Abstract
Preproteins with N-terminal presequences are imported into mitochondria at translocation contact sites that include the translocase of the outer membrane (TOM complex) and the presequence translocase of the inner membrane (TIM23 complex). Little is known about the functional cooperation of these translocases. We have characterized translocation contact sites by a productive TOM–TIM–preprotein supercomplex to address the role of three translocase subunits that expose domains to the intermembrane space (IMS). The IMS domain of the receptor Tom22 is required for stabilization of the translocation contact site supercomplex. Surprisingly, the N-terminal segment of the channel Tim23, which tethers the TIM23 complex to the outer membrane, is dispensable for both protein import and generation of the TOM–TIM supercomplex. Tim50, with its large IMS domain, is crucial for generation but not for stabilization of the supercomplex. Thus, Tim50 functions as a dynamic factor and the IMS domain of Tom22 represents a stabilizing element in formation of a productive translocation contact site supercomplex.
Keywords: contact sites/mitochondria/protein translocation/Saccharomyces cerevisiae
Introduction
Proteins that are imported into the mitochondrial matrix must cross both the outer and inner mitochondrial membranes (Koehler et al., 1999; Bauer et al., 2000; Jensen and Johnson, 2001; Endo and Kohda, 2002; Pfanner and Chacinska, 2002). N-terminal targeting signals (presequences) direct the preproteins to mitochondria and through two protein import complexes, the TOM complex and the TIM23 complex. The TOM complex contains receptors for recognition of preproteins and a general import pore. The two central components of the TOM machinery are the multifunctional receptor Tom22 (van Wilpe et al., 1999) and the pore-forming protein Tom40 (Hill et al., 1998; Künkele et al., 1998). The presequence translocase contains three integral membrane proteins. The pore-forming subunit Tim23 is tightly associated with Tim17 (Dekker et al., 1997; Ryan et al., 1998; Truscott et al., 2001). Tim50 exposes a large domain to the intermembrane space (IMS) (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). The import motor on the matrix side, including the mitochondrial heat shock protein 70 (mtHsp70), drives the completion of protein transport into the matrix.
While considerable information has been accumulated on the composition and function of the individual translocases, little is known about the cooperation of the TOM and TIM23 complexes in protein import. Preproteins in transit can span both mitochondrial membranes at so-called translocation contact sites (Schleyer and Neupert, 1985; Rassow et al., 1989, 1990; Wienhues et al., 1991; Jascur et al., 1992; Kanamori et al., 1997; Schülke et al., 1997). When matrix-targeted preproteins are arrested in translocation contact sites due to a tightly folded domain at the C-terminus, the TOM complex and TIM23 complex can be co-purified (Horst et al., 1995; Dekker et al., 1997; Sirrenberg et al., 1997; Schülke et al., 1999; Geissler et al., 2002). However, it remained open as to which components were required for generation and/or stabilization of the TOM–TIM–preprotein supercomplex. So far, two components that expose domains to the IMS have been suggested to play a role in the TOM–TIM connection. Yeast Tim23 consists of an inner membrane-integrated domain (residues 97–222) and a hydrophilic N-terminal domain that is exposed to the IMS. While the segment formed by amino acid residues 51–96 binds presequences as well as the IMS domain of Tim50 (Bauer et al., 1996; Komiya et al., 1998; Geissler et al., 2002; Yamamoto et al., 2002), a most interesting role has been reported for the N-terminal 50 residues. This segment of Tim23 spans the outer membrane and has been suggested to promote a cooperation of TOM and TIM23 complexes in translocation contact sites and the transfer of preproteins (Donzeau et al., 2000). The IMS domain of Tim50 interacts with preproteins while they are still in contact with the TOM complex and directs the preproteins to the TIM23 import channel (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). A possible function of the IMS domain of Tom22 in formation of a TOM–TIM supercomplex has not been addressed as yet.
We report unexpected roles for the translocase subunits that expose domains to the IMS. The N-terminal outer membrane spanning segment of Tim23 is neither crucial for protein import nor for formation and stability of translocation contact sites. Tim50 is essential for generation but not for stabilization of the translocation contact site supercomplex, while Tom22–IMS forms a stabilizing element.
Results
A productive TOM–TIM–preprotein supercomplex
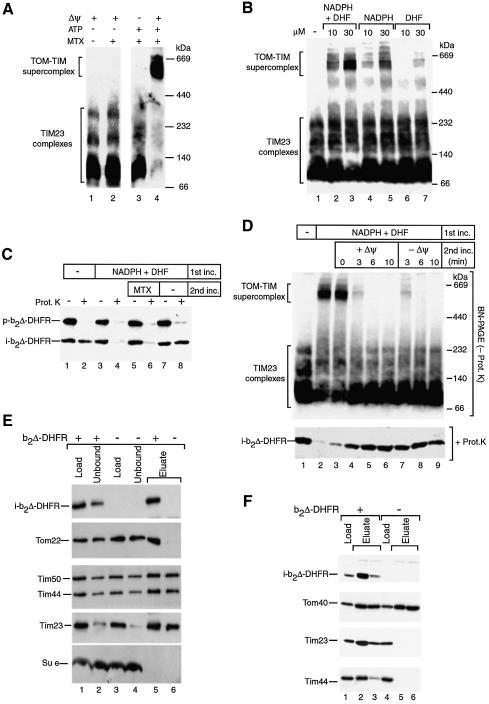
The TOM–TIM supercomplex was generated with the model preprotein b2Δ-DHFR, which consists of an N-terminal matrix-targeted portion of the precursor of cytochrome b2 and dihydrofolate reductase (DHFR) at the C-terminus (Koll et al., 1992; Dekker et al., 1997). In the presence of the substrate analog methotrexate (MTX), DHFR is stably folded, leading to an arrest of b2Δ-DHFR in translocation contact sites such that the b2 portion spans both mitochondrial membranes. We accumulated saturating amounts of purified b2Δ-DHFR/MTX in translocation contact sites of energized yeast mitochondria (Figure 1A, lane 4) (Dekker et al., 1997; Geissler et al., 2002). The mitochondria were lyzed with digitonin and subjected to blue native electrophoresis (BN–PAGE). The supercomplex detected by antibodies against Tim23 migrated at ∼600 kDa, while in the absence of preprotein accumulation, the TIM23 machinery was found in several subcomplexes, a 90 kDa core complex and additional complexes of variable abundance in a range up to 250 kDa (Figure 1A, lanes 1–3) (Dekker et al., 1997).
Fig. 1. The TOM–TIM supercomplex represents a productive translocation intermediate. (A) Formation of the supercomplex. Purified b2Δ-DHFR was incubated with wild-type yeast mitochondria for 15 min at 25°C in the presence or absence of MTX. For sample 3, Δψ was dissipated before the import reaction. For samples 1 and 2, the import assay was depleted of ATP. After the import reaction, the mitochondria were reisolated, lyzed with digitonin and subjected to BN–PAGE. Analysis was performed by immunodecoration with Tim23-specific antibodies. (B) Generation of the supercomplex in the presence of NADPH and DHF. b2Δ-DHFR was imported into isolated mitochondria for 12 min at 25°C, followed by BN–PAGE and immunodecoration for Tim23. (C) Two-step import assay. Purified b2Δ-DHFR was incubated with mitochondria for 15 min at 25°C in the presence or absence of 30 µM NADPH and 30 µM DHF (first incubation). Upon reisolation, the mitochondria of samples 5–8 were resuspended in fresh import buffer and incubated for a further 10 min at 25°C (second incubation). Where indicated, the mitochondria were treated with proteinase K. The samples were separated by SDS–PAGE and immunodecorated with DHFR-specific antibodies. p, precursor; i, processed (intermediate-sized) form of b2Δ-DHFR. (D) Purified b2Δ-DHFR was arrested in mitochondria in the presence or absence of 30 µM NADPH and 30 µM DHF. Upon reisolation, the mitochondria of samples 3–9 were resuspended in fresh import buffer and incubated for the indicated times at 25°C in the presence or absence of a Δψ as indicated. The samples were divided in half. One portion was subjected to BN–PAGE and decorated with Tim23-specific antibodies (upper panel). The other portion was treated with proteinase K, separated by SDS–PAGE and decorated with DHFR-specific antibodies (lower panel). (E) Affinity-purification of the supercomplex via Tim23ProtA. Where indicated, b2Δ-DHFR was accumulated in yeast mitochondria carrying Tim23ProtA in the presence of MTX. The mitochondria were reisolated, lyzed with digitonin, and subjected to IgG affinity chromatography, SDS–PAGE and immunodecoration. Thirty percent of the load and unbound material and 100% of the eluate are shown. (F) Affinity-purification of the supercomplex via Tom22His10. Where indicated, b2Δ-DHFR was accumulated in mitochondria carrying Tom22His10 in the presence of MTX. The reisolated mitochondria were lyzed with digitonin, and subjected to Ni-NTA affinity chromatography, SDS–PAGE and immunodecoration. Ten percent of load and 100% of the eluate fractions are shown.
We asked whether the supercomplex could be dissociated by chasing the accumulated preprotein to its fully imported form. However, when mitochondria containing the supercomplex were reisolated and washed, MTX still bound so tightly to b2Δ-DHFR that a release of MTX and subsequent chase of the protein turned out to be inefficient (not shown). We therefore established a method to reversibly accumulate b2Δ-DHFR by use of the DHFR substrates NADPH and dihydrofolate (DHF) (Figure 1B, lane 3). In a two-step import reaction, b2Δ-DHFR was first accumulated in the supercomplex in the presence of NADPH and DHF (Figure 1C, lanes 3–8). The accumulated b2Δ-DHFR was processed to the i-form by the matrix processing peptidase, yet remained accessible to externally added protease (Figure 1C, lanes 3 and 4). Upon reisolation of the mitochondria, a second incubation was performed in the absence of DHFR ligands (‘chase’), leading to the generation of protease-protected i-b2Δ-DHFR (Figure 1C, lane 8). To monitor the presence of the supercomplex and the formation of protease-protected i-b2Δ-DHFR in parallel, we performed a two-step import-chase assay and divided each sample in half (Figure 1D). In one set, the mitochondria were lyzed with digitonin and analyzed by BN–PAGE, while in the other set, the mitochondria were treated with proteinase K and analyzed by SDS–PAGE. The generation of protease-protected i-b2Δ-DHFR correlated with the disappearence of the TOM–TIM supercomplex (Figure 1D, lanes 4–6). The chase reaction was not blocked by dissipation of the membrane potential Δψ (Figure 1D, lanes 7–9), confirming that the accumulated preprotein in the supercomplex had already passed the Δψ-dependent early import steps. When analyzed by different separation methods (see below), the size and the composition of the supercomplex were independent of which DHFR ligands were used (NADPH/DHFR or MTX). We conclude that the TOM–TIM supercomplex forms a chaseable translocation intermediate, i.e. represents a productive import intermediate.
To analyze the composition of the supercomplex, we used mitochondria derived from yeast strains either carrying a tagged Tim23 (protein A tag) or a tagged Tom22 (His10 tag). The addition of these tags did not impair cellular growth or mitochondrial function (Meisinger et al., 2001; Geissler et al., 2002). Affinity-purification of Tim23 via IgG sepharose led to the co-purification of the other subunits of the TIM23 complex, including Tim50 and Tim44, independently of the presence or absence of the preprotein b2Δ-DHFR (Figure 1E, lanes 5 and 6). The subunit e of the dimeric F0F1-ATPase, also termed Tim11, was used as a non-associated control protein (Arnold et al., 1998). Similarly, affinity-purification of Tom22 via Ni-nitrilotriacetic acid (Ni-NTA) agarose led to a co-purification of other Tom subunits, such as Tom40, independently of the presence of b2Δ-DHFR (Figure 1F) (Geissler et al., 2002). However, subunits of the non-tagged translocase complex were only co-purified when b2Δ-DHFR had been arrested in these mitochondria in the presence of ligand, i.e. co-purification of Tim proteins with tagged Tom22 (Figure 1F, lanes 2 and 3) (Geissler et al., 2002) and co-purification of Tom proteins with tagged Tim23 (Figure 1E, lane 5). Thus, both tagging methods allow for a specific purification of the TOM–TIM supercomplex and demonstrate that the stable association of TOM complex and TIM23 complex depends on the presence of the accumulated preprotein.
The N-terminal segment of Tim23 is not crucial for formation of the supercomplex
We asked which components of the TOM complex or TIM23 complex are involved in formation of the translocation contact site supercomplex, and addressed the role of the three translocase subunits that expose domains to the IMS: Tom22, Tim23 and Tim50.
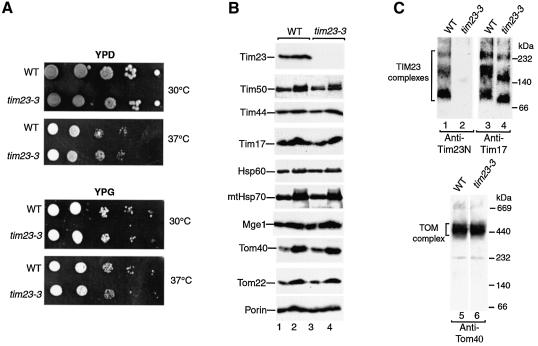
The N-terminal segment of Tim23 (residues 1–50) has been suggested to be important for formation of translocation contact sites (Donzeau et al., 2000). We constructed a yeast strain, tim23-3, that selectively lacked this segment. The strain grew like wild-type yeast under all conditions tested, including fermentable and non-fermentable media (Figure 2A). All marker proteins analyzed in tim23-3 mitochondria were present in wild-type amounts, including Tim17, Tim44 and Tim50, as well as Tom subunits and matrix chaperones (Figure 2B). As expected, Tim23 was no longer detected by an antibody generated against the N-terminal segment (Figure 2B and C). BN–PAGE using antibodies against Tim17 demonstrated that the TIM23 complexes migrated slightly faster than that of wild-type mitochondria, consistent with the lack of the N-terminal segment of Tim23 (Figure 2C, lanes 3 versus 4). The TOM complex was not altered (Figure 2C, lane 6).
Fig. 2. A yeast mutant lacking the N-terminal 50 residues of Tim23. (A) Growth of tim23-3 mutant cells is indistinguishable from wild-type cells. Wild-type (WT) and tim23-3 cells were subjected to consecutive 10-fold dilutions, spotted on fermentable medium (YPD) or non-fermentable medium (YPG) and grown for 3 days at the indicated temperatures. (B) Protein composition of tim23-3 mitochondria. Wild-type mitochondria and tim23-3 mitochondria were subjected to SDS–PAGE and immunodecoration. Odd-numbered and even-numbered samples, 10 and 20 µg mitochondrial protein, respectively. (C) BN–PAGE of digitonin-lyzed wild-type and tim23-3 mitochondria and immunodecoration.
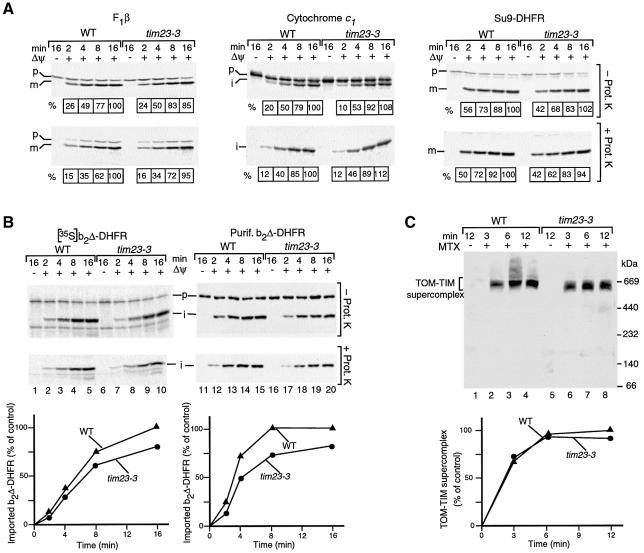
Donzeau et al. (2000) had reported that mutant mitochondria lacking the N-terminal segment of Tim23 were strongly impaired in the import of one presequence-containing preprotein, a fusion protein between the presequence of F0-ATPase subunit 9 and DHFR (Su9-DHFR). We performed a systematic analysis of processing and transport to a protease-protected location of a number of preproteins by tim23-3 mitochondria, including the matrix-targeted protein F1-ATPase subunit β, the inner membrane protein cytochrome c1, as well as the matrix-targeted DHFR fusion proteins Su9-DHFR and b2Δ-DHFR. Surprisingly, we observed only mild import defects for all preproteins analyzed (Figure 3A and B). This mild import defect of tim23-3 mitochondria was similarly observed with small, radiochemical amounts of preprotein (Figure 3B, left panel) and large, saturating amounts of preprotein (Figure 3B, right panel). Why did Donzeau et al. (2000) observe a strong inhibition of protein import into mitochondria lacking the N-terminal Tim23 segment? A likely explanation is that in their import experiments, they used a yeast strain that overexpressed the truncated Tim23 together with a C-terminal His12 tag (Bauer et al., 1996), while the yeast strain tim23-3 used here expressed the N-terminally truncated Tim23 from its own promoter without any further modifications. We conclude that the N-terminal segment of Tim23 plays only a supporting role in dynamic protein import.
Fig. 3. A connection of mitochondrial outer and inner membranes via Tim23 is not critical for formation of the TOM–TIM supercomplex. (A) Import of preproteins into tim23-3 mitochondria. The 35S-labeled precursors of F1-ATPase subunit β (F1β), cytochrome c1 and Su9-DHFR were incubated with wild-type (WT) or tim23-3 mitochondria at 25°C. Where indicated, the mitochondria were treated with proteinase K after the import reaction. The mitochondria were reisolated and analyzed by SDS–PAGE and digital autoradiography. The amount of processed protein in energized wild-type mitochondria after an import time of 16 min was set to 100%, respectively. p, precursor; i, m, processed forms (intermediate-sized, mature) of the protein. (B) 35S-labeled precursor of b2Δ-DHFR (left panel) or purified b2Δ-DHFR (right panel) were imported into wild-type or tim23-3 mitochondria. The amount of protease-protected, processed i-b2Δ-DHFR in energized wild-type mitochondria after an import time of 16 min was set to 100% (control). (C) Formation of the TOM–TIM supercomplex. Purified b2Δ-DHFR was incubated with wild-type or tim23-3 mitochondria in the presence of MTX unless indicated otherwise. The mitochondria were reisolated, lyzed with digitonin and subjected to BN–PAGE and immunodecoration with antibodies directed against DHFR. The amount of supercomplex formed after an incubation of 12 min in wild-type mitochondria was set to 100% (control).
We then addressed the question of whether the N-terminal segment of Tim23 was required for formation of translocation contact sites by analyzing the TOM–TIM supercomplex with BN–PAGE. The tim23-3 mutant mitochondria, however, were able to generate the supercomplex in the presence of accumulated b2Δ-DHFR and MTX (Figure 3C, lanes 6–8). A quantitation revealed that the efficiency of supercomplex formation was only reduced by ∼10% in tim23-3 mitochondria compared with wild-type mitochondria (Figure 3C). Similarly, in a two-step import reaction, the generation of the supercomplex in the presence of NADPH/DHF and the subsequent chase of b2Δ-DHFR into the matrix were only mildly affected (not shown). Thus, the N-terminal segment of Tim23 that connects the TIM23 complexes to the outer membrane is essential neither for dynamic import of preproteins nor for formation of the TOM–TIM supercomplex.
A role for the IMS domain of Tom22 in stabilization of the TOM–TIM supercomplex
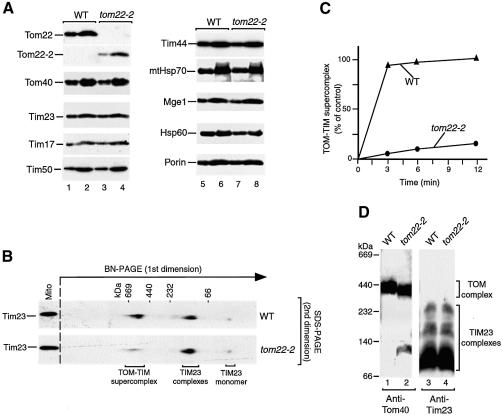
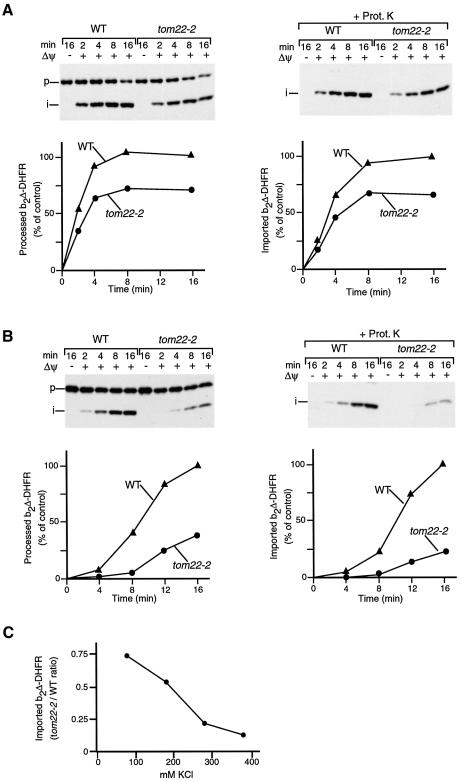
The IMS domain of Tom22 binds mitochondrial presequences and stimulates the import of presequence-containing preproteins (Bolliger et al., 1995; Court et al., 1996; Moczko et al., 1997; Komiya et al., 1998; Kanamori et al., 1999). tom22-2 mitochondria lack this C-terminal IMS domain (Moczko et al., 1997). The steady-state levels of all marker proteins analyzed were comparable between tom22-2 and wild-type mitochondria, including Tom and Tim proteins, as well as matrix chaperones (Figure 4A). The TOM–TIM–preprotein supercomplex, however, was dramatically affected in tom22-2 mitochondria, as shown by BN–PAGE (Figure 4B). In tom22-2 mitochondria, only 5–15% of the yield of supercomplex observed for wild-type mitochondria was obtained (Figure 4C).
Fig. 4. The IMS domain of Tom22 is required for stabilization of the TOM–TIM supercomplex. (A) Protein composition of tom22-2 mitochondria. Wild-type (WT) and tom22-2 mitochondria (odd-numbered and even-numbered samples, 10 and 20 µg protein, respectively) were subjected to SDS–PAGE and immunodecoration. (B) Strong reduction of the supercomplex in tom22-2 mitochondria. Purified b2Δ-DHFR was accumulated in wild-type or tom22-2 mitochondria in the presence of MTX for 15 min at 25°C. The mitochondria were reisolated, lyzed with digitonin and subjected to 2D PAGE (BN–PAGE followed by SDS–PAGE) and immunodecoration with antibodies directed against Tim23. (C) Quantification of supercomplex formation. The import experiment was performed as described for (B) with different import times. The amount of supercomplex formed in wild-type mitochondria after 12 min was set to 100% (control). (D) Stability of TOM and TIM23 complexes in tom22-2 mitochondria. Digitonin-lyzed mitochondria (70 µg protein) were subjected to BN–PAGE and immunodecoration.
Three possibilities were conceivable to explain this strong effect on the TOM–TIM–preprotein supercomplex in tom22-2 mitochondria: (i) a reduced stability of the individual translocase complexes, TOM and TIM23; (ii) a reduced efficiency of preprotein import into the mutant mitochondria; or (iii) a role of the IMS domain of Tom22 in stabilization of the TOM–TIM–preprotein connection. (i) The size of the TOM complex of tom22-2 mitochondria analyzed by BN–PAGE was slightly smaller than that of wild-type mitochondria, as expected due to the shortening of Tom22. The majority of the TOM complex migrated in the high molecular weight range ∼400 kDa, while only a small fraction (<10%) was dissociated to a smaller form of ∼100 kDa (Figure 4D, lanes 1 and 2) (van Wilpe et al., 1999). The TIM23 complexes were indistinguishable between wild-type and tom22-2 mitochondria (Figure 4D, lanes 3 and 4). Thus, the lack of the IMS domain of Tom22 only mildly impairs the stability of the TOM complex. (ii) Moczko et al. (1997) reported a moderate reduction (∼30%) of import of radiochemical amounts of presequence-containing preproteins into tom22-2 mitochondria compared with wild-type mitochondria. However, the import of large amounts of preprotein had not been studied. We therefore analyzed the import of saturating amounts of b2Δ-DHFR into tom22-2 mitochondria under conditions similar to those used for generation of the supercomplex. The efficiency of import, when analyzing either processing to the i-form or transport to a protease-protected location, was reduced by ∼30% in comparison to wild-type mitochondria (Figure 5A). Thus, a lack of the IMS domain of Tom22 moderately impairs the import of preproteins, independently of whether they are added in small, radiochemical amounts or in large, saturating amounts. Taken together, these results demonstrate that the dramatic reduction of supercomplex yield in tom22-2 mitochondria can neither be explained by a reduced stability of the individual TOM and TIM23 complexes nor by a general reduction of protein import. We suggest that the IMS domain of Tom22 is important for stabilization of the TOM–TIM–preprotein connection.
Fig. 5. Protein import into tom22-2 mitochondria. (A) Import of purified b2Δ-DHFR into wild-type (WT) or tom22-2 mitochondria (80 mM KCl in import buffer). Where indicated, the mitochondria were treated with proteinase K after the import reaction. The mitochondria were reisolated and analyzed by SDS–PAGE. The amount of processed (i-form) of b2Δ-DHFR in energized wild-type mitochondria after a 16 min import was set to 100% (control). (B) Import of purified b2Δ-DHFR in the presence of 250 mM KCl. The experiment was performed as described for (A). (C) Dependence of b2Δ-DHFR import on the ionic strength. The experiment was performed as described above at the indicated concentrations of KCl. The ratio of imported protein in tom22-2 mitochondria to wild-type mitochondria is shown.
In order to address under which import conditions into energized mitochondria the stabilizing role of Tom22–IMS was critical for the import of b2Δ-DHFR, we made use of a previous observation for the cytosolic receptor domains of Tom20 and Tom22. Experiments performed at differing ionic strengths revealed a salt-sensitivity in the interaction between preprotein and the cytosolic receptor domain of Tom22, indicating the involvement of ionic interactions, yet a salt-enhanced interaction was observed between preprotein and Tom20, indicating the involvement of hydrophobic-type interactions (Brix et al., 1997). The hydrophobic nature of the Tom20–preprotein interaction was subsequently directly demonstrated by the high resolution structure of the receptor domain (Abe et al., 2000). We imported b2Δ-DHFR at increased ionic strength and found a strong import defect into tom22-2 mitochondria as compared with wild-type mitochondria when 250 mM KCl instead of the usual 80 mM KCl were included (Figure 5B). The import efficiency of b2Δ-DHFR into tom22-2 mitochondria was <20% of that into wild-type mitochondria at higher ionic strength (Figure 5C). Thus the IMS domain of Tom22 is of particular importance for protein import into energized mitochondria under conditions where ionic interactions are disfavored and hydrophobic interactions are favored, resembling the characteristics of Tom20, but not that of the cytosolic domain of Tom22 (Brix et al., 1997; Kanamori et al., 1999; Abe et al., 2000).
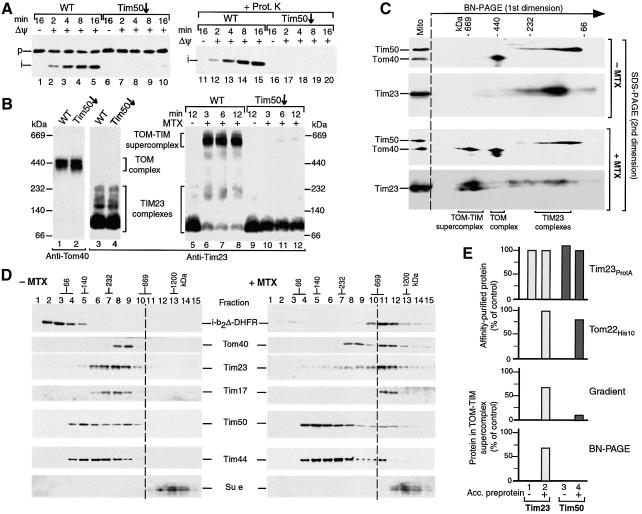
The TOM–TIM supercomplex remains stable after release of Tim50
Tim50 exposes its largest domain of 40 kDa to the IMS and is the first subunit of the TIM23 machinery that contacts polypeptide segments after their passage through the TOM channel (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). Thus, Tim50 represents a likely candidate for a translocase subunit that connects TOM and TIM in the supercomplex. By use of mitochondria isolated from a yeast strain that was depleted of Tim50, it was shown that the import of radiochemical amounts of b2Δ-DHFR required the presence of Tim50 (Geissler et al., 2002). Similarly, the import of saturating amounts of purifed b2Δ-DHFR was strongly inhibited in Tim50-depleted mitochondria (Figure 6A). In addition, the generation of the supercomplex, as determined by BN–PAGE, was blocked in Tim50-depleted mitochondria (Figure 6B, lanes 10–12), while the mobility of TOM and TIM23 complexes on BN–PAGE was unaffected (Figure 6B, lanes 2 and 4). It has been shown that the mutant mitochondria used were selectively depleted of Tim50 and contained wild-type levels of all other Tom, Tim and mitochondrial marker proteins analyzed; moreover, the Tim50-depleted mitochondria were competent in generation of a membrane potential (Geissler et al., 2002). These results indicate that Tim50 is important for dynamic protein import and the generation of the TOM–TIM supercomplex.
Fig. 6. The TOM–TIM supercomplex is not dissociated by a release of Tim50. (A) Import of b2Δ-DHFR is inhibited by depletion of Tim50. Purified b2Δ-DHFR was imported into Tim50-depleted or wild-type (WT) mitochondria. The reisolated mitochondria were separated by SDS–PAGE, followed by immunodecoration with antibodies against DHFR. (B) Generation of the supercomplex is blocked in Tim50-depleted mitochondria. Samples 1–4, isolated wild-type mitochondria and Tim50-depleted mitochondria (70 µg protein each) were lyzed with digitonin and subjected to BN–PAGE and immunodecoration as indicated (absence of preprotein). Samples 5–12, purified b2Δ-DHFR was arrested in wild-type or Tim50-depleted mitochondria (70 µg protein) in the presence of MTX, unless indicated otherwise. The mitochondria were reisolated and subjected to BN–PAGE and immunodecoration with Tim23-specific antibodies. (C) The supercomplex separated by BN–PAGE does not contain Tim50. Purified b2Δ-DHFR was imported into wild-type mitochondria in the presence or absence of MTX for 15 min at 25°C. Protein complexes were separated by 2D PAGE (BN–PAGE followed by SDS–PAGE). Tim50, Tom40 and Tim23 were detected by immunodecoration. (D) Sucrose gradient analysis of the supercomplex. Purified b2Δ-DHFR was imported into wild-type mitochondria in the presence or absence of MTX. The mitochondria were reisolated and lyzed with digitonin. Protein complexes were separated by sucrose gradient centrifugation. Fractions were collected, separated by SDS–PAGE and individual proteins were detected by immunodecoration (15 out of 20 fractions are shown). (E) Quantitative assessment of the presence of Tim50 in the supercomplex. Where indicated, b2Δ-DHFR was accumulated (acc.) in energized mitochondria in the presence of MTX. Top panel, Tim23ProtA mitochondria; second panel, Tom22His10 mitochondria; third and fourth panels, wild-type mitochondria. The mitochondria were reisolated and lyzed with digitonin and subjected to either IgG-affinity chromatography (top panel), Ni-NTA affinity chromatography (second panel), sucrose gradient centrifugation (third panel) or BN–PAGE (fourth panel). The amounts of Tim23 and Tim50 were determined by immunodecoration. First and second panels, the recovery of affinity- purified Tim23 from mitochondria with accumulated b2Δ-DHFR was set to 100% (control). Third and fourth panels, the total amount of solubilized Tim23 and Tim50 was set to 100% (control), respectively.
A surprising observation was made, however, when the composition of the supercomplex of wild-type mitochondria was analyzed by BN–PAGE. While Tim23 and a fraction of Tom40 were shifted to the high molecular weight supercomplex size by the accumulation of b2Δ-DHFR in the presence of MTX, no Tim50 could be detected in the supercomplex (Figure 6C). In contrast, the affinity-purification of the supercomplex via tagged Tom22 led to an efficient co-purification of Tim50 (Geissler et al., 2002) (see below; Figure 6E, second panel). We therefore established sucrose gradient centrifugation as a third independent means to analyze the supercomplex. Wild-type mitochondria were incubated with b2Δ-DHFR, lyzed with digitonin and analyzed by centrifugation on a sucrose gradient. In the presence of MTX, i.e. accumulation of b2Δ-DHFR in translocation contact sites, the preprotein and the bulk of Tim23 and Tim17 were shifted to the high molecular weight range above 600 kDa (Figure 6D, right panel). Only a fraction of Tom40 was shifted, as expected, since the TOM complexes are more abundant than the TIM23 complexes and therefore only a fraction of TOM complexes carry the two-membrane-spanning preprotein when the TIM23 complexes are saturated with preprotein (Dekker et al., 1997; Sirrenberg et al., 1997). Only a relatively small fraction of Tim50, however, was found in the molecular weight range of the supercomplex, comparable to the situation observed for Tim44 (Figure 6D, right panel). As a control, we used subunit e of the F0F1-ATPase, which was not affected by the presence or absence of the two-membrane-spanning preprotein. Thus, two subunits of the TIM23 complex, Tim23 and Tim17, are efficiently shifted to the high molecular weight range of the supercomplex, while the bulk of the other two Tim subunits, Tim50 and Tim44, are not present in the supercomplex after the sucrose gradient separation.
We directly determined the quantitative presence of Tim50 in the supercomplex with the three methods used. Affinity purification via tagged Tom22 led to a recovery of ∼80% of Tim50 molecules in relation to the channel protein Tim23 in the supercomplex (Figure 6E, second panel, columns 2 and 4). After sucrose gradient separation, only ∼10% of Tim50 molecules, but ∼70% of Tim23 molecules, were found in the supercomplex (Figure 6E, third panel, columns 2 and 4). After BN–PAGE, no Tim50 was observed in the supercomplex, i.e. the level was below the detection limit of <1%, while ∼70% of Tim23 molecules were present in the supercomplex (Figure 6E, bottom panel, columns 2 and 4). Since the accumulation of b2Δ-DHFR in translocation contact sites and lysis of mitochondria with digitonin were performed in the identical manner for all three experimental approaches, only the method of separation was different. Thus, the bulk of Tim50 was present in the supercomplex in mitochondria, yet is released by BN–PAGE or sucrose gradient centrifugation, similar to the release of Tim50 from the TIM23 complex that has been observed by various methods (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). Therefore, the TOM–TIM–preprotein connection remains stable under all three methods of separation, independently of whether Tim50 is present or not. Moreover, we did not obtain evidence for a direct interaction between Tom22–IMS and Tim50–IMS (not shown), yet the formation of the supercomplex strictly depended on the presence of the two-membrane-spanning preprotein. Our results indicate that Tim50 is crucial for generation of the supercomplex, but is not an essential structural component once the complex has been formed.
Discussion
The two mitochondrial translocases for matrix-targeted preproteins can function independently of each other. However, during protein import, the TOM complex and TIM23 complex can be stably connected via a precursor polypeptide chain in transit. We show that an accumulated preprotein can be efficiently chased into the matrix, concomitantly with a dissociation of the TOM and TIM23 complexes. Thus the TOM–TIM–preprotein supercomplex represents a productive import intermediate, i.e. is a functional translocation contact site complex. We have characterized the role of three translocase subunits that expose domains to the IMS and separated dynamic and stabilizing elements.
The N-terminal segment of Tim23 spans the outer membrane, tethering the TIM23 complex of the inner membrane to the outer membrane. This two-membrane-spanning topology of Tim23 was proposed to facilitate the formation of translocation contact sites and the transfer of preproteins from the TOM complex to the TIM23 complex (Donzeau et al., 2000). Unexpectedly, we found that deletion of this N-terminal segment of Tim23 only mildly impaired dynamic protein import, as well as formation of the TOM–TIM supercomplex. Since the morphology of the mitochondrial inner membrane is altered in mitochondria lacking the Tim23 segment, including a disappearance of cristae (Donzeau et al., 2000), the two-membrane-spanning topology of Tim23 is apparently crucial for the structural arrangement of the mitochondrial membranes, but not for protein translocation. We conclude that tethering of TIM23 complexes to the outer membrane by the N-terminal segment of Tim23 mildly stimulates protein import, probably by supporting an enrichment of TIM23 complexes in areas of the inner membrane that are close to the outer membrane (Donzeau et al., 2000). However, the formation of translocation contact site supercomplexes can occur in the absence of this N-terminal segment of Tim23.
Tim50 is essential for directing preprotein segments, which have passed through the Tom40 channel of the outer membrane, to the Tim23 channel of the inner membrane (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). The generation of the TOM–TIM supercomplex thus depends strictly on the presence of Tim50. Surprisingly, however, Tim50 can be quantitatively released from the supercomplex, yet the stable interaction between the TOM complex, TIM23 complex and preprotein is not disturbed. When the supercomplex is isolated by a mild affinity purification, Tim50 is present in nearly stoichiometric amounts to Tim23, similar to the situation in the TIM23 complex itself. In a sucrose gradient centrifugation, a major fraction of Tim50 is released and, by BN–PAGE, virtually all Tim50 molecules are released from the supercomplex as well as from the TIM23 complex. These findings also explain the seemingly contradictory results that have been reported on the stoichiometry of Tim50 in the TIM23 complex (Geissler et al., 2002; Yamamoto et al., 2002; Mokranjac et al., 2003). Thus, Tim50 is an essential, but loosely associated, subunit of the TIM23 complex that plays an important dynamic role in protein import, but is not a critical structural part.
In contrast, the IMS domain of Tom22 is of particular importance for the stabilization of the supercomplex. Tom22 is the central receptor protein that is tightly associated with the Tom40 channel and is thus a structural part of the TOM complex as well as the supercomplex. Upon deletion of the IMS domain of Tom22, the stability of the TOM complex is only mildly affected and the TIM23 complex is unaffected. However, the stability of the TOM–TIM–preprotein supercomplex is strongly reduced. The import of preproteins is moderately impaired in the mutant mitochondria, but not blocked (Court et al., 1996; Moczko et al., 1997). Thus, the transfer of preproteins from TOM to TIM23 is still possible as long as all other import conditions are optimal. When further import impairments occur, such as an increased ionic strength or the removal of cytosolic receptor domains, the lack of the IMS domain of Tom22 causes a strong import defect (Moczko et al., 1997; Kanamori et al., 1999; this study). We conclude that the IMS domain of Tom22 promotes an optimal cooperation of the TOM and TIM23 complexes in translocation contact sites by functioning as a stabilizing, structural element of the TOM–TIM–preprotein supercomplex.
In conclusion, mitochondrial translocation contact sites can be isolated as a TOM–TIM23–preprotein supercomplex. In addition to the accumulated preprotein and the channels Tom40 and Tim23 (with the tightly associated Tim17), we found that the IMS domain of Tom22 is a structural element that stabilizes the supercomplex. In contrast, Tim50 is crucial for dynamic protein import and generation of the supercomplex, but does not represent an essential structural element of the supercomplex.
Materials and methods
Yeast strains and growth conditions
The Sacharomyces cerevisiae strains used are listed in Table I. Unless otherwise stated, strains were grown at 30°C on YPG medium (1% yeast extract, 2% bactopeptone and 3% glycerol). To deplete Tim50, the strain AG55Gal carrying genomic TIM50 under control of the GAL1 promoter was precultured on galactose-containing medium at 30°C (1% yeast extract, 2% bactopeptone, 3% lactate, 2% galactose and 1% raffinose pH 5.0) and subsequently shifted to lactate medium (1% yeast extract, 2% bactopeptone and 3% lactate pH 5.0) for 36 h at 30°C (Geissler et al., 2002).
Table I. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| YPH499 (WT) | MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-801 | Sikorski and Hieter (1989) |
| AFY18 (tom22-2) | MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-801 tom22-2 | Moczko et al. (1997); A.E.Frazier, unpublished |
| MR103 (TOM22His10) | MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-801 tom22::TOM22His10-HIS3 | Meisinger et al. (2001) |
| AG55Gal | MATa ade2-101 his3-200 leu2-1 ura3-52 trp1-63 lys2-801 tim50::HIS3-PGAL1-TIM50 | Geissler et al. (2002) |
| PK82 (WT) | MATα ade2-101 leu2-3,112 ura3-52 trp1 lys2 | Gambill et al. (1993) |
| MB29 | MATa ade2 his3 leu2 ura3 trp1 lys2 tim23::LYS2 [YCplac33-TIM23] | Bömer et al. (1997) |
| PRY38 (tim23-3) | MATa ade2 his3 leu2 ura3 trp1 lys2 tim23::LYS2 [pRS413-tim23-3] | Geissler et al. (2002) |
| PRY36 (TIM23ProtA) | MATa ade2 his3 leu2 ura3 trp1 lys2 tim23::LYS2 [pRS414-TIM23ProtA] | Geissler et al. (2002) |
WT, wild type.
Isolation of mitochondria and in vitro import of precursor proteins
Mitochondria were isolated from yeast cells according to established procedures (Ryan et al., 2001). 35S-labeled precursor proteins were synthesized in rabbit reticulocyte lysates. Purified b2Δ-DHFR protein was added to a final concentration of 1 µg per 50 µg of mitochondrial protein (Dekker et al., 1997). Import into isolated yeast mitochondria in the presence of 2 mM NADH, 2 mM ATP and an ATP regenerating system (5 mM creatine phosphate and 0.1 mg/ml creatine kinase) at 25°C and subsequent treatment with proteinase K on ice were performed essentially as described previously (Ryan et al., 2001). The import buffer contained 3% (w/v) bovine serum albumin (BSA), 250 mM sucrose, 80 mM KCl, 5 mM MgCl2, 5 mM KPi, 5 mM methionine and 10 mM MOPS-KOH pH 7.2. The membrane potential was dissipated by addition of 1 µM valinomycin, 8 µM antimycin and 20 µM oligomycin. Samples were subjected to SDS–PAGE analysis, followed by digital autoradiography or immunodecoration with DHFR-specific antibodies.
Generation and purification of the TOM–TIM supercomplex
For the accumulation of preproteins in the import sites, isolated mitochondria were incubated for 15 min at 25°C with recombinant purified b2Δ-DHFR (final concentration: 4 µg of preprotein/100 µg of mitochondria) in the presence of 5 µM MTX, unless otherwise stated. After reisolation, mitochondria were washed with SEM (250 mM sucrose, 1 mM EDTA and 10 mM MOPS-KOH pH 7.2) and lyzed in ice-cold digitonin buffer [1.5% (w/v) digitonin, 20 mM Tri–HCl pH 7.4, 0.1 mM EDTA, 50 mM NaCl, 10% (v/v) glycerol and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. Soluble material was analyzed by BN–PAGE, sucrose gradient centrifugation or subjected to affinity chromatography. To deplete ATP, mitochondria were pretreated with 20 µM oligomycin and 10 U/ml apyrase prior to incubation with b2Δ-DHFR in the presence of 2 mM NADH.
For the two-step import assay (chase experiment), the b2Δ-DHFR precursor was incubated in the presence of 30 µM NADPH and 30 µM DHF in import buffer for 10 min at 25°C prior to addition of mitochondria. The import reaction was performed for 12 min at 25°C. Mitochondria were reisolated and resuspended in import buffer. The chase reaction was performed at 25°C and stopped on ice. The samples were analyzed by immunodecoration following BN–PAGE or SDS–PAGE.
For purification of the supercomplex, Tim23ProtA or Tom22His10 mitochondria (250 µg protein) were incubated with purified b2Δ-DHFR in the presence of 5 µM MTX for 15 min at 25°C. For control reactions, the preprotein was omitted in the import reaction. The mitochondria were reisolated and solubilized with digitonin. The mitochondrial extract (250 µl) was subjected to a clarifying spin at 14 000 g for 15 min. For isolation via Tim23ProtA, the supernatant was incubated for 2 h at 4°C with IgG–Sepharose and then washed with 30 vol. of wash buffer (20 mM Tris–HCl pH 7.4, 1% digitonin, 0.1 mM EDTA, 200 mM NaCl, 10% glycerol and 1 mM PMSF). Proteins were eluted with SDS sample buffer (Geissler et al., 2002). For isolation via Tom22His10, the supernatant was incubated with 0.5 ml Ni-NTA agarose for 30 min on ice in the presence of 5 mM imidazole. After washing with increasing amounts of imidazole (20–50 mM) in 30 vol. wash buffer (20 mM Tris–HCl pH 7.4, 0.2% digitonin, 0.1 mM EDTA, 200 mM NaCl, 10% glycerol and 1 mM PMSF), bound proteins were eluted twice with 200 mM imidazole (Meisinger et al., 2001; Geissler et al., 2002). The samples were separated on SDS–PAGE, blotted onto polyvinylidene fluoride (PVDF) membranes and immunodecorated with various antisera.
BN–PAGE and two-dimensional gel electrophoresis
BN–PAGE was performed essentially as described previously (Schägger and von Jagow, 1991; Dekker et al., 1997). Mitochondrial proteins (70 µg) were solubilized in 50 µl ice-cold 1.5% digitonin-containing buffer and resolved on a 6–13% gradient gel at 4°C. Protein complexes were transferred to PVDF membranes and immunodecorated with specific antibodies. For detection of the TOM–TIM supercomplex, an antibody directed against the N-terminal portion of Tim23 was used in all experiments with the exception of the tim23-3 mutant. In this case, a DHFR-specific antiserum was applied. The High Molecular Weight Calibration Kit for native electrophoresis (Amersham) was used as a molecular weight standard.
For the two-dimensional analysis, mitochondria (100 µg protein) with accumulated b2Δ-DHFR and control mitochondria were lyzed in 70 µl 1.5% digitonin-containing buffer and separated on 6–13% gradient gel. Gel lanes were excised and applied on top of an SDS-polyacrylamide gel. After separation, proteins were blotted onto PVDF membranes and detected by immunodecoration.
Sucrose gradient analysis of the TOM–TIM supercomplex
Linear sucrose gradients were prepared from a solution of 25% (w/v) sucrose, 0.4% (w/v) digitonin, 20 mM Tris–HCl pH 7.4, 50 mM NaCl, 1 mM PMSF and Complete Protease Inhibitor Cocktail (Boehringer) by freezing/thawing at –20°C/room temperature. Mitochondria (1 mg protein) containing accumulated b2Δ-DHFR and control mitochondria were solubilized in 600 µl 1.5% digitonin-containing buffer, loaded onto the top of the gradient and centrifuged for 19 h at 210 000 g. Molecular weight markers (BSA, 66 kDa; bovine heart lactate dehydrogenase, 140 kDa; bovine liver catalase, 232 kDa; porcine thyroid thyroglobulin, 669 kDa; Amersham) were solubilized in 1.5% digitonin-containing buffer and run in a parallel gradient. The collected fractions were precipitated with TCA and separated on SDS–PAGE, followed by immunodecoration.
Miscellaneous
b2Δ-DHFR was expressed and purified as published previously (Dekker et al., 1997). In some figures, non-relevant gel lanes were excised by digital treatment. SDS–PAGE and immunodecoration, using the ECL system (Amersham), were performed according to standard procedures. Quantitation was performed by scanning densitometry or digital autoradiography.
Acknowledgments
Acknowledgements
We thank Dr Rob Jensen for anti-Tim17 antiserum, and Drs Andreas Geissler, Thomas Krimmer and Michiel Meijer for discussion and experimental advice. This work was supported by the Deutsche Forschungsgemeinschaft, Sonderforschungsbereich 388, Max Planck Research Award/Alexander von Humboldt Foundation and the Fonds der Chemischen Industrie/BMBF. A.C. was a recipient of a FEBS long-term fellowship.
References
- Abe Y., Shodai,T., Muto,T., Mihara,K., Torii,H., Nishikawa,S., Endo,T. and Kohda,D. (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell, 100, 551–560. [DOI] [PubMed] [Google Scholar]
- Arnold I., Pfeiffer,K., Neupert,W., Stuart,R.A. and Schägger,H. (1998) Yeast mitochondrial F1F0-ATP synthase exists as a dimer: identification of three dimer-specific subunits. EMBO J., 17, 7170–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer M.F., Sirrenberg,C., Neupert,W. and Brunner,M. (1996) Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell, 87, 33–41. [DOI] [PubMed] [Google Scholar]
- Bauer M.F., Hofmann,S., Neupert,W. and Brunner,M. (2000) Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol., 10, 25–31. [DOI] [PubMed] [Google Scholar]
- Bolliger L., Junne,T., Schatz,G. and Lithgow,T. (1995) Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J., 14, 6318–6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bömer U., Meijer,M., Maarse,A.C., Hönlinger,A., Dekker,P.J.T., Pfanner,N. and Rassow,J. (1997) Multiple interactions of components mediating preprotein translocation across the inner membrane. EMBO J., 16, 2205–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brix J., Dietmeier,K. and Pfanner,N. (1997) Differential recognition of preproteins by the purified cytosolic domains of the mitochondrial import receptors Tom20, Tom22 and Tom70. J. Biol. Chem., 272, 20730–20735. [DOI] [PubMed] [Google Scholar]
- Court D.A., Nargang,F.E., Steiner,H., Hodger,R.S., Neupert,W. and Lill,R. (1996) Role of the intermembrane-space domain of the preprotein receptor Tom22 in protein import into mitochondria. Mol. Cell Biol., 16, 4035–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker P.J., Martin,F., Maarse,A.C., Bömer,U., Müller,H., Guiard,B., Meijer,M., Rassow,J. and Pfanner,N. (1997) The Tim core complex defines the number of mitochondrial translocation contact sites and can hold arrested preproteins in the absence of matrix Hsp70-Tim44. EMBO J., 16, 5408–5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donzeau M., Kaldi,K., Adam,A., Paschen,S., Wanner,G., Guiard,B., Bauer,M.F., Neupert,W. and Brunner,M. (2000) Tim23 links the inner and outer mitochondrial membranes. Cell, 101, 401–412. [DOI] [PubMed] [Google Scholar]
- Endo T. and Kohda,D. (2002) Functions of outer membrane receptors in mitochondrial protein import. Biochim. Biophys Acta, 1592, 3–14. [DOI] [PubMed] [Google Scholar]
- Gambill B.D., Voos,W., Kang,P.J., Miao,B., Langer,T., Craig,E.A. and Pfanner,N. (1993) A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol., 123, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A. et al. (2002) The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell, 111, 507–518. [DOI] [PubMed] [Google Scholar]
- Hill K., Model,K., Ryan,M.T., Dietmeier,K., Martin,F., Wagner,R. and Pfanner,N. (1998) Tom 40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature, 395, 516–521. [DOI] [PubMed] [Google Scholar]
- Horst M., Hilfiker-Rothenfluh,S., Oppliger,W. and Schatz,G. (1995) Dynamic interaction of the protein translocation systems in the inner and outer membranes of yeast mitochondria. EMBO J., 14, 2293–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jascur T., Goldenberg,D.P., Vestweber,D. and Schatz,G. (1992) Sequential translocation of an artificial precursor protein across the two mitochondrial membranes. J. Biol. Chem., 267, 13636–13641. [PubMed] [Google Scholar]
- Jensen R.E. and Johnson,A.E. (2001) Opening the door to mitochondrial protein import. Nat. Struct. Biol., 8, 1008–1010. [DOI] [PubMed] [Google Scholar]
- Kanamori T., Nishikawa,S., Shin,I., Schultz,P.G. and Endo,T. (1997) Probing the environment along the protein import pathways in yeast mitochondria by site-specific photocrosslinking. Proc. Natl Acad. Sci. USA, 94, 485–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori T., Nishikawa,S., Nakai,M., Shin,I., Schultz,P.G. and Endo,T. (1999) Uncoupling of transfer of the presequence and unfolding of the mature domain in precursor translocation across the mitochondrial outer membrane. Proc. Natl Acad. Sci. USA, 96, 3634–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler C.M., Merchant,S. and Schatz,G. (1999) How membrane proteins travel across the mitochondrial intermembrane space. Trends Biochem. Sci., 24, 428–432. [DOI] [PubMed] [Google Scholar]
- Koll H., Guiard,B., Rassow,J., Ostermann,J., Horwich,A.L., Neupert,W. and Hartl,F.U. (1992) Antifolding activity of hsp60 couples protein import into the mitochondrial matrix with export to the intermembrane space. Cell, 68, 1163–1175. [DOI] [PubMed] [Google Scholar]
- Komiya T., Rospert,S., Koehler,C., Looser,R., Schatz,G. and Mihara,K. (1998) Interaction of mitochondrial targeting signals with acidic receptor domains along the protein import pathway: evidence for the ‘acid chain’ hypothesis. EMBO J., 17, 3886–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künkele K.P. et al. (1998) The preprotein translocation channel of the outer membrane of mitochondria. Cell, 93, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Meisinger C. et al. (2001) Protein import channel of the outer mitochondrial membrane: a highly stable Tom40–Tom22 core structure differentially interacts with preproteins, small Tom proteins and import receptors. Mol. Cell. Biol., 21, 2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczko M., Bömer,U., Kübrich,M., Zufall,N., Hönlinger,A. and Pfanner,N. (1997) The intermembrane space domain of mitochondrial Tom22 functions as a trans binding site for preproteins with N-terminal targeting sequences. Mol. Cell. Biol., 17, 6574–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D., Paschen,S.A., Kozany,C., Prokisch,H., Hoppins,S.C., Nargang,F.E., Neupert,W. and Hell,K. (2003) Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J., 22, 816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanner N. and Chacinska,A. (2002) The mitochondrial import machinery: preprotein-conducting channels with binding sites for presequences. Biochim. Biophys Acta, 1592, 15–24. [DOI] [PubMed] [Google Scholar]
- Rassow J., Guiard,B., Wienhues,U., Herzog,V., Hartl,F.U. and Neupert,W. (1989) Translocation arrest by reversible folding of a precursor protein imported into mitochondria: a means to quantitate translocation contact sites. J. Cell Biol., 109, 1421–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J., Hartl,F.U., Guiard,B., Pfanner,N. and Neupert,W. (1990) Polypeptides traverse the mitochondrial envelope in an extended state. FEBS Lett., 275, 190–194. [DOI] [PubMed] [Google Scholar]
- Ryan K.R., Leung,R.S. and Jensen,R.E. (1998) Characterization of the mitochondrial inner membrane translocase complex: the Tim23p hydrophobic domain interacts with Tim17p but not with other Tim23p molecules. Mol. Cell. Biol., 18, 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M.T., Voos,W. and Pfanner,N. (2001) Assaying protein import into mitochondria. Methods Cell Biol., 65, 189–215. [DOI] [PubMed] [Google Scholar]
- Schägger H. and von Jagow,G. (1991) Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem., 199, 223–231. [DOI] [PubMed] [Google Scholar]
- Schleyer M. and Neupert,W. (1985) Transport of proteins into mitochondria: translocational intermediates spanning contact sites between outer and inner membranes. Cell, 43, 339–350. [DOI] [PubMed] [Google Scholar]
- Schülke N., Sepuri,N.B. and Pain,D. (1997) In vivo zippering of inner and outer mitochondrial membranes by a stable translocation intermediate. Proc. Natl Acad. Sci. USA, 94, 7314–7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schülke N., Sepuri,N.B., Gordon,D.M., Saxena,S., Dancis,A. and Pain,D. (1999) A multisubunit complex of outer and inner mitochondrial membrane protein translocases stabilized in vivo by translocation intermediates. J. Biol. Chem., 274, 22847–22854. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg C., Endres,M., Becker,K., Bauer,M.F., Walther,E., Neupert,W. and Brunner,M. (1997) Functional cooperation and stoichiometry of protein translocases of the outer and inner membranes of mitochondria. J. Biol. Chem., 272, 29963–29966. [DOI] [PubMed] [Google Scholar]
- Truscott K.N., Kovermann,P., Geissler,A., Merlin,A., Meijer,M., Driessen,A.J., Rassow,J., Pfanner,N. and Wagner,R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol., 8, 1074–1082. [DOI] [PubMed] [Google Scholar]
- van Wilpe S. et al. (1999) Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature, 401, 485–489. [DOI] [PubMed] [Google Scholar]
- Wienhues U., Becker,K., Schleyer,M., Guiard,B., Tropschug,M., Horwich,A.L., Pfanner,N. and Neupert,W. (1991) Protein folding causes an arrest of preprotein translocation into mitochondria in vivo. J. Cell Biol., 115, 1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Esaki,M., Kanamori,T., Tamura,Y., Nishikawa,S. and Endo,T. (2002) Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes. Cell, 111, 519–528. [DOI] [PubMed] [Google Scholar]