Abstract
Natural hammerhead ribozymes are mostly found in some viroid and viroid-like RNAs and catalyze their cis cleavage during replication. Hammerheads have been manipulated to act in trans and assumed to have a similar catalytic behavior in this artificial context. However, we show here that two natural cis-acting hammerheads self-cleave much faster than trans-acting derivatives and other reported artificial hammerheads. Moreover, modifications of the peripheral loops 1 and 2 of one of these natural hammerheads induced a >100-fold reduction of the self-cleavage constant, whereas engineering a trans-acting artificial hammerhead into a cis derivative by introducing a loop 1 had no effect. These data show that regions external to the central conserved core of natural hammerheads play a role in catalysis, and suggest the existence of tertiary interactions between these peripheral regions. The interactions, determined by the sequence and size of loops 1 and 2 and most likely of helices I and II, must result from natural selection and should be studied in order to better understand the hammerhead requirements in vivo.
Keywords: catalytic RNAs/hammerhead structures/satellite RNAs/viroids
Introduction
Viroids are not supramolecular assemblies such as viruses, but instead are exclusively composed of a small (between 246 and 401 nucleotides, excluding those with sequence repeats), single-stranded, circular, non-coding RNA with the ability to infect certain plants and in most cases incite a pathological disorder (Flores et al., 2000; Diener, 2001). Research on viroids, in addition to viroid discovery itself as the lowest step of the biological scale, has led to other breakthroughs, outstanding among which is the identification of the hammerhead ribozyme in avocado sunblotch viroid (ASBVd) (Hutchins et al., 1986), and in a satellite RNA structurally similar to viroids but functionally dependent on a helper virus (Prody et al., 1986). This finding is regarded as a milestone in molecular virology, with major implications for the replication and evolutionary origin of these RNAs (Forster and Symons, 1987; Diener, 1989). Moreover, owing to its structural simplicity, the hammerhead has sparked considerable basic and applied interest focused on the understanding of its catalytic mechanism (Pley et al., 1994; Scott et al., 1995, 1996) and on its potential use as a therapeutic tool (Rossi and Couture, 1999). This interest promoted since the very beginning the splitting of the hammerhead into two domains: the ribozyme itself, which remains unchanged at the end of the reaction, and the substrate, which it is fragmented into two pieces as a result of a transesterification (Uhlenbeck, 1987). Several splitting formats were assayed (Uhlenbeck, 1987; Haseloff and Gerlach, 1988; Koizumi et al., 1988; Jeffries and Symons, 1989). Such trans-acting designs permitted the study of the reaction kinetics under protein-free conditions and were obligatory for custom-made ribozymes targeted against specific RNAs (Haseloff and Gerlach, 1988). The residues necessary for catalysis have been defined on the basis of detailed analyses (Sheldon and Symons, 1989; Ruffner et al., 1990; for a review see McKay, 1996).
In contrast, kinetic data on natural hammerheads acting in cis, their biological context, are scarce. One of the first studies of this kind, using the (+) hammerhead of the satellite RNA of lucerne transient streak virus (sLTSV), revealed that self-cleavage of the purified RNA was quantitative within 1 min, impeding determination of the catalytic constant under standard conditions (Forster and Symons, 1987). A second study with another cis-acting natural hammerhead, that of the (+) strand of peach latent mosaic viroid (PLMVd) (Hernández and Flores, 1992), also showed quantitative self-cleavage in <1 min (Beaudry et al., 1995). This result was confirmed in a third study (Ambrós and Flores, 1998), in which the self-cleavage rate constant was estimated by a procedure for determining the intramolecular rates of hammerheads during in vitro transcription (Long and Uhlenbeck, 1994). To this aim, the magnesium concentration and the pH of the buffer had to be lowered, but even under these conditions, which affect catalysis negatively (Dahm et al., 1993), the self-cleavage rate constant of the PLMVd (+) hammerhead (Ambrós and Flores, 1998) was higher than the rate constants under standard conditions of two artificial hammerheads acting in cis and in trans (Long and Uhlenbeck, 1994). Although these values were not directly comparable, because the hammerhead formats and the reaction conditions were different, they provided data suggesting that the catalytic properties of natural and artificial hammerheads are not necessarily the same.
A further argument also supports this view. The most efficient hammerheads selected in vitro from a pool of oligoribonucleotides randomized in all positions of the central core contain the motifs strictly conserved in the core of natural hammerheads (Ishizaka et al., 1995; Tang and Breaker, 1997; Eckstein et al., 2001; Salehi-Ashtiani and Szostak, 2001). This indicates that natural hammerheads have been selected through evolution essentially on the basis of optimizing their self-cleaving activity. Therefore, it is conceivable to assume that in addition to the central core, other hammerhead features that include the sequence and size of the adjacent helices and their closing loops may have also been selected to improve the self-cleaving activity of these ribozymes in their cis natural context. Here we provide data showing that this is the case; more specifically that the catalytic behavior of several natural hammerheads in their cis format differs considerably from that of trans-acting derivatives thereof. These results demonstrate that the peripheral regions surrounding the central conserved cores of natural hammerheads play a significant role in catalysis and point out the need of further structural studies of natural cis-acting hammerheads.
Results
Increased yields of uncleaved hammerhead RNAs by transcription in the presence of antisense oligonucleotides
Two approaches have been described to determine the intramolecular self-cleavage rates of hammerheads: isolation of the uncleaved RNA from in vitro transcription reactions and its subsequent incubation into an appropriate buffer (Hutchins et al., 1986; Forster et al., 1988; Miller and Silver, 1991; Pabón-Peña et al., 1991), or direct estimation during in vitro transcription (Long and Uhlenbeck, 1994; Vaish et al., 1997; Ambrós and Flores, 1998). Because most hammerhead RNAs self-cleave efficiently during in vitro transcription, suggesting that the small fraction of uncleaved primary transcripts might be trapped in catalytically inactive conformations (Forster and Symons, 1987) or with mutations affecting self-cleavage (Long and Uhlenbeck, 1994), only those hammerhead RNAs having an intrinsic low self-cleavage rate, e.g. the transcript of the newt satellite 2 DNA (Epstein and Gall, 1987) or the satellite RNA of the cereal yellow dwarf virus-RPV (sCYDV-RPV) (Miller et al., 1991), can be isolated in sufficient amounts to be analyzed under protein-free conditions. Otherwise, when directly determining the self-cleavage rates of natural hammerhead RNAs during in vitro transcription, non-standard conditions, specifically low pH and magnesium concentration, have to be used (Ambrós and Flores, 1998; this work). Such non-standard conditions make further kinetic comparisons between intra- and intermolecular hammerheads difficult.
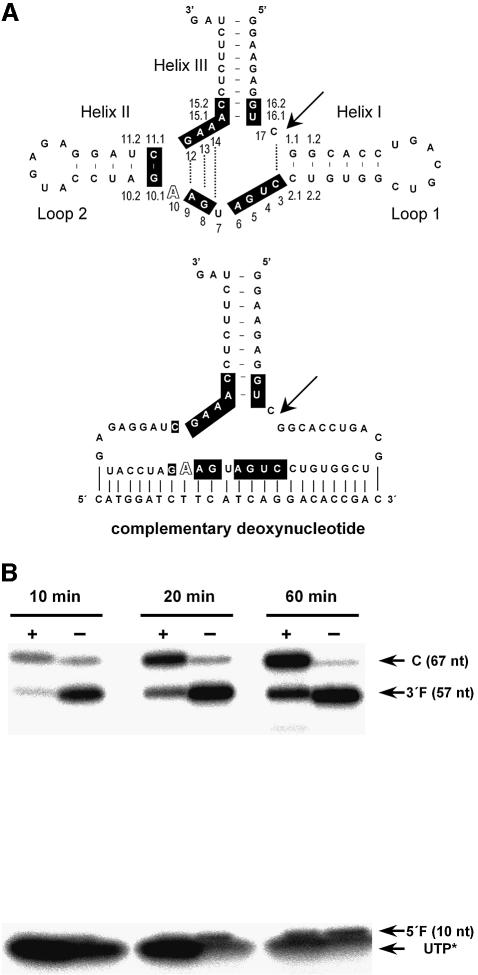
To alleviate this problem we have introduced a modification that permits a comparative analysis, under the same protein-free conditions, of the cleavage rate constants of intra- and intermolecular hammerheads. Extensive self-cleavage of the hammerhead RNA during in vitro transcription was avoided by including a deoxyoligonucleotide complementary to part of the ribozyme, an approach previously used with one of the ribozymes of hepatitis delta virus RNA (Been and Perrotta, 1995) and for in vitro selection of self-cleaving RNAs (Salehi-Ashtiani and Szostak, 2001). The strength of this approach is illustrated by the case of the (+) hammerhead of chrysanthemum chlorotic mottle viroid (CChMVd) (Navarro and Flores, 1997; De la Peña and Flores, 2001), the self-cleavage of which during in vitro transcription decreased from >90% of the total RNA to <25% as a result of incorporating the antisense deoxyoligonucleotide (Figure 1). Transcription levels were not affected by the antisense deoxyoligonucleotide, except for a slight decrease (10–15%) of the total RNA yield at shorter times, and the appearance of some prematurely terminated transcripts (Figure 1). High levels of uncleaved primary transcripts (50–80% of the total RNA) were also obtained in the presence of antisense deoxyoligonucleotides for different mutants of this hammerhead (see below), and for the PLMVd (+) hammerhead, whereas the fraction of the uncleaved primary transcript was lower (<30% of the total RNA) with a cis-derivative of an artificial hammerhead (HH8) (see Figure 2 for the corresponding schemes), probably because the high stability of helices I and II of this hammerhead (rich in GC pairs) impaired efficient annealing of the antisense deoxyribonucleotide. The uncleaved primary transcripts thus obtained were eluted and stored in the absence of divalent cations until further analysis. However, during elution of the uncleaved primary transcript of the PLMVd (+) hammerhead, extensive self-cleavage (up to 60% of the total RNA) was observed even in the presence of high concentrations of EDTA. To reduce this high self-cleavage, gel elution was performed in 40% formamide that decreased considerably the self-cleavage extent (15–25% of the total RNA).
Fig. 1. Effect on self-cleavage during transcription of a deoxyoligonucleotide complementary to part of the hammerhead ribozyme. (A) Schematic representation of the CChMVd (+) hammerhead foldings in the absence (top) and presence (bottom) of the antisense deoxyoligonucleotide. Canonical and non-canonical pairs are indicated by continuous and broken lines, respectively. The nucleotide residues strictly or highly conserved in most natural hammerhead structures are in a black background. Numbering is based on the standard criterion for the consensus hammerhead (Hertel et al., 1992) with the exception of the extra A between residues A9 and G10.1, characteristic of the CChMVd (+) hammerhead, which is referred as number 10 (outlined font). Arrow indicates the self-cleavage site. (B) Self-cleavage kinetics during transcription of the CChMVd (+) hammerhead in the presence (+) or absence (–) of the antisense deoxyoligonucleotide. Reaction aliquots were taken at three different times and analyzed by denaturing (15%) PAGE and autoradiography. Positions of the complete (C) primary transcript and of the resulting self-cleavage 3′ and 5′ fragments (3′F and 5′F, respectively), with their sizes in nucleotides, are indicated on the right.
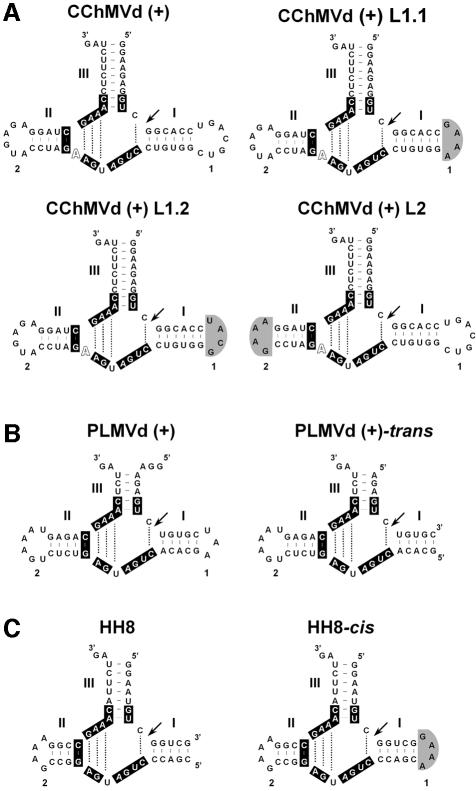
Fig. 2. Schematic representations of the natural and artificial hammerhead structures used in this work. (A) CChMVd (+) hammerhead and three cis-acting derivatives thereof in which loop 1 was substituted by the tetraloops GAAA or UACG [CChMVd (+) L1.1 and CChMVd (+) L1.2, respectively], and loop 2 was substituted by the tetraloop GAAA [CChMVd (+) L2]. (B) PLMVd (+) hammerhead and a trans-acting derivative thereof in which loop 1 was removed. (C) Artificial trans-acting hammerhead 8 (HH8) and a cis-acting derivative thereof in which the 3′-end of the substrate was linked to the 5′-end of the ribozyme through a GAAA tetraloop. The nucleotide residues strictly or highly conserved in most natural hammerhead structures are in a black background. Arrows mark the self-cleavage sites. Gray areas indicate changes affecting loops 1 or 2.
The PLMVd (+) hammerhead self-cleaves under protein-free conditions with a higher catalytic constant than a trans-acting derivative thereof
To confirm that the high self-cleavage rate constant of the PLMVd (+) hammerhead determined previously during transcription (Ambrós and Flores, 1998) was not an artifact, discarding that the proteins present in the transcription reaction could affect the self-cleavage rate constant, this constant was re-evaluated under protein-free conditions. As indicated, elution of the primary transcript was performed in the presence of formamide to limit the extent of self-cleavage. This extent did not change after recovery of the RNA by ethanol precipitation and heating at 95°C for 2 min followed by incubation at 25°C for 10 min, with self-cleavage only occurring after addition of magnesium. Attempts to determine the self-cleavage rate constant under standard conditions (see below) were impeded by the rapid turnover, forcing the modification of these conditions, particularly to decrease the pH taking into account that the logarithm of the cleavage rate constant of a hammerhead increases linearly with the pH (Dahm et al., 1993). More specifically, the trans cleavage rate constants of different artificial hammerheads under standard conditions (50 mM Tris–HCl, pH 7.5, 10 mM Mg2+, 25°C) exhibit a 10-fold reduction at pH 6.5 keeping the other variables unchanged (Clouet-d’Orval and Uhlenbeck, 1996, 1997). Under the new conditions (50 mM PIPES-NaOH, pH 6.5, 10 mM Mg2+, 25°C), the PLMVd (+) hammerhead still showed a high self-cleavage rate constant (3.19 ± 0.35/min), when compared with the catalytic constant of a trans-acting derivative (Figure 2) under the same conditions (0.56 ± 0.03/min). This corroborated the fact that the self-cleavage rate constant of the PLMVd (+) hammerhead determined previously (Ambrós and Flores, 1998) was not the result of specific reaction conditions.
Cis cleavage kinetics during transcription of the CChMVd (+) hammerhead and of its four mutants at position A10
To verify that the high self-cleavage rate constant of the PLMVd (+) hammerhead was not a peculiarity of this ribozyme, the kinetic analysis was extended to another natural hammerhead. Results from a recent study on the CChMVd (+) hammerhead and four mutants thereof have shown that the extra A (A10) between the conserved A9 and the quasi-conserved G10.1 of the wild-type ribozyme (Figure 2) causes a minor decrease of the trans cleavage rate constant with respect to the same hammerhead without this residue, whereas the substitutions A10→C or A10→G have more important effects. In contrast, the A10→U substitution induces a 3- to 4-fold increase (De la Peña and Flores, 2001) (Table I).
Table I. Cis (during transcription) and trans cleavage rate constants of the CChMVd (+) hammerhead ribozyme and its four mutants at position 10.
| Hammerhead | kcat (/min) (cis)a | kcat (/min) (trans)b |
|---|---|---|
| CChMVd-A10 | 1.95 ± 0.12 | 0.80 ± 0.14 |
| CChMVd-Δ10 | 1.59 ± 0.15 | 1.04 ± 0.12 |
| CChMVd-C10 | 2.08 ± 0.44 | 0.18 ± 0.02 |
| CChMVd-G10 | 0.81 ± 0.02c | 0.39 ± 0.02 |
| CChMVd-U10 | 3.79 ± 0.71 | 2.82 ± 0.63 |
aTranscriptions were performed at pH 7 and 6 mM Mg2+ (Ambrós and Flores, 1998).
bFrom De la Peña and Flores (2001), determined at pH 7.5 and 10 mM Mg2+.
cIn the hammerhead ribozyme with the A10→G substitution, adjustment of the experimental data to the proposed equation was difficulted by the significant RNA fraction remaining uncleaved at long reaction times.
For comparative purposes, we studied the effects of the presence of mutations at position 10 of this hammerhead on its cis-acting natural context. In a first experiment series, the self-cleavage rate constants were determined during transcription, as described previously (Long and Uhlenbeck, 1994), with minor modifications (Ambrós and Flores, 1998) (Table I; Supplementary figure 1, available at The EMBO Journal Online). It must be noted that the values of the cis-acting ribozymes, in spite of being quantitatively similar to their trans counterparts, were obtained at lower pH and magnesium concentration, conditions that slow down reaction kinetics and that were necessary to implement in order to obtain a reliable estimation. The cleavage rate constants in cis and trans of the wild-type (A10), the consensus A10→Δ and the A10→G and A10→U CChMVd (+) hammerheads followed the same trend: A10→U > A10 ≈ A10→Δ > A10→G (Table I). In the last case (A10→G), the lower values obtained could reflect the adoption of alternative (catalytically inactive) conformations (De la Peña and Flores, 2001). In the ribozyme with the A10→C substitution the difference between the two cleavage rate constants suggests that the Watson–Crick pair proposed in trans between C10 and G12 (De la Peña and Flores, 2001) would not form during transcription.
Interestingly, a direct comparison with the self-cleavage rate constant of the PLMVd (+) hammerhead (3/min), determined previously under the same conditions (Ambrós and Flores, 1998), revealed that the values of the CChMVd (+) ribozymes were in the same range (Table I). Altogether, these results, and those reported for the sLTSV (+) hammerhead (Forster and Symons, 1987), showed that natural hammerheads (at least these three) display considerably higher cis cleavage activity than their trans-acting derivatives and other artificial hammerheads also acting in trans.
Cis cleavage kinetics under protein-free conditions of the CChMVd (+) hammerhead and of its four mutants at position A10
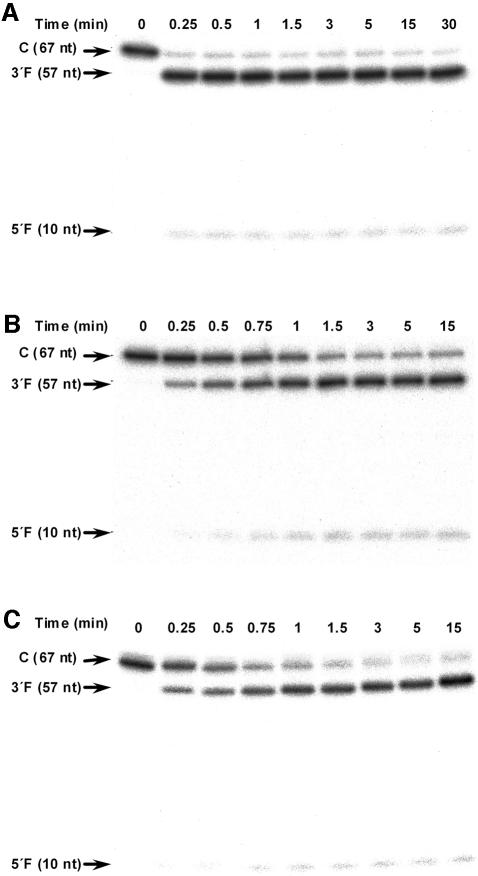
To provide additional support for this view, the self-cleavage rate constants for the CChMVd (+) ribozymes were also determined with the second approach (under protein-free conditions). In a first attempt, uncleaved transcripts of the wild-type (A10) hammerhead ribozyme (Figure 2) were incubated under standard conditions (50 mM Tris–HCl, pH 7.5, 10 mM Mg2+, 25°C). However, >90% of the RNA self-cleaved during the first 15 s, making kinetic analysis by manual pipetting impossible (Figure 3A). To reduce the self-cleavage rate, the pH was brought down to 6.5, as with the PLMVd (+) hammerhead, keeping the other conditions unchanged. The low pH permitted determination of the self-cleavage rate constants for the wild-type CChMVd (+) hammerhead (Figure 3B; Supplementary figure 2A) and for its A10→Δ mutant that were 10- to 20-fold higher than the rate constants of two trans-acting derivatives thereof (Table II), and of other artificial trans-acting hammerheads at the same low pH (∼0.1/min) (Stage-Zimmermann and Uhlenbeck, 1998). The lower values of the ribozymes with the substitutions A10→C and A10→G should be regarded with caution for the reasons stated above.
Fig. 3. Self-cleavage kinetics of the CChMVd (+) hammerhead in protein-free buffers of different composition. (A) Standard buffer (50 mM Tris–HCl, pH 7.5, 10 mM Mg2+, 25°C). (B) Low-pH buffer (50 mM PIPES-NaOH, pH 6.5, 10 mM Mg2+, 25°C). (C) Low- magnesium buffer (40 mM Tris–HCl, pH 7, 0.5 mM Mg2+, 25°C). Positions of the complete (C) primary transcript and of the resulting self-cleavage 3′ and 5′ fragments (3′F and 5′F, respectively), with their sizes in nucleotides are indicated on the left.
Table II. Cis and trans cleavage rate constants under protein-free conditions and low pH of the CChMVd (+) hammerhead ribozyme and its mutants at position 10 and loops 1 and 2.
| Hammerhead | kcat (/min) (cis)a | kcat (/min) (trans)a |
|---|---|---|
| CChMVd-A10 | 2.43 ± 0.52 | 0.13 ± 0.01 |
| CChMVd-Δ10 | 1.41 ± 0.06 | 0.18 ± 0.02 |
| CChMVd-C10 | 0.17 ± 0.02 | ND |
| CChMVd-G10 | 0.060 ± 0.011 | ND |
| CChMVd-U10 | 3.46 ± 0.48 | ND |
| CChMVd-L1.1 | 0.006 ± 0.001 | – |
| CChMVd-L1.2 | 0.002 ± 0.0002 | – |
| CChMVd-L2 | 0.025 ± 0.004 | – |
aReactions were performed in 50 mM PIPES–NaOH, pH 6.5, 10 mM Mg2+, 25°C.
ND, not determined; (–), not applicable.
To extend these observations to other conditions, the kinetic analysis was repeated under the conditions used with another natural cis-cleaving hammerhead (40 mM Tris–HCl, pH 7.0, 10 mM Mg2+, 37°C) (Miller and Silver, 1991; Song et al., 1999). Because the self-cleavage rate was again too fast to be analyzed, the temperature was brought down to 25°C and the magnesium concentration to 0.5 mM (Figure 3C; Supplementary figure 2B), a value within the range estimated for the intracellular concentration of this cation in vivo (Eckstein, 1996), keeping the other conditions unchanged. Under these conditions, the self-cleavage rate constants could be determined (Table III). The trend displayed by the self-cleavage rate constants of the CChMVd (+) hammerheads was similar to that observed under low pH: A10→U > A10 > A10→Δ > A10→C > A10→G. Interestingly, the value of the wild-type CChMVd hammerhead (1.58 ± 0.16/min) was significantly higher than that obtained for its trans-acting derivative (0.034 ± 0.005 min) (Table III), indicating that this, and possibly other cis-acting natural hammerheads, may retain catalytic activity under the in vivo low magnesium concentrations.
Table III. Cis and trans cleavage rate constants under protein-free conditions and low magnesium concentration of the CChMVd (+) hammerhead ribozyme and its four mutants at position 10.
| Hammerhead | kcat (/min) (cis)a | kcat (/min) (trans)a |
|---|---|---|
| CChMVd-A10 | 1.58 ± 0.16 | 0.034 ± 0.005 |
| CChMVd-Δ10 | 0.76 ± 0.06 | ND |
| CChMVd-C10 | 0.17 ± 0.03 | ND |
| CChMVd-G10 | 0.051 ± 0.004 | ND |
| CChMVd-U10 | 3.86 ± 0.51 | ND |
aReactions were performed in 40 mM Tris–HCl, pH 7, 0.5 mM Mg2+, 25°C.
ND, not determined.
Modifications of loops 1 and 2 of the CChMVd (+) hammerhead have profound effects on the self-cleavage rate constant
Because transforming the wild-type cis-acting CChMVd (+) hammerhead and its mutants at position 10 into trans derivatives is accompanied by a significant decrease in their cleavage rate constants, we reasoned that this effect most likely resulted from opening the hammerhead loop 1. Nevertheless, it could not be discarded that a change in the size and sequence of this loop 1 may also affect the self-cleavage rate constant. To assess this possibility, loop 1 of the wild-type CChMVd (+) hammerhead (UGACGUC) was converted by site-directed mutagenesis into a tetraloop (GAAA) (Figure 2). This tetraloop belongs to the GNRA family (where N is any residue and R a purine), which is endowed with a high stability as a consequence of a complex array of non-canonical interactions (Woese et al., 1990; Heus and Pardi, 1991). The substitution of the native loop 1 by the GAAA tetraloop reinforced the stability of the hammerhead domain formed by this loop and its adjacent helix I, thus diminishing the chance of alternative conformations. Supporting this view, the computer-predicted most stable conformation of the hammerhead with the GAAA tetraloop [CChMVd (+) L1.1] was the typical hammerhead structure (Figure 2). However, the extent of self-cleavage of this mutant hammerhead during transcription (in the absence of the antisense deoxyoligonucleotide) was significantly reduced (Figure 4), and self-cleavage rate constant under protein-free conditions was very small (0.006 ± 0.001/min), a value ∼400-fold lower than that obtained for its wild-type counterpart (2.43 ± 0.52/min) under the same reaction conditions (50 mM PIPES-NaOH, pH 6.5, 10 mM Mg2+, 25°C). As expected, increasing the pH of the reaction buffer to 7.5 led to a higher self-cleavage rate constant (0.031 ± 0.007/min) for the hammerhead with the GAAA tetraloop.
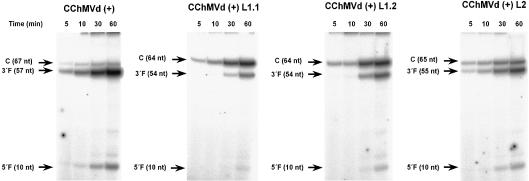
Fig. 4. Self-cleavage during transcription of the CChMVd (+) hammerhead and three derivatives thereof in which loop 1 was substituted by the tetraloops GAAA or UACG [CChMVd (+) L1.1 and CChMVd (+) L1.2, respectively], and loop 2 was substituted by the tetraloop GAAA [CChMVd (+) L2] (see Figure 2). Transcriptions (in 40 mM Tris–HCl pH 8, 6 mM MgCl2, 2 mM spermidine and 10 mM dithiothreitol) were made in the absence of the antisense deoxyoligonucleotide. Reaction aliquots were taken at four different times and analyzed by denaturing (15%) PAGE and autoradio graphy. Positions of the complete (C) primary transcript and of the resulting self-cleavage 3′ and 5′ fragments (3′F and 5′F, respectively), with their sizes in nucleotides are indicated in parentheses on the left.
Comparable data were obtained when loop 1 of the wild-type CChMVd (+) hammerhead was converted by site-directed mutagenesis into another tetraloop (UACG) of the UNCG family, which is also endowed with a high stability (Cheong et al., 1990; Woese et al., 1990). The computer-predicted most stable conformation of the hammerhead with the UACG tetraloop [CChMVd (+) L1.2] was the typical hammerhead structure (Figure 2). The extent of self-cleavage of this mutant hammerhead during transcription (in the absence of the antisense deoxyoligonucleotide) was significantly reduced (Figure 4); the corresponding self-cleavage rate constant under protein-free conditions was 0.0020 ± 0.0002/min at low pH (50 mM PIPES-NaOH, pH 6.5, 10 mM Mg2+, 25°C), and 0.0251 ± 0.002/min when the pH was increased to 7.5.
To explore whether modifications of loop 2 also influenced the catalytic properties, loop 2 of the wild-type CChMVd (+) hammerhead (AUGACA) was converted by site-directed mutagenesis into a tetraloop (GAAA). As with the two mutants affecting loop 1, the computer-predicted most stable conformation of the resulting hammerhead [CChMVd (+) L2] was the typical hammerhead structure (Figure 2). Moreover, the extent of self-cleavage of this mutant hammerhead during transcription (in the absence of the antisense deoxyoligonucleotide) was considerably reduced (Figure 4); the corresponding self-cleavage rate constant under protein-free conditions was 0.025 ± 0.004/min at low pH (50 mM PIPES-NaOH, pH 6.5, 10 mM Mg2+, 25°C), and 0.355 ± 0.035/min when the pH was increased to 7.5, indicating a 100-fold reduction with respect to the rate constant of the wild-type hammerhead under the same conditions. Overall, these results showed that similar cis-acting hammerheads, only differing in loop 1 or 2, exhibited a very distinct catalytic behavior, underlining the strong influence of regions peripheral to the central catalytic core.
Engineering the trans-acting artificial hammerhead HH8 into a cis derivative does not alter the cleavage rate constant
The artificial hammerhead HH8 (Figure 2), has been used previously in studies aimed at defining the catalytic properties of this ribozyme class (Dahm et al., 1993; Long and Uhlenbeck, 1994). The HH8 hammerhead structure has a I/III format and, consequently, lacks loops 1 and 3. In view of the results of the preceding section, we wondered whether engineering this trans-acting hammerhead into a cis derivative by closing helix I with a GAAA tetraloop (Figure 2) would affect its catalytic behavior. When the RNA of this modified HH8 ribozyme, obtained by in vitro transcription in the presence of an antisense oligonucleotide, was kinetically analyzed under protein-free conditions at two pH values (6.5 and 7.5), the cleavage rate constants remained essentially unchanged with respect to their trans counterparts (Table IV). These results showed that modifications that have a strong effect on the cleavage rate constant of a natural hammerhead are catalytically neutral in an artificial context.
Table IV. Cleavage rate constants under protein-free conditions of the trans-acting artificial HH8 hammerhead ribozyme and a cis-acting derivative thereof.
| Hammerhead | kcat (/min)a | kcat (/min)b | kcat (pH 7.5)/kcat (pH 6.5) |
|---|---|---|---|
| HH8 (trans) | 0.20 ± 0.01 | 1.48 ± 0.18 | 7.4 |
| HH8 (cis) | 0.27 ± 0.05 | 1.98 ± 0.20 | 7.3 |
aSelf-cleavage was performed in 50 mM PIPES-NaOH, pH 6.5, 10 mM Mg2+, 25°C.
bSelf-cleavage was performed in 50 mM Tris–HCl, pH 7.5, 10 mM Mg2+, 25°C.
Discussion
Despite the fact that natural hammerheads act in cis, little attention has been paid to the catalytic properties of these ribozymes in their physiological habitat. This can be explained by the technical difficulties that result from their high self-cleavage rates (Forster and Symons, 1987; Beaudry et al., 1995; Ambrós and Flores, 1998), by the interest in trans-acting hammerheads as ‘restriction RNases’ targeted against specific RNAs (Uhlenbeck, 1987; Haseloff and Gerlach, 1988; Rossi and Couture, 1999), and by results indicating that the cleavage rate constant during transcription of a cis-acting hammerhead was virtually the same as that of a closely trans-cleaving derivative of it under protein-free conditions (Long and Uhlenbeck, 1994). However, it should be noted that the cis-acting hammerhead used in this study (HH2) was artificial, and had a design (helices II and III closed by short loops 2 and 3) different from that of most natural hammerheads (in which helices I and II are closed by short loops 1 and 2). In contrast, in similar experiments, the self-cleavage rate constant of the natural PLMVd (+) hammerhead was higher (Ambrós and Flores, 1998). This unexpected behavior raised the question of whether it reflected the singularity of a particular natural hammerhead or had a broader significance, and prompted us to perform a kinetic analysis comparing several natural hammerheads in their cis context (during transcription and under protein-free conditions) with trans-cleaving derivatives thereof.
First of all, we confirmed that under protein-free conditions the PLMVd (+) hammerhead self-cleaves more efficiently than its trans-acting derivative. To obtain enough uncleaved primary transcript for this and for other similar studies, we had to incorporate into the reaction mixture a deoxyoligonucleotide complementary to part of the hammerhead. We then turned our attention to the cis cleavage kinetics of the CChMVd (+) hammerhead, another natural ribozyme for which there are data on the trans cleaving rate constants of derivatives of it and its four mutants at a position (A10) of the central core (De la Peña and Flores, 2001). For proper estimation of the corresponding cis cleaving rate constants during transcription, the pH of the buffer had to be lowered, clearly indicating that the intramolecular rate constants were significantly higher than their intermolecular counterparts. Therefore, the cis self-cleavage behavior during transcription of the CChMVd (+) hammerheads, more specifically of the wild-type (A10) and mutants A10→Δ and A10→U, was comparable to that of the PLMVd (+) hammerhead under the same conditions (Ambrós and Flores, 1998). Similar results for the cis cleaving rate constants of the CChMVd (+) hammerheads were obtained under protein-free conditions in independent experiments at low pH and at low magnesium concentration. These results showed that in these two hammerheads and probably in other natural hammerheads like the sLTSV (+), which also self-cleaves very efficiently (Forster and Symons, 1987), not only the central core but also peripheral regions have been optimized through evolution to increase the catalytic activity under physiological conditions. Further evidence supporting this view was obtained from experiments in which the substitution of the native heptaloop 1 and hexaloop 2 of the wild-type CChMVd (+) hammerhead by the stable GAAA or UACG tetraloops was accompanied by a large decrease of the self-cleavage rate constant. In contrast, transforming the trans-acting artificial hammerhead HH8 into a cis derivative did not alter essentially the cleavage rate constant. It should be pointed out that this hammerhead HH8 has helices I and II, of 5 and 4 bp, respectively, shorter than most natural hammerheads (see below). Altogether, these results demonstrate the strong effect of regions peripheral to the central conserved core on the kinetic properties of a cis-acting natural hammerhead.
There are previous reports on the influence of regions peripheral to the central conserved core on the catalytic behavior of two other natural cis-acting hammerheads, although both with an atypical morphology. Transcripts of the newt satellite 2 DNA, in spite of forming an unstable hammerhead due to a helix I of only 2 bp closed by a 2-nucleotide loop 1, undergo self-cleavage in vitro (Epstein and Gall, 1987). To explain this, a stable double hammerhead that results from the interaction between two separate domains of a newt-like dimeric transcript was proposed (Forster et al., 1988). However, further studies have demonstrated that self-cleavage of some satellite 2 DNA transcripts from the newt, and from two additional families of salamanders, occurs through a single-hammerhead structure that is dependent on sequences external to the conserved hammerhead core, more specifically on an internally looped extension of helix I that must be compatible with helix II (Epstein and Pabón-Peña, 1991; Pabón-Peña et al., 1991; Garrett et al., 1996). The second atypical hammerhead is that of the (+) sCYDV-RPV, which has an extended helix II. In an artificial context, sequences external to the conserved core have a detrimental effect on self-cleavage because of the formation of a pseudoknot between loop 1 and an internal loop of helix II that promotes a catalytically inactive conformation (Miller et al., 1991). In contrast, in a more natural multimeric context containing the full-length sCYDV-RPV RNA and two copies of the hammerhead, self-cleavage is more efficient and occurs via a double hammerhead (Song et al., 1999). Collectively, the results with these two natural cis-acting hammerheads, regardless of their uncommon morphology, underline the influence on the catalytic behavior of factors that operate only in their natural context. Other studies have shown that peripheral elements greatly enhance the catalytic activity of the hairpin and Varkud satellite ribozymes (Murchie et al., 1998; Jones et al., 2001), or that converting a cis-acting into a trans-acting derivative is detrimental for the activity or stability of the ribozymes of hepatitis delta virus RNA (Been and Wickham, 1997).
Considering crystallographic data on the three-dimensional structure of artificial hammerhead–substrate complexes (Pley et al., 1994; Scott et al., 1995, 1996), and previous biochemical and kinetic analyses (McKay, 1996; Stage-Zimmermann and Uhlenbeck, 1998), a plausible explanation consistent with our results is the existence of tertiary interactions between regions peripheral to the central conserved core that would favor a conformation with increased catalytic activity. We propose that in most natural hammerheads these interactions have been shaped by natural selection, are specific, and depend on the sequence and size of loops 1 and 2, and most likely of helices I and II. In line with this view, most natural hammerheads share some common structural features: helices I and II are of 5–7 and 4–5 bp, respectively, and loop 2 is usually a purine-rich tetraloop, with loop 1 having a variable size (3–7 nucleotides and occasionally larger) (Flores et al., 2001). Additional similarities previously noticed in some natural hammerheads, which include the hexanucleotides UGUG(U/C)(G/A) and C(A/G)AAAG forming part of helix I–loop 1 and helix II–loop 2 domains (Flores et al., 2001; Fadda et al., 2003), are also consistent with interactions between these domains. Therefore, the interactions discussed above in the two atypical natural cis-acting hammerheads would be particular cases of a more common situation. Selection pressures on the peripheral regions, like on the central hammerhead core, are expected in general to favor optimization of self-cleavage. However, due to the extreme compression of genetic information in viroid and viroid-like RNAs, residues of certain hammerheads appear to be also compromised in functional properties other than self-cleavage and, on this basis, less than optimal ribozymes have been finally selected (Song et al., 1999; De la Peña and Flores, 2001).
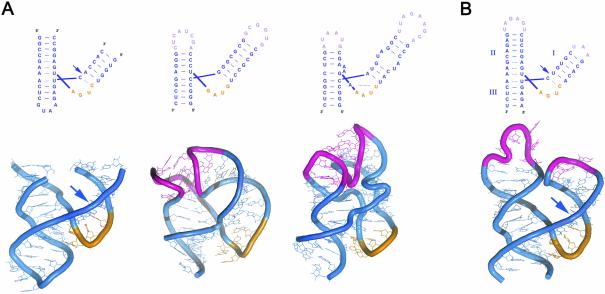
The global architecture that we offer for natural hammerheads resembles those obtained for a fragment of the 23S rRNA bound to ribosomal protein L11 (Wimberly et al., 1999), and for the 5′ domain of SRP RNA bound to the heterodimer of proteins SRP9 and SRP14 in the mammalian signal recognition particle (Weichenrieder et al., 2000). The three-dimensional structures of these three RNAs are similar (Figure 5), with two helical stacks connected by a U-turn (the so-called three helical τ-junction) (Weichenrieder et al., 2000). Interestingly, as we propose for natural hammerheads, a loop–loop interaction has been found in the latter two cases in which the crystallographic structures have been solved, pointing out the need of similar studies for natural hammerheads.
Fig. 5. Schematic representations of the three-helical τ-junctions of different RNAs. (A) Secondary structure (upper panel) and three-dimensional model (lower panel) proposed for the complex of a trans-acting hammerhead and its substrate (left) (Murray et al., 1998), the Alu 5′ domain of the SRP RNA (center) (Weichenrieder et al., 2000) and the L11-binding domain of 23S RNA (right) (Wimberly et al., 1999). The three RNAs show a similar global architecture with two stacked stems (in blue) connected by a U-turn motif (in orange). The peripheral loops of the Alu 5′ and L11- binding domains involved in tertiary interactions are shown in magenta. (B) Secondary structure (upper panel) and a hypothetical three-dimensional model (lower panel) proposed for the PLMVd (+) hammerhead. The three-dimensional model has been built up by superimposing the stems of three different RNA structures: a hexa- and a tri-loop hairpin structures [Protein Data Bank (PDB) accession codes 1JPO and 1I4B, respectively] over the hammerhead (PDB accession code 379D). Note some remarkably similar structural features between these RNAs, particularly the close proximity between phosphate backbones of helices I and II (suggesting the need of a re-arrangement of these regions), and between loops 1 and 2 (facilitating their interaction). Canonical and non-canonical pairs are indicated by continuous and broken lines. Arrows indicate the cleavage sites.
The question remains as to how such interactions influence catalysis. The dramatic increase of the ligation rate constant (without essential changes in the corresponding cleavage constant) reported recently for an artificial hammerhead, in which a covalent cross-link between helices I and II was introduced, has been interpreted as the cross-links reducing the thermal motion of helices of the cleaved rybozyme–product complex or constraining it into a structure that more closely resembles the transition state, thereby increasing the reverse ligation rate (Stage-Zimmermann and Uhlenbeck, 2001; Blount and Uhlenbeck, 2002). In any case, these data show that changes far from the active site of a hammerhead have large effects on its catalytic properties. In natural hammerheads, the inferred tertiary interactions between loops 1 and 2 might help the positioning and rigidity within the active site, thus optimizing the specific interactions of the transition state relative to the ground state and increasing catalysis in a way similar as in protein enzymes (Pauling, 1946; Narlikar and Herschlag, 1997), or in the hairpin and in larger ribozymes (Been and Wickham, 1997; Rupert et al., 2002). Under physiological conditions, these interactions could facilitate the catalytic activity at low magnesium concentration, particularly considering that protein–RNA interactions as those found for the 23S rRNA and the ribosomal protein L11 and for the SRP RNA and the SRP9/SRP14 proteins presumably exist in vivo. In line with this view, recent results have demonstrated an in vivo interaction between a hammerhead viroid and a chloroplastic host protein that facilitates in vitro, and presumably in vivo, the hammerhead-mediated self-cleavage of the viroid RNA (Daròs and Flores, 2002).
Materials and methods
Hammerhead constructs
The cDNAs of the cis-acting CChMVd (+) hammerhead and mutants thereof at position 10 were obtained by PCR amplification with Pfu DNA polymerase of the wild-type cDNA clone CM5 and its derivatives (De la Peña and Flores, 2001), using the sense primer RF-254 (5′-TAAT ACGACTCACTATAGGAAGAGGTCGGCACCTGACG-3′) containing the T7 RNA polymerase promoter (underlined) and the antisense primer RF-255 (5′-CGGTCTAGAAGAGGTTTCGATCCTGTC-3′) with a XbaI site (underlined). The PCR-amplified product was digested with XbaI and cloned in pUC18 opened with SmaI and XbaI. Site-directed mutagenesis in loops 1 and 2 of the wild-type CChMVd (+) hammerhead was performed by PCR, essentially as described previously (Byrappa et al., 1995), using the sense primers RF-437 (5′-GGCACCGAAAGG TGTCCTGATGA-3′) or RF-580 (5′-GGCACCTACGGGTGTCCTGA TGA-3′) and the antisense primer RF-438 (5′-GACCTCTTCCTATA GTGAGTCGTATTA-3′) for mutagenesis of loop 1, and the sense primer RF-542 (5′-GAAAGGATCGAAACCTCTTCTAG-3′) and the antisense primer RF-543 (5′-GGATCTTCATCAGGACACCG-3′) for mutagenesis of loop 2. Cloning and sequencing of the resulting recombinant plasmids confirmed that they contained solely the expected mutations.
The cDNA of the cis-acting PLMVd (+) hammerhead was prepared by PCR amplification of the gds23 cDNA clone (Ambrós and Flores, 1998), using the sense primer RF-89 (5′-TAATACGACTCACTATA GGAAAGAGTCTGTGCTAAGC-3′) and the antisense primer RF-492 (5′-GGGTCTAGAGTTTCGTCTCATTTCAGAGA-3′). The cDNA for the trans-acting PLMVd (+) hammerhead was obtained by PCR amplification after annealing the sense primer RF-490 (5′-GGTAATACGACTCACTATAGGCACACTGATGAGTCTCTGAAA TGA-3′) and the antisense primer RF-492. The substrate RNA (5′-AGAGUCUGUGC-3′) was obtained by chemical synthesis using 2′-orthoester protection (Dharmacon Research, Boulder, CO) and sequentially deprotected with 0.2 M acetic acid and Tris–HCl, pH 8.7. After purification by PAGE in 20% denaturing gels, the substrate RNA was eluted and 5′-labeled with [γ-32P]ATP (3000 Ci/mmol; Amersham) and T4 polynucleotide kinase (Sambrook et al., 1989).
The cDNA of the cis-acting HH8 hammerhead was prepared by PCR-mediated mutagenesis (Byrappa et al., 1995) of a previous clone containing a trans-acting version of this ribozyme (De la Peña and Flores, 2001), using the sense primer RF-435 (5′-TCGGAAACGACCCTGAT GAGGCC-3′) and the antisense primer RF-436 (5′-CCGACATTCC TATAGTGAGTCGTATTGGG-3′).
Secondary RNA structures were predicted with the Mfold program (Zuker, 2003).
In vitro transcriptions
Cis-acting hammerheads were synthesized by in vitro transcription of XbaI-linearized plasmids containing the corresponding cDNA inserts immediately preceded and followed by the promotor of T7 RNA polymerase and the XbaI site, respectively. Transcription reactions (50 µl) contained: 40 mM Tris–HCl, pH 8, 6 mM MgCl2, 2 mM spermidine, 0.5 mg/ml RNase-free bovine serum albumin, 0.1% Triton X-100, 10 mM dithiothreitol, 1 mM each of ATP, CTP and GTP, 0.1 mM UTP plus 0.5 µCi/µl [α-32P]UTP, 2 U/µl of human placental ribonuclease inhibitor, 20 ng/µl of plasmid DNA, 4 U/µl of T7 RNA polymerase and 0.1–1 mM of the blocking deoxyoligonucleotide when indicated. Blocking deoxyoligonucleotides were: RF-122 (5′-GACTCGTCAGTGTGCTTA GCACAGAC-3′), RF-142 (5′-CATGGATCVTCATCAGGACACCGA C-3′) (where V is A, G or C) and RF-309 (5′-CGGCCTCATCAGG GTCGCC-3′) for the PLMVd (+), CChMVd (+) and cis-HH8 hammerheads, respectively. After incubation at 37°C for 1 h, products were fractionated by PAGE in 15% gels with 8 M urea and the uncleaved primary transcripts were eluted by crushing the gel pieces and extracting them with phenol saturated with buffer (Tris–HCl 10 mM, pH 7.5, EDTA 1 mM, SDS 0.1%), recovered by ethanol precipitation and resuspended in deionized and sterile water. For the PLMVd (+) hammerhead, special conditions were used (see Results).
Self-cleavage kinetics during in vitro transcription
Self-cleavage rate constants during in vitro transcription were determined at pH 7 and 6 mM MgCl2, as reported previously (Ambrós and Flores, 1998). Aliquots were removed at appropriate time intervals and quenched with a 5-fold excess of stop solution (8 M urea, 50% formamide, 50 mM EDTA, 0.1% xylene cyanol and bromophenol blue) at 0°C. The uncleaved primary transcripts and their self-cleavage products were separated by PAGE in 15% denaturing gels that were quantitatively scanned with a bioimage analyzer (Fuji BAS1500). Data were fitted to the equation derived previously using a least-squares method (Long and Uhlenbeck, 1994).
Cis and trans cleavage kinetics under protein-free conditions
For determining the cis cleaving rate constants, the uncleaved primary transcripts (from 1 nM to 1 µM) of the PLMVd (+), CChMVd (+) (and its mutants, see Results) and the cis-HH8 hammerheads were incubated in 20 µl of 50 mM PIPES-NaOH, pH 6.5 or 50 mM Tris–HCl, pH 7.5 for 1 min at 95°C and slowly cooled to 25°C for 15 min. After taking a zero-time aliquot, self-cleavage reactions were triggered by adding MgCl2 to 10 mM unless otherwise indicated. The trans cleaving rate constants were determined under single-turnover conditions using an excess of ribozyme (100–1000 nM) and traces of 32P-labeled substrate (<1 nM). The ribozyme and substrate were first annealed by heating at 95°C for 1 min and slowly cooling to 25°C for 15 min in Tris–HCl 50 mM, pH 7.5 (unless otherwise indicated). After taking a zero-time aliquot, cleavage reactions were triggered by adding MgCl2 to 10 mM. In both cases (intra- and intermolecular cleavage reactions), aliquots were removed at appropriate time intervals and quenched with a 5-fold excess of stop solution at 0°C. Substrates and cleavage products were separated by PAGE in 15–20% denaturing gels. The product fraction at different times Ft was determined by quantitative scanning of the corresponding gel bands and fitted to the equation Ft = Fo + F∞ (1 – e–kt), where Fo and F∞ are the product fractions at zero time [which were near to zero except in the case of the cis-cleaving PLMVd (+) hammerhead] and at the reaction endpoint, respectively, and k is the first order rate constant of cleavage (kcat).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank A.Ahuir for excellent technical assistance and Dr C.Hernández for critical reading. This work was supported by grant BMC2002-03694 from the Ministerio de Ciencia y Tecnología (MCyT) (to R.F.). M.D.P. received a pre-doctoral fellowship from the MCyT and S.G. a postdoctoral aid from the Ministerio de Educación, Cultura y Deporte.
Note added in proof
During the review process of this paper, we were informed of an article by Khvorova et al. (Nat. Struct. Biol., advanced online publication July 27, 2003) reporting that natural hammerhead ribozymes require the presence of non-conserved sequence elements outside the conserved catalytic core to enable intracellular activity. These elements may stabilize the hammerhead ribozyme in a catalytically active conformation via tertiary interactions.
References
- Ambrós S. and Flores,R. (1998) In vitro and in vivo self-cleavage of a viroid RNA with a mutation in the hammerhead catalytic pocket. Nucleic Acids Res., 26, 1877–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudry D., Bussiere,F., Laureau,F., Lessard,C. and Perrault,J.P. (1995) The RNA of both polarities of the peach latent mosaic viroid self-cleaves in vitro solely by single hammerhead structures. Nucleic Acids Res., 23, 745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M.D. and Perrotta,A.T. (1995) Optimal self-cleavage activity of the hepatitis delta virus RNA is dependent on a homopurine base pair in the ribozyme core. RNA, 1, 1061–1070. [PMC free article] [PubMed] [Google Scholar]
- Been M.D. and Wickham,G.S. (1997) Self-cleaving ribozymes of hepatitis delta virus RNA. Eur. J. Biochem., 247, 741–753. [DOI] [PubMed] [Google Scholar]
- Blount K.F. and Uhlenbeck,O.C. (2002) Internal equilibrium of the hammerhead ribozyme is altered by the length of certain covalent cross-links. Biochemistry, 41, 6834–6841. [DOI] [PubMed] [Google Scholar]
- Byrappa S., Gavin,D.K. and Gupta,K.C. (1995) A highly efficient procedure for site-specific mutagenesis of full-length plasmids using Vent DNA polymerase. PCR Methods Appl., 5, 404–407. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani,G. and Tinoco,I.,Jr (1990) Solution structure of an unusually stable RNA hairpin, 5′GGAC(UUCG)GUCC3′. Nature, 346, 680–682. [DOI] [PubMed] [Google Scholar]
- Clouet-d’Orval B. and Uhlenbeck,O.C. (1996) Kinetic characterization of two I/II format hammerhead ribozymes. RNA, 2, 483–491. [PMC free article] [PubMed] [Google Scholar]
- Clouet-d’Orval B. and Uhlenbeck,O.C. (1997) Hammerhead ribozymes with a faster cleavage rate. Biochemistry, 36, 9087–9092. [DOI] [PubMed] [Google Scholar]
- Dahm S.C., Derrick,W.B. and Uhlenbeck,O.C. (1993) Evidence for the role of solvated metal hydroxide in the hammerhead cleavage mechanism. Biochemistry, 32, 13040–13045. [DOI] [PubMed] [Google Scholar]
- Daròs J.A. and Flores,R (2002) A chloroplast protein binds a viroid RNA in vivo and facilitates its hammerhead-mediated self-cleavage. EMBO J., 21, 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Peña M. and Flores,R (2001) An extra nucleotide in the consensus catalytic core of a viroid hammerhead ribozyme: implications for the design of more efficient ribozymes. J. Biol. Chem., 276, 34586–34593. [DOI] [PubMed] [Google Scholar]
- Diener T.O. (1989) Circular RNAs: relics of precellular evolution? Proc. Natl Acad. Sci. USA, 86, 9370–9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener T.O. (2001) The viroid: biological oddity or evolutionary fossil? Adv. Virus Res., 57, 137–184. [DOI] [PubMed] [Google Scholar]
- Eckstein F. (1996) The hammerhead ribozyme. Biochem. Soc. Trans, 24, 601–604. [DOI] [PubMed] [Google Scholar]
- Eckstein F., Kore,A.R. and Nakamaye,K.L. (2001) In vitro selection of hammerhead ribozyme sequence variants. Chembiochem, 2, 629–635. [DOI] [PubMed] [Google Scholar]
- Epstein L.M. and Gall,J.G. (1987) Self-cleaving transcripts of satellite DNA from the newt. Cell, 48, 535–543. [DOI] [PubMed] [Google Scholar]
- Epstein L.M. and Pabón-Peña,L.M. (1991) Alternative modes of self-cleavage by newt satellite 2 transcripts. Nucleic Acids Res., 19, 1699–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda Z., Daròs,J.A., Fagoaga,C., Flores,R. and Duran-Vila,N. (2003) Eggplant latent viroid (ELVd): candidate type species for a new genus within family Avsunviroidae (hammerhead viroids). J. Virol., 77, 6528–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R., Randles,J.W., Bar-Joseph,M. and Diener,T.O. (2000) Viroids. In van Regenmortel,M.H.V. et al. (eds), Virus Taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA, pp. 1009–1024. [Google Scholar]
- Flores R., Hernández,C., De la Peña,M., Vera,A. and Daròs,J.A. (2001) Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol., 341, 540–552. [DOI] [PubMed] [Google Scholar]
- Forster A.C. and Symons,R.H. (1987) Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell, 49, 211–220. [DOI] [PubMed] [Google Scholar]
- Forster A.C., Davies,C., Sheldon,C.C., Jeffries,A.C. and Symons,R.H. (1988) Self-cleaving viroid and newt RNAs may only be active as dimers. Nature, 334, 265–267. [DOI] [PubMed] [Google Scholar]
- Garrett T.A., Pabón-Peña,L.M., Gokaldas,N. and Epstein,L.M. (1996) Novel requirements in peripheral structures of the extended satellite 2 hammerhead. RNA, 2, 699–706. [PMC free article] [PubMed] [Google Scholar]
- Haseloff J. and Gerlach,W.L. (1988) Simple RNA enzymes with new and highly specific endoribonuclease activity. Nature, 334, 585–591. [DOI] [PubMed] [Google Scholar]
- Hernández C. and Flores,R. (1992) Plus and minus RNAs of peach latent mosaic viroid self cleave in vitro via hammerhead structures. Proc. Natl Acad. Sci. USA, 89, 3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel K.J. et al. (1992) Numbering system for the hammerhead. Nucleic Acids Res., 20, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus H.A. and Pardi,A. (1991) Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science, 253, 191–194. [DOI] [PubMed] [Google Scholar]
- Hutchins C.J., Rathjen,P.D., Forster,A.C. and Symons,R.H. (1986) Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res., 14, 3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaka M., Ohshima,Y. and Tani,T. (1995) Isolation of active ribozymes from an RNA pool of random sequences using an anchored substrate RNA. Biochem. Biophys. Res. Commun., 214, 403–409. [DOI] [PubMed] [Google Scholar]
- Jeffries A.J. and Symons,R.H. (1989) A catalytic 13-mer ribozyme. Nucleic Acids Res., 17, 1371–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones F.D., Ryder,S.P. and Strobel,S.A. (2001) An efficient ligation reaction promoted by a Varkud satellite ribozyme with extended 5′- and 3′-termini. Nucleic Acids Res., 29, 5115–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi M., Iwai,S. and Ohtsuka,E. (1988) Cleavage of specific sites of RNA by designed ribozymes. FEBS Lett., 239, 285–288. [DOI] [PubMed] [Google Scholar]
- Long D.M. and Uhlenbeck,O.C. (1994) Kinetic characterization of intramolecular and intermolecular hammerhead RNAs with stem II deletions. Proc. Natl Acad. Sci. USA, 91, 6977–6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D.B. (1996) Structure and function of the hammerhead ribozyme: an unfinished story. RNA, 2, 395–403. [PMC free article] [PubMed] [Google Scholar]
- Miller W.A. and Silver,S.L. (1991) Alternative tertiary structure attenuates self-cleavage of the ribozyme in the satellite RNA of barley yellow dwarf virus. Nucleic Acids Res., 19, 5313–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W.A., Hercus,T., Waterhouse,P.M. and Gerlach,W.L. (1991) A satellite RNA of barley yellow dwarf virus contains a novel hammerhead structure in the self-cleavage domain. Virology, 183, 711–720. [DOI] [PubMed] [Google Scholar]
- Murchie A.I., Thomson,J.B., Walter,F. and Lilley,D.M. (1998) Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol. Cell, 1, 873–878. [DOI] [PubMed] [Google Scholar]
- Murray J.B., Terwey,D.P., Maloney,L., Karpeisky,A., Usman,N., Beigelman,L. and Scott,W.G. (1998) The structural basis of hammerhead ribozyme self-cleavage. Cell, 92, 665–673. [DOI] [PubMed] [Google Scholar]
- Narlikar G.J. and Herschlag,D. (1997) Mechanistic aspects of enzymatic catalysis: lessons from comparison of RNA and protein enzymes. Annu. Rev. Biochem., 66, 19–59. [DOI] [PubMed] [Google Scholar]
- Navarro B. and Flores,R. (1997) Chrysanthemum chlorotic mottle viroid: unusual structural properties of a subgroup of viroids with hammerhead ribozymes. Proc. Natl Acad. Sci. USA, 94, 11262–11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabón-Peña L.M., Zhang,Y. and Epstein,L.M. (1991) Newt satellite 2 transcripts self-cleave by using an extended hammerhead structure. Mol. Cell. Biol., 11, 6109–6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling L. (1946) Molecular architecture and biological reactions. Chem. Eng. News, 24, 1375–1377. [Google Scholar]
- Pley H.W., Flaherty,K.M. and McKay,D.B. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature, 372, 68–74. [DOI] [PubMed] [Google Scholar]
- Prody G.A., Bakos,J.T., Buzayan,J.M., Schneider,I.R. and Bruening,G. (1986) Autolytic processing of dimeric plant virus satellite RNA. Science, 231, 1577–1580. [DOI] [PubMed] [Google Scholar]
- Rossi J.J. and Couture,L.A. (1999) Intracellular Ribozyme Applications: Principles and Protocols. Horizon Scientific Press, Wymondham, UK. [Google Scholar]
- Ruffner D.E., Stormo,G.D. and Uhlenbeck,O.C. (1990) Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry, 29, 10695–10702. [DOI] [PubMed] [Google Scholar]
- Rupert P.B., Massey,A.P., Sigurdsson,S.T. and Ferre-D’Amare,A.R. (2002) Transition state stabilization by a catalytic RNA. Science, 298, 1421–1424. [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani K. and Szostak,JW. (2001) In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature, 414, 82–84. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Scott W.G., Finch,J.T. and Klug,A. (1995) The crystal structure of an all-RNA hammerhead ribozyme: a proposed mechanism for RNA catalytic cleavage. Cell, 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- Scott W.G., Murray,J.B., Arnold,J.R.P., Stoddard,B.L. and Klug,A. (1996) Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science, 274, 2065–2069. [DOI] [PubMed] [Google Scholar]
- Sheldon C.C. and Symons,R.H. (1989) Mutagenesis analysis of a self-cleaving RNA. Nucleic Acids Res., 17, 5679–5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S.I., Silver,S.L., Aulik,M.A., Rasochova,L., Mohan,B.R. and Miller,W.A. (1999) Satellite cereal yellow dwarf virus-RPV (satRPV) RNA requires a double hammerhead for self-cleavage and an alternative secondary structure. J. Mol. Biol., 293, 781–793. [DOI] [PubMed] [Google Scholar]
- Stage-Zimmermann T.K. and Uhlenbeck,O.C. (1998) Hammerhead ribozyme kinetics. RNA, 4, 875–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stage-Zimmermann T.K. and Uhlenbeck,O.C. (2001) A covalent crosslink converts the hammerhead ribozyme from a ribonuclease to an RNA ligase. Nat. Struct. Biol., 8, 863–867. [DOI] [PubMed] [Google Scholar]
- Tang J. and Breaker,R.R. (1997) Examination of the catalytic fitness of the hammerhead ribozyme by in vitro selection. RNA, 3, 914–925. [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O.C. (1987) A small catalytic oligonucleotide. Nature, 328, 596–600. [DOI] [PubMed] [Google Scholar]
- Vaish N.K., Heaton,P.A. and Eckstein,F. (1997) Isolation of hammerhead ribozymes with altered core sequences by in vitro selection. Biochemistry, 36, 6495–6501. [DOI] [PubMed] [Google Scholar]
- Weichenrieder O., Wild,K., Strub,K. and Cusack,S. (2000) Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature, 408, 167–173. [DOI] [PubMed] [Google Scholar]
- Wimberly B.T., Guymon,R., McCutcheon,J.P., White,S.W. and Ramakrishnan,V. (1999) A detailed view of a ribosomal active site: the structure of the L11–RNA complex. Cell, 97, 491–502. [DOI] [PubMed] [Google Scholar]
- Woese C.R., Winker,S. and Gutell,R.R. (1990) Architecture of ribosomal RNA: constraints on the sequence of “tetra-loops”. Proc. Natl Acad. Sci. USA, 87, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res., 31, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]