Abstract
We studied the molecular nature of the interaction between the integral membrane protein Sec63p and the lumenal Hsp70 BiP to elucidate their role in the process of precursor transit into the ER of Saccharomyces cerevisiae. A lumenal stretch of Sec63p with homology to the Escherichia coli protein DnaJ is the likely region of interface between Sec63p and BiP. This domain, purified as a fusion protein (63Jp) with glutathione S–transferase (GST), mediated a stable ATP-dependent binding interaction between 63Jp and BiP and stimulated the ATPase activity of BiP. The interaction was highly selective because only BiP was retained on immobilized 63Jp when detergent-solubilized microsomes were mixed with ATP and the fusion protein. GST alone was inactive in these assays. Additionally, a GST fusion containing a point mutation in the lumenal domain of Sec63p did not interact with BiP. Finally, we found that the soluble Sec63p lumenal domain inhibited efficient precursor import into proteoliposomes reconstituted so as to incorporate both BiP and the fusion protein. We conclude that the lumenal domain of Sec63p is sufficient to mediate enzymatic interaction with BiP and that this interaction positioned at the translocation apparatus or translocon at the lumenal face of the ER is vital for protein translocation into the ER.
In the yeast Saccharomyces cerevisiae, molecular chaperones in the cytosol and ER lumen are involved in polypeptide translocation across the ER membrane (for recent reviews on ER translocation see Corsi and Schekman, 1996; Rapoport et al., 1996; Römisch and Corsi, 1996). Molecular chaperones function in a variety of cellular processes by binding unfolded proteins and preventing protein aggregation (Gething and Sambrook, 1992). The ER lumenal protein BiP is a molecular chaperone of the Hsp70 family involved in secretory protein translocation into the ER. BiP is required for efficient import of precursor proteins both in vivo (Vogel et al., 1990; Nguyen et al., 1991) and in vitro (Sanders et al., 1992; Brodsky et al., 1995). Mutations in KAR2, which codes for BiP, cause a block early in translocation before the precursor reaches the translocation pore (Müsch et al., 1992; Sanders et al., 1992) and late in translocation when the precursor is completing transit through the pore (Sanders et al., 1992; Lyman and Schekman, 1995).
As with other Hsp70s, BiP appears to work with a partner protein, Sec63p. In Escherichia coli, the Hsp70, DnaK, functions with a partner protein, DnaJ. Families of both DnaK and DnaJ homologues have been identified in several eukaryotic cellular compartments, including the ER (for review see Caplan et al., 1993; Cyr et al., 1994). Unlike other DnaJ-like proteins identified so far, which are soluble or only peripherally attached to membranes (Caplan et al., 1993; Cyr et al., 1994), Sec63p is an integral membrane protein (Feldheim et al., 1992) spanning the ER membrane three times with a protruding lumenal portion homologous to the J domain, the most highly conserved region of the DnaJ family (Sadler et al., 1989). Sec63p shares 43% identity with DnaJ over a span of 70 amino acids (Sadler et al., 1989; Feldheim et al., 1992). A mutation in a conserved residue (A179T) of this lumenal region of SEC63 (sec63-1; Nelson et al., 1993) causes a defect in precursor translocation across the ER membrane (Rothblatt et al., 1989). This mutation behaves like BiP mutations blocking both early and late phases of translocation in vitro (Sanders et al., 1992; Lyman and Schekman, 1995). sec63-1 and certain kar2 alleles display synthetic lethality, whereas mutations in the cytosolic domain of Sec63p (sec63-101 and sec63-106; Nelson et al., 1993) show no such effect (Scidmore et al., 1993). Additionally, dominant mutations in KAR2 partially suppress the growth and translocation defects of the lumenal sec63-1 mutation (Scidmore et al., 1993). In vitro, these same dominant KAR2 mutations partially relieve the inability of the sec63-1 allele to support complete transit of the precursor through the ER membrane pore (Lyman and Schekman, 1995). Together, these observations provide strong genetic support for an interaction between the J domain of Sec63p and BiP.
A complex of proteins from wild-type yeast membranes has been isolated that includes BiP and Sec63p along with Sec71p and Sec72p (Brodsky and Schekman, 1993), two additional proteins of the translocation apparatus involved in an early step of precursor transit (Feldheim et al., 1993; Kurihara and Silver, 1993; Feldheim and Schekman, 1994; Lyman and Schekman, 1997). This complex and the Sec61p trimeric pore compose the minimal translocation apparatus (or translocon) for in vitro reconstitution of protein import in yeast (Panzner et al., 1995). BiP dissociates from the other members of the complex during isolation of the translocon from sec63-1 membranes (Brodsky and Schekman, 1993). Additionally, in the presence of the nonhydrolyzable analog ATPγS during the last purification step, a significant fraction of BiP dissociates from Sec63p, Sec71p, and Sec72p, implying that BiP interaction with Sec63p is regulated by ATP (Brodsky and Schekman, 1993).
Hsp70s bind and hydrolyze ATP at a site commonly located in the conserved amino-terminal domain resulting in a cycle of ATP hydrolysis that regulates the binding and release of polypeptides at the carboxy-terminal peptide binding domain of the chaperone (for review see Hartl, 1996). The ATPase activity of a number of Hsp70 homologues appears to be regulated by partner proteins often referred to as Hsp40 molecules (Cyr et al., 1994; Rassow et al., 1995). In E. coli, the low intrinsic ATP hydrolysis rate of the Hsp70, DnaK, is stimulated by interaction with its partner protein the Hsp40, DnaJ (Liberek et al., 1991). The stimulation of the ATPase activity of BiP in the ER lumen by a DnaJ homologue located at the membrane would provide an explanation for selective recruitment of BiP to the translocation apparatus in a cycle of ATP hydrolysis.
Neither the molecular determinants of the interaction between Sec63p and BiP nor the effect of Sec63p on the enzymatic activity of BiP is known. To address these issues directly, we constructed a fusion protein with glutathione S–transferase (GST)1 and a portion of the lumenal domain of Sec63p (63Jp), as well as a similar fusion protein with the lumenal domain containing the sec63-1 point mutation (63-1Jp). The 78 amino acids of Sec63p that correspond to the region of homology to DnaJ and the following 23 amino acids of Sec63p up to the third transmembrane domain were joined to the carboxy terminus of GST. The region downstream of the J domain in DnaJ, the glycine/phenylalanine-rich domain, is also required to stimulate the ATPase activity of DnaK (Wall et al., 1994; Karzai and McMacken, 1996; Szabo et al., 1996). Although the corresponding region of Sec63p was not noticeably glycine/phenylalanine rich, the additional amino acids could be important for interaction of the Sec63p lumenal domain with BiP, and thus they were included in our constructs. We discovered that the Sec63p lumenal domain was sufficient to provide specific interaction with BiP and stimulated ATP hydrolysis by BiP. All together, our in vitro studies indicated that the recruitment of BiP to the translocation apparatus through enzymatic interaction with the lumenal domain of Sec63p is necessary for secretory precursor translocation into the ER.
Materials and Methods
Sec63p Lumenal Domain Fusions
Plasmid pGST/63J was made by PCR amplification of pDF41 (Feldheim et al., 1992), which contains the entire coding region of Sec63p. The J domain and the carboxy-terminal DNA sequences that precede the third transmembrane Sec63p domain were amplified by PCR according to the Stratagene (La Jolla, CA) protocol for Pfu Polymerase using the 5′ PCR primer, 5′-CGCGGGGATCCCCACAAAATTATTTGATCCTTA-3′, and the 3′ PCR primer, 5′-CGCGGAATTCTCAGTGGTGGTGGTGGTGGTGTGGAGATGCACTTCCATC-3′. This PCR product made an in-frame fusion with GST in pGEX-3X (Pharmacia LKB Biotechnology, Inc., Piscataway, NJ) at restriction sites BamHI and EcoRI (underlined in the oligos) and introduced six histidines (in italics) at the carboxy terminus just before the stop codon. The resulting plasmid expressed the following chimeric protein (from amino to carboxy terminus): GST fused to the 78 amino acids of the Sec63p J domain plus the 23 successive amino acids of Sec63p with a carboxy-terminal His+ 6-tag.
Plasmid pGST/63-1J was constructed using the PCR primers described above for pGST/63J. The PCR template was genomic DNA prepared from the yeast strain RSY151 (MAT, sec63-1, ura3-52, leu2-3, -112, pep4-3; Rothblatt et al., 1989) according to Hoffman and Winston (1987).
The fidelity of the PCR products generated for constructing pGST/63J, pGST/63-1J, and pAC87-3 (described below) was verified by sequencing the insert regions.
BiP-His+ 6 tag
Full-length BiP without the signal sequence was placed into pRSETB (InVitrogen, San Diego, CA) to provide an amino-terminal His+ 6 sequence for affinity purification from E. coli. The recombinant plasmid, pAC87-3, was made by PCR amplification of KAR2 in two portions using pMR397 (Rose et al., 1989), which contains full-length KAR2 as template DNA. The amino-terminal region of BiP was amplified beginning with the alanine at amino acid position 43, which amino-terminal sequencing showed to be the first amino acid in BiP purified from yeast. Amplification was carried out with the 5′ primer, 5′-CCCGGATCCGGCCGATGATGTAGAAAA-3′, and the 3′ primer, 5′-TTAGCGTCGACTGCAAATG-3′. The 5′ primer contained a BamHI site (underlined) and the 3′ primer introduced a SalI site (underlined) in KAR2, creating a single amino acid change L524V due to changes introduced by the dNTPs indicated in italics. The carboxy-terminal portion of KAR2 was amplified with the 5′ primer, 5′-CATTTGCAGTCGACGCTAA-3′, and 3′ primer, 5′-CCCCGGGCCC/CAGCTGCTACAATTCGTCGTGTT-3′. Restriction sites (underlined) introduced in the carboxy-terminal portion included SalI at the 5′ end and PvuII followed by ApaI at the 3′ end. Each PCR product was first cloned into pBluescript II KS (pBS) (Stratagene) for sequencing. The two halves of KAR2 were subcloned together into pBS by ligating a BamHI/SalI fragment of the amino terminus into pBS containing the carboxy terminus, and then both halves were subcloned together into pRSETB as a BamHI/PvuII fragment.
Purification of GST Fusion Proteins
The lumenal domain of Sec63p was purified from BL21(DE3) E. coli (Studier et al., 1990) expressing pGST/63J. Cells (OD600 1.4) grown in LB plus 100 μg/ml ampicillin at 20°C were induced with 0.2 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) (Sigma Chemical Co., St. Louis, MO) for 75 min. Cells were harvested, washed once in cold 20 mM Hepes, pH 6.8, 150 mM KOAc, 250 mM sorbitol, 2 mM EDTA, and protease inhibitors (0.2 mM ρ-aminobenzamidine, 1 mM ε-amino n-caproic acid, 0.1 mM 4-[2-aminoethyl]-benzenesulfonylfluoride, HCl [AEBSF], and 1 mg/ml leupeptin, pepstatin-A, chymostatin, and aprotinin), and the cell pellet was stored at −20°C. Subsequent steps were carried out at 4°C. Cells (∼3 g/liter) from 1 liter of culture for 63Jp and from 4 liters of culture for 63-1Jp were then resuspended in 4 ml (per liter of cell pellet) of buffer A (PBS, pH 8, 2 mM EDTA, 2 mM β-mercaptoethanol, and protease inhibitors as described above). Because the mutant fusion protein was less soluble, the scale of this purification was increased fourfold. PMSF (1 mM) and 0.1% Triton X-100 were added to the resuspended cells, which were then disrupted with three 30-s bursts (interspersed with 30 s on ice) of sonication using a microtip at 40 W with a Heat Systems Sonicator (Farmingdale, NY). The lysate was centrifuged at 11,000 g (10 min), and the supernatant fraction was subjected to further centrifugation at 100,000 g (30 min). The high-speed supernatant was applied to a 0.5-ml glutathione agarose (Sigma Chemical Co.) column equilibrated with buffer A. The column was washed successively with 40 ml of buffer A, 25 ml of buffer A + 1 M KCl and 0.1% Triton X-100, 50 ml of 50 mM Tris, pH 7.5, 2 mM ATP, 10 mM MgOAc2, 200 mM KOAc (this wash eluted contaminating DnaK), and 20 ml buffer A without EDTA. Bound proteins were eluted successively with 10 ml of PBS, pH 8.0, 20 mM KPi, pH 7, 10 mM reduced glutathione (Sigma Chemical Co.), 4 ml of 50 mM Tris, pH 8, 120 mM KOAc, 20 mM glutathione, and 2 ml 50 mM Tris, pH 8, 200 mM KOAc, 25 mM glutathione, 0.1% Triton X-100. All fractions were pooled and applied to a 0.5-ml Nickel-NTA agarose (QIAGEN, Inc., Chatsworth, CA) column equilibrated with buffer B (50 mM Hepes, pH 6.8, 100 mM KOAc, 5 mM MgOAc2, 5 mM imidazole, pH 7, 2 mM β-mercaptoethanol). The column was washed with 25 ml of 20 mM Hepes, pH 6.8, 500 mM KOAc, 10 mM MgOAc2, 60 mM imidazole, pH 7, followed by 15 ml of buffer C (20 mM Hepes, pH 6.8, 50 mM KOAc, 5 mM MgOAc2, 50 mM imidazole, pH 7). Purified fusion protein was eluted with buffer C containing 300 mM imidazole, pH 7. Peak fractions, as determined by immunoblot, were pooled and dialyzed into buffer 88lK (20 mM Hepes, pH 6.8, 75 mM KOAc, 250 mM sorbitol, 5 mM MgOAc2, 10% glycerol) and frozen in small aliquots in liquid nitrogen. Since the ratio of contaminants to fusion protein was higher for the mutant fusion protein, we were unable to achieve as pure a preparation of 63-1Jp as we obtained for 63Jp. Contaminating proteins in the 63-1Jp did not interfere in our assays. The yield of 63Jp from 2,000 OD600 of cells was ∼750 μg and of 63-1Jp from 8,000 OD600 of cells was ∼400 μg.
GST was purified according to the above protocol except that the Nickel-NTA step was omitted. After elution from glutathione agarose, the purified GST was dialyzed directly into buffer 88lK and frozen. The yield of GST from 2,000 OD600 of cells was ∼8 mg.
Purification of BiP
BL21 (DE3) E. coli cells expressing pAC87-3 were grown at 30°C in LB plus 100 μg/ml ampicillin to OD600 ∼0.8, and BiP expression was induced by adding IPTG to 1 mM. After 2.5 h of induction, cells were harvested and washed once with water, and the cell pellet was stored at −20°C. Cell pellets from 4 liters of culture were resuspended in buffer D (50 mM Hepes, pH 6.8, 400 mM KOAc, 5 mM MgOAc2, 3.5 mM β-mercaptoethanol, 2 mM imidazole, pH 7, plus protease inhibitors except AEBSF as described for buffer A), and the lysate was prepared as for 63Jp above. Subsequent steps were performed at 4°C. The cleared lysate was applied to a 1.5-ml Nickel-NTA agarose column and washed with 20 ml of buffer D, 50 ml of 20 mM Hepes, pH 6.8, 1 M KOAc, 0.1% Triton X-100, 10 mM imidazole, pH 7, 5 mM MgOAc2, 3.5 mM β-mercaptoethanol, and 100 ml buffer E (20 mM Hepes, pH 6.8, 250 mM KOAc, 25 mM imidazole, pH 7, 5 mM MgOAc2, 3.5 mM β-mercaptoethanol). Proteins were eluted with buffer E containing a final concentration of 200 mM imidazole, pH 7. The eluate was pooled and applied to a 1.5-ml Q-Sepharose fast flow (Pharmacia LKB Biotechnology, Inc.) column equilibrated with buffer F (20 mM Hepes, pH 6.8, 375 mM KOAc, 5 mM MgOAc2, 3.5 mM β-mercaptoethanol), and the column was washed with 30 ml of buffer F followed by 100 ml of buffer F + 400 mM KOAc. A highly purified fraction of BiP detected by immunoblot was eluted from the column with 15 ml of buffer F + 500 mM KOAc. The eluate was pooled and dialyzed against buffer G (60 mM KPi, pH 7, 50 mM KOAc, 5 mM MgOAc2, 2 mM β-mercaptoethanol). The dialysate was then passed over a 1-ml hydroxyapatite column (Bernardi, 1971) equilibrated with buffer G and then washed with 40 ml buffer G (80 mM KPi final concentration) followed by 10 ml buffer G (100 mM KPi final concentration). BiP was eluted with 5 ml of 400 mM KPi, pH 7, 50 mM KOAc, 5 mM MgOAc2. The peak of protein, determined by immunoblot, was dialyzed against B88 (buffer 88lK with 150 mM final concentration KOAc) supplemented with 10% (wt/vol) glycerol, and aliquots were frozen in liquid nitrogen. From 8,000 OD600 of cells, we obtained 250 μg of >95% pure BiP. Improved yields were obtained with similar results in our assays if we pooled more column fractions at the expense of purity.
ATPase Assay
63Jp, 63-1Jp, or GST was added as indicated to purified BiP in 50-μl reactions containing 50 mM Hepes, pH 6.8, 50 mM NaCl, 10 mM DTT, 2 mM MgCl2, 100 μM ATP, and 0.5 μCi [γ-32P]ATP (DuPont/NEN, Boston, MA). All reactions were controlled for changes in buffer and salt concentrations due to increasing amounts of added proteins; K+ was adjusted to 20 mM in each assay. Incubations were carried out at 25 or 4°C. At 10-min intervals, 1 μl from each reaction was spotted on polyethyleneimine cellulose thin layer plates (Aldrich Chemical Co., Milwaukee, WI) and chromatographed according to Shlomai and Kornberg (1980). The percentage of 32Pi released (versus the total label added) was determined using a Phosphorimager (Molecular Dynamics, Sunnyvale, CA).
BiP-binding Assay
Glutathione beads (immobilized on cross-linked 4% agarose) (Sigma Chemical Co.) were equilibrated with binding buffer (20 mM Hepes, pH 6.8 [or 20 mM Tris, pH 8, as specified], 100 mM KCl, 5 mM MgCl2, 0.1% NP-40, 2% [wt/vol] glycerol, 1 mM DTT, 0.2 mM AEBSF, and 1 mM EDTA). Reactions contained 20 μl of a 50% suspension of glutathione agarose. 63Jp, 63-1Jp, or GST (3 μg) was added to the beads, the volume was increased to 50 μl with binding buffer, and tubes were rotated at 4°C (1 h). Unbound proteins were collected by centrifugation at 3000 g (2 min) at 4°C followed by two 50-μl washes with binding buffer. Purified BiP (2 μg) and nucleotide as specified was then added in a final volume of 50 μl, and samples were rotated at 4°C (2 h). Unbound protein was collected in a series of four 50-μl washes with binding buffer as above. Proteins remaining on the glutathione beads were solubilized in Laemmli sample buffer, separated by SDS-PAGE, and visualized by staining with Coomassie brilliant blue R-250 (BioRad Labs, Hercules, CA). The amount of BiP associated with 63Jp, 63-1Jp, or GST on the glutathione beads was determined by a comparison with a standard curve of purified BiP using the software Imagequant v1.1 (Molecular Dynamics).
To determine the nucleotide-bound state of BiP associated with 63Jp, we performed standard binding assays adding BiP to the 63Jp or GST glutathione agarose affinity matrices described above with 50 μM ATP and 5 μCi [α-32P]ATP (DuPont/NEN). After a 75-min incubation at 4°C, the radioactivity was observed in the unbound and wash fractions by spotting 1 μl of the bead supernatant to polyethyleneimine cellulose thin layer plates. The BiP that remained bound to the 63Jp or GST matrix after washing was solubilized in 2% SDS for 5 min at 95°C, and the supernatant was also spotted on the thin layer chromotography plates. After chromatography, we determined the nucleotide present in the supernatants in relation to the mobility of nonradioactive ADP and ATP. Reactions containing only nonradioactive ATP were incubated in parallel to observe by SDS-PAGE the amount of BiP bound to the affinity matrices.
For binding of BiP from solubilized membranes, we used glutathione agarose beads linked to GST fusion proteins as described above. Wild-type membranes were solubilized in binding buffer containing 0.5% Triton X-100 (final concentration in reaction), and insoluble material was centrifuged at 100,000 g (30 min). The supernatant was mixed with beads in the presence or absence of 1 mM ATP. Binding reactions performed with solubilized membranes were processed as above except that washes included 0.5% Triton X-100, and SDS–polyacrylamide gels were transferred to nitrocellulose and probed with BiP antibody (Rose et al., 1989) followed by 125I protein A secondary antibody.
Reconstituted Proteoliposome Translocation Assay
Yeast microsomes were prepared from the strain MS137 (MATα, kar2-159, ura 3-52, leu 2-3, -112, ade 2-101; Vogel et al., 1989) as described in Brodsky et al. (1993) based on the Rothblatt and Meyer protocol (1986). Proteoliposomes were formed from a detergent lysate of the kar2-159 membranes according to Brodsky and Schekman (1993) with the following minor modifications: 160 μl of 100,000 g (30 min) supernatant fraction from a 1-ml microsome solubilization reaction was mixed with 12.8 μg of purified BiP and either 63Jp, 63-1Jp, GST, or buffer alone in the amounts indicated. Prepro–α-factor (ppαF) translocation into proteoliposomes was carried out as described in Brodsky and Schekman (1993).
Results
Isolation of the Sec63p Lumenal Domain
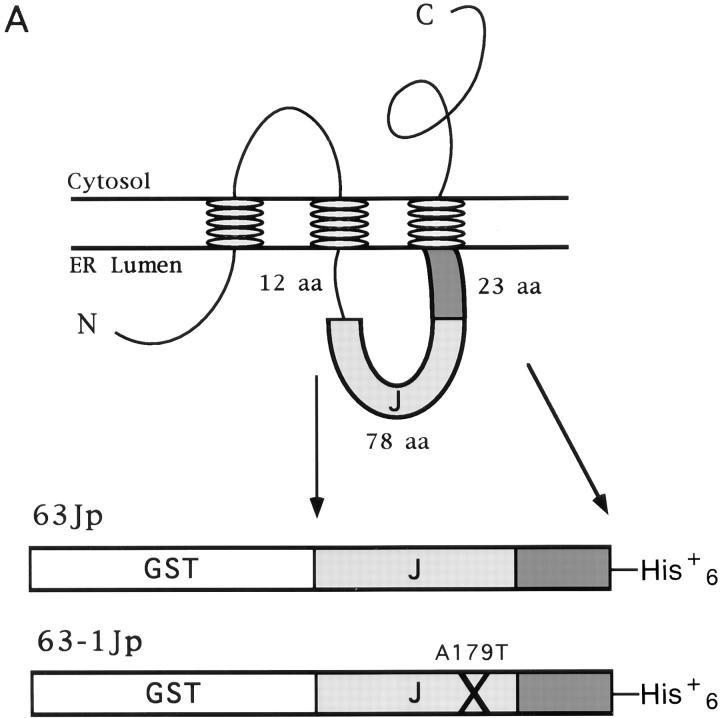
Analysis of the biochemical interaction between Sec63p and BiP has been complicated by the fact that Sec63p is an integral membrane protein and is associated with other integral and peripheral membrane proteins (Brodsky and Schekman, 1993; Panzner et al., 1995). Although previous genetic (Scidmore et al., 1993) and biochemical (Brodsky and Schekman, 1993) data indicated a direct interaction between Sec63p and BiP, neither the precise region of interface nor the effect of Sec63p on the enzymatic activity of BiP has been characterized. To facilitate biochemical studies on the interaction of BiP and the translocon, we isolated the lumenal domain of Sec63p as a GST fusion. We constructed a plasmid that encoded a GST fusion protein (63Jp) containing 101 residues beginning with the first amino acid in Sec63p that corresponds to the start of homology to the amino terminus of DnaJ (Sadler et al., 1989; Feldheim et al., 1992) and continuing up to the third transmembrane domain of Sec63p (Fig. 1 A). To assess the specificity of the Sec63p lumenal domain interaction with BiP, we also constructed a parallel GST fusion protein (63-1Jp) containing the single amino acid change (A179T) resulting in the J domain mutation known as sec63-1 (Nelson et al., 1993) (Fig. 1 A). E. coli strains harboring the pGST/ 63J and pGST/63-1J plasmids were used to express the fusion proteins for purification (see Materials and Methods). Fractions from the two affinity column purification steps (glutathione agarose and Nickel-NTA agarose) containing the peak of protein are shown in Fig. 1 B. As a negative control in our experiments, GST alone was purified using only the glutathione agarose step (not shown).
Figure 1.
Construction and purification of the GST–Sec63p lumenal domain fusion protein. (A) The topology of Sec63p in the ER membrane is shown schematically according to Feldheim et al. (1992). The bottom portion of A depicts GST fused to either a wild-type Sec63p lumenal region (63Jp) or a mutant Sec63-1p lumenal region (63-1Jp) containing the A179T mutation found in sec63-1 (Nelson et al., 1993). (B) Fusions were purified as described in Materials and Methods. The peak fractions from the glutathione agarose and Ni2+-NTA agarose columns are shown on 12.5% SDS–polyacrylamide gels stained with Coomassie brilliant blue R-250.
The Lumenal Domain of Sec63p Stimulates the ATPase Activity of BiP
One of the hallmark interactions of the DnaK–DnaJ partnership is the ability of DnaJ to stimulate the low intrinsic ATPase activity of DnaK (for review see Rassow et al., 1995). In yeast cytosol, it has been shown that the DnaJ-like protein, Ydj1p, stimulates the ATPase activity of the DnaK homologue, Ssa1p (Cyr et al., 1992). We analyzed the effect of 63Jp on the ATPase activity of yeast BiP that was overexpressed and purified from E. coli. Using our assay conditions, the ATPase specific activity of BiP was 2–5 pmol/min/μg. We found that 63Jp reproducibly stimulated the ATPase activity of BiP up to fivefold (Fig. 2). This stimulation was specific to the Sec63p portion of the 63Jp fusion, as GST alone did not stimulate BiP (Fig. 2). Maximum stimulation depended on a functional J domain because the 63-1Jp fusion, which bears a lesion in the J domain, did not increase BiP activity (Fig. 2). Although results varied among preparations, 63-1Jp showed from zero- to twofold stimulation at 25°C. In the instances where stimulation by 63-1Jp occurred at 25°C, no stimulation was observed at 37°C, whereas 63Jp stimulated fivefold at 25°C (Fig. 2) and two- to threefold at 37°C (data not shown). Neither 63Jp, 63-1Jp, nor GST alone exhibited any detectable ATPase activity (data not shown). We conclude that an intact lumenal domain of Sec63p interacts functionally to stimulate ATP hydrolysis by BiP and that the amino acids of the J domain are important for this activity because substitution at an invariant residue abolishes activity.
Figure 2.
The Sec63p lumenal domain stimulates the ATPase of BiP. BiP (0.7 μg) was incubated at 25°C, pH 6.8, with increasing amounts of the Sec63p lumenal domain (63Jp) or the Sec63-1p lumenal domain (63-1Jp) fused to GST, or with GST alone. ATP hydrolysis (%) was measured by quantifying the percentage of 32Pi released from [γ-32P]ATP. All points reported represent 20 min of incubation at 25°C, which was determined to be in the linear range of the reactions.
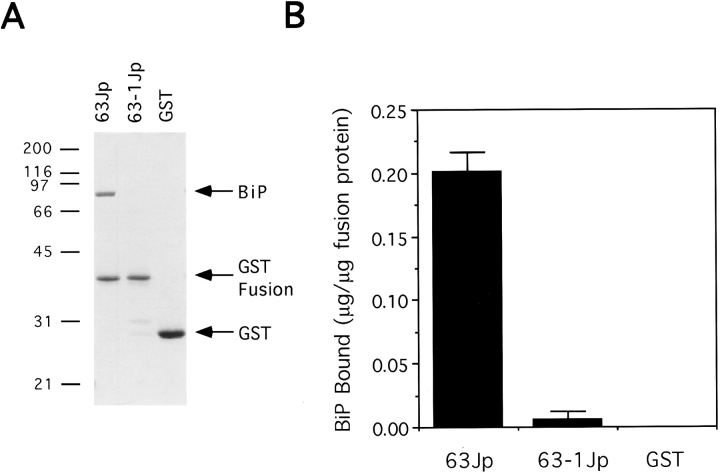
The Lumenal Domain of Sec63p Stably Interacts with BiP
Previous data (Brodsky and Schekman, 1993) suggested that interaction between BiP and Sec63p is mediated by the lumenal domain of Sec63p. However, the complex isolated in these experiments included several other proteins of the ER translocon, and it is not known if the lumenal domain of Sec63p is sufficient for the interaction to occur. To address this question, we examined BiP binding to the GST–Sec63p fusion protein complexed with glutathione beads. Purified BiP was incubated at 4°C with the 63Jp affinity matrix, and any unbound BiP was washed away. Proteins dissociated from the agarose beads by treatment with sample buffer containing SDS were analyzed by SDS-PAGE. Fig. 3 A shows that upon incubation with 1 mM ATP at pH 6.8, a fraction of purified BiP associated with the 63Jp beads. We determined that 0.2 μg of BiP was associated per μg of 63Jp (Fig. 3 B). However, we were not able to calculate a true binding constant because of the uncertain fraction of immobilized 63Jp available and competent for binding to BiP. Fig. 3, A and B, shows that only 0.005 μg of BiP binds to the 63-1Jp matrix, whereas virtually no BiP was found associated with the GST matrix. Thus, as in the ATPase assays, BiP interacts only with an intact J sequence.
Figure 3.
BiP binds to the lumenal domain of Sec63p. (A) Glutathione agarose beads were first incubated with the proteins indicated at the top of the lanes and second with purified BiP in 1 mM ATP at pH 6.8. Beads were then washed as described in Materials and Methods, and the final eluate was resolved on a 12.5% SDS–polyacrylamide gel that was stained with Coomassie brilliant blue R-250. Molecular mass markers (in kD) are indicated at left. (B) BiP was quantified by comparing the signal shown with known amounts (not shown) using a scanner (model ScanMakerIII; Microtek, Redondo Beach, CA) and Imagequant v1.1 software (Molecular Dynamics).
These initial binding experiments were conducted in the presence of 1 mM ATP (a saturating amount; data not shown) to approximate cellular conditions. However, the average K m (ATP) value for Hsp70s is ⩽1 μM (McKay et al., 1994). To investigate the nucleotide requirements for binding of BiP to 63Jp, we assayed binding at lowered ATP levels as well as in the presence of ADP or the nonhydrolyzable analogues ATPγS and AMP-PNP. Fig. 4 A, lane 2, shows that a 10-fold decrease in ATP (100 μM) resulted reproducibly in less than a twofold decrease in BiP binding to 63Jp. No detectable binding occurred in the absence of nucleotide (Fig. 4 A, lane 7) or in the presence of the nonhydrolyzable analogues, ATPγS or AMP-PNP (Fig. 4 A, lanes 5 and 6). To rule out the possibility that ATPγS and AMP-PNP did not bind to BiP, we mixed ATP with the analogues and found that each inhibited BiP-63Jp binding. Fig. 4 A, lanes 9–12, shows that 1 mM ATPγS reduced BiP binding in the presence of 1 mM ATP and 0.1 mM ATP by 2.8 and 16 times, respectively, whereas 1 mM AMP-PNP inhibited 1.8 and 3.4 times, respectively. The omission of Mg2+ from the binding buffer substantially reduced (approximately sevenfold) the amount of BiP bound to the 63Jp affinity matrix (Fig. 4 A, lane 8). ADP afforded a low level of BiP binding (Fig. 4 A, lanes 3 and 4), which we attributed to a low level of contamination by ATP (0.6% in the Sigma ADP preparation). Taken together, these results indicate that ATP hydrolysis is necessary for BiP to bind to the lumenal region of Sec63p.
Figure 4.
The association of BiP and the Sec63p lumenal domain depends on the presence of hydrolyzable ATP. Binding reactions using 63Jp glutathione agarose beads were performed as in Fig. 3. (A) Final concentration of nucleotide in the reaction is indicated. The entire binding reaction depicted in lane 8 was done in the absence of Mg2+ plus 10 mM EDTA. (B) Bound nucleotides associated with glutathione beads (bound to either 63Jp or GST as indicated above the lanes) in the presence (+) or absence (−) of BiP after washing the beads were visualized by thin layer chromatography and a Phosphorimager.
Readily measurable ATP hydrolysis accompanied binding of BiP to 63Jp. Standard ATPase assays conducted at the binding temperature (4°C) reduced the rate of 63Jp-stimulated BiP activity by fivefold in comparison to the rate at 25°C (data not shown). To detect ADP remaining bound to the BiP–63Jp complex, we included [α-32P]ATP in a binding reaction. Significant levels of ADP were associated with glutathione beads only in the presence of both 63Jp and BiP (Fig. 4 B). In an average of four experiments, ADP in the BiP + 63Jp lane was approximately nine times above background in the GST + BiP control. ATP bound nonspecifically to both 63Jp and GST (with or without BiP) affinity matrices.
Next, we examined the specificity with which the Sec63p lumenal domain recruits BiP from solubilized crude yeast membranes. The 63Jp affinity matrix was mixed with a yeast microsomal fraction that had been solubilized with Triton X-100. This experiment, shown in Fig. 5, demonstrated that 63Jp selectively bound BiP from a large pool of microsomal protein, and this binding required ATP. Most of the solubilized ER proteins were recovered in the flow through fraction (Fig. 5, lane FT), and only two prominent proteins remained on the washed beads (Fig. 5, lane B, +ATP). The appearance of the lower mobility band, identified by immunoblot shown in the bottom panel of Fig. 5 as BiP, depended on the presence of ATP, whereas the higher mobility band, identified as 63Jp, remained when ATP was omitted (Fig. 5, lane B, −ATP). 63-1Jp displayed a conditional defect in binding BiP from solubilized microsomes. The experiment in Fig. 6 showed that 63-1Jp bound to yeast microsomal BiP at a neutral pH of 6.8 but not at pH 8.0. Binding of BiP to 63Jp was decreased only marginally at pH 8.0. We interpret these results to mean that in our in vitro binding conditions, BiP is the major ER protein that can associate with 63Jp and that 63-1Jp has reduced but significant binding ability.
Figure 5.
BiP solubilized from yeast microsomes binds to 63Jp in an ATP- dependent manner. Soluble proteins from microsomes solubilized with Triton X-100 were mixed with a glutathione agarose 63Jp affinity matrix either with 1 mM ATP (+ ATP) or no ATP (− ATP) added. Binding reactions were at pH 6.8. The top panel is a 12.5% SDS–polyacrylamide gel stained with Coomassie brilliant blue R-250. Molecular mass markers (in kD) are on the left. P, insoluble microsomal proteins (pellet); FT, proteins not bound to affinity matrix (flow through); W, wash fractions; B, proteins still bound to beads after washing. The bottom panel is the region between 97 and 66 kD of an immunoblot probed with anti-BiP antibody.
Figure 6.
63Jp binds selectively to BiP from detergent-solubilized microsomal membranes, and 63-1Jp associates with BiP from yeast microsomes in a pH-dependent manner. (A) 63Jp or 63-1Jp bound to glutathione agarose beads was incubated with solubilized proteins from a Triton X-100 detergent extract of wild-type yeast membranes in the presence of 1 mM ATP. The entire binding reaction was carried out at pH 6.8 or 8.0, as indicated. Protein profiles are shown on a 12.5% SDS–polyacrylamide gel stained with Coomassie brilliant blue R-250. All fractions are shown for 63Jp binding at pH 6.8; for the other conditions, only the eluate from the beads is shown. Molecular mass markers (in kD) are indicated on the left. L, microsomal detergent extract added to glutathione agarose beads; FT, protein not bound to beads after incubation with detergent lysate; W, material washed off the beads using binding buffer; B, proteins bound to glutathione agarose beads after washes. (B) To quantify the amount of BiP binding from the microsomal extracts, eluate fractions of binding assays were blotted to nitrocellulose and probed with anti-BiP antibody followed by 125I–protein A secondary antibody. The amount of BiP present in the eluate was then compared to a dilution series of the Triton X-100 extracts used in the experiment (not shown) to determine the percentage of BiP bound.
The Soluble Sec63p Lumenal Domain Competes with Sec63p during Precursor Translocation In Vitro
To explore the role of the lumenal domain in the context of the intact Sec63p, we performed reconstituted translocation reactions using proteoliposomes containing 63Jp. Two alternatives were considered to anticipate the influence of the soluble Sec63p lumenal domain on translocation into proteoliposomes. If Sec63p functions principally to regulate the ATPase activity of BiP, then the kinetics of import and perhaps the absolute level of import into proteoliposomes would be stimulated by 63Jp. Alternatively, if Sec63p registers BiP at the Sec61p channel, 63Jp in the proteoliposome may remove BiP from the vicinity of the translocon. To distinguish between these possibilities, we made proteoliposomes from kar2-159 membranes, which do not support translocation of ppαF unless exogenous wild-type BiP is included during the dialysis step used to reconstitute functional vesicles. Translocation was assessed by the appearance of protease-protected, signal- sequence cleaved pro–α-factor (pαF) (Fig. 7 A). When BiP was added to kar2-159 proteoliposomes to a final concentration of 8% of total protein, 12% of the precursor ppαF was translocated (Fig. 7 B). In addition to BiP, when 63Jp was added in the experiment to 0.75 and 1.5% of the total protein concentration (0.5× 63Jp and 1× 63Jp, respectively), ppαF translocation was decreased by approximately four- and ninefold, respectively (Fig. 7, A and B). In contrast, when the same amount of 63-1Jp or GST was added to proteoliposomes along with BiP, no striking inhibition of ppαF translocation was observed (Fig. 7, A and B, right portions of the gel and graph). These data reveal that the Sec63p lumenal domain competes with the intact Sec63p in the membrane to sequester BiP from the translocon, thus impairing the ability of BiP to support translocation.
Figure 7.
The soluble Sec63p lumenal domain inhibits the ability of BiP to support ER translocation. (A) Proteoliposomes were made from kar2-159 microsomes with or without BiP added to 8% of total protein (12.8 μg). As indicated, in addition to BiP some proteoliposomes contained 63Jp added to 0.75% of total protein (0.5×; 1.2 μg) or either 63Jp, 63-1Jp, or GST added to 1.5% of total protein (1×; 2.4 μg). Translocation was assayed using [35S]methionine-labeled ppαF as a substrate. Aliquots from the translocation reactions were untreated (lane 1), treated with trypsin (lane 2), or treated with trypsin plus Triton X-100 (lane 3) and resolved on a 10% SDS–polyacrylamide gel. Translocation efficiency was determined using a Phosphorimager to calculate (pαf [lane 2] − pαf [lane 3])/(pαf + ppαF [lane 1]). Quantification is reported in B.
Discussion
A number of studies have indicated that interaction of the DnaJ homologue Sec63p with the Hsp70 BiP is required for the translocation of secretory precursors across the ER membrane in S. cerevisiae. To facilitate analysis of the BiP and Sec63p interaction, we created a fusion of the lumenal domain of Sec63p to GST (63Jp) and purified this fusion protein for functional assays. We found that the lumenal domain of Sec63p is sufficient to mediate stable interaction between Sec63p and BiP as well as to stimulate the low intrinsic ATPase activity of BiP. In a posttranslational import reaction dependent on exogenously added BiP, the addition of the soluble Sec63p lumenal domain inhibited translocation, presumably by titrating the BiP away from the full-length Sec63p in the translocation apparatus. The ability of the lumenal domain to interact with BiP, stimulate its ATPase activity, and act as an inhibitor of translocation all depended on the wild-type sequences of the J domain. A point mutation in a conserved residue of this domain rendered 63-1Jp ineffective in these assays. Although the importance of the additional 23 amino acids of Sec63p (Fig. 1 A) included in our fusion proteins was not rigorously tested here, our data support Karzai and McMacken's (1996) hypothesis that any unstructured and flexible (though not necessarily glycine/phenylalanine-rich) region adjacent to a J domain will promote DnaK– DnaJ interactions.
Previous data from Brodsky and Schekman (1993) suggested that the interaction between BiP and Sec63p requires the presence of hydrolyzable ATP. We found that ATP was essential for stable association of BiP with the Sec63p lumenal domain (Figs. 4 A and 5). Our experiments suggest that BiP binds to ATP (depicted in model; Fig. 8, step 1) and interaction with the Sec63p lumenal domain stimulates hydrolysis, producing a complex that retains BiP in the ADP-bound form (Fig. 8, step 2). Several observations confirm that BiP and 63Jp represent a DnaK– DnaJ analogue rather than an Hsp70-unfolded protein complex. First, our finding that ATP stimulates whereas ATP analogues inhibit binding of BiP to 63Jp mirrors the findings of Wawrzynów and Zylicz (1995), who observed that DnaK–DnaJ complex formation is stimulated by ATP and inhibited by nonhydrolyzable ATP analogues, whereas DnaK–protein substrate complex formation is inhibited by ATP and stimulated by ATP analogues. Second, the observation that a point mutation in an invariant amino acid in the J domain of Sec63p abolished or destabilized binding indicates a requirement for specific amino acids as opposed to an unfolded region of a protein for a BiP–63Jp interaction. Recent results reported by Holstein et al. (1996) show a similar situation in mammalian cells in the interaction of the DnaJ-like protein auxilin and the cytosolic hsc70 that dissociates clathrin-coated vesicles. Holstein and colleagues observe that a GST fusion protein containing auxilin's carboxy-terminal J domain requires hydrolyzable ATP to bind to hsc70, and hsc70, once bound to the auxilin, is bound to ADP and Pi. The physical interaction between DnaK–DnaJ proteins, therefore, seems likely to occur by a universal mechanism.
Figure 8.
A model for the interaction between Sec63p and BiP in posttranslational translocation across the ER membrane. (1) Our experiments suggest that BiP is recruited to the translocation apparatus by the lumenal domain of Sec63p. (2) The lumenal domain of Sec63p stimulates ATP hydrolysis by BiP to promote stable binding of BiP to the translocon. BiP may continue to associate with Sec63p while binding to the unfolded precursor protein emerging from the Sec61p pore. (3) The precursor or an unidentified protein then would catalyze nucleotide exchange in the ATP binding site of BiP, allowing BiP to undergo another cycle of interaction with Sec63p. This model does not include other precursor interactions with ER chaperones that may execute final secretory protein folding.
An alternative explanation for the nucleotide requirement in our binding assay is that ATP is required to change the oligomeric state of BiP. ATP could create BiP monomers that would be competent to bind to 63Jp rather than to directly affect BiP association with 63Jp. Both Gao et al. (1996) and Benaroudj et al. (1996) have observed that ATP converts cytosolic Hsc70 from oligomers to monomers. We evaluated our purified BiP on native gels according to the protocol described by Freiden et al. (1992). The preparation appeared to be composed almost exclusively of monomers, and incubation with either ATP or ATPγS before loading BiP on the native gel did not alter its mobility (data not shown). Thus, the formation of a BiP–63Jp complex in our experiments seems unlikely to rely on ATP-dependent destabilization of BiP oligomers.
Our results indicate that the interaction between Sec63p and BiP is direct, although the dynamics of the interaction may be modulated by other components of the translocation apparatus. As previously suggested by Brodsky and Schekman (1993), after hydrolyzing ATP, BiP in the ADP-bound form is likely to be anchored to Sec63p near the translocation complex when precursors begin their transit through the pore (Fig. 8, step 2). This prediction is supported by our observation that [α-32P]ADP is retained in the BiP–63Jp complex. Secretory precursor binding to the cytosolic face of the translocon may induce transmembrane structural changes in Sec63p that influence lumenal domain interaction with BiP. Conversely, BiP and ATP cause secretory precursors to disengage from several translocon subunits on the cytosolic face of the ER in preparation for transport through the Sec61p channel (Lyman and Schekman, 1997) and may do so by causing structural changes in the cytosolic portion of Sec63p or other translocon subunits. When the precursor protein emerges on the lumenal side of the ER membrane, the ADP-bound form of BiP may stably bind the exposed hydrophobic regions of unfolded polypeptides (Fig. 8, step 2) as observed for ADP-bound DnaK (Palleros et al., 1994). In analogy to the Hsp70 Ssa1p in the yeast cytosol (Zeigelhoffer et al., 1995) or mammalian cytosolic Hsp70 (Sadis and Hightower, 1992), the unfolded precursor protein may accelerate ATP– ADP exchange. Alternatively, an as yet unidentified protein, acting like GrpE, which enhances nucleotide exchange on DnaK in E. coli (Liberek et al., 1991), may be responsible for nucleotide exchange in the ATP binding site of BiP in the lumen. Once BiP is bound to ATP, it may release the unfolded region of the secretory precursor and be free to interact again with Sec63p (Fig. 8, step 3). The cooperation of BiP and Sec63p in the successive binding and release of precursor is consistent with a “translocation motor” mechanism in which the precursor is pulled through the translocation pore. Such a model has previously been proposed to explain the import of mitochondrial proteins, invoking mitochondrial Hsp70 as the motor that interacts with the peripherally associated inner matrix membrane protein Tim44 (which has very limited homology to DnaJ) (for review see Glick, 1995; for comparison between ER and mitochondrial Hsp70s in protein import see Brodsky, 1996).
Our results are in accord with the suggestion that Sec63p acts as an anchor to localize BiP to the site of translocation (Brodsky and Schekman, 1994) and imply that Sec63p plays an important role in regulating the dynamics of BiP with the translocon by regulating ATP hydrolysis. The sec63-1 allele, which destabilizes interaction between Sec63p and BiP (Brodsky and Schekman, 1993), is defective for translocation in vivo and in vitro (Rothblatt et al., 1989) and is unable to support the complete passage of a precursor protein out of the translocation pore and into the lumen (Lyman and Schekman, 1995). The Sec63-1p mutant protein is labile or deficient both for binding of BiP and for stimulation of ATPase activity (Figs. 2, 3, and 6). We found that although 63-1Jp did not bind appreciably to purified BiP, 63-1Jp bound a fraction of BiP from detergent-solubilized membranes at neutral pH (6.8), but not at pH 8.0 (Fig. 5). Thus, at least at neutral pH, the mutant fusion protein retains some native structure. The mutant protein is essentially inactive in stimulating the ATPase activity of BiP (Fig. 2); therefore, it is unlikely to mimic the effect of unfolded proteins, which increase ATP hydrolysis by BiP (Flynn et al., 1989, 1991; Blond-Elguindi et al., 1993).
The reason for the distinction between BiP from crude membranes and pure BiP in regard to binding 63-1Jp is not obvious. Differences between native and bacterially expressed BiP have been reported (Blond-Elguindi et al., 1993). However, Wei and Hendershot (1995) found that both hamster BiP and bacterially expressed BiP behave identically with respect to oligomeric state, protease digestion patterns, and ATPase properties. Perhaps the presence of full-length Sec63p in the detergent extract stabilizes the interaction by converting BiP to the ADP-bound form, which may then bind to 63-1Jp. This possibility seems unlikely because BiP from wild-type and sec63-1 membranes bound 63-1Jp equally well at pH 6.8 (data not shown). Possibly, other proteins that stabilize a labile interaction between BiP and 63-1Jp are present in the microsomal detergent extract.
The ER lumen contains another DnaJ homologue, Scj1p (Schlenstedt et al., 1995), and another Hsp70 protein, Lhs1p (Craven et al., 1996); however, unlike Sec63p and BiP, Scj1p and Lhs1p are dispensable. Schlenstedt et al. (1995) demonstrated that the Sec63p lumenal domain can be replaced with the corresponding region of Scj1p in vivo, indicating that the lumenal domain may be an interchangeable module that is dependent on its context in the protein for specificity. How Sec63p and BiP may cooperate with Scj1p and Lhs1p to support translocation or assist with protein folding remains unexplored.
The lumenal domain of Sec63p may directly recruit portions of a translocating polypeptide that emerge on the lumenal surface of the ER. DnaJ binds denatured polypeptides (Langer et al., 1992; Wawrzynów et al., 1995; Szabo et al., 1996), and both DnaJ and a yeast cytosolic homologue Ydj1p prevent protein aggregation (Langer et al., 1992; Cyr, 1995). Precursors artificially blocked in transit can be cross-linked to BiP (Müsch et al., 1992; Sanders et al., 1992). However, the orientation of the cross-linked BiP, e.g., bound to Sec63p or free in the lumen, is not known. Binding reactions with the intact translocon, or with subcomplexes of proteins in the translocation apparatus, and denatured precursor may address the location of the translocating precursor in relation to BiP and the Sec63p lumenal domain.
Acknowledgments
The authors wish to thank the following: members of the Schekman lab for helpful advice and discussions, especially Joe Campbell, Susie Lyman for BiP purified from yeast, Jay Chiu for technical assistance, the Rose Lab (Princeton University, Princeton, NJ) for strains and plasmids, and Susie Lyman, Rien Pilon, Meta Kuehn, and Jeff Brodsky for manuscript improvements. We thank Arie Admon for performing the amino-terminal sequencing of yeast BiP. We especially appreciate computer assistance from Jon Bertsch.
This work was supported by National Institutes of Health grant GM26755 and general laboratory assistance was provided by the Howard Hughes Medical Institute.
Abbreviations used in this paper
- 63Jp
fusion protein of Sec63p lumenal domain and GST
- 63-1p
63Jp with sec63-1 mutation
- AEBSF
4-(2-aminoethyl)-benzenesulfonylfluoride, HCl
- GST
glutathione S–transferase
- pαF and ppαF
pro–α-factor and prepro–α-factor
Footnotes
Address all correspondence to R. Schekman, Department of Molecular and Cell Biology, HHMI, Barker Hall, University of California, Berkeley, CA 94720. Tel.: (510) 642-5686. Fax: (510) 642-7846.
References
- Benaroudj N, Triniolles F, Ladjimi MM. Effect of nucleotides, peptides, and unfolded proteins on the self-association of the molecular chaperone HSC70. J Biol Chem. 1996;271:18471–18476. doi: 10.1074/jbc.271.31.18471. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Chromatography of proteins on hydroxyapatite. Methods Enzymol. 1971;22:325–339. doi: 10.1016/s0076-6879(73)27021-0. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MH. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem. 1993;268:12730–12735. [PubMed] [Google Scholar]
- Brodsky JL. Post-translational protein translocation: not all hsc70s are created equal. Trends Biochem Sci. 1996;21:122–126. [PubMed] [Google Scholar]
- Brodsky JL, Schekman R. A Sec63p-BiP complex from yeast is required for protein translocation in a reconstituted proteoliposome. J Cell Biol. 1993;123:1355–1363. doi: 10.1083/jcb.123.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky, J.L., and R. Schekman. 1994. Heat shock cognate proteins and polypeptide translocation across the endoplasmic reticulum membrane. In The Biology of Heat Shock Proteins and Molecular Chaperones. R.I. Morimoto, A. Tissieres, and C. Georgopoulos, editors. Cold Spring Harbor Laboratory Press, Plainview, NY. 85–109.
- Brodsky JL, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic hsc70. J Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Goeckeler J, Schekman R. BiP and Sec63p are required for both co- and post-translational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci USA. 1995;92:9643–9646. doi: 10.1073/pnas.92.21.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. Eukaryotic homologues of Escherichia colidnaJ: a diverse protein family that functions with Hsp70 stress proteins. Mol Biol Cell. 1993;4:555–563. doi: 10.1091/mbc.4.6.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi AK, Schekman R. Mechanism of polypeptide translocation into the endoplasmic reticulum. J Biol Chem. 1996;271:30299–30302. doi: 10.1074/jbc.271.48.30299. [DOI] [PubMed] [Google Scholar]
- Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO (Eur Mol Biol Organ) J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- Cyr DM. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS (Fed Eur Biochem Soc) Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931. [PubMed] [Google Scholar]
- Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-x. [DOI] [PubMed] [Google Scholar]
- Feldheim D, Schekman R. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J Cell Biol. 1994;126:935–943. doi: 10.1083/jcb.126.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim D, Rothblatt J, Schekman R. Topology and functional domains of Sec63p, an ER membrane protein required for secretory protein translocation. Mol Cell Biol. 1992;12:3288–3296. doi: 10.1128/mcb.12.7.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldheim DA, Yoshimura K, Admon A, Schekman R. Structural and functional characterization of Sec66p, a new subunit of the polypeptide translocation apparatus in the yeast ER. Mol Biol Cell. 1993;4:931–939. doi: 10.1091/mbc.4.9.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science (Wash DC) 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature (Lond) 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Freiden PJ, Gaut JR, Hendershot LM. Interconversion of three differentially modified and assembled forms of BiP. EMBO (Eur Mol Biol Organ) J. 1992;11:63–70. doi: 10.1002/j.1460-2075.1992.tb05028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Eisenberg E, Greene L. Effect of constitutive 70-kDa heat shock protein polymerization on its interaction with protein substrate. J Biol Chem. 1996;271:16792–16797. doi: 10.1074/jbc.271.28.16792. [DOI] [PubMed] [Google Scholar]
- Gething M, Sambrook J. Protein folding in the cell. Nature (Lond) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Glick BS. Can Hsp70 proteins act as force-generating motors? . Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- Hartl FU. Molecular chaperones in cellular protein folding. Nature (Lond) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. . Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Holstein SEH, Ungewickell H, Ungewickell E. Mechanism of clathrin basket dissociation: separate functions of protein domains of the DnaJ homologue auxilin. J Cell Biol. 1996;135:925–937. doi: 10.1083/jcb.135.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, McMacken R. A bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia coliDnaK protein. J Biol Chem. 1996;271:11236–11246. doi: 10.1074/jbc.271.19.11236. [DOI] [PubMed] [Google Scholar]
- Kurihara T, Silver P. Suppression of a sec63mutation identifies a novel component of the yeast ER translocation apparatus. Mol Biol Cell. 1993;4:919–930. doi: 10.1091/mbc.4.9.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU. Successive action of DnaK, DnaJ, and GroEL along the pathway of chaperone-mediated protein folding. Nature (Lond) 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coliDnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Interaction between BiP and Sec63p is required for the completion of protein translocation into the ER of Saccharomyces cerevisiae. . J Cell Biol. 1995;131:1163–1171. doi: 10.1083/jcb.131.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman SK, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- McKay, D.B., S.M. Wilbanks, K.M. Flaherty, J.-H. Ha, M.C. O'Brien, and L.L. Shirvanee. 1994. Stress-70 proteins and their interaction with nucleotides. In The Biology of Heat Shock Proteins and Molecular Chaperones. R.I. Morimoto, A. Tissieres, and C. Georgopoulos, editors. Cold Spring Harbor Laboratory Press, Plainview, NY. 153–177.
- Müsch A, Wiedmann M, Rapoport TA. Yeast Sec proteins interact with polypeptides traversing the endoplasmic reticulum membrane. Cell. 1992;69:343–352. doi: 10.1016/0092-8674(92)90414-8. [DOI] [PubMed] [Google Scholar]
- Nelson MK, Kurihara T, Silver PA. Extragenic suppressors of mutations in the cytoplasmic C terminus of SEC63 defines five genes in Saccharomyces cerevisiae. . Genetics. 1993;134:159–173. doi: 10.1093/genetics/134.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Law DTS, Williams DB. Binding protein BiP is required for translocation of secretory proteins into the endoplasmic reticulum in Saccharomyces cerevisiae. . Proc Natl Acad Sci USA. 1991;88:1565–1569. doi: 10.1073/pnas.88.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palleros DR, Shi L, Reid KL, Fink AL. Hsp70-protein complexes: complex stability and conformation of bound substrate protein. J Biol Chem. 1994;269:13107–13114. [PubMed] [Google Scholar]
- Panzner S, Dreier L, Hartmann E, Kostka S, Rapoport TA. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell. 1995;81:562–570. doi: 10.1016/0092-8674(95)90077-2. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Kutay U. Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Rassow J, Voos W, Pfanner N. Partner proteins determine multiple functions of Hsp70. Trends Cell Biol. 1995;5:207–212. doi: 10.1016/s0962-8924(00)89001-7. [DOI] [PubMed] [Google Scholar]
- Römisch, K., and A. Corsi. 1996. Protein translocation into the endoplasmic reticulum. In Protein Targeting. S.M. Hurtley, editor. Oxford University Press, Oxford, England. 101–122.
- Rose MD, Misra LM, Vogel JP. KAR2, a karyogymy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Meyer DI. Secretion in yeast: reconstitution of the translocation and glycosylation of α-factor and invertase in a homologous cell-free system. Cell. 1986;44:619–628. doi: 10.1016/0092-8674(86)90271-0. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;109:2641–2652. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadis S, Hightower LE. Unfolded proteins stimulate molecular chaperone Hsc70 ATPase by accelerating ADP/ATP exchange. Biochemistry. 1992;31:9406–9412. doi: 10.1021/bi00154a012. [DOI] [PubMed] [Google Scholar]
- Sadler IA, Chiang A, Kurihara T, Rothblatt J, Way J, Silver P. A yeast gene essential for protein assembly in the endoplasmic reticulum and the nucleus has homology to DnaJ, an E. coliheat shock protein. J Cell Biol. 1989;109:2665–2675. doi: 10.1083/jcb.109.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Harris S, Risse B, Lill R, Silver PA. A yeast DnaJ homologue, Scj1p, can function in the endoplasmic reticulum with BiP/ Kar2p via a conserved domain that specifies interaction with Hsp70s. J Cell Biol. 1995;129:979–988. doi: 10.1083/jcb.129.4.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scidmore MA, Okamura HH, Rose MD. Genetic interactions between KAR2 and SEC63, encoding eukaryotic homologues of DnaK and DnaJ in the endoplasmic reticulum. Mol Biol Cell. 1993;4:1145–1159. doi: 10.1091/mbc.4.11.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J, Kornberg A. A prepriming DNA replication enzyme of Escherichia coli.I. Purification of protein n′: a sequence-specific, DNA- dependent ATPase. J Biol Chem. 1980;255:6789–6793. [PubMed] [Google Scholar]
- Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Szabo A, Korszun R, Hartl FU, Flanagan J. A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO (Eur Mol Biol Organ) J. 1996;15:408–417. [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Misra LM, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coliDnaJ protein stimulate the ATPase activity of DnaK and are sufficient for λ replication. J Biol Chem. 1994;269:5446–5451. [PubMed] [Google Scholar]
- Wawrzynów A, Zylicz M. Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J Biol Chem. 1995;270:19300–19306. doi: 10.1074/jbc.270.33.19300. [DOI] [PubMed] [Google Scholar]
- Wawrzynów A, Banecki B, Wall D, Liberek K, Georgopoulos C, Zylicz M. ATP hydrolysis is required for the DnaJ-dependent activation of DnaK chaperone for binding to both native and denatured protein substrates. J Biol Chem. 1995;270:19307–19311. doi: 10.1074/jbc.270.33.19307. [DOI] [PubMed] [Google Scholar]
- Wei J, Hendershot LM. Characterization of the nucleotide binding properties and ATPase activity of recombinant hamster BiP purified from bacteria. J Biol Chem. 1995;270:26670–26676. doi: 10.1074/jbc.270.44.26670. [DOI] [PubMed] [Google Scholar]
- Zeigelhoffer T, Lopez-Buesa P, Craig E. The dissociation of ATP from hsp70 of Saccharomyces cerevisiaeis stimulated by both Ydj1p and peptide substrates. J Biol Chem. 1995;270:10412–10419. doi: 10.1074/jbc.270.18.10412. [DOI] [PubMed] [Google Scholar]