Abstract
Membrane traffic in eukaryotic cells requires that specific v-SNAREs on transport vesicles interact with specific t-SNAREs on target membranes. We identified a novel Saccharomyces cerevisiae v-SNARE (Vti1p) encoded by the essential gene, VTI1. Vti1p interacts with the prevacuolar t-SNARE Pep12p to direct Golgi to prevacuolar traffic. vti1-1 mutant cells missorted and secreted the soluble vacuolar hydrolase carboxypeptidase Y (CPY) rapidly and reversibly when vti1-1 cells were shifted to the restrictive temperature. However, overexpression of Pep12p suppressed the CPY secretion defect exhibited by vti1-1 cells at 36°C. Characterization of a second vti1 mutant, vti1-11, revealed that Vti1p also plays a role in membrane traffic at a cis-Golgi stage. vti1-11 mutant cells displayed a growth defect and accumulated the ER and early Golgi forms of both CPY and the secreted protein invertase at the nonpermissive temperature. Overexpression of the yeast cis-Golgi t-SNARE Sed5p suppressed the accumulation of the ER form of CPY but did not lead to CPY transport to the vacuole in vti1-11 cells. Overexpression of Sed5p allowed growth in the absence of Vti1p. In vitro binding and coimmunoprecipitation studies revealed that Vti1p interacts directly with the two t-SNAREs, Sed5p and Pep12p. These data suggest that Vti1p plays a role in cis-Golgi membrane traffic, which is essential for yeast viability, and a nonessential role in the fusion of Golgi-derived vesicles with the prevacuolar compartment. Therefore, a single v-SNARE can interact functionally with two different t-SNAREs in directing membrane traffic in yeast.
Eukaryotic cells contain numerous membrane compartments, each of which maintains distinct protein and lipid compositions for specific tasks. Trafficking between different membrane compartments occurs via transport vesicles. Transport of proteins along the secretory pathway starts with protein translocation into the endoplasmic reticulum (ER)1. Proteins are transported via the intermediate compartment to the Golgi apparatus, which can be subdivided into cis-, medial-, and trans-Golgi compartments. These Golgi compartments are well characterized at the morphological level in mammalian cells (Farquhar and Palade, 1981). The Golgi compartments of yeast are defined only by the glycosylation and the processing enzymes they contain (Franzusoff and Schekman, 1989).
The SNARE hypothesis (Söllner et al., 1993) proposes that a unique set of proteins on the transport vesicle (v-SNARE) and proteins on the target membrane (t-SNARE) provide specific recognition between both membranes (Ferro-Novick and Jahn, 1994; Rothman, 1994). v- and t-SNAREs have structural similarities. They contain a single COOH-terminal transmembrane domain and regions that are predicted to form coiled coil domains. The yeast proteins Sec22p (Sly2p), Bet1p (Sly12p), Bos1p, and possibly Ykt6p have been identified as v-SNAREs in ER to Golgi transport (Newman et al., 1990; Ossig et al., 1991; Sogaard et al., 1994). Their partner is the t-SNARE Sed5p, which is localized on the first Golgi compartment in yeast (Hardwick and Pelham, 1992). In addition, Sed5p interacts with the v-SNARE Sft1p, which is localized on a more distal Golgi compartment and is therefore involved in retrograde transport (Banfield et al., 1995). Recently, Ufe1p, a t-SNARE localized in the ER and involved in retrograde transport to the ER, has been identified (Lewis and Pelham, 1996). Mutations inhibiting retrograde traffic to the ER or to the cis-Golgi compartment lead to a very rapid blockage of forward traffic, making it difficult to distinguish by phenotypic analysis whether a protein is involved in anterograde or retrograde transport. The tight coupling of anterograde and retrograde membrane traffic suggests that components needed for forward transport have to be recycled (Letourneur et al., 1994; Lewis and Pelham, 1996).
The yeast late Golgi compartment, which corresponds to the mammalian TGN, functions as a sorting compartment (Pryer et al., 1992; Nothwehr and Stevens, 1994; Stack et al., 1995). Over 50 VPS (vacuolar protein sorting) and PEP (peptidase deficient) genes have been identified that are involved in the transport of the soluble vacuolar protease carboxypeptidase Y (CPY) from the Golgi apparatus to the vacuole (Jones, 1977; Bankaitis et al., 1986; Rothman and Stevens, 1986), and none of these genes is essential for cell growth (Stack et al., 1995). CPY binds to the CPY sorting receptor, Vps10p, in the late Golgi compartment (Marcusson et al., 1994), and this complex is directed away from the secretory pathway by packaging into transport vesicles that eventually fuse with the prevacuolar or endosomal compartment (Raymond et al., 1992b ; Vida et al., 1993; Piper et al., 1995; Stack et al., 1995). Whereas CPY is released and is transported on to the vacuole, the CPY receptor Vps10p recycles to the Golgi apparatus (Cereghino et al., 1995; Piper et al., 1995; Cooper and Stevens, 1996).
Recently, the t-SNARE Pep12p was found to be localized to the yeast prevacuolar compartment (Becherer et al., 1996). Deletion of the PEP12 gene leads to the accumulation of small vesicles, suggesting that it is necessary for fusion of Golgi-derived transport vesicles with the prevacuolar compartment.
Here we report the identification of a novel yeast v-SNARE, Vti1p, that interacts with Pep12p and is involved in membrane traffic from the Golgi apparatus to the prevacuolar compartment. VTI1 was found to be an essential gene, and its essential function is related to the interaction of Vti1p and the cis-Golgi t-SNARE Sed5p. Therefore, Vti1p is a v-SNARE that interacts functionally with two t-SNAREs, Pep12p localized to the prevacuolar compartment and Sed5p localized to the cis-Golgi compartment.
Materials and Methods
Materials
Reagents were used from the following sources: enzymes for DNA manipulation from New England Biolabs (Beverly, MA) and Boehringher Mannheim Biochemicals (Indianapolis, IN), secondary antibodies from Promega Biotech. (Madison, WI), Amersham Corp. (Arlington Heights, IL), and Jackson ImmunoResearch Labs (West Grove, PA); 35S-Express label and ECL solution from New England Nuclear (Boston, MA); fixed Staphylococcus aureus cells (IgGsorb) from The Enzyme Center (Malden, MA); Oxalyticase from Enzogenetics (Corvallis, OR); protein G–Sepharose from Pharmacia Fine Chemicals (Uppsala, Sweden); and the plasmid pQE30 and Ni-NTA resin from Qiagen (Hilden, Germany). All other reagents were purchased from Sigma Chemical Co. (St. Louis, MO).
Plasmid manipulations were performed in the Escherichia coli strains MC1061 or XL1Blue using standard media. Yeast strains (Table I) were grown in rich media (1% yeast extract, 1% peptone, 2% dextrose, YEPD) or standard minimal medium (SD) with appropriate supplements. To induce expression from the GAL1 promoter, dextrose was replaced by 2% raffinose and 2% galactose.
Table I.
Yeast Strains Used in this Study
| Strain | Genotype | Reference | ||
|---|---|---|---|---|
| CTY10-5D | MATa leu2-3,112 his3-Δ200 trp1-Δ901 gal4 gal80 URA3::lexAop-LacZ | (Bartel and Fields, 1995) | ||
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ mel- | (Robinson et al., 1988) | ||
| SEY6211 | MATa leu2-3,112 ura3-52 his3-Δ200 ade2-101 trp1-Δ901 suc2-Δ9 mel- | (Robinson et al., 1988) | ||
| SEY5186 | MATα leu2-3,112 ura3-52 gal2 sec18-1 | (Emr et al., 1984) | ||
| FvMY6 | MATα leu2-3,112 ura3-52 ade2-101 trp1-Δ901 suc2-Δ9 mel- vti1::ΔHIS3 | This study | ||
| FvMY7 | MATa leu2-3,112 ura3-52 his3-Δ200 ade2-101 trp1-Δ901 suc2-Δ9 mel- vti1-1 | This study |
Plasmids and Strains
The plasmids used in this study are listed in Table II. pSN206 was constructed by PCR amplifying the VPS10 tail (codons 1413–1579) with EcoRI–BamHI ends and cloning it into the 2μ-HIS3 vector pEG202, which makes LexA fusion proteins under the control of an ADH promoter (Golemis et al., 1996). A 1.1-kb fragment starting at the SacI site 150 bp upstream of the VTI1-ATG and adding an XhoI site was amplified from genomic DNA and cloned into pBluescript KS+. For the disruption construct pSN258 the VTI1 coding region between the HindIII and NdeI sites was replaced by the HIS3 gene. pFvM7 consists of VTI1 (codons 1–194) PCR-amplified with BamHI–SmaI ends in pGex2T (Smith and Johnson, 1988). A HindIII site was introduced upstream of the VTI1 initiation codon and the HindIII–XhoI fragment was cloned together with GAL1promoter DNA into pRS314 (Sikorski and Hieter, 1989) to yield pFvM17. pFvM16 is identical to pFvM17 except for the presence of a single HA tag in the BglII site. The MBP–VTI1::HA fusion construct was derived by PCR amplification of VTI1 (codons 1–194) with BamHI–HindIII ends and cloning into pMal-p2 (Maina et al., 1988). pFvM28, pFvM29, and pFvM32 consist of a 1.8-kb fragment encoding VTI1 between a SacI and an XhoI site in pRS314, pRS316, or YEp352 (Hill et al., 1986). VTI1 (codons 1–194) was PCR amplified with BamHI–SmaI ends and put into pQE30 to produce pFvM38. pFvM40 was constructed by PCR amplification of SED5 (codons 1–324; Hardwick and Pelham, 1992) with EcoRI and BamHI ends and subcloning this fragment into pGex2T. pFvM89 contains a PCR-amplified 1.8-kb fragment encoding SED5 starting 400 bp upstream of the ATG with BamHI and SacI ends in YEp351. PCR mutagenesis was performed as described (Muhlrad et al., 1992) using PvuII-digested pFvM28 as template and oligonucleotides annealing to nucleotides 1823–1844 and 2074– 2053 of pRS314. In the PCR reaction, 3 mM MgCl2, 0.25 mM MnCl2 and 25 μM dATP, 250 μM dCTP, dGTP, and dTTP, or 25 μM dGTP, 250 μM dATP, dCTP, and dTTP were used. The PCR fragments were cotransformed with XhoI–SacI digested pRS314 into FvMY6 pFvM29. The transformants were pooled, the plasmid pFvM29 removed by plating on 5-fluoroorotic acid (5-FOA) (Boeke et al., 1984), and colonies screened for CPY secretion at 37°C by colony overlay (Rothman et al., 1986) or for growth at 22° and 37°C. Mutant plasmids were rescued, retransformed, and the plasmid pFvM29 removed to allow isolation of pFvM92 and pFvM93. pFvM103 consists of a PCR-amplified fragment of SSO2 (codons 1–269; Aalto et al., 1993) with BamHI and EcoRI ends subcloned into pGex2T. pFvM104 was constructed by subcloning a 2.9-kb EcoRI–SacI fragment from pRB58 (Carlson and Botstein, 1982) encoding SUC2 into pRS316.
Table II.
Plasmids Used in this Study
| Plasmid | Description | Reference | ||
|---|---|---|---|---|
| pSN206 | ADH1-LexA-VPS10(tail) fusion in pEG202 (2μ-HIS3) | This study | ||
| p2H-1 | 5 kb genomic DNA with VTI1 (codons 106-217) in pGAD3 | This study | ||
| pSN258 | vti1Δ::HIS3 disruption construct in pRS305 | This study | ||
| pFvM7 | E. coli expression vector pGex2T encoding GST-Vtilp (aal-194) | This study | ||
| pFvM16 | GAL1-VTI1::HA in pRS314 (CEN6-TRP1) | This study | ||
| pFvM17 | GAL1-VTI1 in pRS314 (CEN6-TRP1) | This study | ||
| pFvM20 | E. coli expression vector pMal-p2 encoding MBP-Vtilp::HA (aal-194) | This study | ||
| pFvM28 | 1.8 kb containing VTI1 in pRS314 (CEN6-TRP1) | This study | ||
| pFvM29 | 1.8 kb containing VTI1 in pRS316 (CEN6-URA3) | This study | ||
| pFvM32 | 1.8 kb containing VTI1 in YEp352 (2μ-URA3) | This study | ||
| pFvM38 | E. coli expression vector pQE30 encoding 6His-Vtilp (aal-194) | This study | ||
| pFvM40 | E. coli expression vector pGEX2T encoding GST-Sed5p (aa1-324) | This study | ||
| pFvM47 | 7.7 kb Chrom XII (nt190000-197700) containing SED5 in YEp24 (2μ-URA3) | This study | ||
| pFvM89 | SED5 in YEp351 (2μ-LEU2) | This study | ||
| pFvM92 | vti1-11 in pRS314 (CEN6-TRP1) | This study | ||
| pFvM93 | vti1-2 in pRS314 (CEN6-TRP1) | This study | ||
| pFvM103 | E. coli expression vector pGex2T encoding GST-Sso2p (aal-269) | This study | ||
| pFvM104 | 2.9 kb EcoRI-SnaBI DNA from pRB58 with SUC2 in pRS316 | This study | ||
| pRCP59 | E. coli expression vector pGex3X encoding GST-Pep12p (aal-263) | R.C. Piper*/T.H. Stevens | ||
| pRCP110 | PEP12 in YEp351 (2μ-LEU2) | R.C. Piper*/T.H. Stevens |
Institute of Molecular Biology, University of Oregon, Eugene, OR
To delete VTI1, a yeast diploid strain (SEY6210 × SEY6211) was transformed with SacI–XbaI-digested pSN258 and selected for His+ colonies. A vti1Δ::HIS3 knockout was confirmed by PCR, and the strain was transformed with a VTI1 covering plasmid. After sporulation, tetrads were dissected and vti1Δ cells identified (FvMY6, Table I). For hydroxylamine mutagenesis, 250 μg pFvM28 purified on a CsCl-gradient was incubated with 2 ml 0.4 M hydroxylamine in 150 mM KPO4, pH 6.0, at 75°C for 60 min (Schauer et al., 1985). The DNA was cycled through E. coli and transformed into FvMY6 containing plasmid pFvM29. The transformants were pooled, the plasmid pFvM29 removed by plating on 5-FOA plates, and colonies screened for CPY secretion at 37°C by colony overlay (Rothman et al., 1986). A mutant plasmid was recovered, retested, and the vti1-1 DNA subcloned into the integration vector pRS306 (Sikorski and Hieter, 1989). FvMY7 was constructed by integration of this linearized DNA into SEY6211 and looping out the wild-type VTI1 on 5-FOA plates (Boeke et al., 1984).
Two Hybrid Screen
The strain CTY10-5D carrying the plasmid pSN206 expressing the LexA-VPS10(tail) fusion protein was transformed with the libraries pGAD1, pGAD2, and pGAD3 (Chien et al., 1991; Bartel and Fields, 1995; Golemis et al., 1996). About 30,000 colonies were screened for expression of the lacZ gene. The rescued library plasmid containing the GAL4 activation domain fused to codons 109–217 of Vti1p, p2H-1, did not interact with the LexA protein alone.
Production of Vti1p and Pep12p Antisera
An antiserum was raised in rabbits against a fusion protein containing glutathione–S-transferase and amino acids 1–194 of Vti1p purified from E. coli. The antiserum was affinity purified with an Affigel 10 (BioRad Laboratories, Hercules, CA) column with covalently bound maltose-binding protein Vti1p (amino acids 1–194) fusion protein purified from E. coli. The Pep12p antiserum was raised in rabbits against a purified fusion protein containing glutathione–S-transferase and amino acids 1–263 of Pep12p.
Immunofluorescence Microscopy
Indirect immunofluorescence microscopy was performed with SEY6211 cells as previously described (Raymond et al., 1992a ). The following antibody combination was used: 1:50 diluted affinity-purified Vti1p antiserum, 1:100 diluted biotin-conjugated goat anti–rabbit IgG (H + L), and 1:100 diluted rhodamine-conjugated streptavidin.
SDS-PAGE and Western Blotting
Yeast extracts were prepared and the proteins subjected to SDS-PAGE and electroblotting as described (Laemmli, 1970; Towbin et al., 1979). Affinity-purified rabbit antisera were used for the detection of Vti1p, Pep12p, Vps10p (Piper et al., 1995), and dipeptidyl aminopeptidase A (DPAP A; Roberts et al., 1992). Rabbit antiserum was used for PGK and monoclonal antibodies for Vph1p (Molecular Probes Inc., Eugene, OR), Sac1p (Mayinger et al., 1995), and Dpm1p (Molecular Probes Inc.). An incubation with HRP-conjugated secondary antibodies was followed by visualization with ECL reagents.
Subcellular Fractionation
Membrane fractionation by differential centrifugation was performed as described (Paravicini et al., 1992). SEY6211 spheroplasts were lysed, centrifuged at 500 g (5 min) to remove debris, at 13,000 g (10 min) to yield P13 and S13, and at 200,000 g (20 min) to produce P200 and S200.
The S13 fraction was further separated by sucrose gradient centrifugation (Becherer et al., 1996). The gradient consisted of the following steps: 0.5 ml 60%, 1 ml 37%, 1.5 ml 34%, 2 ml 32%, 2 ml 29%, 1 ml 27%, 1.5 ml 22%, and 1 ml 19% (wt/wt) sucrose in 10 mM Hepes-NaOH, pH 7.6, and was centrifuged for 17 h at 170,000 g in an SW41 rotor. 13 fractions were collected from the top. Sucrose density was measured with a refractometer. Fractions were either concentrated by TCA precipitation or directly analyzed by SDS-PAGE and immunoblotting.
Immunoprecipitation of 35S-labeled Proteins
The procedure for CPY immunoprecipitation was described earlier (Vater et al., 1992). Briefly, log-phase growing yeast cells were labeled for 10 min with 35S-Express label (10 μl/0.5 OD of cells) followed by a 30-min chase (chase was initiated by addition of 500 μg/ml methionine and cysteine). Invertase immunoprecipitation and a secondary immunoprecipitation of proteins containing α1,6-mannose linkages were performed as described (Franzusoff and Schekman, 1989). Invertase was derepressed by an incubation in minimal medium containing 0.1% glucose, 50 mM KPO4, and 1 mg/ml BSA for 45 min at 22°C or 30 min at 22°C plus 15 min at 37°C. Pulse labeling with 35S-Express label (10 μl/0.5 OD of cells) was done for 7 min, an aliquot removed (in some experiments), and the remaining sample chased for 30 min. Cells were pelleted, the medium was removed, and the cells were spheroplasted. The spheroplasts were pelleted and lysed giving rise to the intracellular fraction. The supernatant was combined with the medium fraction to yield the extracellular fraction. Immunoprecipitates were analyzed by SDS-PAGE and autoradiography.
Suppressor Screen
To identify multicopy suppressors that allow for growth in the absence of Vti1p, the strain FvMY6/pFvM16 (vti1Δ/pCEN-GAL1-VTI1) was used. FvMY6/pFvM16 cells were transformed with a YEp24 2μ library (Carlson and Botstein, 1982) and plated on SD-Ura plates, conditions that turn off the expression of VTI1 (due to the repression of the GAL1 promoter by dextrose). Colonies that grew were tested for the absence of Vti1p by immunoblot analysis and the inability to grow on 5-FOA plates to determine if the suppression was plasmid linked. Plasmids were recovered, retransformed, and colonies, which lost the pFvM16 plasmid, selected to confirm the suppression. Portions of the inserts were subcloned into YEp351 to determine the minimal DNA fragment needed for suppression.
Native Immunoprecipitation
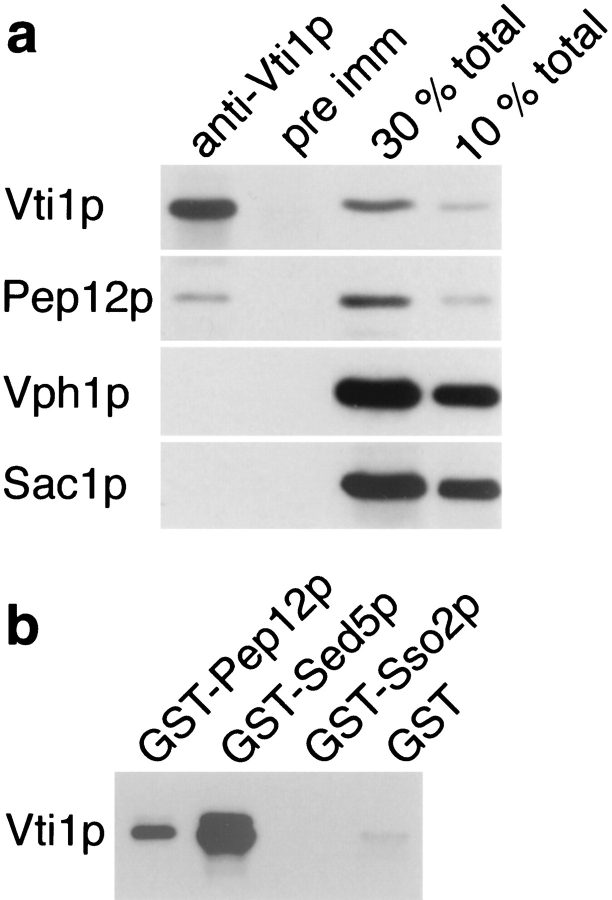
1 OD600nm (∼107 cells) of wild-type yeast cells were spheroplasted, solubilized with 700 μl 0.5% Triton X-100 in PBS, and centrifuged at 12,000 g for 10 min. The supernatant fractions were incubated with anti-Vti1p antiserum or pre-immune serum for 2 h at 4°C and then with protein G-sepharose for an additional 2 h at 4°C. The sepharose beads were washed three times for 5 min with 0.5% Triton X-100 in PBS. The proteins bound to the beads were analyzed by SDS-PAGE and immunoblotting with antibodies against Vti1p, Pep12p, Vph1p, and Sac1p. To quantify how much of each protein was immunoprecipitated, the amount of supernatant fraction corresponding to 30 and 10% of the total protein present in the immunoprecipitations were analyzed by SDS-PAGE and immunoblotting.
In vitro Binding Assay
Recombinant fusion proteins were purified from E. coli extracts. Specifically, bacteria were induced in mid-log phase with 0.5 mM IPTG for 6 h at 30°C to express 6-His–Vti1p (amino acids 1–194). The bacterial pellet was resuspended in 50 mM NaPO4, pH 8.0, 300 mM NaCl, frozen, and lysed by sonification. After centrifugation for 10 min with 12,000 g, the supernatant was incubated with Ni-NTA resin with mixing for 1 h at 4°C. The resin was loaded in a column, washed with 50 mM NaPO4, pH 6.0, 300 mM NaCl, and 10% glycerol, followed by washes with 20 and 100 mM imidazole in the same wash buffer. 6-His–Vti1p was eluted with 500 mM imidazole and dialyzed against PBS. Bacteria were induced with 0.5 mM IPTG for 4 h at 37°C to express GST, GST-Pep12p, GST-Sed5p, and GST-Sso2p. Bacterial pellets were resuspended in PBS, frozen, and lysed by sonification. Debris was removed by centrifugation at 12,000 g for 10 min. The supernatant was incubated with glutathione–agarose for 15 min. The beads were washed three times and the amount of bound protein estimated after separation by SDS-PAGE and visualization by Coomassie staining. About 1 μg of these immobilized GST-fusion proteins was incubated with 9 μg of 6-His–Vti1p in 0.5% Triton X-100 in PBS with mixing for 2 h at 4°C. The beads were washed three times with 0.5% Triton X-100 in PBS and the bound protein eluted and analyzed by SDS-PAGE and immunoblotting with anti-Vti1p antibodies.
Electron Microscopy
Electron microscopy experiments were performed as described (Kaiser and Schekman, 1990; Piper et al., 1994).
To quantify the number of vesicles, 12–20 cells for each condition were photographed. The negatives were scanned, vesicle-like dark structures were counted, and the cytoplasmic area was determined using the NIH Image program. The number of vesicles per unit cytoplasmic area was compared for different conditions. The length of membrane sheets that were not adjacent to the cell wall or around the nucleus was measured using the NIH Image program and divided by the cytoplasmic area as a measure for ER accumulation.
Results
Identification of VTI1
We were interested in identifying proteins that are involved in trafficking of the CPY sorting receptor Vps10p (Marcusson et al., 1994). Since the cytosolic tail of Vps10p directs its recycling (Cereghino et al., 1995; Cooper and Stevens, 1996), this portion of the protein was fused to the LexA DNA-binding domain to create a “bait” protein to screen for interacting proteins in the yeast two-hybrid system (Brent and Ptashne, 1985; Chien et al., 1991). An activating plasmid containing the VTI1 gene (Vps ten interacting) was isolated and the insert sequenced. The sequence matched an open reading frame upstream of CIK1 (Page and Snyder, 1992), which was also sequenced by the Genome Sequencing Project on chromosome XIII and designated open reading frame YMR197c. VTI1 is predicted to encode a protein of 217 amino acids with a COOH-terminal transmembrane domain (Fig. 1).
Figure 1.
Amino acid sequence of yeast Vti1p. The predicted transmembrane domain is boxed. The predicted coiled coil domain is underlined (amino acids 159–188 with a P = 0.285 using the paircoil program [Berger et al., 1995]; amino acids 149–169 with a P = 0.58 using the coil program with a window of 21 [Lupas et al., 1991]). These sequence data are available from GenBank under accession number AF006074.
The regions adjacent to the transmembrane domain are predicted to fold into an amphipathic α-helical coiled coil domain. These structural elements are characteristic of SNARE proteins (Ferro-Novick and Jahn, 1994; Rothman, 1994). In pairwise sequence comparisons with SNARE proteins, Vti1p shares the highest sequence similarity with synaptobrevin (VAMP; 28% amino acid identity) and with Sft1p (32% amino acid identity), which are much smaller proteins. Sft1p is a yeast v-SNARE involved in retrograde transport to the cis-Golgi compartment (Banfield et al., 1995), and synaptobrevin is a v-SNARE found on synaptic vesicles. Domains close to the NH2 terminus of Vti1p (amino acids 39–63) and of synaptobrevin (amino acids 29–56) are predicted to adopt the same structure as viral fusion peptides with charged amino acids on one side of the α-helix and bulky hydrophobic amino acids on the other. The presence of a domain of this sort close to the NH2 terminus is characteristic for v-SNAREs (Dascher et al., 1991; White, 1992; Jahn and Südhof, 1994). In general, members of the v-SNARE family share common structural elements but exhibit low sequence identities in pairwise combinations (18–35% amino acid identities), and Vti1p fits this pattern.
t-SNAREs display significant amino acid sequence identity in the coiled coil domain adjacent to the transmembrane domain, and these regions of identity do not extend to v-SNAREs such as Vti1p. Most t-SNAREs contain nine invariant amino acid residues in this region (Pelham, 1993; Lewis and Pelham, 1996). The coiled coil domain in Vti1p does not share sequence identity with this coiled coil domain in different t-SNAREs (8% compared to Pep12p, 13% compared to Sed5p, and 17% compared to syntaxin3 and syntaxin5). Only two of the invariant nine amino acid residues are present in Vti1p. Therefore, by all of the above criteria, Vti1p belongs to the v-SNARE family of proteins.
Vti1p Localizes to Golgi and Prevacuolar Membranes
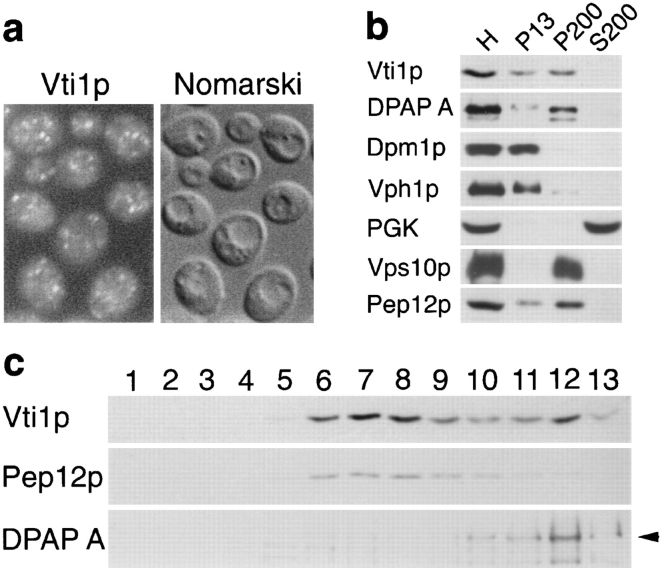
A rabbit antiserum was generated against Vti1p. Anti-Vti1p antibodies identified a single 27-kD band in protein extracts from wild-type yeast cells analyzed by SDS-PAGE and immunoblotting. This band was more intense in a strain carrying a high copy VTI1 plasmid and absent in vti1Δ mutant cells (data not shown). Vti1p was immunolocalized by indirect immunofluorescence in wild-type cells. Vti1p antibodies labeled numerous punctate structures throughout the cytosol (Fig. 2 a), some of which were adjacent to the vacuole, but vacuolar and ER membranes were not stained.
Figure 2.
Vti1p localization. (a) Immunofluorescence microscopy of Vti1p. Anti-Vti1p antibodies reveal a punctate staining pattern distinct from the vacuoles seen by Nomarski optics. (b) Differential centrifugation of total yeast homogenate (H). Vti1p was predominantly found in the 200,000 g pellet (P200) with lower amounts in the 13,000 g pellet (P13). The syntaxin-related protein Pep12p had a similar distribution. (c) Sucrose density centrifugation of the S13 fraction. Vti1p migrated in two peaks, one cofractionating with the prevacuolar protein Pep12p and the other with the Golgi protein DPAP A. Fractions were collected from the top of the gradient (fraction 1) and analyzed by SDS-PAGE and immunoblotting.
Differential centrifugation of wild-type yeast lysates followed by SDS-PAGE and immunoblot analysis (Fig. 2 b) revealed that Vti1p was predominantly localized in the 200,000 g membrane pellet (P200), which contained small vesicles and Golgi membranes, as defined by the enrichment of the proteins DPAP A and Vps10p (Paravicini et al., 1992; Roberts et al., 1992; Marcusson et al., 1994). Lower amounts of Vti1p were present in the 13,000 g pellet (P13), which included vacuolar (Vph1p [Manolson et al., 1992]), ER (Dpm1p [Orlean, 1990]), and mitochondrial membranes. The prevacuolar t-SNARE Pep12p was most abundant in the P200 fraction with lower amounts in the P13 fraction, consistent with previous results (Becherer et al., 1996). As expected, the cytosolic protein PGK fractionated with the S200 soluble protein fraction.
To study the localization of Vti1p in more detail, the 13,000 g supernatant (S13) was separated on a 19–37% sucrose density equilibrium gradient (Becherer et al., 1996). Fractions were collected from the top and analyzed by SDS-PAGE and immunoblot analysis (Fig. 2 c). Vti1p was found at two different buoyant densities: in fractions 6–8 (at 29.5% sucrose) and in fraction 12 (36.4% sucrose). As observed previously (Becherer et al., 1996), Golgi membranes (as indicated by the presence of DPAP A) migrated close to the bottom of the gradient and cofractionated with the higher density Vti1p peak. By contrast, Pep12p was found in a single peak together with the lower density Vti1p peak. These data indicate that a smaller portion of Vti1p is localized in the Golgi apparatus, whereas the majority is colocalized with Pep12p in the prevacuolar compartment.
VTI1 Is an Essential Gene Required for CPY Sorting
The VTI1 gene was disrupted in yeast to determine the functional role of Vti1p. One of the copies of VTI1 in a diploid strain (SEY6210 × SEY6211) was disrupted by replacement with the HIS3 gene (vti1Δ::HIS3). Tetrad dissections did not yield viable His+ spores, revealing that VTI1 is an essential gene. His+ spores derived from tetrad dissection of FvMY4 after transformation with a VTI1 plasmid were only viable in the presence of the VTI1-containing plasmid. vti1Δ::HIS3 cells carrying the VTI1–URA3 plasmid could not grow on 5-FOA because plasmid loss was lethal, confirming that VTI1 is an essential gene. By contrast, none of the VPS or PEP genes are essential for haploid yeast cell growth (Stack et al., 1995).
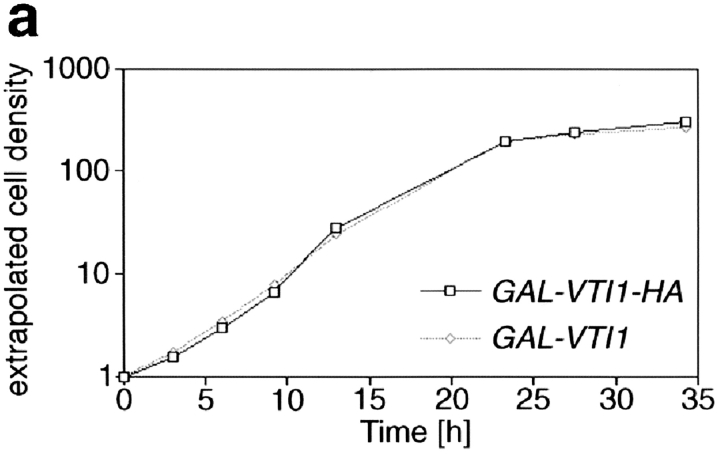
The phenotypic consequences of the loss of Vti1p were investigated by placing VTI1 expression under the control of the GAL1 promoter. vti1Δ cells carrying the GAL1– VTI1 plasmid were maintained on galactose-containing medium. Approximately 20 h after shifting these cells to glucose-containing medium (about six cell divisions) the cells stopped dividing (Fig. 3 a). Immunoblot analysis revealed that VTI1 expression under control of the GAL1 promoter was ∼10-fold higher in the presence of galactose than expression from the VTI1 promoter. Wild-type levels of Vti1p were reached ∼12 h after shift to glucose medium (data not shown).
Figure 3.
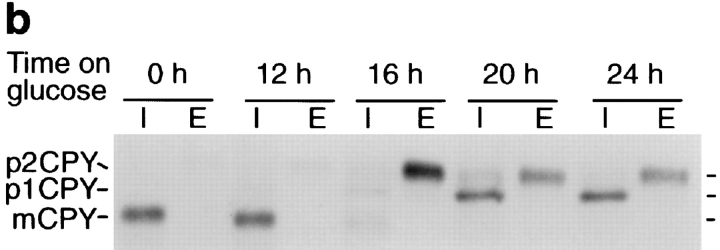
Depletion of Vti1p leads to growth arrest and CPY mislocalization. (a) Growth curves of the GAL1–VTI1 and GAL1– VTI1-HA strains in glucose-containing medium. Cells of both strains displayed equivalent growth properties and stopped dividing after 22 h. Cell density was measured by optical density at 600 nm. The cultures were diluted when they reached an optical density of about one to keep cells growing in logarithmic phase. Taking the dilution factor into account, the extrapolated cell densities were calculated from the measured optical densities. (b) CPY sorting in the GAL1–VTI1-HA strain grown in galactose and for different amounts of time in glucose medium. CPY sorting was followed by pulse–chase labeling and CPY immunoprecipitation from cellular extracts (I, intracellular) and medium (E, extracellular). For cells cultured in galactose, CPY reached the vacuole (presence of mCPY). After 16 h in glucose most of the CPY was secreted as the late Golgi precursor form, p2CPY. After longer time periods in glucose, the ER and early Golgi forms of CPY (p1CPY) accumulated within the cell.
To determine if sorting of the soluble vacuolar hydrolase CPY was affected by lowered Vti1p levels, the fate of newly synthesized CPY was monitored in GAL1–VTI1 cells grown on galactose medium or after shift to glucose medium for different periods of time (Fig. 3 b). Pulse– chase immunoprecipitations of CPY in GAL1–VTI1 cells grown on galactose medium or grown on glucose medium for 12 h revealed that CPY was found only in the mature, vacuolar form in the intracellular fraction, reflecting correct sorting and delivery of CPY to the vacuole. After growth on glucose medium for 16 h, hardly any newly synthesized CPY reached the vacuole. Instead, the Golgi-modified precursor form of CPY (p2CPY [Stevens et al., 1982]) was secreted. These data indicate that Vti1p is necessary for sorting of the soluble vacuolar hydrolase CPY. This missorting of CPY occurred before the growth defect became apparent, suggesting that it was not an indirect effect. After the GAL1–VTI1 cells had grown on glucose medium for 20 to 24 h, the ER and early-Golgi form of CPY (p1CPY [Stevens et al., 1982]) was found to accumulate. These data imply that transport through the Golgi apparatus was slowed or blocked upon severe depletion of Vti1p. However, it remains unclear whether p1CPY accumulation was caused directly by Vti1p depletion or an indirect effect due to the severe growth defect.
vti1 Mutants Are Defective in Two Vesicle Traffic Steps
The genes identified as being required for vacuolar protein sorting (VPS) are all nonessential in yeast, yet VTI1 function is required for CPY sorting, and the gene is essential. This suggests that VTI1 has a second function in addition to vacuolar protein sorting. Temperature-sensitive vti1 mutants were generated to elucidate the role of Vti1p in Golgi to vacuolar traffic and the essential function for this protein. Using shuttle mutagenesis with hydroxylamine-treated VTI1 on a CEN-based plasmid, a mutant was identified (vti1-1) that secreted CPY at 37°C yet grew with nearly a wild-type doubling time at this temperature. The vti1-1 mutation was integrated into the genome for further analysis. Additional vti1 alleles (vti1-2 and vti1-11) were generated by PCR mutagenesis. vti1-11 grew normally at 20°C but grew very slowly at 37°C. The doubling time of vti1-11 at 37°C was about 10 h compared to 2.5 h for wild-type yeast cells. The vti1-2 mutant cells had a slight growth defect at 37°C.
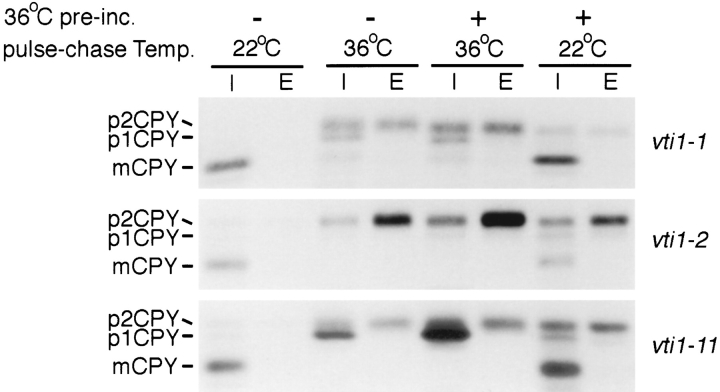
The effects of the vti1 mutations on CPY sorting were studied by CPY immunoprecipitations after pulse–chase labeling. CPY was correctly transported to the vacuole at 22°C in the vti1-1 mutant (Fig. 4, top row). If 35S label was added immediately upon shift to 36°C, very little CPY reached the vacuole, and instead the Golgi-modified p2CPY was either secreted or accumulated intracellularly. The amount of p1CPY detected was small and varied somewhat between experiments. In cells that were labeled at 22°C after a preincubation at 36°C (30 min), the majority of CPY reached the vacuole (as evidenced by the presence of mature CPY; mCPY), indicating that the vti1-1 temperature-sensitive mutation is rapidly reversible. This extremely fast onset of the CPY sorting defect and fast recovery demonstrate that Vti1p is directly involved in CPY transport from the Golgi apparatus to the vacuole. The mutant vti1-2 also exhibited a fast onset of CPY secretion upon shift to 36°C (Fig. 4, middle row), however, the recovery was somewhat slower. The extent of CPY secretion was greater in vti1-2 mutant cells than in vti1-1 mutant cells upon shift to 36°C; some p2CPY accumulated intracellularly but p1CPY did not.
Figure 4.
CPY sorting in vti1-ts mutants. CPY sorting was followed in the vti1-1, vti1-2, and vti1-11 strains by pulse–chase labeling and CPY immunoprecipitations from cellular extracts (I, intracellular) and medium (E, extracellular). vti1-1 cells were preincubated for 30 min at 36°C and vti1-2 and vti1-11 cells for 15 min. In the vti1-1 and vti1-2 cells most CPY was secreted or accumulated intracellularly as p2CPY at the restrictive temperature. In vti1-11 cells, predominantly the ER or early Golgi form p1CPY accumulated intracellularly at the restrictive temperature.
In the vti1-11 mutant, which was temperature sensitive for growth, CPY reached the vacuole and was processed normally at 22°C (Fig. 4, bottom row). p1CPY was the predominant form of the protein found when vti1-11 cells were labeled at the restrictive temperature (36°C). Lower amounts of intracellular and secreted p2CPY were also present, but mature CPY was not. After return to the permissive temperature, CPY sorting to the vacuole recovered quickly in vti1-11 cells, indicating that the vti1-11 temperature-sensitive mutation is rapidly reversible. These experiments indicate that in the vti1-11 cells, CPY accumulated either in the ER or in the cis-Golgi compartment (as p1CPY) with a rapid onset upon shift to the restrictive temperature. CPY that reached the trans-Golgi compartment was not targeted to the vacuole but instead secreted, suggesting that Golgi to vacuole traffic is also defective in vti1-11 mutant cells.
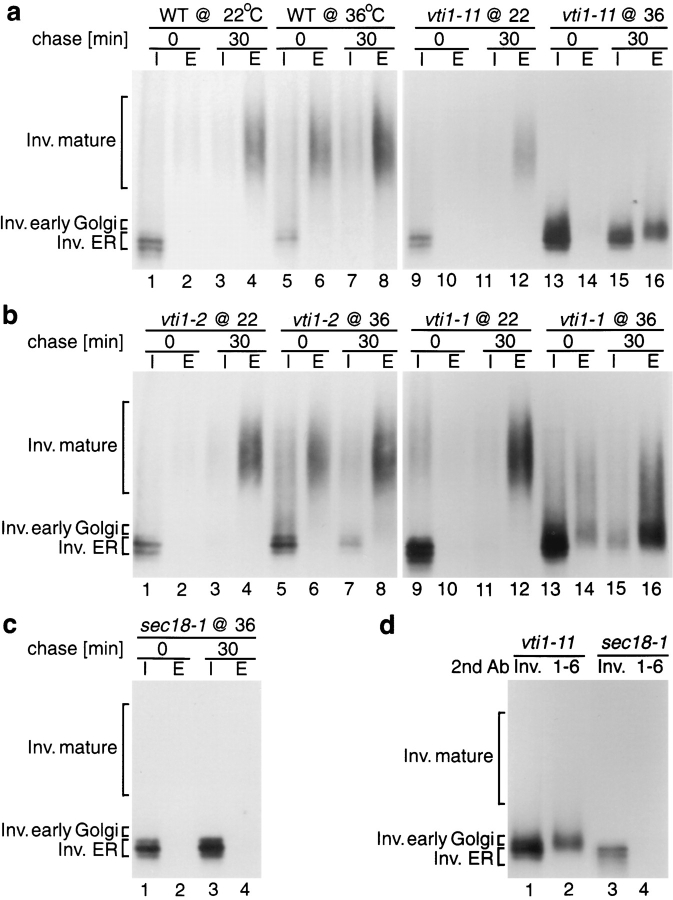
To further characterize these mutants, trafficking of the secretory protein invertase was studied (Fig. 5). In wild-type yeast cells the core glycosylated ER form of invertase seen before the chase period received outer chain glycosylation and was secreted during the chase at 22° (Fig. 5 a, lanes 1–4) and at 36°C (lanes 5–9). As expected, sec18-1 cells, which are defective in ER to Golgi vesicle traffic (Esmon et al., 1981), accumulated the core glycosylated ER form of invertase at the restrictive temperature (Fig. 5 c, lanes 1–4). Invertase glycosylation and transport were normal in all of the vti1 mutants at 22°C. In the vti1-11 mutant labeled at 36°C, the ER form and a slightly larger form of invertase accumulated intracellularly (Fig. 5 a, lane 15). Invertase molecules that were secreted by vti1-11 mutant cells were severely underglycosylated (Fig. 5 a, lane 16). By contrast, nearly all invertase was secreted by vti1-1 cells, but it was somewhat underglycosylated at the restrictive temperature (Fig 5 b, lanes 15 and 16). Glycosylation and secretion of invertase were nearly normal in the vti1-2 cells at the nonpermissive temperature (Fig. 5 b, lanes 7 and 8), indicating that in the vti1-2 mutant only transport from the late-Golgi compartment to the vacuole was blocked.
Figure 5.
Invertase sorting in vti1-ts mutants. (a–c) Invertase immunoprecipitations from intracellular (I) and extracellular (E) fractions were performed directly after the pulse or after a 30-min chase. Wild-type (WT), vti1, and sec18-1 cells were labeled at 22°C or after a 15-min preincubation at 36°C. Invertase was processed and secreted correctly in vti1-2 strains at either temperature. In vti1-1 cells, partially glycosylated invertase was secreted at 36°C. In vti1-11 cells, the ER and early Golgi forms of invertase accumulated, and partially glycosylated invertase was secreted at 36°C. (d) Invertase was immunoprecipitated from the intracellular fraction of vti1-11 and sec18-1 cells, which were incubated at 36°C and chased for 30 min. Secondary immunoprecipitations were performed with antiserum against invertase (Inv.) or against α1, 6–linked mannose (1-6). vti1-11 cells accumulated the ER form of invertase, which had not received α1,6–linked mannose, and the slightly larger cis-Golgi form, which could be precipitated with antiserum against α1,6–linked mannose.
Glycosylation occurs in a characteristic order in Saccharomyces cerevisiae because glycosylation enzymes are localized in separate Golgi subcompartments (Franzusoff and Schekman, 1989; Graham and Emr, 1991). In the first Golgi compartment, one to four α1-6 mannose residues are added to Asn-linked carbohydrate chains. Further α1-6 mannose outer chain elongation follows in the next compartment. The next modification is addition of α1-2 and α1-3 mannose. Secondary immunoprecipitation with antiserum against α1,6-mannose linkages was used to determine whether the internal invertase that accumulated in vti1 mutant cells had received this modification after chase at the restrictive temperature. The lower molecular weight fraction of invertase that accumulated in vti1-11 cells (Fig. 5 d, lane 1) had not received α1,6-linked mannose and was similar to the core glycosylated, ER-localized invertase found in sec18-1 mutants (Fig. 5 d, lanes 3 and 4), which could not be immunoprecipitated with antiserum against α1-6 mannose linkages. The slightly larger molecular weight fraction of invertase in the vti1-11 cells (Fig. 5 d, lane 2) contained α1,6-linked mannose and had therefore reached the cis-Golgi compartment. These data indicate that ER to Golgi membrane traffic is drastically slowed in vti1-11 mutant cells shifted to the nonpermissive temperature. Secretion of under-glycosylated invertase still occurred slowly, suggesting that the vti1-11 allele was “leaky” and that the Golgi apparatus had lost the ability to glycosylate invertase normally.
The behavior of vti1-11 mutant cells is reminiscent of sec22Δ/sly2Δ cells at nonpermissive temperature (Ossig et al., 1991) and ypt1-1 mutant cells (Segev et al., 1988). Sec22p/ Sly2p is a v-SNARE necessary for ER to cis-Golgi membrane traffic. Ypt1p, a rab-related small GTPase, is involved in ER to cis-Golgi and cis- to medial-Golgi vesicle fusion (Jedd et al., 1995). As with vti1-11 cells, ypt1-1 mutant cells slowly secrete underglycosylated invertase (Segev et al., 1988). These data indicate that Vti1p is required both for membrane traffic at the cis-Golgi compartment and for sorting CPY from the Golgi apparatus to the vacuole.
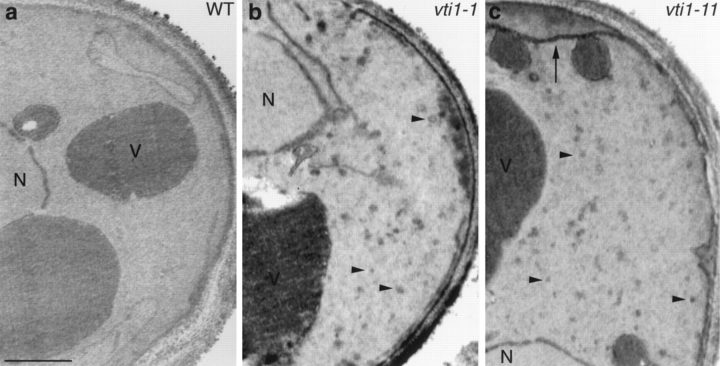
Electron microscopy was used to investigate whether vti1 mutant cells accumulated membrane organelles. Wild-type, vti1-1, and vti1-11 cells were grown at 22°C, incubated for 1 h at 37°C, and prepared for electron microscopy (Fig. 6). The vti1-1 and vti1-11 cells contained about three times the number of small vesicles relative to isogenic wild-type cells. In addition, vti1-11 cells accumulated more ER membranes than wild-type cells, but vti1-11 cells contained similar amounts of ER membranes at both 22° and 37°C. The accumulation of vesicles in vti1-1 and vti1-11 mutant cells is consistent with a proposed role for Vti1p as a v-SNARE in vesicle docking and membrane fusion.
Figure 6.
Electron microscopy of wild-type and vti1 mutant cells. Cells were grown at 22°C and transferred to 37°C for 1 h. In vti1-1 cells (b), small vesicles (arrowheads) accumulated. In vti1-11 cells (c), small vesicles and ER membranes (arrow) accumulated. N, nucleus; V, vacuole. Bar, 500 nm.
SED5 Overexpression Suppresses vti1Δ Lethality
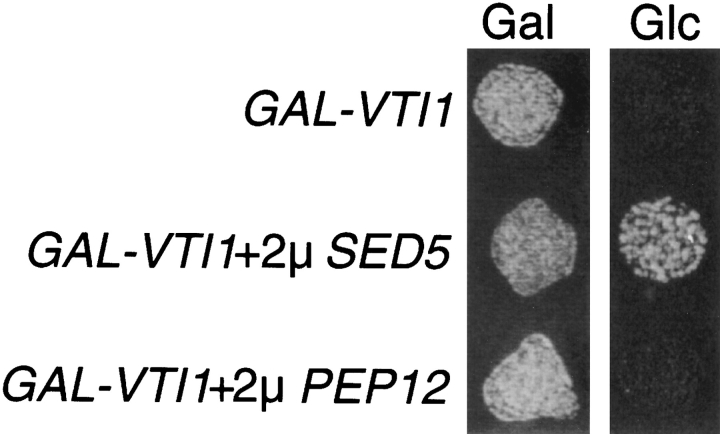
To elucidate further the essential function of VTI1, we screened for multicopy suppressors that allow yeast cells to grow in the absence of Vti1p. A vti1Δ strain harboring the GAL1–VTI1 plasmid was transformed with a multicopy yeast library (Carlson and Botstein, 1982) and transformants plated on glucose plates to switch off expression of VTI1. Yeast transformants capable of forming a colony on glucose media were tested for the absence of Vti1p by immunoblotting and for the inability to lose the multicopy plasmid to confirm that the suppression was linked to the library plasmid. In addition, the library plasmid was recovered, retransformed, and a vti1Δ strain isolated that did not contain the GAL1–VTI1 plasmid. A plasmid containing a 7.7-kb DNA insert was identified that supported growth in the absence of Vti1p. The minimal complementing fragment (2.7 kb) contained the gene SED5, which encodes a t-SNARE involved in anterograde transport from the ER to the early-Golgi compartment as well as retrograde transport from the medial- to the early-Golgi compartment (Hardwick and Pelham, 1992; Banfield et al., 1995). To confirm that SED5 was a vti1Δ suppressor, a fragment encoding Sed5p plus 400 bp 5′ and 400 bp 3′ of the coding region was amplified by PCR and cloned into a multicopy yeast vector. The 2μ-SED5 plasmid (pFvM89) was transformed into vti1Δ cells that carried a CEN-based URA3–VTI1 plasmid. The URA3–VTI1 plasmid was removed by plating cells on 5-FOA plates, resulting in vti1Δ cells containing the 2μ-SED5 plasmid that were able to grow with a doubling time of 5 h compared to a doubling time of 3 h for VTI1 wild-type yeast cells. Immunoblot analysis demonstrated that these cells were not expressing Vti1p.
To investigate whether overexpression of the prevacuolar t-SNARE Pep12p could suppress lethality due to the vti1Δ mutation, vti1Δ cells carrying the URA3–VTI1 plasmid were transformed with a multicopy PEP12 plasmid (2μ-PEP12). Overexpression of Pep12p was confirmed by Western blot analysis (data not shown). vti1Δ cells carrying the 2μ-PEP12 and URA3-VTI1 plasmids failed to grow on 5-FOA, indicating that these cells could not lose the VTI1 plasmid. It was also determined whether vti1Δ cells carrying the 2μ-PEP12 or the 2μ-SED5 plasmid and expressing VTI1 from the GAL1 promoter could grow when Vti1p expression was repressed on glucose medium. Whereas the 2μ-SED5 plasmid could support growth of vti1Δ cells after shut off of GAL1–VTI1 expression (Glc medium), the multicopy PEP12 plasmid could not (Fig. 7). These results demonstrate that VTI1 and SED5 interact genetically, providing further evidence that the essential function of VTI1 involves membrane traffic in to or out of the early-Golgi compartment.
Figure 7.
SED5 overexpression suppresses vti1Δ lethality. Growth of the GAL1–VTI1 strain alone or overexpressing Sed5p or Pep12p on plates containing galactose (Gal) or glucose (Glc). The GAL1–VTI1 strain could not grow on glucose after the expression of VTI1 was shut off. Overexpression of the early Golgi t-SNARE Sed5p allowed growth in the absence of Vti1p. Overexpression of the prevacuolar-localized t-SNARE Pep12p did not support growth of vti1Δ cells.
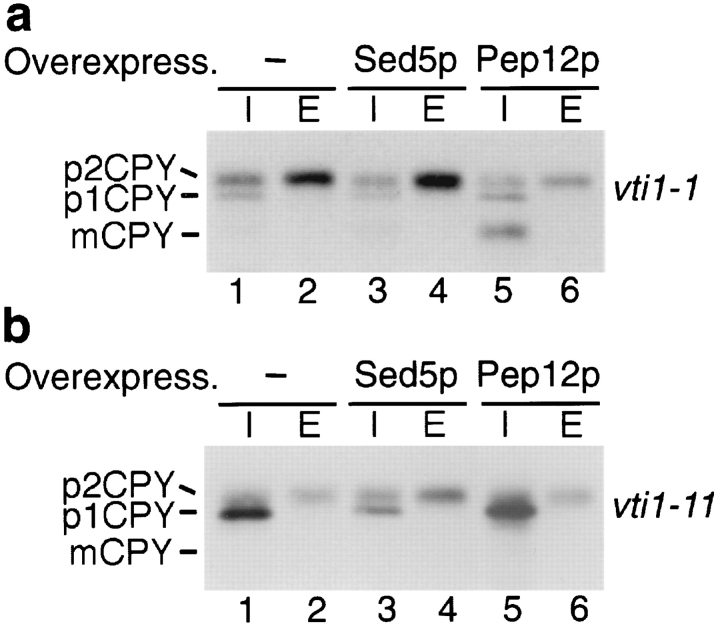
Overexpression of Pep12p or Sed5p Restores CPY Transport
The function of Vti1p was investigated further by monitoring the effects of overexpression of the t-SNAREs Pep12p and Sed5p on trafficking of CPY in the various vti1-ts mutants (Fig. 8). Whereas vti1-1 mutant cells secreted 44% of CPY at 36°C (average of four experiments; Fig. 8 a, lane 2) and sorted only 6% of CPY to the vacuole, overexpression of Pep12p led to a significant decrease in CPY secretion (26%, lane 6) and an increase in intracellular mature CPY (30%, lane 5). This dramatic increase in intracellular CPY (6–30%) due to Pep12p overexpression reflects a highly significant suppression of the CPY sorting defect in vti1-1 mutant cells. This suppression was allele specific because overexpression of Pep12p did not suppress the sorting defect observed in vti1-2 mutant cells (data not shown). By contrast, overexpression of Sed5p did not have an effect on CPY sorting in vti1-1 cells (Fig. 8 a, lanes 3 and 4). These data indicate that in vti1-1 mutant cells, which are defective in transport of CPY from the late-Golgi apparatus to the vacuole, the overexpression of the prevacuolar-localized t-SNARE Pep12p restored the ability of vesicles to dock and fuse with the prevacuolar compartment.
Figure 8.
CPY sorting in vti1-1 (a) and vti1-11 (b) cells overexpressing Sed5p and Pep12p. Overexpression of Pep12p in vti1-1 cells partially restored CPY sorting to the vacuole at the restrictive temperature (36°C). Overexpression of Sed5p in vti1-11 mutant cells reduced p1CPY accumulation at the restrictive temperature.
We also investigated whether overexpression of either Pep12p or Sed5p could suppress the membrane traffic defect at the cis-Golgi compartment in vti1-11 mutant cells. Overexpression of Sed5p in the vti1-11 strain reduced the amount of p1CPY that accumulated at restrictive temperature and increased the amount of secreted p2CPY (Fig. 8 b, lanes 3 and 4). Whereas 56% of the total CPY was found as p1CPY and 21% was secreted as p2CPY in vti1-11 mutant cells at the restrictive temperature (Fig. 8 b, lanes 1 and 2), overexpression of Sed5p led to a decrease in p1CPY accumulation to 28% and an increase in secreted p2CPY to 44% (average of two experiments). By contrast, overexpression of Pep12p in vti1-11 cells did not suppress the CPY missorting defect or the accumulation of p1CPY (Fig. 8 b, lanes 5 and 6). These results indicate that in vti1-11 mutant cells, which are blocked in traffic to the early-Golgi compartment, overexpression of the early-Golgi t-SNARE Sed5p (but not Pep12p) promoted CPY transport from the ER to the Golgi compartment but had no effect on transport of CPY from the late-Golgi compartment to the vacuole.
Vti1p Interacts with the Cytosolic Domains of Pep12p and Sed5p
To determine whether Vti1p and Pep12p interact directly, Vti1p was immunoprecipitated from yeast lysates solubilized with 0.5% Triton X-100. The native immunoprecipitation samples were analyzed by SDS-PAGE and immunoblotted for Vti1p, Pep12p, and two control membrane proteins. Approximately 10% of the total Pep12p was coprecipitated with Vti1p (Fig. 9 a). Importantly, preimmune serum did not precipitate Pep12p, and anti-Vti1p antibodies did not coprecipitate two other membrane proteins, Vph1p (a vacuolar membrane protein [Manolson et al., 1992]) or Sac1p (ER and Golgi membrane protein [Mayinger et al., 1995]). These data indicate that Vti1p and Pep12p interact specifically.
Figure 9.
Physical interaction of Vti1p with Sed5p and Pep12p. (a) Coimmunoprecipitation of Pep12p with Vti1p. Yeast spheroplasts were solubilized with 0.5% Triton X-100. Vti1p was quantitatively precipitated from the Triton extract by native immunoprecipitation with anti-Vti1p antibodies, and ∼10% of the total Pep12p was coimmunoprecipitated. In contrast, the membrane proteins Vph1p (vacuole) and Sac1p (ER and Golgi apparatus) were not coimmunoprecipitated, and pre-immune serum did not precipitate any of these proteins. For quantitative comparison, Triton extracts equivalent to 30 and 10% of total protein present in the immunoprecipitations were analyzed in lanes 3 and 4. The proteins bound to the beads and the starting extracts were analyzed by SDS-PAGE and immunoblotting with antibodies against Vti1p, Pep12p, Vph1p, and Sac1p. (b) In vitro binding of the soluble domain of Vti1p to immobilized GST fusion proteins containing the soluble domains of different yeast t-SNAREs. Vti1p interacted with GST-Sed5p and GST-Pep12p but not with GST-Sso2p or GST alone. Equal amounts of immobilized GST fusion proteins were incubated with 6-His–Vti1p. The bound proteins were analyzed by SDS-PAGE and immunoblotting with anti-Vti1p antibodies.
Because Vti1p was identified in a two-hybrid screen with the cytosolic domain of Vps10p as “bait,” we investigated whether Vti1p and Vps10p interacted physically. Only very low levels of the CPY receptor, Vps10p (<2% of the total Vps10p), coprecipitated with Vti1p under native immunoprecipitation conditions (data not shown), and additional experiments suggest that this effect may be nonspecific. Specific binding between Vps10p and Vti1p could not be detected using either chemical crosslinking followed by immunoprecipitation or in vitro binding assays with the soluble cytosolic domains. This may reflect a low degree of colocalization between Vps10p and Vti1p or that these proteins interact only transiently. Whether Vps10p and Vti1p interact functionally remains unclear.
An in vitro binding assay was used to study further the interaction of Vti1p with different yeast t-SNAREs. The soluble cytosolic domain of Vti1p was expressed in E. coli and purified as a 6-His–fusion protein. GST-fusion proteins with the soluble domains of the yeast t-SNAREs Pep12p (prevacuolar compartment [Becherer et al., 1996]), Sed5p (cis-Golgi compartment [Hardwick and Pelham, 1992]), and Sso2p (plasma membrane [Aalto et al., 1993]) were expressed in E. coli and purified. Each of the GST-fusion proteins immobilized on glutathione agarose was incubated with a constant amount of the purified 6-His– Vti1p (see Materials and Methods). The pellets were washed and the bound protein eluted, separated by SDS-PAGE, and then analyzed by immunoblotting for bound Vti1p (Fig. 9 b). High levels of Vti1p bound to the GST–Sed5p fusion. Consistent with the coimmunoprecipitation data, Vti1p also bound to GST–Pep12p. By contrast, Vti1p did not bind to GST–Sso2p or to GST alone. These data indicate that Vti1p interacts specifically with a prevacuolar t-SNARE (Pep12p) and a cis-Golgi t-SNARE (Sed5p) but not with a t-SNARE (Sso2p) involved in a pathway (secretory vesicle fusion with the plasma membrane) that is unaffected in vti1 mutant cells.
Discussion
We have identified a new v-SNARE, Vti1p, that functions in membrane traffic to the yeast vacuole. More importantly, these studies have revealed that a single v-SNARE can function in two separate vesicle trafficking steps. The yeast v-SNARE Vti1p was found to interact with two t-SNAREs, one found on the cis-Golgi compartment (Sed5p) and the second on the prevacuolar compartment (Pep12p). The specificity of the v- and t-SNAREs for each other must necessarily require the intimate involvement of additional proteins (e.g., Rab/Ypt proteins, Sec1 protein family members, and/or other proteins) so that a single v-SNARE can control the fusion of two different classes of vesicles.
Role of Vti1p in Golgi to Prevacuole Membrane Traffic
Upon depletion of Vti1p, yeast cells secreted the vacuolar hydrolase CPY, a phenotype characteristic of vacuolar protein sorting (vps) mutants (Stack et al., 1995). This result was confirmed by the isolation of the temperature-sensitive vti1 mutants, vti1-1 and vti1-2, that displayed CPY secretion with rapid onset upon shift to restrictive temperature as well as rapid recovery upon return to permissive temperature. Therefore, Vti1p is directly involved in transport of CPY from the late Golgi compartment to the vacuole.
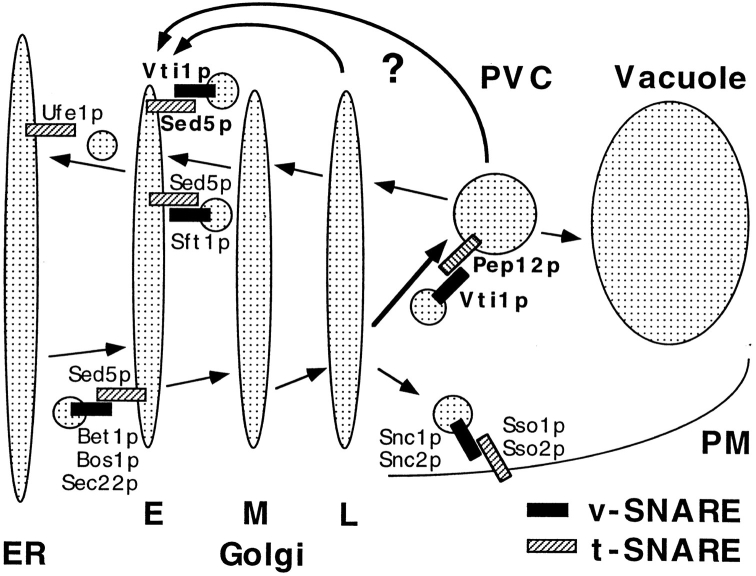
Recently, Pep12p was identified as a t-SNARE involved in sorting CPY, and this protein was localized to the prevacuolar compartment, suggesting that it acts in docking and fusion of Golgi-derived transport vesicles (Becherer et al., 1996). Therefore, we tested whether Vti1p is the v-SNARE partner of Pep12p. Overexpression of Pep12p in the vti1-1 strain suppressed the CPY sorting defect significantly at the restrictive temperature. This suppression did not represent a general bypass of the Vti1p-dependent step because it was allele specific. Native immunoprecipitation and an in vitro binding assay demonstrated physical interaction between Vti1p and Pep12p. In addition, Vti1p localized both to the Golgi apparatus and the prevacuolar compartment, consistent with a role in Golgi to prevacuolar traffic. vti1-1 cells were found to accumulate small vesicles at the restrictive temperature. Taken together, these data indicate that Vti1p–Pep12p SNARE complex formation is required for consumption of Golgi-derived transport vesicles at the prevacuolar compartment (Fig. 10).
Figure 10.
Model for Vti1p function in vesicle docking and fusion. Yeast Golgi- derived vesicles contain the v-SNARE Vti1p on the membrane allowing for its interaction with the t-SNARE Pep12p on the surface of the prevacuolar compartment (PVC). This interaction provides part of the specificity for fusion of Golgi-derived vesicles with the prevacuole. In addition, Vti1p interacts with the early Golgi t-SNARE Sed5p. We propose that Vti1p is involved in retrograde traffic from either the prevacuolar compartment or the late Golgi compartment back to the early Golgi compartment. Other v-SNAREs and t-SNAREs functioning in yeast ER, Golgi, and plasma membrane (PM) traffic are included. E, early Golgi; M, medial Golgi; L, late Golgi; PVC, prevacuolar compartment; PM, plasma membrane.
Role of Vti1p in Early-Golgi Membrane Traffic
In contrast to all VPS genes (Stack et al., 1995), VTI1 is an essential yeast gene, suggesting that it may have a second function in addition to regulating membrane traffic to the vacuole. The mutant vti1-11 grew very slowly and rapidly accumulated the ER and early-Golgi form of proCPY (p1CPY) upon shift to the restrictive temperature. CPY sorting recovered quickly after return to the permissive temperature. The core glycosylated ER form and the slightly larger α1,6-linked mannose-containing, cis-Golgi form of the secreted protein invertase accumulated intracellularly in the vti1-11 strain at the restrictive temperature. The fact that some invertase had received the initial α1,6-mannose modification indicates that glycosylation in the cis-Golgi compartment is normal. The remainder of the invertase was secreted but was severely underglycosylated. This suggests that invertase failed to receive α1,6-mannose chain elongation, which occurs in the medial-Golgi compartment. These defects in invertase secretion and processing are reminiscent of the phenotype of the ypt1-1 mutant (Segev et al., 1988). Ypt1p acts in ER to early-Golgi and in early- to medial-Golgi vesicle fusion steps (Jedd et al., 1995). An accumulation of ER core glycosylated invertase and secretion of partially glycosylated invertase was also observed at the nonpermissive temperature in sly2/sec22Δ cells (Ossig et al., 1991). Sec22p/Sly2p is a v-SNARE involved in vesicle transport from the ER to the early-Golgi compartment. These similarities indicate that trafficking into and/or out of the early-Golgi compartment is blocked or severely slowed down in vti1-11 mutant cells. The mechanisms causing underglycosylation are unclear. Possible explanations could be indirect effects due to failure to retrieve glycosylation enzymes from the TGN or the prevacuolar compartment, or that conditions in the Golgi lumen are unfavorable for the glycosylation enzymes.
The cytosolic domains of Vti1p and of the early-Golgi t-SNARE Sed5p (Hardwick and Pelham, 1992) were found to interact in an in vitro binding assay. The binding data suggest that Vti1p and Sed5p form a SNARE complex required for membrane transport to the cis-Golgi compartment (Fig. 10). Genetic analysis revealed a strong interaction between the VTI1 and the SED5 genes. Strains in which VTI1 was deleted were able to grow if Sed5p was overexpressed, indicating that SED5 is a bypass suppressor of lethality due to the vti1Δ mutation. In addition, overexpression of Sed5p in vti1-11 cells suppressed the accumulation of p1CPY, indicating that excess Sed5p overcame the cis-Golgi transport block in these cells. However, while Sed5p overexpression could suppress the cis-Golgi transport block of vti1-11 cells, Sed5p overexpression had no effect on the vacuolar protein sorting defect of any vti1 mutant (vti1-1, vti1-11, or vti1Δ/2μ-SED5). These data are not inconsistent with the Vti1p–Sed5p in vitro binding data. SED5 may be able to act as a bypass suppressor of a VTI1 deletion if a second protein on the transport vesicles interacts with Vti1p and facilitates the formation of the Sed5p–Vti1p SNARE-complex. This protein could allow a less efficient fusion of transport vesicles if high levels of Sed5p are present on the target membrane. In fact, it has been demonstrated that a complex of the v-SNAREs Bos1p and Sec22p in ER-derived transport vesicles is necessary for efficient formation of a SNARE complex with Sed5p in ER to cis-Golgi transport (Lian et al., 1994; Jiang et al., 1995).
It is important to note that a block in ER to Golgi traffic does not necessarily implicate Vti1p in this exact trafficking step. It was reported recently that a block in retrograde traffic from the Golgi apparatus to the ER leads to a rapid block in anterograde traffic from the ER to the Golgi apparatus (Letourneur et al., 1994; Lewis and Pelham, 1996). In fact there were no differences in the lag before p1CPY accumulated in cells conditionally defective for proteins involved in anterograde traffic (sec18, sec12) or retrograde traffic (ufe1, sec20, sec21). It is unlikely that Vti1p acts together with the v-SNAREs Sec22p/Sly2p, Bet1p/Sly12p, and Bos1p in ER to cis-Golgi traffic, because Sec22p, Bet1p, and Bos1p were localized predominantly to the ER by a combination of immunofluorescence and subcellular fractionation (Newman et al., 1992; Jiang et al., 1995), whereas Vti1p was not detected in the ER.
We propose that Vti1p is involved in retrograde traffic to the cis-Golgi compartment in yeast. Whereas anterograde Golgi traffic is studied extensively, not much is known about retrograde transport. Because v-SNAREs bud with transport vesicles from the donor compartment and are incorporated into the target membrane there has to be a mechanism by which v-SNAREs and other components of the transport machinery recycle to the donor compartment. Recently, evidence for retrograde traffic from the medial- to the cis-Golgi compartment in yeast was reported (Banfield et al., 1995). It was also reported that the cis-Golgi enzyme Och1p rapidly reaches the TGN and returns to the cis-Golgi compartment by retrograde transport (Harris and Waters, 1996). The localization of the medial-Golgi protein Emp47p involves retrograde transport to the ER (Schröder et al., 1995). Recycling of late-Golgi proteins from the prevacuolar compartment back to the Golgi apparatus has also been demonstrated (Cereghino et al., 1995; Piper et al., 1995; Cooper and Stevens, 1996; Nothwehr et al., 1996), but it is unclear which Golgi stack serves as the acceptor membrane. In all examples it is also not clear whether retrograde traffic moves back from compartment to compartment, mirroring anterograde flow, or whether retrograde transport vesicles from different compartments have a common target, e.g., the ER or the cis-Golgi compartment (Rothman and Wieland, 1996).
Vti1p is probably not involved in retrograde traffic from the medial- to the early-Golgi compartment together with the v-SNARE Sft1p, because the phenotypes resulting from vti1 and sft1 mutations are different. At the restrictive temperature, sft1-ts mutants accumulated large amounts of Golgi membranes and the early-Golgi but not the ER form of invertase (Banfield et al., 1995). An attractive possibility is that Vti1p is involved in retrograde vesicle traffic from the prevacuolar compartment back to the cis-Golgi compartment. Alternatively, Vti1p could control retrograde traffic from the late-Golgi to the cis-Golgi compartment, a possible pathway for Och1p retrieval in yeast (Harris and Waters, 1996). Future efforts will be directed at identifying the putative proteins whose retrieval to the cis-Golgi compartment is required for secretion in yeast.
Implications for the SNARE Hypothesis
In its original form, the SNARE hypothesis proposed that a specific set of v-SNAREs on transport vesicles and a specific set of t-SNAREs on target membranes serve as identifying markers. Their specific pairwise recognition would ensure that transport vesicles would fuse only with the correct target organelle (Söllner et al., 1993; Ferro-Novick and Jahn, 1994; Rothman, 1994). Recently an extension of the original SNARE hypothesis was made necessary by the discovery that Sed5p could interact not only with the v-SNAREs Sec22p/Sly2p, Bet1p/Sly12p, and Bos1p that are required for ER to early Golgi traffic but also with the v-SNARE Sft1p, which is involved in retrograde traffic from a later Golgi to the early-Golgi compartment (Banfield et al., 1995). Vti1p-containing vesicles would represent a third kind of vesicle fusing with the early-Golgi compartment. This could mean that a t-SNARE defines a target organelle and can interact with v-SNAREs of all incoming vesicles to that organelle, instead of being restricted to a specific vesicle fusion event.
We demonstrate here by genetic and biochemical studies that a single v-SNARE can interact functionally with two different t-SNAREs in distinct compartments. A model to explain these data is that a v-SNARE functioning in anterograde trafficking to a target organelle also plays a critical role in docking and fusion of retrograde vesicles originating from the target organelle, while the v-SNARE itself is recycling. Alternatively, if Vti1p directs both anterograde and retrograde traffic from the late-Golgi compartment this could mean that a v-SNARE is present on different vesicle populations budding from a single donor compartment and may define the donor membranes. To interact specifically with either the anterograde or the retrograde t-SNARE, this v-SNARE must either be able to assume two different conformations, two modified states, or form complexes with additional proteins that modify the action of the v-SNARE. A protein able to fulfill the latter type of function has been identified in mammalian neurons. The v-SNARE synaptobrevin forms a complex with the synaptic vesicle protein synaptophysin, which excludes the t-SNARE syntaxin 1 (Edelmann et al., 1995), suggesting a role for synaptophysin in control of synaptobrevin activity. Therefore, it seems likely that the activity and possibly the specificity of v-SNAREs are regulated by interactions with additional proteins so that a single v-SNARE can direct vesicle fusion of two different classes of vesicles.
Acknowledgments
We thank J. Selker for superb electron microscopy work and G. Hamilton for help with the two-hybrid screen. We acknowledge R.C. Piper and S.R. Gerrard for providing PEP12 plasmids and anti-Pep12p antiserum. N.J. Bryant, R.C. Piper, W. Voos, E. Conibear, G. Flynn, and G.F. Sprague, Jr. are thanked for stimulating discussions and critical reading of the manuscript. We also thank M. Snyder for supplying a CIK1 plasmid, R. Brent and S. Fields for two-hybrid plasmids and strains, R. Schekman for the antiserum against α1,6-linked mannose, and P. Orlean for anti-Dpm1p antibodies and DPM1 plasmids.
This work was supported by grants from the Deutsche Forschungsgemeinschaft, from the American Cancer Society (S.F. Nothwehr; PF-3608), and from the National Institutes of Health (T.H. Stevens; GM32448).
Footnotes
1. Abbreviations used in this paper: CPY, carboxypeptidase Y; ER, endoplasmic reticulum; FOA, fluoroorotic acid.
Please address all correspondence to Tom H. Stevens, Institute of Molecular Biology, University of Oregon, Eugene, OR 97403-1229. Tel.: (541) 346-5884; Fax: (541) 346-4854; E-mail: stevens@molbio.uoregon.edu
S.F. Nothwehr's present address is Division of Biological Sciences, University of Missouri, Columbia, MO 65211
References
- Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO (Eur Mol Biol Organ) J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Pelham HR. A SNARE-like protein required for traffic through the Golgi complex. Nature (Lond) 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Johnson LM, Emr SD. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc Natl Acad Sci USA. 1986;83:9075–9079. doi: 10.1073/pnas.83.23.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel PL, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. doi: 10.1016/0076-6879(95)54018-0. [DOI] [PubMed] [Google Scholar]
- Becherer KA, Rieder SE, Emr SD, Jones EW. Novel syntaxin homolog, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J.D., F. LaCroute, and G.R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoroorotic acid resistance. Mol. Gen. Genet. 197. [DOI] [PubMed]
- Brent R, Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted proteins with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPSgene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AA, Stevens TH. Vps10p cycles between the late-Golgi and prevacuolar compartments in its function as the sorting receptor for multiple yeast vacuolar hydrolases. J Cell Biol. 1996;133:529–541. doi: 10.1083/jcb.133.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher C, Ossig R, Gallwitz D, Schmitt HD. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RASsuperfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann L, Hanson PI, Chapman ER, Jahn R. Synaptobrevin binding to synaptophysin: a potential mechanism for controlling the exocytotic fusion machine. EMBO (Eur Mol Biol Organ) J. 1995;14:224–231. doi: 10.1002/j.1460-2075.1995.tb06995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmon B, Novick P, Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981;25:451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- Farquhar MG, Palade GE. The Golgi apparatus (complex)-(1954– 1981)-from artifact to center stage. J Cell Biol. 1981;91:77S–103S. doi: 10.1083/jcb.91.3.77s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature (Lond) 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Franzusoff A, Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7mutation. EMBO (Eur Mol Biol Organ) J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemis, E., J. Gyuris, and R. Brent. 1996. Interaction trap/two-hybrid systems to identify interacting proteins. In Current Protocols in Molecular Biology. F.M. Ausubel, editor. John Wiley & Sons, New York. 20.1.1–20.1.28.
- Graham TR, Emr SD. Compartmental organization of Golgi-specific protein modification and vacuolar protein sorting events defined in a yeast sec18(NSF) mutant. J Cell Biol. 1991;114:207–218. doi: 10.1083/jcb.114.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagologg A. E. colishuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Jahn R, Südhof TC. Synaptic vesicles and exocytosis. Annu Rev Neurosci. 1994;17:219–246. doi: 10.1146/annurev.ne.17.030194.001251. [DOI] [PubMed] [Google Scholar]
- Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Y., M. Sacher, B. Singer-Krüger, J.P. Lian, S. Stone, and S. Ferro-Novick. 1995. Factors mediating the late stages of ER-to-Golgi transport in yeast. Cold Spring Harbor Symp. Quant. Biol. LX, 119–126. [DOI] [PubMed]
- Jones EW. Proteinase mutants of Saccharomyces cerevisiae. . Genetics. 1977;85:23–33. doi: 10.1093/genetics/85.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CA, Schekman R. Distinct sets of SECgenes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, D'Emolliere C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- Lian JP, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Ypt1p implicated in v-SNARE activation. Nature. 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science (Lond) 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Maina CV, Riggs PD, Grandea AGD, Slatko BE. An Escherichia colivector to express and purify foreign proteins by fusion to and separation from maltose-binding protein. Gene. 1988;74:365–373. doi: 10.1016/0378-1119(88)90170-9. [DOI] [PubMed] [Google Scholar]
- Manolson MF, Proteau D, Preston RA, Stenbit A, Roberts BT, Hoyt MA, Preuss D, Mulholland J, Botstein D, Jones EW. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivoassembly and activity of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Mayinger P, Bankaitis VA, Meyer DI. Sac1p mediates the adenosine triphosphate transport into yeast endoplasmic reticulum that is required for protein translocation. J Cell Biol. 1995;131:1377–1386. doi: 10.1083/jcb.131.6.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Groesch ME, Ferro-Novick S. Bos1p, a membrane protein required for ER to Golgi transport in yeast, co-purifies with the carrier vesicles and with Bet1p and the ER membrane. EMBO (Eur Mol Biol Organ) J. 1992;11:3609–3617. doi: 10.1002/j.1460-2075.1992.tb05445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Stevens TH. Sorting of membrane proteins in the yeast secretory pathway. J Biol Chem. 1994;269:10185–10188. [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRDgenes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidylinositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. . Mol Cell Biol. 1990;10:5796–5805. doi: 10.1128/mcb.10.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossig R, Dascher C, Trepte HH, Schmitt HD, Gallwitz D. The yeast SLY gene products, suppressors of defects in the essential GTP-binding Ypt1protein, may act in endoplasmic reticulum-to-Golgi transport. Mol Cell Biol. 1991;11:2980–2993. doi: 10.1128/mcb.11.6.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. . Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Paravicini G, Horazdovsky BF, Emr SD. Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol Biol Cell. 1992;3:415–427. doi: 10.1091/mbc.3.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham HRB. Is epimorphin involved in vesicular transport? . Cell. 1993;73:425–426. doi: 10.1016/0092-8674(93)90128-d. [DOI] [PubMed] [Google Scholar]
- Piper RC, Whitters EA, Stevens TH. Yeast Vps45p is a Sec1p-like protein required for the consumption of vacuole-targeted, post-Golgi transport vesicles. Euro J Cell Biol. 1994;65:305–318. [PubMed] [Google Scholar]
- Piper RC, Cooper AA, Hong Y, Stevens TH. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. . J Cell Biol. 1995;131:603–617. doi: 10.1083/jcb.131.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Wuestehube LJ, Schekman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vpsmutants. Mol Biol Cell. 1992a;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CK, Roberts CJ, Moore KE, Howald-Stevenson I, Stevens TH. Biogenesis of the vacuole in Saccharomyces cerevisiae. . Int Rev Cytol. 1992b;139:59–120. doi: 10.1016/s0074-7696(08)61410-2. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nothwehr SF, Stevens TH. Membrane protein sorting in the yeast secretory pathway: evidence that the vacuole may be the default compartment. J Cell Biol. 1992;119:69–83. doi: 10.1083/jcb.119.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J. Mechanism of intracellular protein transport. Nature (Lond) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- Rothman JH, Stevens TH. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT. Protein sorting by transport vesicles. Science (Wash DC) 1996;227:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- Schauer I, Emr S, Gross C, Schekman R. Invertase signal and mature sequence substitutions that delay intercompartmental transport of active enzyme. J Cell Biol. 1985;100:1664–1675. doi: 10.1083/jcb.100.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder S, Schimmöller F, Singer-Krüger B, Riezman H. The Golgi-localization of yeast Emp47p depends on its di-lysine motif but is not affected by the ret1-1 mutation in α-COP. J Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia colias fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Sogaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Sollner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Söllner T, Whiteheart SW, Brunner M, ErdjumentBromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature (Lond) 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stack JH, Horazdovsky BF, Emr SD. Receptor-mediated protein sorting to the vacuole in yeast: Roles for a protein kinase, a lipid kinase and GTP-binding proteins. Annu Rev Cell Dev Biol. 1995;11:1–33. doi: 10.1146/annurev.cb.11.110195.000245. [DOI] [PubMed] [Google Scholar]
- Stevens T, Esmon B, Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vater CA, Raymond CK, Ekena K, Howald-Stevenson I, Stevens TH. The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J Cell Biol. 1992;119:773–786. doi: 10.1083/jcb.119.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida TA, Huyer G, Emr SD. Yeast vacuolar proenzymes are sorted in the late Golgi complex and transported to the vacuole via a prevacuolar endosome-like compartment. J Cell Biol. 1993;121:1245–1256. doi: 10.1083/jcb.121.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JM. Membrane fusion. Science (Wash DC) 1992;258:917–924. doi: 10.1126/science.1439803. [DOI] [PubMed] [Google Scholar]