Abstract
In mammalian cells, extracellular signals can regulate the delivery of particular proteins to the plasma membrane. We have discovered a novel example of regulated protein sorting in the late secretory pathway of Saccharomyces cerevisiae. In yeast cells grown on either ammonia or urea medium, the general amino acid permease (Gap1p) is transported from the Golgi complex to the plasma membrane, whereas, in cells grown on glutamate medium, Gap1p is transported from the Golgi to the vacuole. We have also found that sorting of Gap1p in the Golgi is controlled by SEC13, a gene previously shown to encode a component of the COPII vesicle coat. In sec13 mutants grown on ammonia, Gap1p is transported from the Golgi to the vacuole, instead of to the plasma membrane. Deletion of PEP12, a gene required for vesicular transport from the Golgi to the prevacuolar compartment, counteracts the effect of the sec13 mutation and partially restores Gap1p transport to the plasma membrane. Together, these studies demonstrate that both a nitrogen-sensing mechanism and Sec13p control Gap1p transport from the Golgi to the plasma membrane.
The trans-Golgi is a major branch point in the secretory pathway, where proteins directed to the cell surface are segregated from proteins that are destined for the lysosome. In some specialized mammalian cells, particular cargo proteins are segregated at the trans-Golgi into compartments that allow regulated delivery to the plasma membrane. For example, in pancreatic secretory cells and islet cells, secreted enzymes and hormones are packaged into regulated secretory vesicles known as secretory granules (Burgess and Kelly, 1987). In adipose cells, the GLUT-4 glucose transporter is segregated into a specialized post-Golgi compartment that allows its delivery to the cell surface to be regulated by insulin (Haney and Mueckler, 1994). Similarly, in renal duct cells, water channels are delivered from a post-Golgi compartment to the plasma membrane in response to vasopressin (Nielsen et al., 1995), and in stomach epithelial cells the H+–K+ ATPase is delivered to the plasma membrane in response to gastrin (Urushidani and Forte, 1987). These regulated trafficking processes allow the protein composition of the plasma membrane and the release of proteins into the extracellular environment to be modulated in response to extracellular signals. An understanding of how different cargo proteins are segregated in the trans-Golgi and how extracellular signals control vesicle function is actively being sought, but the underlying molecular mechanisms remain poorly understood.
The trans-Golgi compartment is also a major branch point in the secretory pathway of Saccharomyces cerevisiae. At least three different kinds of vesicles bud from the yeast trans-Golgi: one vesicle type carries soluble and membrane vacuolar proteins to the pre-vacuolar compartment (Horazadovsky et al., 1995; Conibear and Stevens, 1995), and two separable vesicle types carry different sets of proteins to the plasma membrane (Harsay and Bretscher, 1995). Transport by all three post-Golgi mechanisms appears to be constitutive, and regulation of protein delivery to the plasma membrane has not been observed in yeast. As a possible subject for regulated transport in yeast, we have explored the mechanism by which the activity of certain amino acid permeases is regulated by the nitrogen source.
The amino acid permeases of S. cerevisiae constitute a family of >20 related proteins that can be divided into two classes according to their regulation and function (André, 1995). The constitutive permeases are high affinity permeases that transport either specific amino acids, or a set of chemically related amino acids (Horak, 1986; Grenson, 1992). The best characterized permeases in this class include the histidine permease, Hip1p (Tanaka and Fink, 1985); the lysine permease, Lyp1p (Syrchova and Chevallier, 1993); and the basic amino acid permease, Can1p (Hoffman, 1985). This class of permease is thought to transport amino acids primarily for use in protein synthesis. The regulated permeases include Gap1p, which can transport all naturally occurring amino acids (Grenson et al., 1970; Jauniaux and Grenson, 1990); and Put4p, which transports proline (Lasko and Brandriss, 1981; Vandenbol et al., 1989). These permeases have a high capacity and are coordinately induced by growth on poor nitrogen sources, implying that they function in the acquisition of amino acids for use by the cell as a source of nitrogen (Courchesne and Magasanik, 1993; Grenson, 1992).
Gap1p is thought to play a pivotal role in the control of nitrogen metabolism since the amino acids that Gap1p transports are both the substrates for, and the inducers of, amino acid utilization pathways (Magasanik, 1992). Accordingly, Gap1p activity is strictly regulated in response to the type of nitrogen source that is available in the growth medium. The GLN3, NIL1, and URE2 genes coordinately control transcription of a number of nitrogen- responsive genes. Under the control of these transcription factors, GAP1 mRNA is expressed in cells grown on urea or glutamate, but not in cells grown on glutamine as a nitrogen source (Stanbrough and Magasanik, 1995). Posttranslational regulation of Gap1p is also evident when Gap1 protein levels and permease activity are compared in cells grown on different nitrogen sources. For example, Gap1 protein levels are similar in cells grown on either urea or glutamate, but Gap1p activity in cells grown on urea is 100-fold higher than in cells grown on glutamate (Stanbrough and Magasanik, 1995).
Here we follow the intracellular location of Gap1p in cells grown on different nitrogen sources and find that in glutamate-grown cells, Gap1p is transported to the vacuole rather than to the plasma membrane. Thus, one of the underlying mechanisms in posttranslational regulation of Gap1p is regulated protein sorting at the trans-Golgi by a process that appears to be similar to regulated delivery of proteins to the plasma membrane in mammalian cells. In addition, we find that certain alleles of SEC13 selectively disrupt the transport of nitrogen-regulated permeases to the cell surface, but do not affect the constitutive permeases. Together, our studies indicate that there is a specialized transport step for delivery of Gap1p and Put4p to the plasma membrane that requires Sec13p function and is regulated by nitrogen source.
Materials and Methods
Strains, Plasmids, and Media
The yeast strains listed in Table I are all in the S288C genetic background that carries a loss of function allele at the per1 locus and therefore expresses high levels of Gap1p and Put4p permeases when ammonia is used as a nitrogen source (Courchesne and Magasanik, 1983). pPL257 is the vector pRS316 with the GAP1 gene that has been tagged with the nine– amino acid hemagglutinin 1 (HA1)1 epitope inserted between amino acids 62 and 63 of Gap1p (Ljungdahl et al., 1992). pMS29 is the pBL101 vector with a GAP1–lacZ fusion at amino acid 53 of Gap1p (Stanbrough and Magasanik, 1995). Minimal media are composed of Difco yeast nitrogen base without amino acids and without ammonium sulfate (Difco Laboratories Inc., Detroit, MI), 2% glucose, and a nitrogen source: 0.1% glutamate, 0.1% glutamine, 0.2% urea, or 0.5% ammonium sulfate. For SFD (nitrogen-free media) no nitrogen source was added. Minimal media were adjusted to pH 4.0 with either NaOH or HCl.
Table I.
Saccharomyces cerevisiae Strains
| Strain | Genotype | Reference or source | ||
|---|---|---|---|---|
| PLY134 | MATa ade2 ade3 leu2-3, 112 lys2Δ201 ura3-52 | Ljungdahl et al., 1992 | ||
| gap1Δ1::LEU2 shr3Δ1::URA3 [pPL257] | ||||
| CKY443 | MATa (prototroph) | |||
| CKY444 | MATα sec13-1 | |||
| CKY445 | MATα leu2-3, 112 gap1Δ1::LEU2 | |||
| CKY455 | MATa sec13-1 pep12Δ::TRP1 | |||
| CKY456 | MATa sec13-1 ends3-1 | |||
| CKY457 | MATa sec13-1 pep4Δ::LEU2 leu2-3, 112 | |||
| CKY465 | MATα leu2-3, 112 ura3-52 gap1Δ1::LEU2 [pPL257] | |||
| CKY466 | MATα sec13-1 leu2-3, 112 ura3-52 gap1Δ1::LEU2 [pPL257] | |||
| CKY467 | MATα sec13-1 pep4Δ::LEU2 leu2-3, 112 ura3-52 [pPL257] | |||
| CKY468 | MATα sec13-1 pep12::TRP1 leu2-3, 112 ura3-52 gap1::LEU2 [pPL257] | |||
| CKY517 | MATa sec6-4 | |||
| CKY518 | MATα sec6-4 leu2-3, 112 ura3-52 [pPL257; pNV31(CEN LEU2 TPI1-SUC2] | |||
| CKY519 | MATa pep4Δ::LEU2 leu2-3, 112 ura3-52 [pPL257] | |||
| CKY520 | MATα ura3-52 [pPL257] | |||
| CKY521 | MATα sec13-1 ura3-52 [pPL257] | |||
| CKY522 | MATα ura3-52 | |||
| CKY523 | MATα sec13-1 ura3-52 |
All strains are from this study where not otherwise indicated.
Assays for Amino Acid Uptake
Strains to be assayed were cultured to 8 × 106 cells/ml in minimal medium. Cells collected by filtration on a 0.45-μM nitrocellulose filter (Millipore Corp., Bedford, MA) were suspended in SFD medium. Cell density was adjusted according to the amino acid being assayed so that the rate of uptake was linear during the assay. The assay was initiated by addition of a 14C-radiolabeled amino acid to a final concentration of 4 μM and 100-μl samples were taken at five intervals, 30 s apart. For some citrulline uptake assays it was necessary to shorten this interval to keep the assay linear. Cells were collected by filtration through a No. 30 glass fiber filter (Schleicher & Schuell Inc., Keene, NH), and were washed free of labeled amino acid with 30 ml of chilled water. The filters were dried and intracellular labeled amino acid was determined by liquid scintillation counting. The activity was then determined from the velocity of uptake for at least two independent samples. The specific activities of the amino acids used were 312 mCi/ mmol for histidine, 125 mCi/mmol for lysine, leucine, and proline, 57.7 mCi/mmol for citrulline, and 51.8 mCi/mmol for tryptophan. Labeled amino acids were obtained from: Amersham Corp., Arlington Heights, IL; Dupont-NEN, Boston, MA; and Moravek Biochemicals, Inc., Brea, CA.
For transfer of cells from one nitrogen source to another, cells were grown in minimal glutamate or glutamine medium to a density of 8 × 106 cells/ml. Cells were harvested on 0.45 μM nitrocellulose filter and suspended in urea medium, or urea medium with 1.5 μg/ml cycloheximide. Citrulline uptake assays were then performed at 0, 5, 10, 15, 20, and 30 min after the transfer. For experiments with the sec6 mutants, glutamate grown cultures were shifted to 36°C for 10 min before the medium transfer.
Membrane Protein Preparation, Western Blotting, and Antibodies
Cultures were grown to exponential phase and 4 × 108 cells were collected and washed once in STE10 (10% [wt/wt] sucrose, 10 mM tris-HCl [pH 7.6], 10 mM EDTA). Cell pellets were resuspended in 20 μl of STE10 with protease inhibitors (1 mM PMSF, 0.5 mg/ml leupeptin, 0.7 mg/ml pepstatin; Boehringer Mannheim Biochemicals, Indianapolis, IN) and lysed by agitation with glass beads. Cell lysates were diluted to 1 ml with STE10 and centrifuged for 2 min at 300 g. The supernatants were then centrifuged at 150,000 g for 1 h, membrane pellets were resuspended in 100 μl STE10, and total protein was determined using the DC protein assay (Bio Rad Laboratories, Hercules, CA). Membranes were solubilized by adding 10 μl of membranes to 90 μl 2× sample buffer (4% SDS, 0.1 M tris-HCl [pH 6.8], 4 mM EDTA, 20% glycerol, 2% 2-mercaptoethanol, 0.02% bromophenol blue).
Proteins were resolved by SDS-PAGE (Laemmli, 1970). Western blotting was performed using standard methods (Harlow and Lane, 1988). Antibodies were used as follows: anti-HA antibody, 12CA5 (BAbCO, Richmond, CA), at 1:1,000 dilution; rabbit anti–Sec61p at 1:1,000 dilution; rabbit anti–Pma1p (gift of A. Chang, Albert Einstein College of Medicine, New York) at 1:500 dilution; and HRP-coupled sheep anti–rabbit and HRP-coupled sheep anti–mouse (both Amersham Corp.) at 1:10,000 dilution. Blots were developed by chemiluminescence (ECL kit; Amersham Corp.).
Cell Fractionation and Equilibrium Density Centrifugation
Cell organelles were fractionated on equilibrium density gradients essentially as described by Kölling and Hollenberg (1994). Exponential cultures were arrested by the addition of sodium azide and potassium fluoride to 10 mM. Cells were washed once in 10 mM sodium azide, 10 mM potassium fluoride, 5 mM tris-HCl (pH 7.6). For fractionation of membranes in the absence of Mg2+, cells were pelleted and resuspended in 0.5 ml STE10 with protease inhibitors. Glass beads were added to the meniscus and cells were disrupted by rigorous vortexing for 2 min. After the addition of 1 ml STE10, the extract was transferred to a fresh tube and cleared of unbroken cells and cell walls by centrifugation at 300 g for 2 min. 0.3 ml of cleared extracts was layered on top of a 5-ml, 20–60% linear sucrose gradient made up in 10 mM tris-HCl (pH 7.6), 10 mM EDTA. For fractionation of membranes in the presence of Mg2+, all solutions contained either 1 or 2 mM MgCl2 as indicated, and no EDTA. Samples were centrifuged for 18 h at 100,000 g in a SW50.1 rotor (Beckman Instrs., Fullerton, CA). 300 μl fractions were collected from the top of the gradient using a micropipette. Fractions were diluted to 1 ml with water and protein was precipitated by the addition of 100 μl of 0.15% deoxycholate and 100 μl of 72% TCA. After a 1-h incubation on ice, proteins were pelleted by centrifugation, and reprecipitated with cold acetone. Protein pellets were dried and suspended in 2× sample buffer. Gap1p-HA, Pma1p, and Sec61p were resolved by SDS-PAGE and were detected by Western blotting. The quantity of each protein in cell fractions was determined by densitometry using an Ultroscan (model 2202; LKB Instruments, Inc., Gaithersburg, MD). GDPase activity was assayed in gradient fractions before protein precipitation using standard methods (Abeijon et al., 1989).
Radiolabeling and Immunoprecipitation
Gap1p-HA stability was examined by pulse–chase experiments. Cells for labeling were grown in minimal medium to a density of 8 × 106 cells/ml in glutamate or 1.6 × 107 cells/ml in ammonia. Cells were collected by filtration and resuspended in 1.6 ml of growth medium at a density of 1.7 × 108 cells/ml. Labeling of cellular protein was initiated by the addition of 300 μCi of [35S]methionine and [35S]cysteine (Dupont-NEN). After 7 min, the chase was initiated by diluting labeled cells into 26 ml of growth medium with 5.9 μM methionine and 5.5 μM cysteine. In experiments with sec13-1 mutants, the strains were labeled for 10 min and chased with 268 μM methionine and 250 μM cysteine because of reduced uptake of amino acids in sec13-1 mutants. 5-ml samples were removed from labeled cultures and added to 100 μl of 50× stop mix (0.5 M sodium azide, 0.5 M potassium fluoride, 50 mM tris-HCl [pH 7.4]). Cells were lysed in 20 μl of STE10 with protease inhibitors by agitation with glass beads. Further steps were essentially as described by Silve et al. (1991). Extracts were diluted with 1 ml of IP buffer (50 mM tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 1% Triton X-100), containing 0.5% fixed Staphylococcus aureus cells, incubated for 10 min, and cleared by centrifugation. 900 μl of the cleared supernatant was added to 100 μl of IP buffer containing 1 μl of the 12CA5 monoclonal antibody. Samples were incubated at room temperature for 1 h after which 100 μl of 5% protein A–Sepharose beads (Pharmacia LKB Biotechnology Inc., Piscataway, NJ) were added and the incubation was continued for an additional hour. Protein A–Sepharose pellets were washed three times with IP buffer and once with IP buffer without Triton X-100. Proteins were solubilized in 20 μl sample buffer and resolved by SDS-PAGE. Gap1p-HA was imaged by fluorography with a phosphoimager (Molecular Dynamics Inc., Sunnyvale CA). The amount of Gap1p-HA was determined using ImageQuaNT software and standard protocols (Molecular Dynamics).
The kinetics of carboxypeptidase Y (CPY) maturation was evaluated by pulse–chase experiments as previously described (Gimeno et al., 1995).
β-Galactosidase Assay
CKY522 (ura3-52) and CKY523 (sec13-1 ura3-52) were transformed with the plasmid pMS29 carrying a gap1–lacZ fusion. Fresh transformants were grown overnight in ammonium medium to a cell density of 1.6 × 107 cells/ml. Cells were permeabilized by chloroform–SDS treatment and β-galactosidase activity was assayed as described by Guarente (1983). Enzymatic activities were normalized to OD600. Five independent transformants were assayed for each determination.
Immunofluorescence Microscopy
Cellular Gap1p-HA was visualized by indirect immunofluorescence using standard methods for fixation and immunostaining (Pringle et al., 1991). Strains were grown in SD medium to a density of 1.6 × 107 cells/ml. Cells were fixed in 3.7% formaldehyde for 1 h at room temperature, washed in 100 mM potassium phosphate (pH 7.2), and spheroplasted by lyticase treatment at 30°C for 30 min (100 U lyticase in 0.1 M potassium phosphate, pH 7.2; 28 mM β-mercaptoethanol). Primary and secondary antibody incubations were for 1 h at 25°C. The 12CA5 monoclonal antibody was used at a 1:5,000 dilution (BAbCO). The anti-Kar2p polyclonal antibody (a gift from M. Rose, Princeton University, Princeton, NJ) was used at a dilution of 1:1,000. FITC-coupled goat anti–mouse IgG (Boehringer Mannheim Biochemicals) and rhodamine-coupled donkey anti–rabbit IgG (Amersham Corp.) were used at a 1:200 dilution. Samples were viewed with a Zeiss axioscope equipped for epifluorescence (Carl Zeiss, Thornwood, NY). Images were recorded with Kodak T-Max 400 film (Eastman Kodak Co., Rochester NY).
Results
Regulation of Amino Acid Permeases by Growth on Glutamate
We evaluated the effect of the nitrogen source on Gap1p regulation in our wild-type genetic background by measuring both Gap1 protein levels and Gap1p activity in cells grown on different nitrogen sources. Gap1 protein was measured using a strain expressing Gap1p fused to the HA1 epitope of influenza hemagglutinin (Ljungdahl et al., 1992). High levels of Gap1p-HA were detected in cells grown on ammonia or urea as the sole nitrogen source, somewhat lower levels were detected in cells grown on glutamate, and no Gap1p-HA was detected in cells grown on glutamine (Fig. 1 A). Gap1p permease activity was measured by uptake of [14C]citrulline, an amino acid that enters cells only through Gap1p (Grenson et al., 1970). Gap1p activity was high in cells grown on either ammonia or urea, and was low in cells grown on glutamate or glutamine (Stanbrough and Magasanik, 1995) (Fig. 1 B). These findings reproduce the observation that although Gap1p is expressed in cells grown on glutamate, Gap1p experiences some form of posttranslational inhibition and the permease activity is low.
Figure 1.
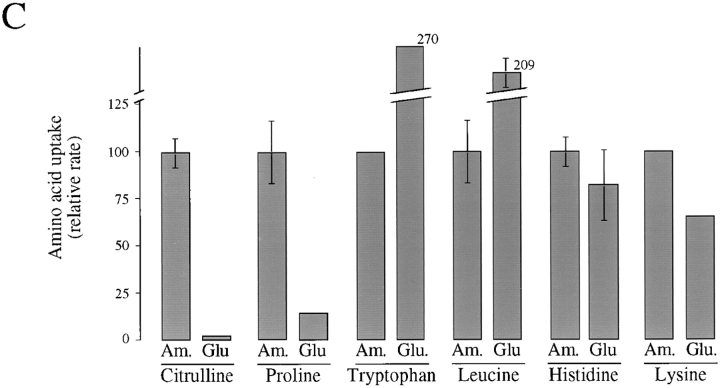
Gap1 protein is expressed but permease activity is low in cells grown on glutamate as a nitrogen source. (A) Wild-type cells expressing GAP1-HA (CKY465) were grown exponentially in synthetic medium with either ammonium sulfate, urea, glutamate, or glutamine as the only nitrogen source. The amount of tagged Gap1p in protein extracts was determined by SDS-PAGE and Western blotting with anti-HA antibody. Equal quantities of each extract as determined by protein assay were loaded onto the gel. (B) Wild-type cells (CKY443) were grown exponentially in synthetic medium with different nitrogen sources. Cells were harvested by filtration and suspended in SFD, and 4 μM [14C]citrulline was added. Five aliquots were removed at 30-s intervals, and label associated with the cell bodies was determined. Permease activity is expressed as the rate of citrulline uptake relative to the rate for cells grown on ammonia. The absolute rate of citrulline uptake on ammonia was 31 pmol/min/OD600. (C) Gap1p-independent uptake of amino acids was determined for a gap1-Δ1 mutant (CKY445) after growth on synthetic medium supplemented with either ammonium sulfate or glutamate. (Citrulline uptake was determined for a wild-type strain as described above.) The rate of uptake of [14C]-labeled amino acids proline, tryptophan, leucine, histidine, and lysine was determined as above. The absolute rate of uptake for each amino acid, in ammonia-grown cells, was as follows: proline, 4.7 pmol/min/OD600; tryptophan, 2.0 pmol/min/OD600; leucine, 2.4 pmol/min/OD600; histidine, 18 pmol/min/OD600; and lysine, 36 pmol/min/OD600.
The effect of glutamate on the activity of other amino acid permeases was also examined by measuring the rate of uptake in different radiolabeled amino acids. For these experiments, the contribution of Gap1p to the uptake of specific amino acids was eliminated by use of a gap1Δ mutant. The uptake of proline was 10-fold lower in cells grown on glutamate than on ammonia, indicating that Put4p activity was also inhibited on glutamate. In contrast, the uptake of tryptophan, leucine, histidine, and lysine was either the same or greater in cells grown on glutamate than on ammonia, indicating that the activities of at least four specific permeases are not inhibited by growth on glutamate (Fig. 1 C). Thus, the inhibitory effect of growth on glutamate appears to be specific for Gap1p and Put4p.
Gap1p Is Rapidly Delivered to the Plasma Membrane on Transfer From Glutamate to Urea
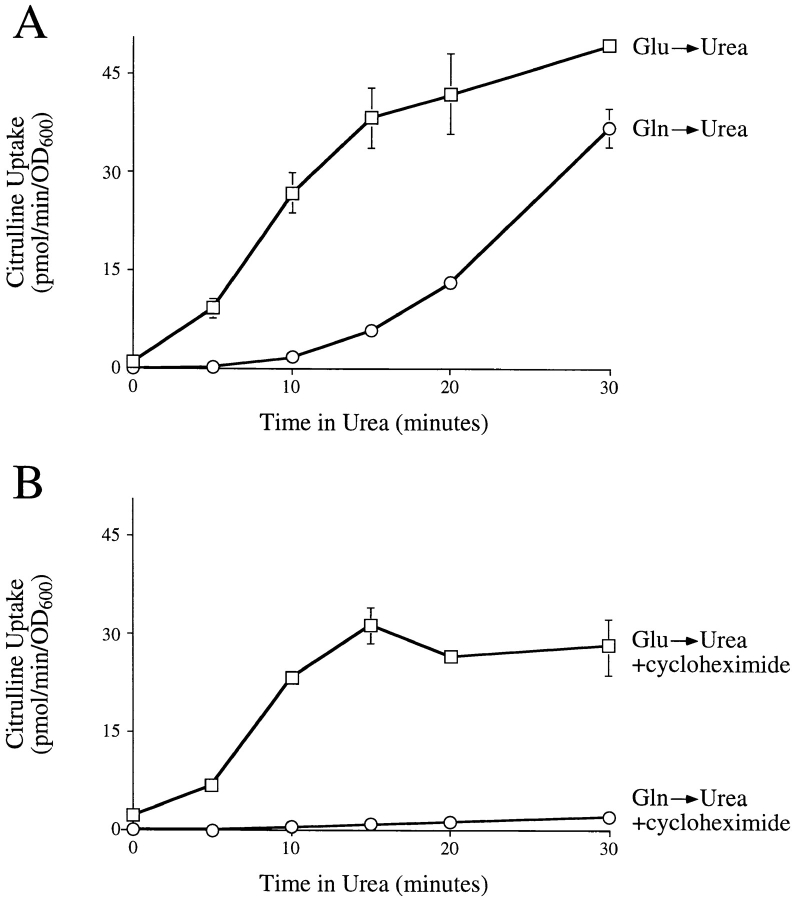
To determine how rapidly the Gap1 protein in cells grown on glutamate could become active for amino acid uptake, the kinetics of Gap1p induction were followed by measuring Gap1p activity after a shift in nitrogen source. When cells grown on glutamate were transferred to urea medium, Gap1p permease activity was induced rapidly, increasing 10-fold after 5 min, and 100-fold after 20 min (Fig. 2 A). When cells were grown on glutamine before transfer to urea medium, Gap1p activity increased only after a delay of about 15 min. This lag period probably represented the minimum time required to induce synthesis and to transport new Gap1 protein to the cell surface. The rapid onset of Gap1p activity after transfer from glutamate to urea suggested that the cellular pool of Gap1p present in cells grown on glutamate was available for rapid activation after cells were transferred to urea medium.
Figure 2.
(A) Gap1p is rapidly activated after glutamate-grown cells are transferred to urea medium. Wild-type cells (CKY443) were grown on either glutamate or glutamine and the kinetics of induction of permease activity was followed after transfer to urea. Permease activity was measured by the rate of [14C]citrulline uptake as described above. (B) Wild-type cells were treated as for A, but were shifted to urea medium containing 1.5 μg/ml cycloheximide.
To test whether the Gap1p in cells grown on glutamate could be activated without further protein synthesis, cells grown on glutamate were transferred to urea medium containing cycloheximide. Under these conditions, Gap1p activity increased as rapidly as in cells transferred to urea medium without cycloheximide, but the increase in Gap1p activity was not sustained after 15 min (Fig. 2 B). When cells grown on glutamine were transferred to urea containing cycloheximide, a substantial increase in Gap1p activity was not detected, confirming that treatment with cycloheximide blocked new Gap1p synthesis. These results show that the Gap1 protein present in cells grown in glutamate can be activated upon a shift to urea medium.
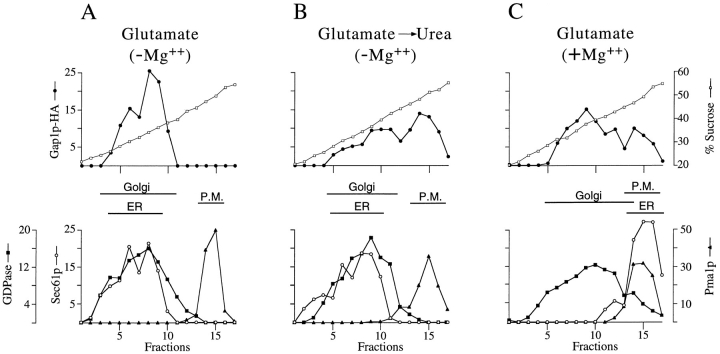
We considered the possibility that the initial rapid activation of Gap1p upon transfer from glutamate to urea could be the consequence of a redistribution of Gap1p from an intracellular compartment to the cell surface. Extracts from cells grown on glutamate, or grown on glutamate and then shifted to urea for 20 min, were fractionated on sucrose density gradients in the presence of EDTA. Under these conditions, where Mg2+ was removed from the lysate by chelation, ER and Golgi membranes had a low buoyant density and were well resolved from the more dense plasma membrane. Gap1p-HA from cells grown on glutamate fractionated in one peak coincident with the intracellular markers, Sec61p (ER) and GDPase activity (Golgi), but was not detected in fractions containing the plasma membrane marker, Pma1p (Fig. 3 A). In contrast, Gap1p-HA from cells shifted to urea fractionated as two peaks—one coincident with the intracellular markers, and the other with the plasma membrane marker (Fig. 3 B). Thus, the low level of Gap1p activity observed in glutamate-grown cells correlates with the absence of detectable levels of Gap1p in the plasma membrane. Moreover, after transfer to urea medium the 100-fold increase in Gap1p activity was accompanied by the appearance of Gap1 protein in the plasma membrane.
Figure 3.
In cells grown on glutamate, Gap1p-HA is in the ER and Golgi but not the plasma membrane. (A) Cell lysates from wild-type (CKY465) grown on glutamate, were fractionated on density gradients of 20–60% sucrose containing 10 mM EDTA. Relative levels of Gap1p-HA, Pma1p (plasma membrane marker), and Sec61p (ER marker) in each fraction were quantitated by Western blotting and densitometry. GDPase (Golgi marker) was determined by enzymatic assay of each fraction. The position of the peak fractions for each marker are indicated (these fractions contain at least 80% of the total marker on the gradient). (B) Cell lysates from wild-type (CKY465) grown on glutamate and then transferred to urea medium were fractionated on density gradients containing 10 mM EDTA. (C) Cell lysates from wild-type (CKY465) grown on glutamate were fractionated as above but on density gradients containing 2 mM Mg2+.
ER and Golgi fractions overlap extensively in sucrose density gradients when cell extracts have been prepared in the presence of EDTA. To further define the subcellular distribution of Gap1p in cells grown on glutamate, extracts were prepared with MgCl2 and no EDTA. These conditions produce ER membranes with a much higher buoyant density than Golgi membranes, possibly because a Mg2+-dependent association of ribosomes with ER membranes is preserved (Dobrota and Hinton, 1992). On gradients containing Mg2+, Gap1p-HA from glutamate-grown cells resolved in two peaks, one coincident with the Golgi and the other with the ER (Fig. 3 C). Thus, in cells grown on glutamate, Gap1p-HA was found in membranes of both the ER and Golgi compartments, but not in the plasma membrane.
Regulated Delivery of Gap1p to the Plasma Membrane Requires Vesicular Transport
The role of vesicular transport in the redistribution of Gap1p to the plasma membrane after transfer from glutamate to urea medium was tested by examining Gap1p activity in mutants that block transport in the late secretory pathway. SEC6 is 1 of 10 genes required for the fusion of post-Golgi secretory vesicles with the plasma membrane. In a sec6 temperature-sensitive mutant at 36°C, transport from the Golgi to the plasma membrane is blocked and secretory vesicles accumulate (Novick et al., 1981). No induction of Gap1p permease activity was observed in a sec6 mutant when transfer from glutamate to urea was preceded by a 10-min shift from 24°C to 36°C (Fig. 4 A). In an isogenic wild-type strain under the same conditions, Gap1p activity increased up to 25-fold after 15 min. The effect of the sec6 mutation on the subcellular distribution of Gap1p-HA was also examined by cell fractionation on a sucrose density gradient containing EDTA. No Gap1p-HA was detected in the plasma membrane fractions from an extract prepared from a sec6 mutant grown on glutamate at the permissive temperature, and then transferred to urea medium at the restrictive temperature for 20 min (Fig. 4 B). Based on these results, we conclude that post-Golgi vesicular transport was required for the redistribution of Gap1p to the plasma membrane upon a shift from glutamate to urea.
Figure 4.
Transport of Gap1p to the plasma membrane requires secretory vesicles. (A) Wild-type (CKY443) and sec6-1 mutant (CKY517) strains were grown in glutamate at 24°C, shifted to 36°C for 10 min, and then transferred to prewarmed urea medium. Gap1p activity was measured by the rate of [14C]citrulline uptake. (B) A sec6 mutant (CKY518) was grown in glutamate at 24°C, shifted to 36°C for 10 min, and then shifted to prewarmed urea medium for 20 min. A lysate prepared from this culture was fractionated on a density gradient of 20–60% sucrose containing 10 mM EDTA. Gap1p-HA and markers for different subcellular compartments were evaluated for each fraction as described in Fig. 2.
Gap1p Is Transported from the Golgi to the Vacuole in Glutamate-grown Cells
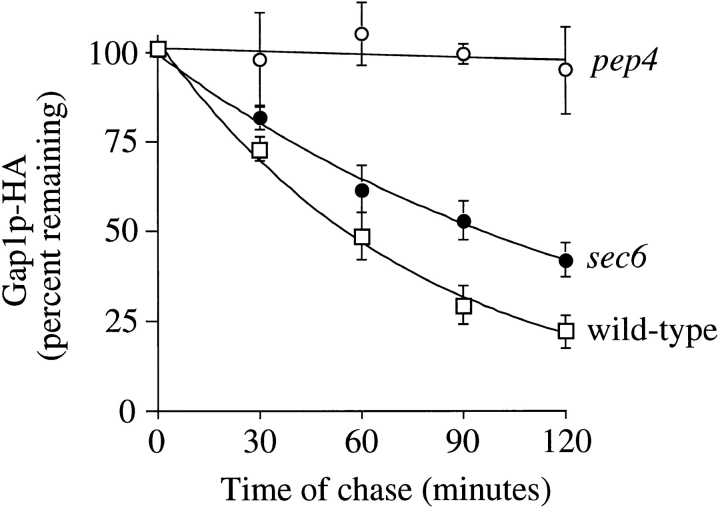
Gap1p was found in the ER and the Golgi membranes of glutamate-grown cells; however, it did not appear to accumulate in either organelle, which suggests that Gap1p was continuously turned over. In pulse–chase experiments to examine the stability of Gap1p-HA in glutamate-grown cells, Gap1p-HA was found to be degraded with a half-time of 60 min (Fig. 5). This degradation on glutamate was found to occur in the vacuole since Gap1p-HA was stabilized by pep4Δ, a mutation that prevents the enzymatic activation of vacuolar proteases (Jones et al., 1982).
Figure 5.
Gap1p stability in pep4, sec6, and wild-type strains grown in glutamate. Isogenic wild-type (CKY520), sec6 (CKY518), and pep4Δ (CKY519) strains expressing GAP1-HA were grown in glutamate at 24°C. Cultures were shifted to 36°C for 10 min, and cell proteins were radioactively labeled during a 7-min pulse of [35S]methionine and [35S]cysteine followed by a chase of excess unlabeled methionine and cysteine. Gap1p-HA was immunoprecipitated and the fraction of labeled Gap1p-HA remaining after different chase times was determined with a phosphoimager.
Studies of membrane protein delivery to the yeast vacuole have demonstrated that there are two possible transport pathways from the Golgi to the vacuole. Usually, vacuolar proteins follow an intracellular route via the prevacuolar compartment (Horazdovsky et al., 1995; Conibear and Stevens, 1995). However, when normal transport to the prevacuolar compartment is blocked, proteins can be transported first to the plasma membrane and then to the vacuole via the endocytic pathway (Nothwehr et al., 1995; Redding et al., 1996). To determine which pathway is followed by Gap1p in glutamate-grown cells, we evaluated the effect on Gap1p degradation of a block in transport from the Golgi to the plasma membrane. In pulse– chase experiments with glutamate-grown cells shifted to 36°C, Gap1p-HA was turned over at similar rates in isogenic sec6 mutant and wild-type strains, demonstrating that Gap1p transport to the vacuole did not require transport from the Golgi to the plasma membrane (Fig. 5). If Gap1p were delivered to the vacuole via the plasma membrane, then a sec6 mutation should have caused Gap1p to be sequestered in post-Golgi vesicles, preventing its degradation. Thus, in cells grown on glutamate, Gap1p traverses the early part of the secretory pathway and then appears to be transported directly from the Golgi to the vacuole via the prevacuolar compartment.
To confirm that transport of Gap1p to the vacuole in glutamate-grown cells was not a result of endocytosis from the plasma membrane, the effect of end3-1 and end4-1 mutations on Gap1p activity was evaluated. In these mutants, endocytosis of the integral membrane proteins, Ste2p, Ste6p, and the uracil permease, is blocked at the restrictive temperature (Schandel and Jenness, 1994; Berkower et al., 1994; Volland et al., 1994). We expected that if the low level of Gap1p activity observed in cells grown on glutamate was a consequence of endocytosis, these mutants would exhibit relatively high levels of Gap1p activity on glutamate at the restrictive temperature. Assays of [14C]citrulline uptake by cells grown on glutamate and shifted to 36°C for 1 h demonstrated that Gap1p activity was actually at least twofold lower in end3-1 and end4-1 mutants than in a wild-type strain under identical conditions. The inability of endocytosis mutants to restore Gap1p activity to cells grown on glutamate supports our conclusion that the low Gap1p activity in wild-type cells grown on glutamate results from decreased transport of Gap1p to the plasma membrane rather than enhanced endocytosis.
SEC13 Mutants Affect Gap1p Activity on Ammonia but Not on Glutamate
We were interested in identifying cellular factors that are required for Gap1p transport to the cell surface. A simple hypothesis to explain how Gap1p localization is regulated in response to the external nitrogen source is that growth on glutamate inhibits the selective transport of Gap1p to the plasma membrane. Accordingly, mutations in genes required for this selective transport step should exhibit an intracellular distribution of Gap1p on ammonia that is comparable to the distribution of Gap1p in wild-type cells grown on glutamate: namely Gap1p should be present in the Golgi but not in the plasma membrane. In the process of studying mutants that genetically interact with SEC13, a gene previously recognized to encode an essential component of the COPII vesicle coat, we found that the activities of Put4p and Gap1p were particularly sensitive to sec13 mutations. This finding led us to investigate the relationship between SEC13 function and nitrogen-regulated delivery of Gap1p to the plasma membrane.
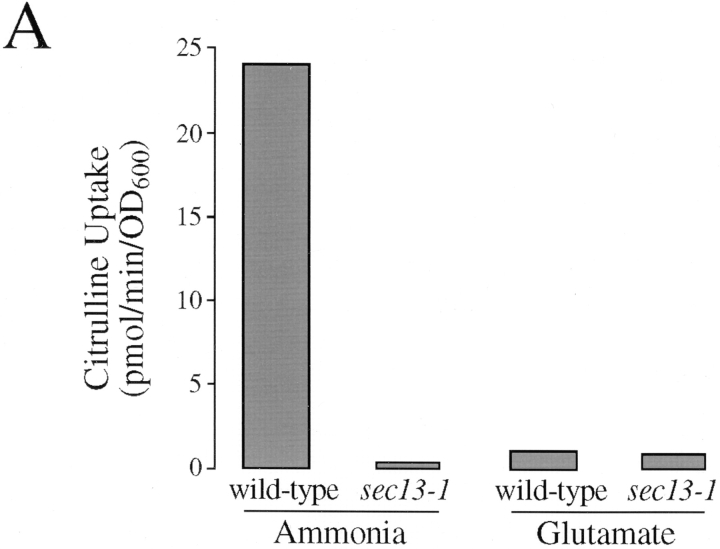
Because Gap1p activity is highest in wild-type cells grown on ammonia (Fig. 1 B), analysis of the effect of sec13-1 on Gap1p activity was conducted on cells grown on ammonia as the only nitrogen source. When a sec13-1 mutant was grown at 24°C on ammonia, its rate of [14C] citrulline uptake was 50 to 100-fold lower than that of an isogenic wild-type strain (Fig. 6 A). Similarly, [14C]proline uptake was 20-fold lower in a sec13-1 mutant than in wild type (Roberg, K., and C. Kaiser, unpublished data). These effects on amino acid uptake cosegregated with the sec13-1 mutation in genetic crosses and could be complemented by a wild-type copy of the SEC13 gene. Uptake of tryptophan, leucine, histidine, and lysine were not significantly altered by the sec13-1 mutation. Thus, like growth on glutamate, the sec13-1 mutation affects the activities of Gap1p and Put4p, but not Hip1p, Lyp1p, or permeases required for the uptake of either tryptophan or leucine.
Figure 6.
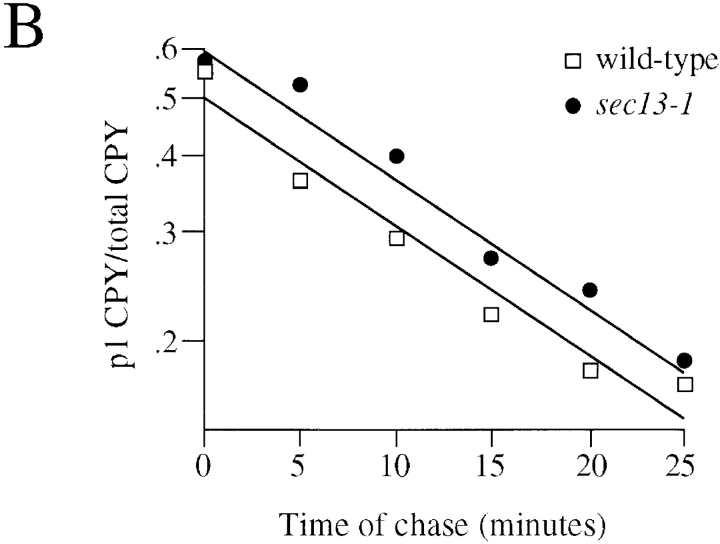
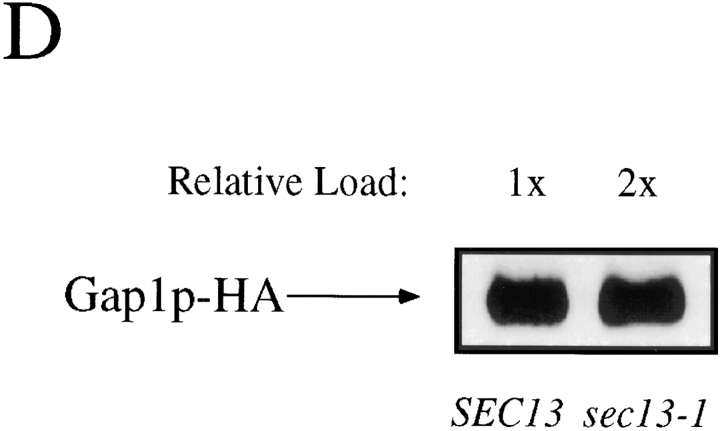
sec13-1 mutants greatly reduce Gap1p activity but not protein levels. (A) Isogenic wild-type (CKY443) and sec13-1 (CKY 444) strains were grown at 24°C in synthetic medium supplemented with either ammonium sulfate or glutamate as the only nitrogen source. Gap1p activity was determined by the rate of [14C] citrulline uptake. (B) ER to Golgi transport was assayed by the kinetics of CPY maturation. Wild-type (CKY443) and sec13-1 (CKY444) strains were grown at 24°C in minimal medium with ammonia. Cultures were labeled with [35S]methionine and [35S]cysteine for 5 min and chased by the addition of an excess of unlabeled methionine and cysteine. CPY was immunoprecipitated from labeled extracts and resolved by SDS-PAGE. The quantity of p1 CPY and total CPY was determined with a phosphorimager. Conversion of p1 CPY to the p2 and M forms gives the rate of transport out of the ER. (C) Expression of Gap1p in sec13-1 was determined by β-galactosidase expression from a gap1–lacZ reporter fusion (pMS29) in isogenic wild-type (CKY522) and sec13-1 (CKY523) strains grown in ammonia medium at 24°C. (D) Gap1p-HA protein levels in isogenic wild-type (CKY465) and sec13-1 (CKY466) strains grown in ammonia medium at 24°C were determined by Western blotting with anti-HA antibody. The amount of extract loaded was adjusted to give equivalent intensities of the Gap1p-HA band.
Importantly, the sec13-1 mutation had no significant effect on Gap1p activity in cells grown on glutamate. Both wild-type and sec13-1 strains had similar low levels of Gap1p activity (0.64 ± 0.18 pmol/min/OD600 for wild-type; 0.45 ± 0.04 pmol/min/OD600 for sec13-1) (Fig. 6 A). If it were the case that the sec13-1 mutation and growth on glutamate reduced permease activity for different reasons, then the effect of sec13-1 combined with the effect produced by growth on glutamate should have reduced activity even further. The absence of such a synergistic effect indicates that growth on glutamate and the sec13-1 mutation act on the same process to reduce permease activity.
The Effect of sec13-1 on Gap1p Activity Is Not Due to a Defect in General ER to Golgi Transport
As the sec13-1 mutation was originally recognized to block ER to Golgi transport at restrictive temperatures, it seemed possible that the permease defect in sec13-1 could be related to an unanticipated link between general ER to Golgi transport and transport of specific permeases from the ER. We could eliminate this possibility, however, since a sec13-1 mutant grows at the same rate as wild-type at 24°C (doubling time of 3 h in minimal medium) and therefore must have a normal rate of secretion-coupled cell surface growth. As a specific test for the rate of ER to Golgi transport, pulse–chase experiments were performed to follow the rate of maturation of CPY from the ER form (p1) to the Golgi (p2) and vacuolar (M) forms of the enzyme. In these experiments, the rate of transport of CPY from the ER was the same in sec13-1 and wild-type at 24°C (Fig. 6 B). These data show that sec13-1 does not have a general ER to Golgi defect at 24°C and the effect of sec13-1 on permease transport cannot be explained as a consequence of slowed ER to Golgi transport.
We also examined the possibility that sec13-1 reduces expression of Gap1p. Both wild-type and sec13-1 mutants exhibited similar levels of expression of a gap1–lacZ gene fusion (Fig. 6 C). In addition, Gap1 protein levels in strains expressing Gap1p-HA were quantified by immunoblotting. The levels of Gap1p-HA in a sec13-1 mutant were approximately twofold lower than in wild-type (Fig. 6 D); a reduction in protein level insufficient to account for the 100-fold reduction in Gap1p activity.
Gap1p-HA Is Not Transported to the Cell Surface in sec13-1 Mutants
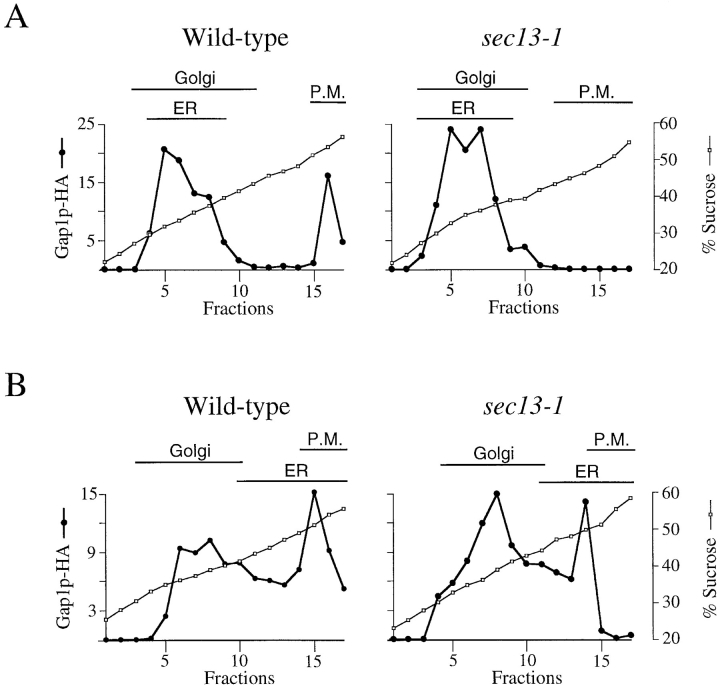
To determine if the effect of sec13-1 on Gap1p activity was due to altered trafficking of the permease, we compared the subcellular distribution of Gap1p-HA in wild-type and a sec13-1 mutant. Lysates were prepared from cells grown on ammonia at 24°C and fractionated on sucrose density gradients containing EDTA. Consistent with a defect in transport to the plasma membrane, Gap1p-HA from sec13-1 cells was found in a single peak coincident with the ER-and Golgi-containing fractions, and no Gap1p-HA could be detected in the plasma membrane fractions (Fig. 7 A). As expected, Gap1p-HA from wild-type cells was detected in two distinct peaks: one coinciding with both the ER marker Sec61p and Golgi GDPase, and the other coinciding with the plasma membrane marker Pma1p. The finding that, in wild-type cells grown on ammonia, less than half of the total Gap1p-HA is in the plasma membrane at steady state is likely the consequence of a slow rate of transport of Gap1p to the plasma membrane.
Figure 7.
In sec13-1 mutants, Gap1p exits the ER but is not transported to the plasma membrane. (A) Wild-type (CKY465) and sec13-1 (CKY466) strains expressing Gap1p-HA were grown in ammonia medium at 24°C, and cell lysates were fractionated on density gradients of 20–60% sucrose containing 10 mM EDTA. Gap1p-HA and markers for different subcellular compartments were evaluated for each fraction as described in Fig. 3. (B) Wild-type (CKY465) and sec13-1 (CKY466) strains expressing Gap1p-HA were grown in ammonia medium at 24°C, and cell lysates were fractionated on density gradients of 20–60% sucrose containing 1 mM Mg2+.
To determine if Gap1p is transported from the ER in a sec13-1 mutant at 24°C, the ER and Golgi from a sec13-1 mutant were resolved on sucrose gradients containing Mg2+. Under these conditions, Gap1p-HA from both sec13-1 and wild-type cells resolved in two peaks. One peak coincided with the ER, and the other with the Golgi (Fig. 7 B), thereby demonstrating that in the sec13-1 mutant, Gap1p resided in both the ER and Golgi compartments.
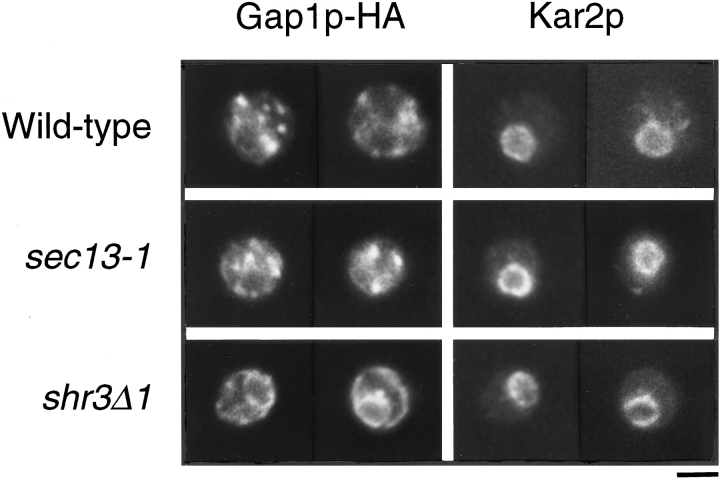
That the sec13-1 mutation did not block Gap1p at the level of the ER at 24°C was confirmed by indirect immunofluorescence microscopy using an anti-HA antibody to detect Gap1p-HA. Punctate staining, clearly distinct from the ER, was observed at the cell periphery in both wild-type and sec13-1 cells. Only a small fraction of the Gap1p-HA was coincident with the perinuclear ER compartment, as marked by the ER-resident protein Kar2p in double- labeling experiments (Fig. 8). A shr3Δ mutant was used as a control to show the staining pattern for Gap1p-HA that had been retained in the ER. Mutations in SHR3 cause a block in transport of Gap1p-HA from the ER (Ljungdahl et al., 1992). As expected, the distribution of Gap1p-HA in the shr3Δ mutant was perinuclear and coincident with Kar2p staining. Together, the localization experiments show that in sec13-1 cells Gap1p is located in both the ER and Golgi compartments, but not the plasma membrane. The absence of a clear accumulation of Gap1p in the ER in sec13-1 reinforces the conclusion that the permease defect in sec13-1 occurs at a late stage of the secretory pathway and is not the result of a failure to transport Gap1p out of the ER.
Figure 8.
Localization of Gap1p-HA by indirect immunofluorescence microscopy in sec13-1, shr3Δ, and wild-type strains. Wild-type (CKY465), sec13-1 (CKY466), and shr3Δ (PLY134) strains expressing Gap1p-HA from a centromeric plasmid were grown in ammonia medium, fixed in formaldehyde, and double labeled with anti–HA and anti–Kar2p antibodies. Mouse anti-HA was visualized with FITC-coupled secondary antibody and rabbit anti–Kar2p was visualized with rhodamine-coupled secondary antibody. Bar, 2.5 μm.
SEC13 Is Required for Transport of Gap1p from the Golgi to the Plasma Membrane
We examined the possibility that Gap1p was transported to the vacuole in sec13-1 mutants grown on ammonia. In pulse–chase experiments to examine protein turnover in cells growing at 24°C, Gap1p-HA had a half-life of 55 min in a wild-type strain and 35 min in a sec13-1 mutant (Fig. 9 A). Degradation of Gap1p-HA in the sec13-1 mutant occurred in the vacuole since Gap1p-HA was not degraded in a sec13-1 pep4Δ double mutant. Thus, we conclude that both for wild-type cells grown on glutamate and for sec13-1 mutants grown on ammonia, Gap1p was transported to the vacuole.
Figure 9.

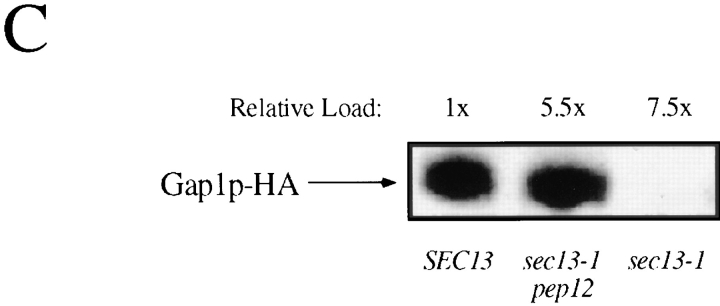
Gap1p is transported from the Golgi to the vacuole in sec13-1 mutants. (A) Wild-type (CKY520), sec13-1 (CKY521), and sec13-1 pep4Δ (CKY467) strains were grown at 24°C to exponential phase. Cultures were labeled with [35S]methionine and [35S]cysteine for 10 min and chased by the addition of an excess of unlabeled methionine and cysteine. Gap1p-HA was immunoprecipitated from labeled extracts, resolved by SDS-PAGE, and was quantified using a phosphorimager. (B) Gap1p activity was assayed by the rate of [14C]citrulline uptake in wild-type (CKY443), sec13-1 (CKY444), sec13-1 pep12Δ::TRP1 (CKY455), sec13-1 end3-1 (CKY456), and sec13-1 pep4Δ::LEU2 (CKY457) strains. (C) Cell lysates from wild-type (CKY465), sec13-1 (CKY466), and sec13-1 pep12Δ::TRP1 (CKY468) strains were fractionated on density gradients of 20%–60% sucrose containing 10 mM EDTA. Fractions containing Pma1p were pooled, and Gap1p-HA in these fractions was detected by Western blotting with anti-HA antibody. The relative amount of plasma membrane in each extract was determined by quantitation of Pma1p. For A–C, all strains were grown in ammonia medium.
To strengthen the argument that SEC13 is required for the regulated transport of Gap1p to the plasma membrane, we wished to define the route taken by Gap1p to the vacuole in sec13-1 cells grown on ammonia. We utilized a pep12Δ mutation that blocks transport of proteins from the Golgi to the prevacuolar compartment at 24°C (Becherer et al., 1996). If Gap1p arrived at the vacuole via the prevacuolar compartment, we reasoned that a blockade of transport along this pathway could result in the accumulation of Gap1p at the Golgi. Furthermore, this accumulation might result in nonspecific packaging of Gap1p into vesicles by a SEC13-independent process, leading to the transport of Gap1p from the Golgi to the plasma membrane. Consistent with this hypothesis, the rate of [14C]citrulline uptake was 15-fold greater for a sec13-1 pep12Δ double mutant than for an isogenic sec13-1 single mutant (Fig. 9 B). In control experiments, we found no significant difference between the rate of citrulline uptake in sec13-1 and sec13-1 pep4 mutants, showing that suppression of the sec13-1 defect by pep12Δ was not simply due to a loss of vacuolar protease activity.
We confirmed the ability of pep12Δ to suppress the Gap1p transport defect of sec13-1 mutants by directly comparing the amount of Gap1p in the plasma membrane of sec13-1 and sec13-1 pep12Δ strains. Cell extracts from wild-type, sec13-1, and sec13-1 pep12Δ strains were fractionated on density gradients and the plasma membrane–containing fractions, marked by the presence of Pma1p, were collected. The amount of Gap1p-HA in the pooled plasma membrane fractions from sec13-1 pep12Δ double mutants was ∼20% of that found in wild-type cells, consistent with the relative Gap1p activity in the two strains (Fig. 9 C). In contrast, Gap1p-HA could not be detected in the plasma membrane of a sec13-1 single mutant.
We also examined the possibility that decreased endocytosis could restore Gap1p activity in sec13-1 mutants. The ability of pep12Δ to partially restore Gap1p activity in sec13-1 could not be explained by an effect of pep12Δ on endocytosis, since isogenic pep12Δ and wild-type strains showed similar capacity to take up the endocytic marker Lucifer yellow (data not shown). Moreover, citrulline uptake by a sec13-1 end3-1 double mutant was only twofold greater than for an isogenic sec13-1 single mutant (Fig. 9 B), demonstrating that an end3-1 mutation could not restore Gap1p activity to a sec13-1 mutant.
These results demonstrate that Gap1p was transported to the vacuole via the prevacuolar compartment in a sec13-1 mutant, and that when this route was blocked by pep12Δ Gap1p transport to the plasma membrane was partially restored. In addition, these results strengthen the conclusion that the sec13-1 mutation, like growth on glutamate, affects Gap1p sorting at the trans-Golgi.
Discussion
One way that S. cerevisiae cells adapt to different growth conditions is to alter the capacity of the plasma membrane to transport small molecules. The amino acid permeases Gap1p and Put4p are regulated according to nitrogen source, and their induction on poor nitrogen sources increases the cells' capacity to extract amino acids from the extracellular environment for use as a nitrogen source (Magasanik, 1992). Regulation of Gap1p by nitrogen source was previously shown to be accomplished by transcriptional control of GAP1 mRNA synthesis and by hitherto poorly understood posttranslational regulatory mechanisms. In this paper, we have followed Gap1p localization within yeast cells grown on different nitrogen sources and found that the posttranslational regulation of Gap1p activity is a consequence of regulated sorting of Gap1 protein in the secretory pathway. In cells grown on either urea or ammonia as a nitrogen source, Gap1p can be detected in the plasma membrane and the permease activity is high. In cells grown on glutamate as the nitrogen source, Gap1p is absent from the plasma membrane and permease activity is low. In glutamate grown cells, Gap1p appears to be transported to the vacuole from a late-Golgi compartment.
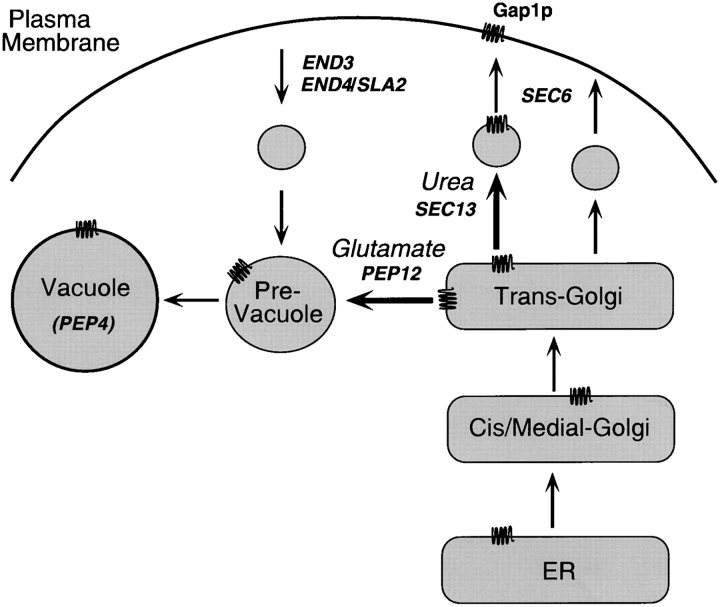
These findings are most easily explained if a special type of secretory vesicle, or a special cargo-loading step into a general vesicle, is required for the transport of Gap1p from the late-Golgi to the cell surface (Fig. 10). According to this idea, a regulatory mechanism sensitive to growth on glutamate inhibits the packaging of Gap1p into vesicles destined for the plasma membrane, thereby causing Gap1p to be transported to the vacuole where the protein is degraded. Examination of mutants of a variety of membrane proteins has led to the view that transport from the Golgi to the vacuole is the default condition for membrane proteins that are not correctly targeted to the plasma membrane, or not correctly retained in the Golgi or ER. For example, mutant forms of the plasma membrane ATPase are not transported to the cell surface, but instead are diverted to the vacuole (Chang and Fink, 1995). In addition, mutant forms of the Golgi dipeptidyl aminopeptidase that lack a Golgi retention signal are transported to the vacuole, but not via the plasma membrane (Roberts et al., 1992). Finally, fusion proteins containing a domain of a resident ER membrane protein are transported to the vacuole when these proteins escape the ER-retention mechanisms (Gaynor et al., 1994). These results imply that transport of membrane proteins from the Golgi to the plasma membrane involves signal-mediated sorting into secretory vesicles, and membrane proteins that lack an appropriate signal are transported to the vacuole by default. Our finding that the regulated permeases, Gap1p and Put4p, have different requirements for their transport to the cell surface than do the constitutive amino acid permeases implies the two classes of permeases carry different sorting signals. Comparison of permease sequences did not reveal any obvious features that are unique to Gap1p and Put4p (André, 1995), and it remains to be determined how these proteins are selected for a nitrogen-regulated transport step resulting in delivery to the plasma membrane.
Figure 10.
A model for the alternative routes of transport of Gap1p from the late-Golgi controlled physiologically by nitrogen source, and genetically by SEC13 and PEP12. In cells grown on either ammonia or urea, Gap1p is packaged into Golgi-derived vesicles destined for the plasma membrane. On glutamate, or in a sec13-1 mutant, Gap1p is diverted to the vacuole. This model is based on the idea that a specialized class of vesicle carries Gap1p to the plasma membrane and that unregulated permeases and other plasma membrane proteins are transported by a general class of vesicles. Alternatively, loading of Gap1p into general secretory vesicles could be regulated by both Sec13p and in response to the nitrogen source.
The localization of Gap1p has also allowed us to evaluate mutants for an effect on the mechanism of Gap1p localization. Alleles of SEC13 seem to produce the same effect on Gap1p sorting in the Golgi as that produced by growth on glutamate. In sec13-1 mutants growing on ammonia as a nitrogen source, Gap1p does not appear in the plasma membrane but is transported to the vacuole instead. Moreover, in sec13-1 mutants growing on glutamate, the reduction in Gap1p activity is no greater than for wild-type cells growing on glutamate, indicating that the sec13-1 allele affects the same physiological process that is regulated by glutamate. Finally, sec13-1 appears to show the same specificity for Gap1p and Put4p as does nitrogen-regulated transport.
Sec13p is a component of the COPII vesicle coat that acts in ER to Golgi transport (Kaiser and Schekman, 1990; Pryer et al., 1993). The simplest explanation for the specific requirement for SEC13 in the transport of Gap1p from the Golgi to the plasma membrane is that Sec13p is an essential component of the secretory vesicles that carry Gap1p to the plasma membrane. Two different types of secretory vesicles—with different physical characteristics and carrying different cargo molecules—have been identified in yeast (Harsay and Bretscher, 1995). Although both classes of secretory vesicles share a common machinery for fusion with the plasma membrane, it is not known whether these vesicles have coats, nor is anything known of the specific genetic or biochemical requirements for their budding. We propose that there is a third class of post-Golgi vesicle that is specialized to carry Gap1p and Put4p to the plasma membrane, and that Sec13p functions as a component of the coat for these vesicles. An alternative possibility is that Sec13p is required for loading of Gap1p and Put4p into one of the general classes of secretory vesicle. These two models can be properly addressed once Gap1p-containing secretory vesicles have been purified and characterized. In either case, the functional studies presented in this work provide strong evidence that Sec13p participates in the formation of two different types of vesicles: Gap1p containing vesicles that bud from the late-Golgi, as well as COPII vesicles that bud from the ER.
Although the requirement for SEC13 in the proper trafficking of nitrogen-regulated permeases implies that Sec13p participates directly in vesicle budding from the trans-Golgi, we cannot rule out the possibility that the role of SEC13 in this process is indirect. One speculative, indirect effect could involve an as yet unidentified chaperone protein which follows the secretory pathway and is required in the trans-Golgi for proper transport of Gap1p to the plasma membrane. If such a protein existed, the effect of sec13 mutations on Gap1p transport could be explained if we further postulate that transport of the chaperone out of the ER were inhibited by sec13 mutants under conditions where general ER to Golgi transport was not affected. Alternatively, since some Sec13p has been shown to be associated with nuclear pore complexes (Siniossoglou et al., 1996), it is possible that sec13 mutants have a subtle effect on the nuclear pore and block import of a factor specifically required for expression of a Gap1p-sorting factor. It will be possible to more directly address these issues once we are able to test for the physical association of Sec13p with Gap1p-containing vesicles.
We have explored the possibility that COPII proteins other than Sec13p could also be required for transport of Gap1p from the late-Golgi. Mutations in the COPII genes, SEC12, SEC16, SEC23, and SEC31, had no effect on Gap1p activity (Roberg, K., and C. Kaiser, unpublished data). Given the sensitivity and reproducibility of the permease assay, and the complete absence of a measurable effect of other COPII mutations, we think it is likely that Sec13p is the only known COPII protein that participates in Gap1p sorting into post-Golgi vesicles. More likely candidates for additional components of a Gap1p carrying vesicle were identified in a screen for mutations that are synthetically lethal with sec13-1. Three new genes isolated in this screen, LST4, LST7, and LST8, are also required for Gap1p activity. In lst4, lst7, and lst8 mutants, Gap1p and Put4p activity is reduced to levels similar to those in sec13-1. Preliminary data also suggest that Gap1p is not transported to the plasma membrane in lst4, lst7, and lst8 mutants (Roberg, K., and C. Kaiser, unpublished data).
When the pathway for transport from the Golgi to the prevacuolar compartment is blocked, vacuolar membrane proteins can be transported to the cell surface instead (Nothwehr et al., 1995). A suggestion that the levels of amino acid permeases in the plasma membrane can be increased by decreasing transport to the vacuole came from the early finding that some vacuolar protein sorting mutants could suppress certain missense alleles of CAN1, the gene encoding the arginine permease (Jones, 1983). In particular, mutations in PEP12, a gene encoding a putative t-SNARE for protein trafficking from the trans-Golgi to the prevacuolar compartment (Becherer et al., 1996), are capable of suppressing can1 mutations. In pep12Δ, the absence of a functional transport pathway from the Golgi to the vacuole presumably allows the concentration of membrane proteins that would otherwise be transported to the vacuole. At sufficiently high concentrations in the Golgi, we presume that even membrane proteins that lack appropriate sorting signals, such as mutant Can1p, will enter vesicles destined for the plasma membrane. We have taken advantage of this phenomenon to show that pep12Δ can partially compensate for reduced Gap1p transport to the plasma membrane caused by sec13-1.
We propose that for Gap1p and Put4p, there is a hierarchy of routes out of the Golgi. On ammonia or urea, these permeases are preferentially loaded into a class of vesicles that depends on Sec13p, and are destined for the plasma membrane. When the pathway to the cell surface is not available, in cells grown on glutamate, or in sec13-1 cells grown on ammonia, the alternative pathway taken by the permeases appears to be first, transport to the prevacuolar compartment, and then to the vacuole. In a pep12Δ mutant, this alternative pathway is blocked and transport to the plasma membrane is restored, presumably because the only route out of the Golgi for Gap1p and Put4p is by constitutive secretory vesicles destined for the plasma membrane.
The regulation of permease activity, by sorting in the secretory pathway, raises the interesting question of why cells grown on glutamate would express a large quantity of Gap1p that is degraded in the vacuole without appearing at the cell surface. One possible benefit might be that regulation of Gap1p activity by sorting would allow rapid adaptation to a changing environment. The availability of glutamate as a nitrogen source could signal an impending need for Gap1p permease activity. On the other hand, glutamate as a nitrogen source might also be a condition where it would be undesirable to have Gap1p and Put4p permeases in the plasma membrane because of their potential to cause needless induction of amino acid utilization pathways. Accordingly, cells grown on glutamate would produce an internal pool of Gap1p to populate the early compartments of the secretory pathway that would be available for rapid delivery to the plasma membrane once glutamate had been consumed. Indeed, during growth on glutamate, Gap1p activity can be induced within 5 min after a change in nitrogen source; this interval is considerably shorter than the induction time for cells grown on glutamine, where de novo synthesis of Gap1p is required (Fig. 2).
It is also possible that Gap1p may perform an important function in the vacuole. In the experiments reported here, Gap1p delivered to the vacuole is degraded. However, it is not known how long the permease remains active in the vacuolar membrane before proteolysis. Gap1p is an H+– amino acid symporter (Grenson, 1992); if Gap1p were transiently active in the vacuolar membrane, its activity should produce a flux of amino acids from the vacuolar lumen to the cytosol. The vacuole is a storage compartment for a number of amino acids, particularly arginine (Cooper, 1982). The membrane pumps responsible for concentrating amino acids in the vacuole have been detected as biochemical activities, but the corresponding genes have not yet been identified (Ohsumi and Anraku, 1981; Sato et al., 1984). Conditions that cause amino acids stored in the vacuole to appear in the cytosol include nitrogen starvation and growth on glutamate (Cooper, 1982; Courchesne and Magasanik, 1983). We propose that Gap1p delivered to the vacuole in cells grown on glutamate may be one of the agents responsible for release of amino acids from the vacuole, and we are currently examining the role of Gap1p in this process. Regulated sorting of Gap1p in the late Golgi could be thought of as a decision either to send Gap1p to the plasma membrane to extract amino acids from the extracellular environment, or to send Gap1p to the vacuole to utilize the internally stored amino acids.
Finally, it is worth noting the parallels between the regulation of Gap1p localization in yeast and the regulation of the GLUT-4 glucose transporter in insulin-responsive mammalian cells. One way fat and muscle cells respond to insulin, a signal of high levels of glucose in the blood, is to increase their capacity to take up glucose. This is accomplished by a specialized insulin-induced protein trafficking step that delivers GLUT-4 transporter to the cell surface from an intracellular compartment (Haney and Mueckler, 1994). Dysfunction of the cell's ability to redistribute GLUT-4 to the cell surface compromises glucose homeostasis and could be the underlying cause of certain forms of non–insulin-dependent diabetes (James and Piper, 1994). This form of control of carbon source metabolism by mammalian cells is strikingly similar to the mechanism we propose here for control of nitrogen metabolism by yeast cells. Here we show one way that the capacity of a yeast cell to utilize amino acids as a nitrogen source is regulated by the controlled delivery of the Gap1p permease to the cell surface. When Gap1p is not delivered to the cell surface, it is eventually degraded in the vacuole after slow transit through the ER and Golgi compartments. In the parallel case of unstimulated mammalian cells, intracellular GLUT-4 protein is found in intracellular compartments thought to be related to either the trans-Golgi or the endosome (Haney and Mueckler, 1994). For yeast cells the functional equivalent of a storage compartment for the permease might be the slow transit of Gap1p through the ER and Golgi. Since little is known of the molecular mechanisms that underlie GLUT-4 trafficking, it is our hope that study of Gap1p transport in yeast will lead to the identification of factors required for regulated transport in a variety of organisms.
Acknowledgments
We thank B. Jones for providing the pep12Δ strain. We also thank A. Chang, M. Rose, and R. Schekman for their generous gifts of antibodies, and the members of the Kaiser lab for their technical assistance, advice, and encouragement, with special thanks to A. Frand and K. Lemon for critically reading parts of this manuscript. We are especially grateful to M. Stanbrough and B. Magasanik for helpful discussions throughout the course of this work.
This work was supported by a grant from the National Institute of General Medical Sciences and the Searle Scholars Program (to C.A. Kaiser), a National Institutes of Health predoctoral traineeship (to K.J. Roberg), and a Human Frontiers International postdoctoral fellowship (to N. Rowley). C.A. Kaiser is a Lucille P. Markey Scholar, and this work was funded in part by the Lucille P. Markey Charitable Trust.
Footnotes
1. Abbreviations used in this paper: CPY, carboxypeptidase Y; HA, hemagglutinin.
Please address all correspondence to Chris A. Kaiser, Department of Biology, Room 68-533, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139. Tel.: (617) 253-9804. Fax: (617) 253-8699. e-mail: ckaiser@mit.edu
References
- Abeijon C, Orlean P, Robbins PW, Hirschberg CB. Topography of glycosylation in yeast: characterization of GDP mannose transport and lumenal guanosine diphospatase activities in Golgi-like vesicles. Proc Nat Acad Sci USA. 1989;86:6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. . Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homologue, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkower C, Loayza D, Michaelis S. Metabolic instability and constitutive endocytosis of STE6, the a-factor transporter of Saccharomyces cerevisiae. . Mol Biol Cell. 1994;5:1185–1198. doi: 10.1091/mbc.5.11.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess TL, Kelly RB. Constitutive and regulated secretion of proteins. Annu Rev Cell Biol. 1987;3:243–293. doi: 10.1146/annurev.cb.03.110187.001331. [DOI] [PubMed] [Google Scholar]
- Chang A, Fink GR. Targeting of the yeast plasma membrane [H+]ATPase: a novel gene AST1prevents mislocalization of mutant ATPase to the vacuole. J Cell Biol. 1995;128:39–49. doi: 10.1083/jcb.128.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vacuolar biogenesis in yeast: sorting out the sorting proteins. Cell. 1995;83:513–516. doi: 10.1016/0092-8674(95)90088-8. [DOI] [PubMed] [Google Scholar]
- Cooper, T.G. 1982. Nitrogen metabolism in Saccharomyces cerevisiae. In The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression. J.N. Strathern, E.W. Jones, and J.R. Broach, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 39–100.
- Courchesne WE, Magasanik B. Ammonia regulation of amino acid permeases in Saccharomyces cerevisiae. . Mol Cell Biol. 1983;3:672–683. doi: 10.1128/mcb.3.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrota, M., and R. Hinton. 1992. Conditions for density gradient separations. In Preparative Centrifugation: A Practical Approach. D. Rickwood, editor. Oxford University Press, Oxford, England. 77–142.
- Gaynor EC, te Heesen S, Graham TR, Aebi M, Emr SD. Signal-mediated retrieval of a membrane protein from the Golgi to the ER in yeast. J Cell Biol. 1994;127:653–665. doi: 10.1083/jcb.127.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno RE, Espenshade P, Kaiser CA. SED4encodes a yeast endoplasmic reticulum protein that binds Sec16p and participates in vesicle formation. J Cell Biol. 1996;131:325–338. doi: 10.1083/jcb.131.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson, M. 1992. Amino acid transport in yeast: structure, function and regulation. In Molecular Aspects of Transport Proteins. J.J.H.H.M. de Pont, editor. Elsevier Science Publishers B.V., Amsterdam. 219–245.
- Grenson M, Hou C, Crabeel M. Multiplicity of the amino acid permeases in Saccharomyces cerevisiae.IV. Evidence for a general amino acid permease. J Bacteriol. 1970;103:770–777. doi: 10.1128/jb.103.3.770-777.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZfusions designed to study expression of cloned genes in yeast. Meth Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Haney PM, Mueckler M. Subcellular targeting and regulation of glucose transporters. Curr Top Membr. 1994;41:89–107. [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Harsay E, Bretscher A. Parallel secretory pathways to the cell surface in yeast. J Cell Biol. 1995;131:297–310. doi: 10.1083/jcb.131.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman W. Molecular characterization of the CAN1 locus in Saccharomyces cerevisiae.A transmembrane protein without a N-terminal signal sequence. J Biol Chem. 1985;260:11831–11837. [PubMed] [Google Scholar]
- Horak J. Amino acid transport in eukaryotic microorganisms. Biochim Biophys Acta. 1986;864:223–256. doi: 10.1016/0304-4157(86)90001-8. [DOI] [PubMed] [Google Scholar]
- Horazadovsky BF, DeWald DB, Emr S. Protein transport to the yeast vacuole. Curr Opin Cell Biol. 1995;7:544–551. doi: 10.1016/0955-0674(95)80012-3. [DOI] [PubMed] [Google Scholar]
- Jauniaux JC, Grenson M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae: nucleotide sequence, protein similarity with other baker's yeast amino acid permeases, and nitrogen catabolite repression. Eur J Biochem. 1990;190:39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- James DE, Piper RC. Insulin resistance, diabetes, and the insulin-regulated transport of GLUT-4. J Cell Biol. 1994;126:1123–1126. doi: 10.1083/jcb.126.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, E.W. 1983. Genetic approaches to the study of protease function and proteolysis in Saccharomyces cerevisiae. In Yeast Genetics: Fundamental and Applied Aspects. J.F.T. Spencer, D. Spencer, and A.W. Smith, editors. Springer-Verlag New York, Inc., New York. 167–203.
- Jones EW, Zubenko GS, Parker RR. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. . Genetics. 1982;102:665–677. doi: 10.1093/genetics/102.4.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R. Distinct sets of SECgenes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kölling R, Hollenberg CP. The ABC–transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO (Eur Mol Biol Organ) J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Brandriss MC. Proline transport in Saccharomyces cerevisiae. . J Bacteriol. 1981;148:241–247. doi: 10.1128/jb.148.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl PO, Gimeno CJ, Styles CA, Fink GR. SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell. 1992;71:463–478. doi: 10.1016/0092-8674(92)90515-e. [DOI] [PubMed] [Google Scholar]
- Magasanik, B. 1992. Regulation of nitrogen utilization. In The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Gene Expression. J.R. Broach, E.W. Jones, and J.R. Pringle, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 283–317.
- Nielsen S, Chou CL, Marples D, Christensen EI, Kishore BK, Knepper MA. Vasopressin increases water permeability of kidney collecting duct by inducing translocation of aquaporin-CD water channels to plasma membrane. Proc Natl Acad Sci USA. 1995;92:1013–1017. doi: 10.1073/pnas.92.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Ohsumi Y, Anraku Y. Active transport of basic amino acids driven by a proton motive force in vacuolar membrane vesicles of Saccharomyces cerevisiae. . J Biol Chem. 1981;256:2079–2082. [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immuno-fluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pryer NK, Salama NR, Schekman R, Kaiser CA. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Seeger M, Payne GS, Fuller RS. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol Cell Biol. 1996;7:1667–1677. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Ohsumi Y, Anraku Y. Substrate specificities of active transport for amino acids in vacuolar-membrane vesicles of Saccharomyces cerevisiae.Evidence of seven independent proton/amino acid antiport systems. J Biol Chem. 1984;259:11505–11508. [PubMed] [Google Scholar]
- Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silve S, Volland C, Garnier C, Jund R, Chevallier MR, Haguenhauer-Tsapis R. Membrane association of uracil permease, a polytopic plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- Stanbrough M, Magasanik B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. . J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrchova H, Chevallier MR. Cloning and sequencing of the Saccharomyces cerevisiae gene LYP1coding for the lysine permease. Yeast. 1993;9:771–782. doi: 10.1002/yea.320090711. [DOI] [PubMed] [Google Scholar]
- Tanaka J, Fink G. The histidine permease (HIP1) of Saccharomyces cerevisiae. . Gene. 1985;38:205–214. doi: 10.1016/0378-1119(85)90219-7. [DOI] [PubMed] [Google Scholar]
- Urushidani T, Forte JG. Stimulation-associated redistribution of H+, K+-ATPase activity in isolated gastric glands. Am J Physiol. 1987;252:G458–G465. doi: 10.1152/ajpgi.1987.252.4.G458. [DOI] [PubMed] [Google Scholar]
- Vandenbol M, Jauniaux JC, Grenson M. Nucleotide sequence of the Saccharomyces cerevisiae PUT4proline–permease-encoding gene: similarities between CAN1, HIP1, and PUT4 permeases. Gene. 1989;83:153–159. doi: 10.1016/0378-1119(89)90413-7. [DOI] [PubMed] [Google Scholar]
- Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]