completely, indicating that all molecules are mobile. In addition, the recovery time increases with increasing spot size.
A motion-derived origin for the partial recovery is also suggested by results from two additional experiments; see Fig. 4, a and b. First, when samples are dried down and immobilized, the recovery largely disappears. Second, when hydrated samples are deoxygenated, the recovery time is essentially unaltered, which indicates that the initial recovery does not arise from reversible photobleaching (see Discussion). It is important to eliminate this latter possibility because the effects of reversible photobleaching have previously been observed in FRAP experiments on DNA and chromatin (Scalettar et al., 1990; Selvin et al., 1990).
Figure 4.
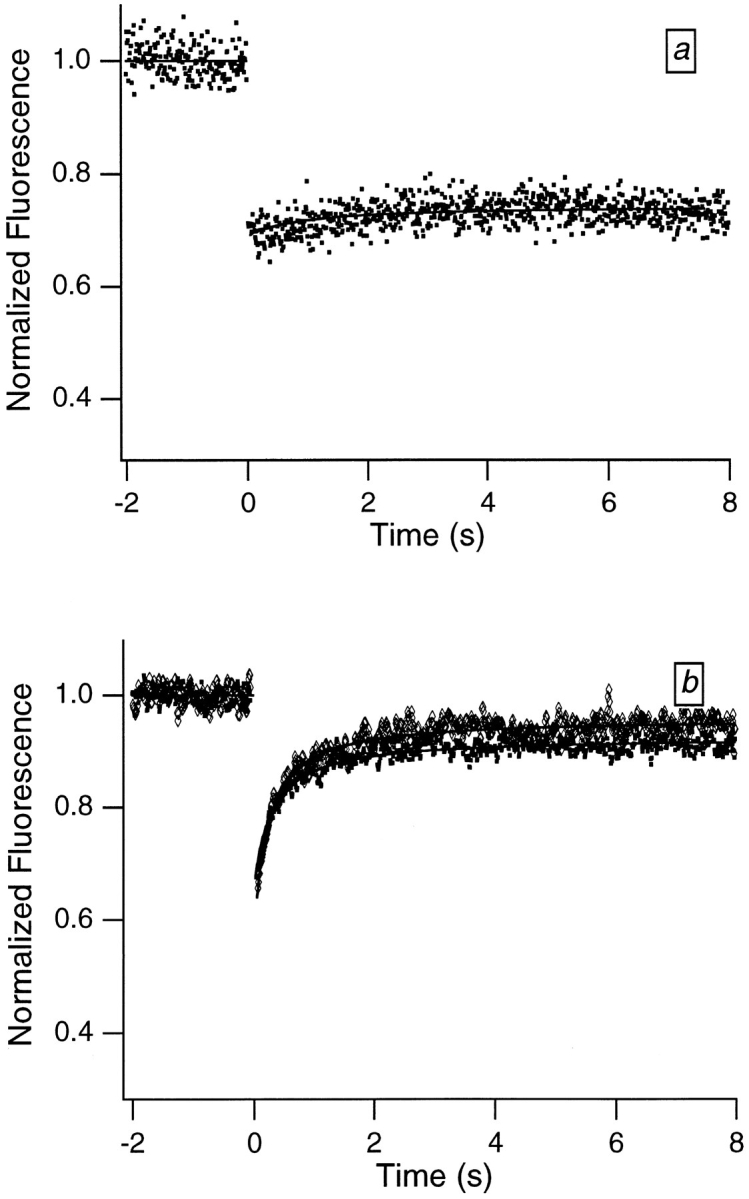
Typical FRAP curves obtained from Swiss 3T3 cells that were labeled with dihydroethidium and then (a) dried down or (b) placed in a deoxygenated buffer (diamonds) or an atmosphere-equilibrated buffer (squares). Bleach and data collection times were 10 ms. The virtual absence of the initial recovery in a sample immobilized by drying (a) supports the idea that the recovery is motion derived. The failure of the initial relaxation time to vary appreciably with oxygen concentration in the hydrated samples (b) indicates that the initial relaxation does not represent reversible photobleaching. Six times more power was required to achieve bleaching in the deoxygenated sample comparable to that achieved in the nondeoxygenated sample. This reflects the greater difficulty of doing irreversible bleaching in the absence of oxygen, and shows that the deoxygenation was successful.
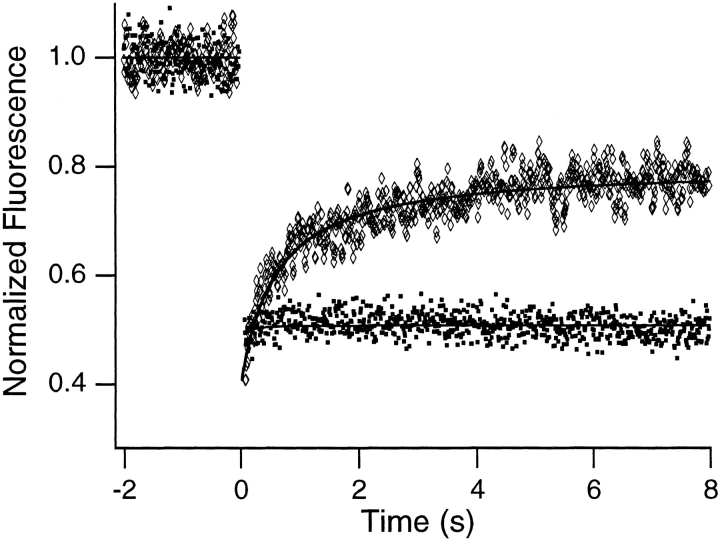
The mobile fraction could represent chromatin, RNA, or a subpopulation of the ethidium bromide that transiently comes off the chromatin. To distinguish these possibilities, FRAP control experiments were performed on cells fixed and labeled with either ethidium bromide or ethidium homodimer-1, which binds essentially irreversibly to DNA and RNA (Gaugain et al., 1978; Markovits et al., 1985; Rye and Glazer, 1995). Fig. 5 shows that the mobile fraction is present in the fixed ethidium bromide–labeled samples but is absent from the fixed ethidium homodimer-1–labeled samples. Specifically, the results of curve fits for the fixed ethidium bromide-labeled samples are 29 ± 6% immobile, 71 ± 6% mobile, and D = 7.1 (± 0.1) × 10−10 cm2/s. The results for the fixed ethidium homodimer-1– labeled samples are essentially 0% mobile, 100% immobile, and D ≈ 0 cm2/s. Fixation should largely inhibit motion of protein-containing macromolecules on the FRAP distance scale. This, together with the fact that only the nucleic acid label differs between the two control samples, strongly suggests that the mobile fraction is a subpopulation of the ethidium bromide that transiently comes off the chromatin and diffuses freely (see Discussion). The mobile chromatin fraction is thus small (or zero).
Figure 5.
Typical FRAP curves obtained from fixed ethidium bromide–labeled (diamonds) and ethidium homodimer-1–labeled (squares) Swiss 3T3 cells. Note that the initial component is absent when the homodimer is used as a label.
A potential artifact in interpretation could arise if the lack of full fluorescence recovery is due to depletion of the finite reservoir of fluorescence in the nucleus, and not to an immobile chromatin fraction. However, the photographs show that the volume of the bleached region is small, indicating that the bleach is unlikely to significantly deplete the total nuclear fluorescence. Moreover, if the “immobile fraction” simply reflected depletion of that fraction of the total nuclear fluorescence, a few bleaches would render the nucleus essentially nonfluorescent. This was not found to be the case. Finally, the long-lived spots shown in the photographs demonstrate that the nucleus does not become uniformly less fluorescent with time after the bleach.
Artifacts in interpretation could also arise if the cells were not viable or were damaged during the FRAP experiments. For this reason, cell viability during and after photobleaching experiments was tested using the two methods described under Materials and Methods. No leakage of the viability dye calcein from bleached or unbleached cells was observed during the ∼1-h period that the cells remained on the microscope stage, demonstrating viability during the experiments. In contrast, substantial leakage of calcein occurred <2 min after cells were placed in buffer containing 0.1% Triton X-100. Moreover, bleached cells in an easily identified (scratched) region of the coverslip were observed to divide and proliferate after being returned to the incubator. Both of these results strongly suggest that the FRAP data reflect interphase chromatin dynamics in living and unperturbed Swiss 3T3 and HeLa cells.
Several other results, observations, and aspects of the experimental protocol argue against light-induced artifacts in the FRAP experiments. First, the mobility results were independent of light intensity and bleaching time. Second, only a very small fraction of the nuclear volume was typically exposed to light. Finally, ethidium was excited through its visible absorption band, using green light; this is likely to result in significantly less nuclear damage than would excitation of ethidium through its UV absorption band.
Discussion
The experiments described here were directed at (a) measuring the translational mobility of chromatin, especially euchromatin, in living interphase cells using FRAP; and (b) relating the results to current ideas about chromatin function and organization during interphase.
Visually Constrained Motion
Even in the absence of photobleaching data, it is evident that there are some constraints on chromatin motion in living interphase Swiss 3T3 cells. For example, the photographs in Fig. 1 show that heterochromatin is largely immobile, since the bright heterochromatic spots remain in approximately the same place in the nucleus during long periods of observation.
The mobility of euchromatin in living Swiss 3T3 cells, and of chromatin in general in small HeLa cells, is more difficult to assay visually. Hence, we turned to photobleaching techniques to give us more complete information on chromatin mobility in interphase cells. When we bleached the fluorescence in euchromatic regions of Swiss 3T3 nuclei, the subsequent temporal dependence of the fluorescence could be monitored both photographically (Fig. 1) and through photobleaching recovery curves (Fig. 2). Both types of data show that it is possible to bleach a long-lived spot in Swiss 3T3 euchromatin, indicating that a substantial fraction of Swiss 3T3 chromatin is immobile over distance scales comparable to, or larger than, the size (∼0.25 μm) of a chromatin looped domain (Nelson et al., 1986). Similar results were obtained after photobleaching a spectrum of regions in HeLa nuclei, indicating that chromatin is similarly immobile in this human cell line. This immobility of interphase chromatin is in marked contrast to the known mobility of ethidium bromide and ethidium bromide–stained naked DNA in isolated samples (Icenogle and Elson, 1983a , b ; Scalettar et al., 1989).
Origin of Initial Recovery
Although long-lived, the bleached spots quickly refill partially, as manifest in the partial recoveries in Fig. 2. We have studied the properties of this initial recovery in detail to verify that it is attributable to molecular motion. This is essential because there are mechanisms that can lead to recovery in FRAP experiments, such as reversible photobleaching, which are not mobility derived (Velez and Axelrod, 1988; Scalettar et al., 1990; Selvin et al., 1990).
In our case, several lines of evidence suggest that the initial recovery arises from motion and not effects such as reversible bleaching. First, the relaxation time associated with mobility-derived recoveries is expected to increase with increasing size of the illuminated spot, an effect which can be seen experimentally in the data in Fig. 2. Also, the relaxation time associated with reversible recoveries is expected to be independent of spot size but dependent on oxygen concentration, effects that we do not observe experimentally (Figs. 2 and 4). (The dependence on oxygen concentration is expected because reversible recovery reflects a long-lived residence in a triplet state, and triplet lifetimes are very sensitive to oxygen concentration; Scalettar et al., 1990; Selvin et al., 1990.) Second, the relaxation time associated with reversible photobleaching of ethidium bromide when bound to DNA or chromatin is <2 ms (Scalettar et al., 1990), about two orders of magnitude shorter than that of the initial recovery observed here. Finally, the initial recovery disappears when the molecules are dried down; drying leads to molecular immobilization but does not eliminate reversible bleaching (Scalettar et al., 1988b ).
Effects of High and Low Affinity Binding of Ethidium Bromide
To identify the molecular species giving rise to the partial recovery, control experiments were performed on fixed samples that were labeled with either ethidium bromide or ethidium homodimer-1, which binds to naked DNA about 1,000 times more tightly than does ethidium bromide (Gaugain et al., 1978). Fixed samples were used because neither dye is membrane-permeant; however, fixation in formaldehyde was not expected to immobilize the dye, because formaldehyde cross-links amino groups (Baker, 1958) and because the dye was added after fixation. This expectation was borne out by the fact that the mobile fraction was still present in fixed, ethidium bromide–labeled samples and in dihydroethidium-labeled samples that were fixed after labeling.
In contrast, the mobile fraction was absent from fixed, ethidium homodimer-1–labeled samples. Because fixing and labeling with ethidium bromide leaves the mobile fraction unaltered, whereas fixing and labeling with the more tightly binding ethidium homodimer-1 eliminates the mobile fraction, we have attributed the initial recovery to independent motion of ethidium bromide arising from its weaker binding to DNA. In addition, the effects of on/ off binding of ethidium bromide have been previously observed in FRAP data (Icenogle and Elson, 1983a , b ).
Over the time-scale we have probed, only a subpopulation of the ethidium molecules is transiently moving independently of the chromatin. If all ethidium molecules were moving independently, the FRAP curves could be described by a single recovery time reflecting the fraction of ethidiums bound and the diffusion coefficient of ethidium alone (Icenogle and Elson, 1983a , b ; Kao et al., 1993). Our data show two recovery times (one approximately several hundred milliseconds and one around infinity), indicating that not all ethidiums are moving independently.
Previous studies (Lawrence and Daune, 1976) of the interaction of ethidium bromide with chromatin suggest a mechanism that could give rise to two ethidium populations in our samples. These studies identified high and low affinity ethidium binding sites, which differ by close to three orders of magnitude in binding constant (Lawrence and Daune, 1976). Moreover, the low affinity sites constitute the dominant fraction (∼82%) of the ethidium sites in native chromatin and are present in formaldehyde-fixed chromatin. It thus seems likely that the mobile species observed here is ethidium that diffuses after release from low affinity sites. This interpretation is strengthened by the fact that our mobile fractions (∼64–73%) are in reasonable agreement with the fraction of sites in native chromatin that have a low affinity for ethidium bromide.
This interpretation is further strengthened by the quantitative agreement between mobile fractions in living dihydroethidium-labeled samples and fixed ethidium bromide–labeled samples. The mobile fractions in the living dihydroethidium labeled Swiss 3T3 samples were 73 ± 4% and in the fixed ethidium bromide–labeled Swiss 3T3 samples were 71 ± 6%. Thus, the entire mobile fraction can be attributed to transiently unbound ethidium bromide, and the FRAP data show that interphase chromatin is immobile on the ⩾0.4 μm distance scale.
Comparison with Previous Studies of Chromosome Organization and Dynamics
Results obtained in a number of studies suggest that chromosome substructures can be nonrandomly organized and that chromosomes are confined to domains within interphase nuclei. From a dynamic perspective, these results suggest that there are restrictions on the translational mobility of interphase chromatin, as observed here.
Examples of nonrandom organization and confinement to domains are numerous. For example, a nuclear membrane–apposed/peripheral distribution has been observed for inactivated X chromosomes in fibroblasts, centromeres in some types of cells, and centromeres and telomeres of polytene chromosomes in Drosophila salivary gland nuclei (Moroi et al., 1981; Mathog et al., 1984; Manuelidis and Borden, 1988; Dyer et al., 1989). Similarly, chromosomal confinement to domains has been observed or deduced from in situ hybridization studies and DNA irradiation studies (Cremer et al., 1982; Hens et al., 1983; Schardin et al., 1985; Cremer et al., 1988; Lichter et al., 1988; Manuelidis and Borden, 1988; Pinkel et al., 1988; Leitch et al., 1990; Popp et al., 1990). Finally, active genes can exhibit nonrandom organization, concentrating in the nuclear periphery of mouse L and P19 embryonal carcinoma cells and near the borders of condensed chromatin in nucleated newt erythrocytes (Hutchison and Weintraub, 1985; de Graaf et al., 1990). These examples all suggest constraints on chromosome lateral mobility during interphase.
More direct evidence for constraints on chromosome lateral mobility during interphase comes from particularly pertinent recent studies of the mobility of centromeres in living HeLa cells (Shelby et al., 1996). Motion of centromeres was tracked by labeling centromeres with a fusion protein consisting of green fluorescent protein and CENP-B, a centromeric satellite DNA–binding protein. Most centromeres were found to remain motionless for a time period of up to 2 h.
In contrast, some studies have indicated that chromatin can be mobile during interphase. For example, in living interphase neurons the position of heterochromatin has been shown to change in tandem with changes in nucleoli position (DeBoni and Mintz, 1986). Similarly, centromeres have been observed to relocate in a variety of interphase cells (Manuelidis, 1985; Bartholdi, 1991; Ferguson and Ward, 1992; Janevski et al., 1995; Shelby et al., 1996), as have X chromosome centromeres in certain pathological states (Borden and Manuelidis, 1988). Finally, chromosome arms in polytene nuclei (Hochstrasser and Sedat, 1987) and neuronal cells (Manuelidis and Borden, 1988) can exhibit some randomness in configuration and position.
Our work is in basic agreement with the body of work suggesting the existence of constraints on interphase chromatin mobility. A unique feature here is that the results reflect the mobility of interphase chromatin in general, rather than the mobility of a particular chromosome or chromosome substructure. The results thus give a global view of chromatin mobility while also defining a distance scale for chromatin immobilization. In addition, the results were obtained from living cells, unlike many of those discussed above.
Chromatin in Transformed and Cultured Cells
Swiss 3T3 and HeLa cells are commonly used in studies of chromatin organization and nuclear structure. Nevertheless, there are potential differences between nuclear structure in these cell lines and nuclear structure in untransformed, uncultured cells. For example, transformed cells may have less heterochromatin and a somewhat altered nuclear matrix protein composition when compared with untransformed cells (for review see Manuelidis, 1990; Nickerson et al., 1995). Similarly, chromatin in cultured cells may partially expand to a more euchromatic state (Manuelidis, 1990). However, because these potential changes in chromatin structure would probably tend to “loosen up” chromatin and increase its mobility, our results should represent a lower bound on the immobility of interphase chromatin in cells in general.
Origin of Chromatin Immobilization on the ⩾0.4 μm Distance Scale
There are two fundamentally different types of interactions that could act alone or in combination to produce the chromatin immobilization observed here. These are (a) outright attachment of chromatin to nuclear substructures, such as nucleoli, the nuclear lamina, or the nuclear matrix; and (b) motion-impeding steric interactions inherent in a highly concentrated milieu such as the nucleus. We consider each of these possibilities, in turn.
Interphase chromatin is known to be attached to nucleoli and the nuclear envelope (Gerace and Burke, 1988; Manuelidis, 1990; Taniura et al., 1995). Such attachment will certainly hinder chromatin motion; moreover, it might completely immobilize chromatin if the attachment is extensive enough. Alternatively, less extensive attachment might also lead to immobilization if interphase chromatin is relatively rigid, and segmental motion between attachment sites thus cannot occur on the ⩾0.4 μm distance scale. Indeed, some data suggest that interphase chromatin is quite rigid (Selvin et al., 1990), making the latter mechanism more plausible.
Interphase chromatin may also be attached to the nuclear matrix (Berezney et al., 1995). In the matrix model, interphase chromatin is organized into looped domains on the order of 0.25 μm in size and containing 5–200 kb of DNA (Nelson et al., 1986; Davie, 1995). Because these tethered loops are smaller than the FRAP spot, segmental motion of the loops would not be expected to produce a FRAP recovery. Thus, the immobility observed in the FRAP experiments is also consistent with the tethering postulated in the matrix model.
Finally, chromatin will interact sterically with the broad spectrum of macromolecules found in the congested nuclear milieu (Livolant and Maestre, 1988; Selvin et al., 1990), including other chromatin. These steric interactions will also impede mobility (Lang et al., 1986; Scalettar et al., 1989); however, it has been shown experimentally (Lang et al., 1986; Scalettar et al., 1989; Hou et al., 1990) and theoretically (Scalettar et al., 1988a ; Abney et al., 1989) in a variety of systems that steric interactions usually do not lead to complete immobilization of an untethered species. For example, dextrans in nuclei (Lang et al., 1986), tracer particles in the cytoplasm of Swiss 3T3 cells (Luby-Phelps et al., 1987; Kao et al., 1993), and proteins in biological membranes that are not attached to cytoskeletal components (for review see Jacobson et al., 1987) all diffuse translationally, albeit more slowly than in dilute solution. These examples all suggest that steric interactions, in the absence of some attachment, are unlikely to produce complete molecular immobilization.
Similarly, large DNAs in concentrated (“semi-dilute”) solution are significantly more mobile than interphase chromatin. For DNA molecules with radii of gyration on the order of 0.5 μm, diffusion is somewhat slower in semi-dilute solution than in dilute solution and diffusion coefficients range from 10−9 cm2/s to 10−8 cm2/s (Thomas et al., 1980; Scalettar et al., 1989). Associated FRAP recovery times are on the order of 100 ms, assuming the spot radius is 0.4 μm. In contrast, the FRAP recovery times reported here for interphase chromatin are ⩾1 h. This and the overall immobility of the FRAP spot imply that interphase chromosomes, which occupy discrete micron-sized domains, diffuse at least four orders of magnitude more slowly than comparably sized DNA molecules.
Steric interactions alone can produce such major inhibition of mobility in highly entangled polymer solutions (Luby-Phelps et al., 1987, 1988). Significantly, individual chromosomes appear not to be entwined (for reviews see Manuelidis, 1990; Jackson, 1991; van Driel et al., 1995), suggesting that entanglement does not have a major effect on chromatin motion. Steric interactions can also produce such major inhibition of mobility if the system contains a very high concentration of immobile obstacles that block all potential paths for motion (Luby-Phelps et al., 1987, 1988; Saxton, 1993). However, if such a situation exists in interphase nuclei, it would once again suggest that immobile nuclear substructures extensively bind nuclear constituents, thereby producing a high concentration of immobile obstacles.
Conclusions
We have shown that interphase euchromatin and heterochromatin in living Swiss 3T3 fibroblasts and HeLa cells are translationally immobile over distance scales ⩾0.4 μm. This immobilization may reflect motion-impeding steric interactions that arise in the highly concentrated nuclear milieu or outright attachment of the chromatin to underlying nuclear substructures, such as nucleoli, the nuclear lamina, or the nuclear matrix. However, independent of its origin, this immobilization ensures that spatial organization of chromosomes within interphase nuclei is maintained, as postulated in current models of a highly ordered interphase nucleus.
Acknowledgments
We thank Dr. John Langmore of the University of Michigan for a critical reading of the manuscript.
This work was supported by grants CC3819 from Research Corporation (to B.A. Scalettar), BIR-9510226 from the National Science Foundation (to B.A. Scalettar), and MCB-9405298 from the National Science Foundation (to D. Axelrod).
Footnotes
1. Abbreviation used in this paper: D, diffusion coefficient.
Please address all correspondence to Bethe A. Scalettar, Department of Physics, Lewis & Clark College, Portland, OR 97219. Tel.: (503) 768-7585. Fax: (503) 768-7369. e-mail: bethe@lclark.edu
James R. Abney's current address is Kolisch Hartwell Dickinson McCormack & Heuser, PC, Patent, Trademark and Copyright Attorneys, 520 S.W. Yamhill Street, Suite 200, Portland, OR 97204.
References
- Abney JR, Scalettar BA, Owicki JC. Self diffusion of interacting membrane proteins. Biophys J. 1989;55:817–833. doi: 10.1016/S0006-3495(89)82882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SP, Rieder CL. Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol. 1991;113:805–815. doi: 10.1083/jcb.113.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, J.R. 1958. Principles of Biological Microtechnique. John Wiley & Sons Inc., New York. 357 pp.
- Bartholdi MF. Nuclear distribution of centromeres during the cell cycle of human diploid fibroblasts. J Cell Sci. 1991;99:255–263. doi: 10.1242/jcs.99.2.255. [DOI] [PubMed] [Google Scholar]
- Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int Rev Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- Blonk JCG, Don A, Van Aalst H, Birmingham JJ. Fluorescence photobleaching recovery in the confocal scanning light microscope. J Microsc (Oxf) 1993;169:363–374. [Google Scholar]
- Borden J, Manuelidis L. Movement of the X chromosome in epilepsy. Science (Wash DC) 1988;242:1687–1691. doi: 10.1126/science.3201257. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C, Baumann H, Luedtke EK, Sperling K, Teuber V, Zorn C. Rabl's model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser–UV-microbeam experiments. Hum Genet. 1982;60:46–56. doi: 10.1007/BF00281263. [DOI] [PubMed] [Google Scholar]
- Cremer T, Lichter P, Borden J, Ward DC, Manuelidis L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situhybridization using chromosome-specific library probes. Hum Genet. 1988;80:235–246. doi: 10.1007/BF01790091. [DOI] [PubMed] [Google Scholar]
- Davie JR. The nuclear matrix and the regulation of chromatin organization and function. Int Rev Cytol. 1995;162A:191–250. doi: 10.1016/s0074-7696(08)61232-2. [DOI] [PubMed] [Google Scholar]
- De Boni U, Mintz AH. Curvilinear, three-dimensional motion of chromatin domains and nucleoli in neuronal interphase cells. Science (Wash DC) 1986;234:863–866. doi: 10.1126/science.3775367. [DOI] [PubMed] [Google Scholar]
- de Graaf A, van Hemert F, Linnemans WA, Brakenhoff GJ, de Jong L, van Renswoude J, van Driel R. Three-dimensional distribution of DNase I-sensitive chromatin regions in interphase nuclei of embryonal carcinoma cells. Eur J Cell Biol. 1990;52:135–141. [PubMed] [Google Scholar]
- Dyer KA, Canfield TK, Gartler SM. Molecular cytological differentiation of active from inactive X domains in interphase: implications for X chromosome inactivation. Cyogenet Cell Genet. 1989;50:116–120. doi: 10.1159/000132736. [DOI] [PubMed] [Google Scholar]
- Eils R, Dietzel S, Bertin E, Schrock E, Speicher MR, Reid T, Robert-Nicoud M, Cremer C, Cremer T. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol. 1996;135:1427–1440. doi: 10.1083/jcb.135.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M, Ward DC. Cell cycle dependent chromosomal movement in pre-mitotic human T-lymphocyte nuclei. Chromosoma. 1992;101:557–565. doi: 10.1007/BF00660315. [DOI] [PubMed] [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)-containing and RNP- depleted matrices analyzed by sequential fractionation and resinless section microscopy. J Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle–dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugain B, Barbet J, Capelle N, Roques BP, Le Pecq J-B. DNA bifunctional intercalators. 2. Fluorescence properties and DNA binding interaction of an ethidium homodimer and an acridine ethidium heterodimer. Biochemistry. 1978;17:5078–5088. doi: 10.1021/bi00617a002. [DOI] [PubMed] [Google Scholar]
- Gerace L, Burke B. Functional organization of the nuclear envelope. Annu Rev Cell Biol. 1988;4:335–374. doi: 10.1146/annurev.cb.04.110188.002003. [DOI] [PubMed] [Google Scholar]
- Haugland, R.P. 1996. Handbook of Fluorescent Probes and Research Chemicals. Molecular Probes, Inc., Eugene, OR. 679 pp.
- He D, Nickerson JA, Penman S. Core filaments of the nuclear matrix. J Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hens L, Baumann H, Cremer T, Sutter A, Cornelis JJ, Cremer C. Immunocytochemical localization of chromatin regions UV-microirradiated in S phase or anaphase. Evidence for a territorial organization of chromosomes during cell cycle of cultured Chinese hamster cells. Exp Cell Res. 1983;149:257–269. doi: 10.1016/0014-4827(83)90397-x. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Minden JS, Swedlow JR, Sedat JW, Agard DA. Focal points for chromosome condensation and decondensation revealed by three-dimensional in vivotime-lapse microscopy. Nature (Lond) 1989;342:293–296. doi: 10.1038/342293a0. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M, Sedat JW. Three-dimensional organization of Drosophila melanogasterinterphase nuclei. II. Chromosome spatial organization and gene regulation. J Cell Biol. 1987;106:1471–1483. doi: 10.1083/jcb.104.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Lanni F, Luby-Phelps K. Tracer diffusion in F-actin and ficoll mixtures: toward a model for cytoplasm. Biophys J. 1990;58:31–43. doi: 10.1016/S0006-3495(90)82351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison N, Weintraub H. Localization of DNase I-sensitive sequences to specific regions of interphase nuclei. Cell. 1985;43:471–482. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- Icenogle RD, Elson EL. Fluorescence correlation spectroscopy and photobleaching recovery of multiple binding reactions. I. Theory and FCS measurements. Biopolymers. 1983a;22:1919–1948. doi: 10.1002/bip.360220808. [DOI] [PubMed] [Google Scholar]
- Icenogle RD, Elson EL. Fluorescence correlation spectroscopy and photobleaching recovery of multiple binding reactions. II. FPR and FCS measurements at low and high DNA concentrations. Biopolymers. 1983b;22:1949–1966. doi: 10.1002/bip.360220809. [DOI] [PubMed] [Google Scholar]
- Jackson DA. Structure–function relationships in eukaryotic nuclei. BioEssays. 1991;13:1–10. doi: 10.1002/bies.950130102. [DOI] [PubMed] [Google Scholar]
- Jacobson K, Ishihara A, Inman R. Lateral diffusion of proteins in membranes. Annu Rev Physiol. 1987;49:163–175. doi: 10.1146/annurev.ph.49.030187.001115. [DOI] [PubMed] [Google Scholar]
- Janevski J, Park PC, De Boni U. Organization of centromeric domains in hepatocyte nuclei: rearrangement associated with de novo activation of the vitellogenin gene family in Xenopus laevis. . Exp Cell Res. 1995;217:227–239. doi: 10.1006/excr.1995.1082. [DOI] [PubMed] [Google Scholar]
- Kao HP, Abney JR, Verkman AS. Determinants of the translational mobility of a small solute in cell cytoplasm. J Cell Biol. 1993;120:175–184. doi: 10.1083/jcb.120.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang I, Scholz M, Peters R. Molecular mobility and nucleocytoplasmic flux in hepatoma cells. J Cell Biol. 1986;102:1183–1190. doi: 10.1083/jcb.102.4.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Lalande M. Homologous association of oppositely imprinted chromosomal domains. Science (Wash DC) 1996;272:725–728. doi: 10.1126/science.272.5262.725. [DOI] [PubMed] [Google Scholar]
- Lawrence JJ, Daune M. Ethidium bromide as a probe of conformational heterogeneity of DNA in chromatin. The role of histone H1 . Biochemistry. 1976;15:3301–3307. doi: 10.1021/bi00660a021. [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH, Marselle LM. Highly localized tracks of specific transcripts within interphase nuclei visualized by in situhybridization. Cell. 1989;57:493–502. doi: 10.1016/0092-8674(89)90924-0. [DOI] [PubMed] [Google Scholar]
- Leitch AR, Mosgoller W, Schwarzacher T, Bennett MD, Heslop-Harrison JS. Genomic in situhybridization to sectioned nuclei shows chromosome domains in grass hybrids. J Cell Sci. 1990;95:335–341. doi: 10.1242/jcs.95.3.335. [DOI] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC. Delineation of individual human chromosomes in metaphase and interphase cells by in situhybridization using recombinant DNA libraries. Hum Genet. 1988;80:224–234. doi: 10.1007/BF01790090. [DOI] [PubMed] [Google Scholar]
- Livolant F, Maestre MF. Circular dichroism microscopy of compact forms of DNA and chromatin in vivo and in vitro: cholesteric liquid-crystalline phases of DNA and single dinoflagellate nuclei. Biochemistry. 1988;27:3056–3068. doi: 10.1021/bi00408a058. [DOI] [PubMed] [Google Scholar]
- Luby-Phelps K, Castle PE, Taylor DL, Lanni F. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc Natl Acad Sci USA. 1987;84:4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luby-Phelps K, Lanni F, Taylor DL. The submicroscopic properties of cytoplasm as a determinant of cellular function. Annu Rev Biophys Biophys Chem. 1988;17:369–396. doi: 10.1146/annurev.bb.17.060188.002101. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Different central nervous system cell types display distinct and nonrandom arrangements of satellite DNA sequences. Proc Natl Acad Sci USA. 1984;81:3123–3127. doi: 10.1073/pnas.81.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis L. Indications of centromere movement during interphase and differentiation. Ann NY Acad Sci. 1985;450:205–221. doi: 10.1111/j.1749-6632.1985.tb21494.x. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A view of interphase chromosomes. Science (Wash DC) 1990;250:1533–1540. doi: 10.1126/science.2274784. [DOI] [PubMed] [Google Scholar]
- Manuelidis L, Borden J. Reproducible compartmentalization of individual chromosome domains in human CNS cells revealed by in situhybridization and three-dimensional reconstruction. Chromosoma (Berl) 1988;96:397–410. doi: 10.1007/BF00303033. [DOI] [PubMed] [Google Scholar]
- Markovits J, Ramstein J, Roques BP, Le Pecq JB. Effect of B–Z transition and nucleic acid structure on the conformational dynamics of bound ethidium dimer measured by hydrogen deuterium exchange kinetics. Nucleic Acids Res. 1985;13:3773–3788. doi: 10.1093/nar/13.10.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathog D, Hochstrasser M, Gruenbaum Y, Saumweber H, Sedat J. Characteristic folding pattern of polytene chromosomes in Drosophilasalivary gland nuclei. Nature (Lond) 1984;308:414–421. doi: 10.1038/308414a0. [DOI] [PubMed] [Google Scholar]
- Moroi Y, Hartman AL, Nakane PK, Tan EM. Distribution of kinetochore (centromere) antigen in mammalian cell nuclei. J Cell Biol. 1981;90:254–259. doi: 10.1083/jcb.90.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WG, Pienta KJ, Barrack ER, Coffey DS. The role of the nuclear matrix in the organization and function of DNA. Annu Rev Biophys Biophys Chem. 1986;15:457–475. doi: 10.1146/annurev.bb.15.060186.002325. [DOI] [PubMed] [Google Scholar]
- Nickerson JA, Blencowe BJ, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J. Fluorescence in situhybridization with human chromosome– specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci USA. 1988;85:9138–9142. doi: 10.1073/pnas.85.23.9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science (Wash DC) 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Popp S, Scholl HP, Loos P, Jauch A, Stelzer E, Cremer C, Cremer T. Distribution of chromosome 18 and X centric heterochromatin in the interphase nucleus of cultured human cells. Exp Cell Res. 1990;189:1–12. doi: 10.1016/0014-4827(90)90249-a. [DOI] [PubMed] [Google Scholar]
- Rye HS, Glazer AN. Interaction of dimeric intercalating dyes with single-stranded DNA. Nucleic Acids Res. 1995;23:1215–1222. doi: 10.1093/nar/23.7.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitzky A, Golay MJE. Smoothing and differentiation of data by simplified least squares procedures. Anal Chem. 1964;36:1627–1639. [Google Scholar]
- Saxton MJ. Lateral diffusion in an archipelago. Dependence on tracer size. Biophys J. 1993;64:1053–1062. doi: 10.1016/S0006-3495(93)81471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalettar BA, Abney JR, Owicki JC. Theoretical comparison of the self diffusion and mutual diffusion of interacting membrane proteins. Proc Natl Acad Sci USA. 1988a;85:6726–6730. doi: 10.1073/pnas.85.18.6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalettar BA, Selvin PR, Axelrod D, Hearst JE, Klein MP. A fluorescence photobleaching study of the microsecond reorientational motions of DNA. Biophys J. 1988b;53:215–226. doi: 10.1016/S0006-3495(88)83083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalettar BA, Hearst JE, Klein MP. FRAP and FCS studies of self-diffusion and mutual diffusion in entangled DNA solutions. Macromolecules. 1989;22:4550–4559. [Google Scholar]
- Scalettar BA, Selvin PR, Axelrod D, Klein MP, Hearst JE. A polarized photobleaching study of DNA reorientation in agarose gels. Biochemistry. 1990;29:4790–4798. doi: 10.1021/bi00472a007. [DOI] [PubMed] [Google Scholar]
- Scalettar BA, Swedlow JR, Sedat JW, Agard DA. Dispersion, aberration and deconvolution in multi-wavelength fluorescence images. J Microsc (Oxf) 1996;182:50–60. doi: 10.1046/j.1365-2818.1996.122402.x. [DOI] [PubMed] [Google Scholar]
- Schardin M, Cremer T, Hager HD, Lang M. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum Genet. 1985;71:281–287. doi: 10.1007/BF00388452. [DOI] [PubMed] [Google Scholar]
- Selvin PR, Scalettar BA, Langmore JP, Axelrod D, Klein MP, Hearst JE. A polarized photobleaching study of chromatin reorientation in intact nuclei. J Mol Biol. 1990;214:911–922. doi: 10.1016/0022-2836(90)90345-M. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of α-satellite DNA domains visualized in situin living human cells. J Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniura H, Glass C, Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JC, Allison SA, Schurr JM. Dynamic light scattering studies of internal motions in DNA. II. Clean viral DNAs. Biopolymers. 1980;19:1451–1474. doi: 10.1002/bip.1980.360190804. [DOI] [PubMed] [Google Scholar]
- van Driel R, Wansink DG, van Steensel B, Grande MA, Schul W, de Jong L. Nuclear domains and the nuclear matrix. Int Rev Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- Velez M, Axelrod D. Polarized fluorescence photobleaching recovery for measuring rotational diffusion in solutions and membranes. Biophys J. 1988;53:575–591. doi: 10.1016/S0006-3495(88)83137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Axelrod D. Subnanosecond polarized fluorescence photobleaching: rotational diffusion of acetylcholine receptors on developing muscle cells. Biophys J. 1995;69:690–700. doi: 10.1016/S0006-3495(95)79944-1. [DOI] [PMC free article] [PubMed] [Google Scholar]