Abstract
Molecular motors undergo conformational changes to produce force and move along cytoskeletal filaments. Structural changes have been detected in kinesin motors; however, further changes are expected because previous crystal structures are in the same or closely related conformations. We report here a 2.5 Å crystal structure of the minus-end kinesin, Ncd, with the coiled-coil stalk/neck and one head rotated by ∼75° relative to the other head. The two heads are asymmetrically positioned with respect to the stalk and show asymmetry of nucleotide state: one head is fully occupied, but the other is unstably bound to ADP. Unlike previous structures, our new atomic model can be fit into cryoelectron microscopy density maps of the motor attached to microtubules, where it appears to resemble a one-head-bound motor with the stalk rotated towards the minus end. Interactions between neck and motor core residues, observed in the head that moves with the stalk, are disrupted in the other head, permitting rotation of the stalk/neck. The rotation could represent a force-producing stroke that directs the motor to the minus end.
Keywords: force production/kinesin microtubule motors/molecular motors/Ncd/stalk rotation
Introduction
Molecular motors bind to and hydrolyze ATP, and use the energy from nucleotide hydrolysis to produce force and move along cytoskeletal filaments. The mechanism by which motors capture the energy from ATP hydrolysis and convert it into work is as yet unknown. Motors are thought to couple nucleotide binding, hydrolysis or release of hydrolysis products to force-producing conformational changes, undergoing changes in binding affinity for their filament as they move along the filament. Identifying the structural elements that move and the force-producing conformational changes of the motor will be essential to understand how these motors work.
The kinesin microtubule motors consist of a motor domain with highly conserved nucleotide- and microtubule-binding motifs, usually joined to an α-helical coiled-coil stalk and a tail. The tail is thought to bind to cellular cargo, e.g. vesicles or organelles, for transport along microtubules. The first discovered or conventional kinesin is a plus-end motor that moves towards the fast polymerizing/depolymerizing ends of microtubules, but Ncd and a group of related kinesin motors move in the opposite direction, towards the more stable minus ends. Analysis of chimeric and mutant motors has localized the region of the motor that determines directionality to the neck, just adjacent to the conserved motor domain (Case et al., 1997; Henningsen and Schliwa, 1997; Endow and Waligora, 1998; Endow and Higuchi, 2000). The neck of conventional kinesin consists of the end of the coiled-coil stalk and two neck linkers, which join the stalk to the motor domains, while the neck of Ncd consists only of the end of the coiled-coil stalk.
Over the past several years much has been learned about how kinesin works from studies using X-ray crystallography and cryoelectron microscopy (cryoEM). For example, the first crystal structures of the motor domains of conventional kinesin and Ncd showed a remarkable similarity, providing the first evidence that the opposite directionality of movement along microtubules of the two motors resides in features outside the conserved motor domain (Kull et al., 1996; Sablin et al., 1996). The neck linkers of conventional kinesin were identified by X-ray crystallography (Kozielski et al., 1997) and have been implicated in kinesin motility based on cryoEM, together with biochemical and biophysical evidence (Rice et al., 1999). A signaling pathway linking the microtubule-binding site to the active site of the kinesin motors was discovered from crystal structures (Sablin et al., 1998; Yun et al., 2001) and mutant analysis (Song and Endow, 1998; Yun et al., 2001), and helix α4 at the microtubule-binding site of the motor has been identified as a key element in this pathway by analysis of crystal structures (Kikkawa et al., 2001).
Unfortunately, the atomic structures of conventional kinesin and other plus-end kinesin motors available to date include only ADP structures (Sack et al., 1999; Kull and Endow, 2002) and one ATP-like structure (Kikkawa et al., 2001). The ATP-like structure shows only small changes compared with the ADP structures, and probably represents a collision ATP complex—a first step in ATP binding that does not produce major conformational changes (Cooke, 1986). Only ADP structures are available for the minus-end kinesin motor, Ncd, and nucleotide-free structures of the kinesin motors have not yet been reported.
The absence of crystal structures showing new conformations of the kinesin motors represents an obstacle to understanding the motor mechanism of function. One approach to obtaining new crystal forms is to weaken the ADP state by mutating residues required for its stabilization. Here, we use a mutant of a kinesin motor that decouples microtubule binding from nucleotide hydrolysis (Song and Endow, 1998) and may destabilize the ADP form of the minus-end kinesin motor, Ncd. The crystal structure of the mutant motor shows a striking new conformation that can also be observed, although at lower resolution, for the wild-type motor. The new conformation may explain how the Ncd motor generates force and why the motor moves to the microtubule minus end. Unlike previous crystal structures, the new atomic structure of Ncd can be fit into the electron density corresponding to the motor in complexes with microtubules observed by cryoEM, where it appears to resemble a motor bound by one head to a microtubule.
Results
Decoupling mutant
Residue N600 of Ncd is present in the microtubule-binding region of the motor domain and is highly conserved in the kinesin proteins (Hoyt et al., 1993). Mutation of N600 to K causes the motor to bind tightly to microtubules (Song and Endow, 1998). The mutant has basal ATPase activity, but is completely blocked in ATPase activation by microtubules, which is essential for motor movement along microtubules. The tight binding to microtubules, but absence of ATPase activation by microtubules, indicates that signaling between the microtubule- and nucleotide-binding sites of the motor is disrupted, decoupling microtubule and nucleotide binding by the motor.
We tested dimeric NcdN600K protein in single turnover assays without microtubules and found a small but significant increase in the dissociation rate of mant·ADP, a fluorescent nucleotide analog, compared with the corresponding wild-type motor. The dissociation rate constants for the mutant and wild type were 0.00245 ± 0.00026/s (n = 11) and 0.00155 ± 0.00021/s (n = 11), respectively, when 500 µM ATP was added to 0.5 or 1 µM motor-mant·ADP. The ∼1.6-fold increase in ADP dissociation rate from the mutant compared with wild type indicates that the N600K mutant may bind ADP less stably than wild type. This differs from the results we reported previously for monomeric NcdN600K, which showed no significant difference in the dissociation rate of mant·ADP compared with the monomeric wild-type motor (Song and Endow, 1998).
A new conformation of the Ncd motor
The X-ray crystal structure of dimeric NcdN600K was determined to 2.5 Å resolution (Table I). Under the same crystallization conditions, the corresponding wild-type Ncd protein crystallized in the same space group as the mutant, although the crystals did not diffract to as high resolution, indicating that the mutation may stabilize the new structure. A refined model of the wild-type Ncd protein, determined to ∼3.5 Å resolution, matched the mutant structure [root mean square deviation (r.m.s.d.) = 0.9 Å, including both heads and the stalk/neck], demonstrating that the wild-type Ncd structure was the same conformation as the mutant (see Supplementary data, available at The EMBO Journal Online). The higher resolution NcdN600K mutant structure was used for structural interpretation.
Table I. X-ray diffraction data collection and refinement statistics.
| Data collection |
|
| Resolution (Å) | 20.0–2.5 |
| Measured reflections | 570 918 |
| Unique reflections | 33 852 |
| Completeness (%, > –1σ)a | 97.7 (96.0) |
| Rsym (%)a,b | 5.3 (46.4) |
| Average I/σ(I)a |
19.47 (1.9) |
| Refinement |
|
| Rworkc | 0.260 |
| Rfreec | 0.302 |
| Number of reflections [F > 2σ (F)] | 28 067 |
| Number of protein atoms | 5549 |
| Number of non-protein atoms | 337 |
| R.m.s.d.d bond length (Å) | 0.009 |
| R.m.s.d.d bond angles (°) | 1.47 |
| Average B-factors (Å2, main chain/side chain) | 61.5/62.3 |
| Ramachandran analysis most favored/allowed | 82.8/17.2 |
aLast shell (2.59–2.5 Å resolution) value is in parentheses.
bRsym = Σhkl[Σi|Ihkl,i – <Ihkl>|]/Σhkl,i<Ihkl>, where Ihkl,i is the intensity of an individual measurement of the reflection with Miller indices h, k and l, and <Ihkl> is the mean intensity of the reflection.
cRwork = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factor amplitudes. Rfree is equivalent to Rwork, except 5% of total reflections were set aside to test the progress of refinement.
dR.m.s.d., root mean square deviation from ideal geometry.
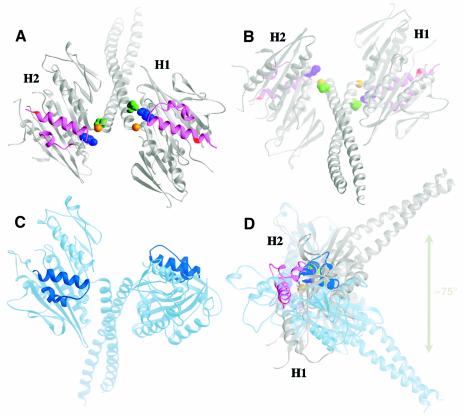
Remarkably, the new crystal structure shows that the coiled-coil stalk/neck (residues H293–R346) and one head, H1, are rotated by ∼75° when the other head, H2, is held in the same position as one of the two heads of a previous atomic model (Figure 1A, C and D). The pivot point of stalk/neck rotation is G347 at the junction between the coiled coil and conserved motor domain. The mutated residue N600K of head H2 is at the N-terminal end of helix α4, but lies internal to helix α4 of head H1, which is longer by one turn than in head H2. A network of interactions between the coiled coil and the two heads, observed in previous Ncd dimer structures (Sablin et al., 1998; Kozielski et al., 1999), are present in head H1, which has rotated with the stalk, but are disrupted in head H2, permitting the stalk/neck and interacting head H1 to move relative to head H2. These include interaction of N340 with K640 (Figure 1A and B), which may be required to stabilize the head against the stalk. The rotation of the coiled coil and head H1 alters the symmetry of the heads relative to the coiled coil—the two heads of the motor in the new structure show twofold rotational symmetry, but are asymmetrically positioned with respect to the coiled coil, differing from previous Ncd dimer crystal structures, which show actual or approximate twofold rotational symmetry of the heads around the axis of the coiled coil (Sablin et al., 1998; Kozielski et al., 1999) (Figure 1C). The rotation of head H1with the stalk/neck causes the tubulin-binding elements of the two heads, helices α4 and α5 and loops L11 and L12, to be present on the same face of the motor (Figure 1A), rather than on opposite faces, as in previous Ncd dimer crystal structures (Figure 1C), and changes the orientation of head H2 relative to the stalk and head H1, which may be important for functional interactions of the motor with microtubules.
Fig. 1. New Ncd crystal structure. The new structure shows a large rotation of the coiled-coil stalk/neck and asymmetry of the two heads relative to the coiled coil. (A) Microtubule-binding face with the coiled coil tilted back (tubulin-interacting elements L11–α4–L12–α5, magenta; N600K, red). Neck residue N340 (green) touches motor core residue K640 (purple) in ADP-bound head H1, but the residues are separated in unstably occupied head H2. Stalk rotation occurs at G347 (gold) of H2. (B) View 180° from (A) with the coiled coil tilted forward. (C) A previous Ncd structure (PDB accession code 1CZ7) (Kozielski et al., 1999) (L11-α4-L12-α5, dark blue) with left head in same orientation as H2 of (A). (D) Side view of (A) and (C) superposed structures rotated 90° vertically, showing the ∼75° rotation of the NcdN600K coiled coil relative to the previous structure. Figure prepared using RIBBONS (Carson, 1997).
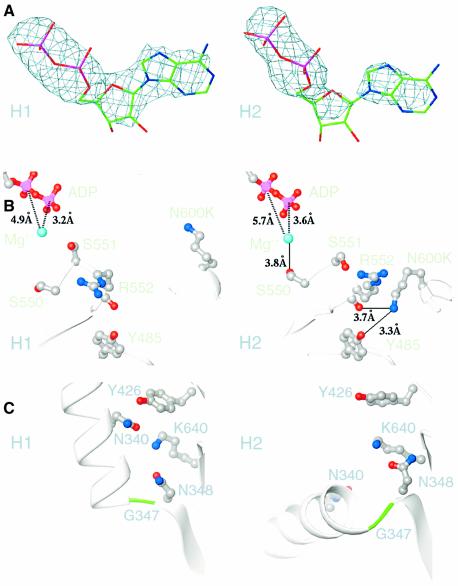
The two heads of the motor in the new structure are both complexed with Mg·ADP, but the ADP bound to head H2 is reduced in density compared with the ADP of head H1. Fo – Fc difference maps show clear density for the ADP bound to the active site of head H1, whereas the pyrophosphate density is clear in head H2, but the adenosine density is reduced (Figure 2A). This indicates that the nucleotide bound to head H2 is mobile or unstable, consistent with the small but reproducible increase in ADP dissociation rate that we observe for the mutant compared with the wild type in biochemical assays.
Fig. 2. Structural features of the two heads in the new structure. (A) Weighted Fo – Fc difference maps of the ADP bound to heads H1 and H2. The ADP bound to head H2 shows reduced density of the adenine base (right) and pentose ring (center) compared with the ADP of head H1, indicating that the adenosine is mobile or unstably bound. Map contoured at 3.5σ. (B) The mutated residue, N600K, of head H2 has moved toward R552 and forms a hydrogen bond (solid line) to R552 and Y485, causing S550 to hydrogen bond to the Mg2+ (cyan), destabilizing the bound ADP (ADP pyrophosphate, pink/red). This interaction is not observed in head H1. Distances, broken lines. (C) Movement of N600K towards R552 in head H2 weakens interactions between neck residue N340 and head H2 residues K640, N348 and Y426, permitting the coiled coil to rotate at G347 of head H2, while interactions of N340 with these residues in head H1 cause head H1 to rotate with the coiled coil.
Destabilization of the nucleotide bound to head H2 can be explained by changes in the switch I and II regions, which are structurally homologous to G protein regions that move or change in conformation upon nucleotide binding or release (Sablin et al., 1996). Switch I (SSRSH, residues 550–554) and switch II (LAGSE, residues 581–585) are visible in both heads, but N600K in head H2 is hydrogen bonded to R552 of switch I and the SSR has moved by an average of 1.5 Å around the axis of strand β6, allowing S550 to hydrogen bond to the magnesium ion, which coordinates the bound ADP (Figure 2B). This interaction is not observed in head H1 or previous Ncd dimer structures and probably weakens nucleotide coordination by the magnesium ion, destabilizing the bound ADP and explaining the instability of the nucleotide bound to head H2. In the wild-type structure, the conformation of N600 in head H1 resembles that of N600K in head H1 of the NcdN600K mutant, while the side chain of N600 in head H2 points toward R552, but does not form a hydrogen bond with the residue.
To interact with R552, N600K moves toward switch I by 2.8 Å, causing the switch II helix, α4, together with helices α5 and α6, to move towards the nucleotide-binding cleft by an average of 1 Å. This movement weakens the network of interactions between neck and motor core residues that include N340, K640, N348 and Y426, which are observed in head H1, but are disrupted in head H2 (Figure 2C). These interactions stabilize head H1 interactions with the stalk/neck, causing head H1 to rotate with the coiled coil, whereas disrupting these interactions in head H2 allows the coiled coil to move. The rotation of the stalk/neck is thus caused by the same interaction of N600K with R552 in head H2 that results in instability of the ADP bound to head H2.
The instability of the nucleotide bound to head H2, coupled with changes in nucleotide coordination at the active site, indicates that the new structure may represent a transition away from the ADP state towards a no-nucleotide state. Other differences between the two heads include disordered loops L8, L10 and L12 in head H2, which are visible in head H1, and visible C-termini of loops L2 and L9 in head H2, which are disordered in head H1. The salt bridge between switch I and II (Kull and Endow, 2002) is not present in either head and the C-terminus of the switch II loop, L11, is disordered in both heads.
Functional tests
We performed several functional tests of the new model, first, by mutating the residue at the pivot point of stalk/neck rotation, G347, to determine the effects on motility. Gly is a residue that can adopt many different conformations and is therefore likely to be important for the coiled-coil rotation. Missense mutants with changes of G347 to A or T were constructed, expressed in bacteria with glutathione S-transferase (GST) fused to 138 residues of the coiled coil, followed by the motor domain (Chandra et al., 1993), and assayed in coverslip gliding assays. The G347A and G347T point mutations caused the motor to bind poorly to microtubules in gliding assays and reduced microtubule gliding velocity to 8.7 ± 0.3 µm/min (n = 15) and 7.4 ± 0.2 µm/min (n = 11), respectively, compared with 12.3 ± 0.3 µm/min (n = 32) for the corresponding wild-type GST/Ncd motor. The change of G347T was more severe than G347A with respect to both microtubule binding and gliding velocity, consistent with the greater constraint on the conformations Thr can adopt compared with Ala or Gly. The sensitivity of microtubule binding and motor velocity to changes in G347 supports the idea that the residue is important and may serve as the pivot point for movements of the stalk/neck that are required for motor binding to microtubules and movement along the microtubule.
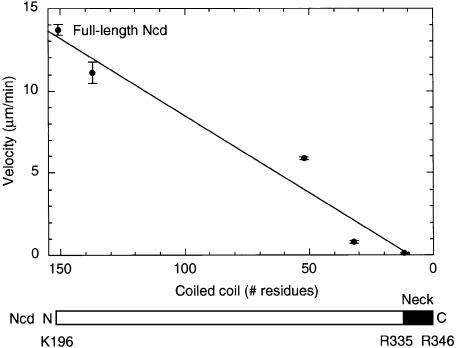
We then asked whether truncation of the coiled coil affects motor velocity, as predicted if the coiled-coil rotation constitutes a force-producing stroke of the motor. A series of GST/Ncd motors with truncations of the coiled coil has been reported previously by ourselves and others (Chandra et al., 1993; Stewart et al., 1993). We analyzed the dependence of motor velocity in gliding assays on the length of the coiled coil and found a linear relationship with slower velocities for motors containing shorter coiled coils (Figure 3). The dependence of motor velocity on the length of the coiled coil is consistent with a swinging lever arm model in which the size of the displacement produced at each stroke, and consequently the gliding velocity, is proportional to the length of the lever arm (Uyeda et al., 1996). The relatively small number of truncated motors analyzed here, however, leaves room for other interpretations of the effects of the truncations on motor velocity. This means that further experiments will be required to demonstrate the possible role of the Ncd stalk in force generation by the motor.
Fig. 3. Dependence of gliding velocity on coiled-coil length. The gliding velocity of stalk-truncated GST/Ncd motors decreases linearly with the length of the coiled coil. The velocity of full-length Ncd is shown for comparison. The coiled coil of full-length Ncd is predicted to begin at K196 and extends to R346 in the new crystal structure. The Ncd neck consists of residues R335–R346 (Sablin et al., 1998), with G347 serving as the pivot point for stalk/neck rotation (this study). Data for gliding velocities were from Stewart et al. (1993).
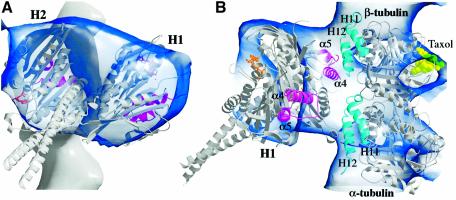
We next asked whether our new atomic structure could be docked into three-dimensional reconstructions made from cryoEM density maps of dimeric Ncd bound to microtubules, and thus represents a functional motor. Previous dimer crystal structures of kinesin motors (Kozielski et al., 1997; Sablin et al., 1998; Kozielski et al., 1999) fit poorly into cryoEM density maps of motor–microtubule complexes, largely because the rotational symmetry of the heads relative to the coiled coil causes the heads to be positioned relative to one another and to the coiled coil so that they do not fit well into the electron density (see Supplementary data). Because the two heads of the motor in our new structure are oriented with one of the heads (H2) changed in position relative to the coiled coil and the other head (H1), we were able to dock our new atomic structure into cryoEM density maps of dimeric Ncd bound to microtubules (Hirose et al., 1998, 1999), where it accounted for almost all of the density corresponding to the motor (Figure 4).
Fig. 4. Docking into a motor–microtubule cryoEM map. The new atomic structure of Ncd is shown docked into a cryoEM density map of nucleotide-free Ncd bound to microtubules (Hirose et al., 1998, 1999). (A) View perpendicular to the microtubule axis (similar view of motor as Figure 1B but rotated to the left). A microtubule protofilament is shown at the back with the plus end at the top (motor elements L11–α4–L12–α5, magenta; ADP, gold). (B) View from the side of the unattached head H1. An α/β tubulin atomic model (PDB accession code 1TUB) (Nogales et al., 1998) is also shown docked into the cryoEM map (helices H11 and H12, cyan; taxol, light green). Figure prepared with BOBSCRIPT (Esnouf, 1997).
The new crystal structure fit into the nucleotide-free cryoEM density map with head H2, to which nucleotide is less stably bound, interacting with the microtubule, and head H1 unattached (Figure 4). The last visible residues at the C-terminus of each motor domain in the crystal structure lie between the two heads, thus the empty density between the two heads in the cryoEM density map would be occupied by the 30 (H1) or 33 (H2) C-terminal residues, which are not resolved in the crystal structure. The coiled-coil stalk of the atomic structure is not visible in the cryoEM density map because of its low density and possibly its mobility; consequently, the coiled coil of the atomic model protrudes from the surface contour map. The docking is consistent with 3D models that show the Ncd motor bound by one head to the microtubule and the other head unattached (Hirose et al., 1996, 1998). It shows the coiled-coil stalk/neck rotated towards the microtubule minus end. The structural elements of head H2 that touch tubulin in the docking, helices α4 and α5 and loops L11 and L12, are thought from previous work to be at the motor–microtubule interface (Woehlke et al., 1997; Alonso et al., 1998) (Figure 4B). Head H2 elements α4–L12–α5 interact with β-tubulin helices 11 and 12. The disordered loop L8 of head H2, when stabilized by tubulin, may also interact with β-tubulin helices 11 and 12, and the visible portion of loop L11 in head H2 is located near the loop between helices 11 and 12 in the adjacent α-tubulin subunit towards the minus end, while loop L2 interacts with α-tubulin helices 11 and 12.
The crystal structure fit best into the nucleotide-free map, prepared in the presence of apyrase, compared with the motor-AMP·PNP and motor-ADP maps (see Supplementary data), consistent with the instability of the nucleotide bound to head H2, which interacts with the microtubule in the docking. The favorable fit suggests that the new crystal structure may resemble the motor as it binds to a microtubule by one head and releases ADP from the bound head.
Discussion
Features of the new Ncd crystal structure
We report here a new 2.5 Å crystal structure of a minus-end kinesin motor, Ncd, that shows a strikingly different conformation compared with previous crystal structures. The new structure is of a mutant, NcdN600K, but the same conformation was observed for the wild-type motor (see Supplementary data); however, the crystals were less well ordered and diffracted only to ∼3.5 Å. This suggests that the N600K mutation, which alters a residue in the microtubule-binding region of the motor, may stabilize the new conformation. The new crystal structure shows the coiled-coil stalk/neck and one head rotated by ∼75° relative to the other head, a conformation that has not been observed previously in crystal structures of the kinesin motors. Rotation of the coiled-coil stalk/neck and one head (H1) is caused by movement of the mutated residue N600K towards R552 of switch I in the head that does not move with the stalk/neck (H2), resulting in subsequent movements that disrupt interactions between neck and motor core residues, including interaction of N340 with K640, permitting the stalk/neck to rotate.
This same movement of N600K towards R552 of switch I results in a movement of switch I that disrupts coordination of the bound ADP by the magnesium ion, destabilizing the ADP and causing the two heads of the motor in the new structure to differ in nucleotide stability: the head that rotates with the stalk/neck, head H1, shows stably bound ADP, but the ADP bound to the other head, H2, shows reduced density, indicating that it is mobile or unstably bound. The instability of the nucleotide bound to head H2, coupled to changes in nucleotide coordination at the active site, suggests that the motor may represent a transition away from the ADP state towards a no-nucleotide state. This is consistent with the small, but significant increase in ADP dissociation from the mutant motor compared with wild type and better fit of the atomic structure into the nucleotide-free cryoEM density map than the AMP·PNP or ADP maps.
Rotation of the stalk/neck and motor directionality
Disruption of neck–motor core interactions in the head that does not move with the stalk/neck, including neck residue N340 with motor core residue K640, permits the coiled coil to rotate. N340–K640 may be a key interaction in positioning the head against the stalk/neck in the ADP conformation. Weakening the interactions between the stalk/neck and motor core by mutating N340 or K640 might allow the stalk/neck movement to occur in either direction. This would account for gliding of the mutants in either direction in coverslip assays and movement in either direction of a conformational or angle change detected in laser-trap assays that is biased towards the minus end in wild-type Ncd (Endow and Higuchi, 2000). The conformational or angle change detected in laser-trap assays depends on interaction of neck residue N340 with motor core residue K640 and directs the motor towards the minus end. The stalk/neck rotation in the new crystal structure could correspond to the conformational or angle change observed in laser-trap assays, representing a stroke of the motor that directs the motor towards the microtubule minus end.
Ncd function and stalk/neck rotation
Mutants that affect the pivot point of coiled-coil rotation or its length reduce motor velocity of movement on microtubules, consistent with the possibility that the coiled coil functions like a lever arm in force production by the motor. Mutation of the residue at the pivot point of stalk/neck rotation, G347, reduced both the binding of the motor to microtubules and its gliding velocity in coverslip assays; thus, rotation of the stalk/neck may occur when the motor binds to microtubules and may be needed for the motor to translocate microtubules in gliding assays. Ncd motors with truncations of the stalk show decreased motor velocity with decreasing length of the coiled coil, providing tentative evidence that the coiled coil may act like a lever to amplify force produced by the motor, resulting in a working stroke of the motor.
Rotation of the coiled-coil stalk/neck and one head in the new structure causes the tubulin-binding elements of the two heads to be on the same face of the motor, and one of the two heads to be in a different orientation relative to the coiled coil than in previous kinesin and Ncd dimer crystal structures. This allowed us to fit our new atomic structure into cryoEM density maps of Ncd motor–microtubule complexes, where it appears to resemble a motor with one head bound to the microtubule and the other head unattached, and the stalk/neck rotated towards the minus end. The head that touches the microtubule in the docking is head H2, to which ADP is less stably bound, thus the crystal structure may resemble a motor bound by one head to a microtubule, releasing ADP from the bound head, with the coiled coil tilted towards the microtubule minus end.
This interpretation differs from the proposal by others that the coiled-coil stalk/neck rotates when Ncd binds ATP, inferred from lower resolution cryoEM data using a density marker, SH3, at the end of a truncated coiled-coil stalk/neck to visualize the position of the coiled coil (Wendt et al., 2002). The density corresponding to the SH3 marker was not connected to the motor density in the nucleotide-free maps, however, and was not visible in maps of the motor bound to microtubules in the presence of the ATP analog, AMP·PNP, presumably because of its mobility. Thus, the angle change of the stalk/neck was not directly visualized in these studies and the nucleotide state of the motor at the time it was inferred to occur may have been incorrectly identified. Docking our new Ncd atomic structure into the coordinates of SH3-Ncd bound to microtubules without nucleotide (Wendt et al., 2002) shows the coiled coil touching a region of SH3 density positioned towards the microtubule minus end, rather than one towards the plus end, as observed for a previous Ncd dimer crystal structure (Wendt et al., 2002) (see Supplementary data). However, the density corresponding to the unattached head of the motor in the SH3 map is much smaller compared with previous nucleotide-free Ncd cryoEM maps (Hirose et al., 1998, 1999) (Figure 4); thus, both the previous structure and our new structure show a poor fit of the unbound head into the electron density. Further studies will be required to distinguish between the hypotheses that a rotation occurs when the motor releases ADP, as proposed here, or when the motor binds ATP, as proposed by Wendt et al. (2002).
The new Ncd dimer crystal structure shows that the coiled-coil domain of the motor can undergo a large rotational movement that could represent a force- producing stroke of the motor. If so, the role of ATP binding and hydrolysis may be to unbind the motor from the microtubule, which has been estimated to require as much as half the free energy of ATP hydrolysis for interactions of myosin with actin (for a review see Kinosita et al., 1998). Remarkably, the acute movement of the coiled-coil stalk in our new structure is not accompanied by melting of the neck. Instead, the stalk/neck behaves like a rigid domain and may act like a lever arm, resembling the rod of myosin (Rayment et al., 1993; Uyeda et al., 1996; Suzuki et al., 1998). In contrast to minus-end Ncd, current models for motility of conventional kinesin and other plus-end kinesin motors propose a molecular rachet mechanism, rather than a lever arm mechanism (Rice et al., 1999; Turner et al., 2001). This could be a fundamental difference between the plus- and minus-end kinesin motors that may be related not only to motor directionality, but also to processivity of motor movement along the microtubule.
Materials and methods
Protein purification
Plasmids encoding wild-type or mutant proteins were constructed using conventional methods, including overlap extension PCR (Ho et al., 1989). PCR-synthesized DNA regions were confirmed by DNA sequence analysis. Dimeric NcdN600K protein (MGSM-H293-K700; 412 residues) was purified by chromatography on SP-Sepharose FF (Pharmacia), followed by FPLC on Superose 12 and/or MonoQ (Pharmacia) (Song and Endow, 1997; Sablin et al., 1998).
Single turnover ADP release assays
Single turnover mant·ADP release experiments were performed in 50 mM Tris–acetate pH 7.4, 2.5 mM MgCl2, 1.5 mM dithiothreitol (DTT), 5 mM HEPES, 0.5 mM EGTA and 94 mM NaCl at room temperature in a SPEX FluoroMax spectrofluorometer. Four micromolar mant·ATP [2′(3′)-O-(N-methyl-anthraniloyl)-adenine 5′-triphosphate] was incubated with 0.5 or 1 µM Ncd or NcdN600K for ≥1 h on ice in the dark, then warmed to room temperature for 5–10 min immediately before addition of 500 µM ATP. Fluorescence (excitation, 356 nm; emission, 446 nm) was recorded for 600–1000 s. Data were fit to a single exponential using Kaleidagraph v. 3.0.8 to obtain dissociation rate constants.
Gliding assays
Gliding assays of GST/Ncd fusion proteins were performed using lysates prepared from induced bacterial cells (Song et al., 1997). Lysates were analyzed on western blots probed with anti-GST antibodies to ensure that the amount of motor in the lysate was the same for the G347A or G347T mutant and corresponding wild-type GST/Ncd control, GST/MC1 (Chandra et al., 1993). Microtubules gliding on the motor bound to glass coverslips were imaged by VE-DIC microscopy (Walker et al., 1990) and recorded onto videotape. Velocities were determined using a custom tracking program (a gift from E.D.Salmon and N.R.Gliksman).
Protein crystallization
Protein for crystallization was concentrated to 17.0 mg/ml in 20 mM HEPES pH 7.4, 200 mM NaCl, 10 mM MgCl2 and 2 mM DTT, and was pre-incubated with 4 mM AMP·PNP or ATP for 2 h. Crystals grew in 11.0% PEG 8000, 0.8 M NaCl, 50 mM Na2HPO4/NaH2PO4 (pH 6.8) and 7 mM DTT at 18°C and were flash-frozen in liquid N2 in cryoprotectant (45% ethylene glycol, 2.5 mM MgCl2) containing 0.5 mM AMP·PNP or ATP, depending on the nucleotide used for pre-incubation. Diffraction data were measured under cryoconditions (100°K) at the macromolecular beamline at Pohang Accelerator Laboratory, Korea, equipped with DIP image plate detectors. The dataset was scaled by the program DENZO/SCALEPACK (Otwinowski and Minor, 1997). NcdN600K crystallized in space group C2, differing from the P6122 and C2221 of previous Ncd dimer structures (Sablin et al., 1998; Kozielski et al., 1999), with unit cell dimensions of a = 162.6 Å, b = 66.6 Å, c = 94.8 Å, β = 98.0°. The NcdN600K structure reported here was obtained after adding 4 mM AMP·PNP to the protein prior to crystallization, but the same crystal form was obtained when either 4 mM AMP·PNP or ATP was added. The model is based on 20.0–2.5 Å resolution data and has an Rfree of 0.302 and Rwork of 0.260. The wild-type Ncd protein, pre-incubated with 4 mM AMP·PNP, crystallized in space group C2 with unit cell dimensions of a = 162.1 Å, b = 67.4 Å, c = 94.6 Å, β = 98.5°. Refinement of a model based on 20 to ∼3.5 Å resolution data gave an Rfree of 0.337 and Rwork of 0.296.
Structure determination
The structure was determined by molecular replacement using the head domain (residues 350–670) from an Ncd dimer structure [Protein Data Bank (PDB) accession code 2NCD] (Sablin et al., 1998) as a search model. Cross-rotation and translation functions calculated using the program CNS (Brunger et al., 1998) and data from 15–4.0 Å gave two solutions that corresponded to two monomers. A model containing the two head domains was subjected to rigid body refinement at 6.0–4.0 Å resolution, followed by conjugate gradient minimization and individual B-factor refinement at 20.0–2.5 Å resolution, giving an Rwork of 36.8% and Rfree of 43.2%. Reflections at low resolution (to 20.0 Å) were included in the refinement after taking into consideration the solvent contribution to their intensities using the structure factor amplitudes of solvent molecules in the crystal calculated by the bulk correction routine in CNS. The resulting difference map revealed electron density for the stalk/neck region (residues 293–349) and one Mg·ADP that was absent from the search model, suggesting the correct molecular replacement solution. The stalk/neck and Mg·ADP were fit into this density. A few iterations of model building in the program O (Jones et al., 1991) and refinement in CNS improved the map quality, showing electron density for the second Mg·ADP. The final refinement statistics, after including the second Mg·ADP, are given (Table I). The refinement included simulated annealing, conjugate gradient minimization and individual B-factor refinement at 20.0–2.5 Å resolution. The coordinates for the NcdN600K structure have been deposited into the PDB under accession code 1N6M.
Docking
Manual docking of the new structure was performed using the program O (Jones et al., 1991), and cryoEM density maps of dimeric Ncd bound to microtubules in the absence of nucleotide (with added apyrase), or presence of AMP·PNP or ADP (Hirose et al., 1998, 1999). Head H2 of the NcdN600K atomic structure was fit into the attached head density while keeping head H1 within the unattached head density. Reversing the heads gave a poor fit. The α/β tubulin atomic structure (PDB accession code 1TUB) (Nogales et al., 1998) was matched with the corresponding microtubule density. Dockings of a previous Ncd structure into an ADP map (Hirose et al., 1998, 1999) and of the previous and new structures into the SH3-Ncd no-nucleotide map (Wendt et al., 2002) were performed by fitting the motor and tubulin structures into the density to maintain interactions of the motor tubulin-binding elements L11–α4–L12–α5 with H11 and H12 of tubulin.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank H.S.Lee and the staff at the Pohang Accelerator Laboratory, Pohang, Korea, for help with data collection, Linda Amos (MRC Laboratory of Molecular Biology, Cambridge, UK) for cryoEM coordinates and Steve Rosenfeld (University of Alabama, Birmingham, AL) for the gift of mant·ATP. This work was supported by St Jude Children’s Research Hospital Cancer Center Grant and American Lebanese Syrian Associated Charities (H.-W.P.) and grants from the National Institutes of Health and Human Frontiers Science Program to S.A.E.
References
- Alonso M.C., van Damme,J., Vandekerckhove,J. and Cross,R.A. (1998) Proteolytic mapping of kinesin/ncd–microtubule interface: nucleotide-dependent conformational changes in the loops L8 and L12. EMBO J., 17, 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) Ribbons. Methods Enzymol., 277, 493–505. [PubMed] [Google Scholar]
- Case R.B., Pierce,D.W., Hom-Booher,N., Hart,C.L. and Vale,R.D. (1997) The directional preference of kinesin motors is specified by an element outside of the motor catalytic domain. Cell, 90, 959–966. [DOI] [PubMed] [Google Scholar]
- Chandra R., Salmon,E.D., Erickson,H.P., Lockhart,A. and Endow,S.A. (1993) Structural and functional domains of the Drosophila ncd microtubule motor protein. J. Biol. Chem., 268, 9005–9013. [PubMed] [Google Scholar]
- Cooke R. (1986) The mechanism of muscle contraction. CRC Crit. Rev. Biochem., 21, 53–118. [DOI] [PubMed] [Google Scholar]
- Endow S.A. and Waligora,K.W. (1998) Determinants of kinesin motor polarity. Science, 281, 1200–1202. [DOI] [PubMed] [Google Scholar]
- Endow S.A. and Higuchi,H. (2000) A mutant of the motor protein kinesin that moves in both directions on microtubules. Nature, 406, 913–916. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes generally enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–134. [DOI] [PubMed] [Google Scholar]
- Henningsen U. and Schliwa,M. (1997) Reversal in the direction of movement of a molecular motor. Nature, 389, 93–96. [DOI] [PubMed] [Google Scholar]
- Hirose K., Lockhart,A., Cross,R.A. and Amos,L.A. (1996) Three-dimensional cryoelectron microscopy of dimeric kinesin and ncd motor domains on microtubules. Proc. Natl Acad. Sci. USA, 93, 9539–9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose K., Cross,R.A. and Amos,L.A. (1998) Nucleotide-dependent structural changes in dimeric NCD molecules complexed to microtubules. J. Mol. Biol., 278, 389–400. [DOI] [PubMed] [Google Scholar]
- Hirose K., Löwe,J., Alonso,M., Cross,R.A. and Amos,L.A. (1999) Congruent docking of dimeric kinesin and ncd into three-dimensional electron cryomicroscopy maps of microtubule–motor ADP complexes. Mol. Biol. Cell, 10, 2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S.N., Hunt,H.D., Horton,R.M., Pullen,J.K. and Pease,L.R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene, 77, 51–59. [DOI] [PubMed] [Google Scholar]
- Hoyt M.A., He,L., Totis,L. and Saunders,W.S. (1993) Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics, 135, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) O. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kikkawa M., Sablin,E.P., Okada,Y., Yajima,H., Fletterick,R.J. and Hirokawa,N. (2001) Switch-based mechanism of kinesin motors. Nature, 411, 439–445. [DOI] [PubMed] [Google Scholar]
- Kinosita J.K., Yasuda,R., Noji,H., Ishiwata,S. and Yoshida,M. (1998) F1-ATPase: a rotary motor made of a single molecule. Cell, 93, 21–24. [DOI] [PubMed] [Google Scholar]
- Kozielski F., Sack,S., Marx,A., Thormählen,M., Schönbrunn,E., Biou,V., Thompson,A., Mandelkow,E.-M. and Mandelkow,E. (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell, 91, 985–994. [DOI] [PubMed] [Google Scholar]
- Kozielski F., De Bonis,S., Burmeister,W.P., Cohen-Addad,C. and Wade,R.H. (1999) The crystal structure of the minus-end-directed microtubule motor protein ncd reveals variable dimer conformations. Structure, 7, 1407–1416. [DOI] [PubMed] [Google Scholar]
- Kull F.J. and Endow,S.A. (2002) Kinesin: switch I & II and the motor mechanism. J. Cell Sci., 115, 15–23. [DOI] [PubMed] [Google Scholar]
- Kull F.J., Sablin,E.P., Lau,R., Fletterick,R.J. and Vale,R.D. (1996) Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature, 380, 550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E., Wolf,S.G. and Downing,K.H. (1998) Structure of the αβ tubulin dimer by electron crystallography. Nature, 391, 199–203. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Rayment I., Rypniewski,W.R., Schmidt-Base,K., Smith,R., Tomchick,D.R., Benning,M.M., Winkelmann,D.A., Wesenberg,G. and Holden,H.M. (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor. Science, 261, 50–58. [DOI] [PubMed] [Google Scholar]
- Rice S. et al. (1999) A structural change in the kinesin motor protein that drives motility. Nature, 402, 778–784. [DOI] [PubMed] [Google Scholar]
- Sablin E.P., Kull,F.J., Cooke,R., Vale,R.D. and Fletterick,R.J. (1996) Crystal structure of the motor domain of the kinesin-related motor ncd. Nature, 380, 555–559. [DOI] [PubMed] [Google Scholar]
- Sablin E.P., Case,R.B., Dai,S.C., Hart,C.L., Ruby,A., Vale,R.D. and Fletterick,R.J. (1998) Direction determination in the minus-end-directed kinesin motor ncd. Nature, 395, 813–816. [DOI] [PubMed] [Google Scholar]
- Sack S., Kull,F.J. and Mandelkow,E. (1999) Motor proteins of the kinesin family. Structure, variations and nucleotide binding sites. Eur. J. Biochem., 262, 1–11. [DOI] [PubMed] [Google Scholar]
- Song H. and Endow,S.A. (1997) Rapid purification of microtubule motor domain proteins expressed in bacteria. BioTechniques, 22, 82–85. [DOI] [PubMed] [Google Scholar]
- Song H. and Endow,S.A. (1998) Decoupling of nucleotide- and microtubule-binding in a kinesin mutant. Nature, 396, 587–590. [DOI] [PubMed] [Google Scholar]
- Song H., Golovkin,M., Reddy,A.S.N. and Endow,S.A. (1997) In vitro motility of AtKCBP, a calmodulin-binding kinesin protein of Arabidopsis. Proc. Natl Acad. Sci. USA, 94, 322–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R.J., Thaler,J.P. and Goldstein,L.S.B. (1993) Direction of microtubule movement is an intrinsic property of the motor domains of kinesin heavy chain and Drosophila ncd protein. Proc. Natl Acad. Sci. USA, 90, 5209–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Yasunaga,T., Ohkura,R., Wakabayashi,T. and Sutoh,K. (1998) Swing of the lever arm of a myosin motor at the isomerization and phosphate-release steps. Nature, 396, 380–383. [DOI] [PubMed] [Google Scholar]
- Turner J., Anderson,R., Guo,J., Beraud,C., Fletterick,R. and Sakowicz,R. (2001) Crystal structure of the mitotic spindle kinesin Eg5 reveals a novel conformation of the neck-linker. J. Biol. Chem., 276, 25496–25502. [DOI] [PubMed] [Google Scholar]
- Uyeda T.Q.P., Abramson,P.D. and Spudich,J.A. (1996) The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc. Natl Acad. Sci. USA, 93, 4459–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R.A., Gliksman,N.R. and Salmon,E.D. (1990) Using video-enhanced differential interference contrast microscopy to analyze the assembly dynamics of individual microtubules in real time. In Herman,B. and Jacobson,K. (eds), Optical Microscopy for Biology. Wiley-Liss, New York, NY, pp. 395–407. [Google Scholar]
- Wendt T.G., Volkmann,N., Skiniotis,G., Goldie,K.N., Müller,J., Mandelkow,E. and Hoenger,A. (2002) Microscopic evidence for a minus-end-directed power stroke in the kinesin motor ncd. EMBO J., 21, 5969–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G., Ruby,A.K., Hart,C.L., Ly,B., Hom-Booher,N. and Vale,R.D. (1997) Microtubule interaction site of the kinesin motor. Cell, 90, 207–216. [DOI] [PubMed] [Google Scholar]
- Yun M., Zhang,X., Park,C.-G., Park,H.-W. and Endow,S.A. (2001) A structural pathway for activation of the kinesin motor ATPase. EMBO J., 20, 2611–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]