Abstract
The NF-κB transcription factors consist of dimeric proteins of the Rel homology family. They activate many promoters containing highly divergent κB-site sequences. We have generated cell lines lacking individual and multiple NF-κB proteins and used them to establish interactions between components of the NF-κB–IκB signaling system. Functional compensation within the family of dimers was evident in knockout cell lines. Analysis of transiently transfected genes gave an impression of promiscuity that was not borne out by analysis of endogenous genes. Using TNFα as an inducer, a panel of endogenous genes showed a wide range of subunit specificities as well as highly variable kinetics of induction. Comparing the function and subunit specificity of genes with the sequence of the κB DNA-binding site we found little correlation, indicating that NF-κB family member specificity for endogenous promoters is not solely encoded by the κB site sequence itself.
Keywords: compensation/NF-κB/Rel/specificity/transcription
Introduction
Cell signaling molecules such as transcription factors are encoded in gene families whose members have distinct yet overlapping functions. The fact that increasingly complex genomes contain a greater number of gene family members rather than novel families (Lander et al., 2001) suggests that diversity within a gene family may provide for specificity and versatility in the regulation of cellular responses. Characterizing the specific functions of the members of transcription factor gene families contributes to our understanding of gene regulatory networks and to the development of specific therapeutic strategies. For example, biochemical studies of the nuclear hormone receptor family have shown that the spacing between two tandem cognate DNA half-sites determines which family members bind and function (Rastinejad, 2001). Similarly within the large zinc finger transcription factor family, biochemical and biophysical characterization of DNA–protein interactions are the basis for synthetic transcription factor engineering (Wolfe et al., 2000). However, for many other transcription factor families, such as the C/EBP (Ramji and Foka, 2002), E2F (Trimarchi and Lees, 2002), AP-1 (Chinenov and Kerppola, 2001; Mechta-Grigoriou et al., 2001) and NF-κB families, marked DNA-binding specificity rules have not been identified. Indeed, the raison d’être of multiple members within signal transduction protein families may not be molecular interaction specificities but could be found in their own differential regulation. Considering the immediate regulators of NF-κB activity, the IκB protein family, for example, the difference between family members that is of primary physiological importance appears to be regulation of synthesis, as demonstrated by the fact that IκBα knockout lethality was rescued by placing IκBβ under the control of the IκBα promoter (Cheng et al., 1998). A quantitative analysis of the signaling characteristics of IκB family members has supported that conclusion (Hoffmann et al., 2002).
The NF-κB (also called NF-κB/Rel) family of dimeric transcription factors mediates cellular responses to a wide variety of different stimuli by regulating the expression of a large number of genes of highly diverse functions (Pahl, 1999). Mouse knockout studies have revealed this pleiotropic signal transducer to be involved in inflammatory and stress responses, the control of the cell cycle, apoptosis, growth and proliferation, cell–cell communication, lymphocyte development and maturation, and neuronal learning (Gerondakis et al., 1999; Meffert et al., 2003). The functional transcription factors consist of pairs taken from the NF-κB protein family (p50, p52, p65, cRel, RelB), proteins that utilize the Rel homology domain (RHD) for DNA binding and dimerization. Their cognate DNA-binding element, the κB-site, is found in the promoters of NF-κB responsive genes but displays a remarkably loose consensus, often described as G–5G–4G–3R–2N–1N0Y+1Y+1C+2C+3 (Ghosh et al., 1998). The heterogeneity of κB-sites has been thought to confer specificity of regulation mediated by differential affinities of NF-κB isoforms. Purified recombinant RHD protein dimers could indeed be shown to select differential optimal DNA-binding motifs from a pool of random oligonucleotides (Kunsch et al., 1992). Similarly, one study with transfected promoter–reporter constructs showed that multimerized κB-sites from one of three different promoters (Igκ, MHC H2 and IFN-β) exhibited differential responses when co-transfected with p50 and/or p65 expression plasmids (Fujita et al., 1992).
However, more recent structural studies did not reveal evidence for highly sequence-specific DNA–NF-κB interactions, with base-specific contacts within the p50 homodimer structure involving primarily 5′-G–5G–4G–3 (Ghosh et al., 1995; Muller et al., 1995) and contacts within the p65 homodimer structure involving primarily 5′-G–4G–3 (Chen et al., 1998). In fact, these studies have not revealed sufficient DNA base contacts to rationalize κB-site sequence-specific binding. Though homodimers of p65 or cRel require only a 9 bp sequence for binding, this may be contained within certain 10 bp consensus-conforming κB-sites. X-ray structures of the p50:p65 heterodimer bound to different κB-sites, such as those derived from the immunoglobulin and HIV enhancers G–5G–4G–3A–2C–1T0T+1T+2C+3C+4 (Chen et al., 1998), the IFN-β enhancer G–5G–4G–3A–2A-1A0T+1T+2C+3C+4 (Berkowitz et al., 2002; Escalante et al., 2002), and the urokinase plasminogen activator gene promoter G–5G–4G–3A–2A-1A0G+1T+2A+3C+4 (Chen-Park et al., 2002) demonstrate that NF-κB binds a variety of sequences. Furthermore, the two RHDs contained in each dimer constituent relate to each other differently in different structures and lead to different degrees of DNA bending (Chen and Ghosh, 1999), emphasizing the remarkable permissiveness of NF-κB–DNA interactions.
Despite the difficulty in discerning specificity at the level of DNA–protein interactions, mice deficient in a single RHD protein show specific phenotypes. rela–/– animals are embryonic lethal with massive hepatocyte apoptosis (Beg et al., 1995), while crel–/– mice develop normally but have defects in lymphocyte proliferation (Kontgen et al., 1995). In part, family member-specific functions can be inferred by tissue-specific expression. While p65 appears to be ubiquitous, cRel is expressed constitutively primarily in mature monocytic and lymphocytic lineages (Liou et al., 1994), while in others, such as fibroblasts, only following stimulation (Grumont and Gerondakis, 1990). In addition, there are differences in the molecular characteristics of RHD family members: transfection studies have identified activation domains in p65 and cRel, but not in p50 and p52 (Ghosh et al., 1998). DNA-binding assays were used to identify dimerization rules: p50 (and presumably p52) can partner with all RHD family members including themselves, while RelB and cRel do not appear to engage in homotypic interactions. RelB, furthermore, does not appear to dimerize with p65 or cRel. Little is known about how the results from in vitro biochemical characterizations of RHD proteins can serve to explain physiologically relevant functional specificity in gene regulation, because selectivity of endogenous κB-site containing promoters remains largely unexplored. In fact, in the case of the IL-12 promoter, results from cell-free or extra-chromosomal templates are shown not to recapitulate endogenous promoter specificity (Sanjabi et al., 2000).
Genetic approaches are suitable to address questions of in vivo mechanistic specificity, but must take into account the possibility of interdependent regulation of factors in mammalian signaling networks; resulting compensatory mechanisms among functionally related molecules may lend increased robustness to signaling systems and affect the phenotype of genetic deletions. Here, we have undertaken a genetic analysis of RHD proteins with respect to NF-κB-dependent gene activation by creating a panel of single and double knockout cell lines. We have biochemically characterized NF-κB activation within them, and measured the TNFα responsiveness of a diverse set of NF-κB target genes. Our results reveal that RHD protein family members exhibit differential target gene specificities and demonstrate that a combination of genetic and biochemical analyses can be applied to decode functional specificity rules of gene promoters in mammalian cells.
Results
Cross-regulation and compensation within the NF-κB–IκB regulatory system
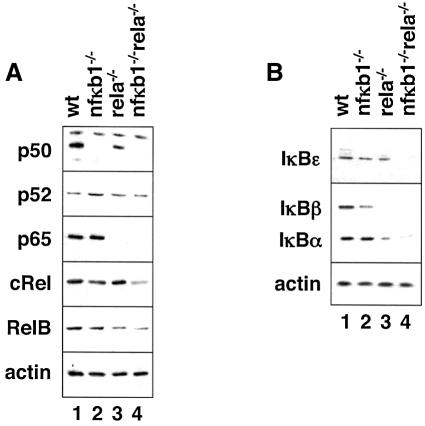
The dominant κB-binding activity induced by TNFα in fibroblasts is composed of p50 and p65, the mature gene products of the nfκb1 and rela genes. Microarray studies with nfκb1–/–rela–/– doubly deficient cells have confirmed their key role in activating most TNFα-responsive genes; fibroblasts lacking both factors show no activation of genes controlled by NF-κB (A.Hoffmann and D.Baltimore, in preparation). To address the specific roles of p50 and p65 in NF-κB-dependent gene expression, we generated fibroblast cell lines from nfκb1–/– (Sha et al., 1995), rela–/– (Beg et al., 1995) and nfκb1–/–rela–/– embryos (Horwitz et al., 1997). Western blots confirmed that p50 and p65 proteins were absent in respective knockout cell lines (Figure 1A), while other RHD proteins remained, though at somewhat altered levels. In particular, p52 levels were enhanced in nfκb1–/– cells, cRel levels were increased in rela–/– cells, and cRel as well as RelB levels were somewhat reduced in doubly deficient cells. Strikingly, rela–/– cells contained markedly reduced levels of IκBα and IκBβ proteins, while all three IκB proteins were reduced to almost undetectable levels in doubly deficient cells (Figure 1B).
Fig. 1. RHD proteins in NF-κB knockout cells. Western blots of cells derived from wild-type, nfκb1–/–, rela–/– and nfκb1–/–/rela–/– mouse embryos with antibodies directed against p50, p52, p65, cRel, RelB and actin (A), and IκBε, -β, -α and actin (B) as indicated. Specific bands are shown, but additional bands were detectable with some antibodies.
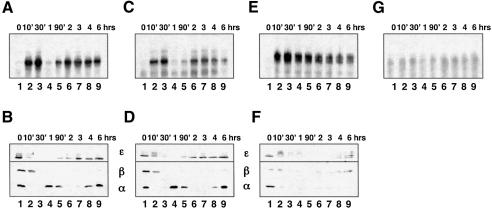
To investigate the result of genetic knockouts within the RHD family on the activated NF-κB transcription factor, we examined the induction of nuclear κB-binding activity in response to TNFα stimulation in wild-type and mutant cell lines (Figure 2). Gel shift and western blot analyses of the nuclear and cytoplasmic compartments of wild-type cells revealed a characteristic biphasic induction of nuclear NF-κB (Figure 2A) that results from the coordinated degradation and synthesis of IκB family members (Figure 2B). Cells lacking p50 showed only a marginal reduction in κB-binding activity (Figure 2C) with no alteration in the temporal regulation. In p65-deficient cells, however, induction produced two distinct κB-binding activities that did not undergo post-induction repression (Figure 2E), correlating with defective IκBα protein synthesis (Figure 2F). Finally, no significant κB-binding activity was induced at any time point within a 6 h TNFα time course in doubly deficient cells (Figure 2G). These results confirm that p50 and p65 make up the major constituents of NF-κB in fibroblasts. However, neither p50 nor p65 single knockout cells were deficient in κB-binding activity. In addition, the temporal regulation of nuclear NF-κB is altered in rela–/– cells.
Fig. 2. TNFα induction of κB-binding activity in NF-κB knockout cells. EMSA was used to monitor nuclear NF-κB in wild-type (A), nfκb1–/– (C), rela–/– (E) and nfκb1–/–rela–/– (G) cells in the indicated (in min and h) time course following the onset of TNFα stimulation. The cytoplasmic portion from wild-type (B), nfκb1–/– (D), relA–/– (F) was probed in western blots to monitor concurrent degradation and synthesis of IκB proteins, IκBα and IκBβ (bottom panels) and IκBε (top panels).
Compensation and specificity on non-chromosomal promoter DNA
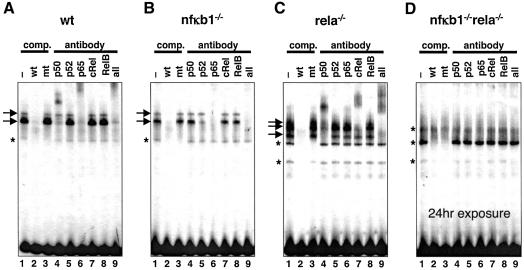
To characterize the molecular composition of induced κB-binding complexes, we employed antibodies specific for each Rel protein in gel mobility supershift analyses of nuclear extracts made after 30 min of TNFα stimulation. As expected, nuclear κB-binding activity in wild-type cells (Figure 3A) was sensitive to antibodies specific to p50 and p65; indeed distinct complexes comprising dimers of p65 (upper arrow), p50:p65 (lower arrow) and p50 (data not shown) were identified. The lower complex (indicated by an asterisk) appeared to be non-specific as revealed by competition analysis with wild-type and mutant double-stranded oligonucleotides (lanes 2 and 3 in each panel). Not surprisingly, complexes apparent at long exposure in extracts from nfκb1–/–rela–/– double knockout cells (Figure 3D) were not recognized by any of the NF-κB family antibodies and were not specifically competed by double-stranded oligonucleotides.
Fig. 3. Molecular composition of κB-binding activity in NF-κB knockout cells. Immediate early protein–DNA complexes induced by TNFα in wild-type (A), nfκb1–/– (B), rela–/– (C) and nfκb1–/–rela–/– (D) cells and detected by EMSA in Figure 2 are indicated by arrows in lanes 1, while constitutive complexes are indicated by asterisks. They are specific for κB-sites as shown by competition with double-stranded wild-type and mutant oligonucleotides (lanes 2 and 3) and are probed with antibodies directed against the indicated RHD proteins (lanes 4–9). This results in ‘supershift’ or ablation of the characteristic protein–DNA complex. We conclude that the κB-binding activity in wild-type cells consists of p50:p65 heterodimer and p65 homodimer, in nfκb1–/– cells p52:p65 heterodimer and p65 homodimer, in rela–/– cells p50 homodimer, cRel:p50 heterodimer, and most likely, but not unambiguously cRel homodimer. While experiments shown in panels A, B and C were exposed to film for 6 h, panel D shows a 24 h exposure.
Turning to single knockouts, anti-p50 antibodies, as expected, did not recognize κB-binding complexes in extracts derived from nfκb1–/– cells (Figure 3B), nor did anti-p65 antibodies recognize κB-binding complexes derived from rela–/– cells (Figure 3C). Instead, antibodies directed against p52 ablated the nfκb1–/– complex of the same mobility as the p50:p65 complex found in wild-type cells. Similarly, antibodies directed against cRel supershifted the analogous rela–/– κB-binding complex, as well as a much weaker, slower migrating complex that may correspond to a cRel homodimer. These results strikingly revealed molecular compensation within the NF-κB/Rel family; however, not every family member functioned interchangeably with one another. Molecular compensation rules appear to reflect previously noted homology relationships, and the results from molecular characterization studies that inferred structural and functional similarities between p50 and p52, and p65 and cRel proteins (Ghosh et al., 1998).
These results suggest multiple mechanisms of cross-regulation between the RHD and IκB protein family members. One known mechanism is transcriptional feedback: cRel (Grumont et al., 1993), RelB (Bren et al., 2001) and IκBα (Scott et al., 1993) are known NF-κB target genes, which presumably explains observed decreases in protein levels in p50 and p65 doubly deficient cells, as well as the absence of post-induction repression of nuclear NF-κB in rela–/– cells. Another cross-regulation mechanism may be based on the protein stability differential between uncomplexed and complexed polypeptides; indeed reduced IκB protein levels in mutant cells may be the result of the loss of NF-κB dimers available for complex formation. Similar reasoning might explain the increased levels of p52 and cRel-containing complexes in nfκb1–/– and rela–/– cells by respective substitution in dimer formation.
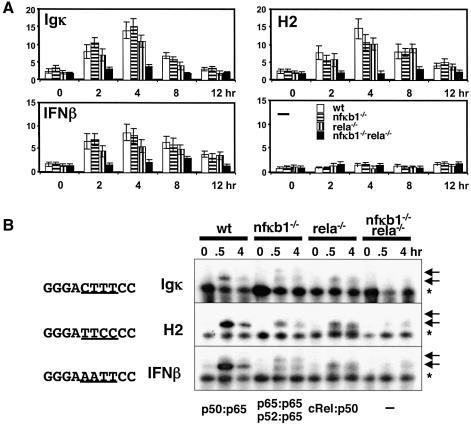
To address the transcriptional activity of p50- or p65-deficient NF-κB complexes, we utilized transient transfections with promoter–reporter constructs driven by multimerized κB-sites. Each construct contained κB-binding site sequences taken from one of three different promoters: Igκ, MHC H2 or IFN-β. These constructs had previously been shown to have differential responses to co-transfected p50 and p65 expression plasmids (Fujita et al., 1992). We observed significant transcriptional activation of κB-site-containing reporters in wild-type fibroblasts following TNFα stimulation (Figure 4A, white bars). Such activation was dependent on the presence of κB-sites (lower right panel) and was minimal in cells deficient in both p50 and p65 (black bars). However, fibroblasts lacking only one family member, either p50 (horizontally striped bars) or p65 (vertically striped bars), still showed significant levels of activation, with some reduction observed (about half) in rela–/– cells. Surprisingly, the results were similar for all three promoter constructs. Those κB-sites also revealed little difference in NF-κB complexes when employed in a gel shift assay with nuclear extracts from the above-described TNFα-stimulated mutant cells lines. Each probe revealed p50:p65 complexes in wild-type cells, p52:p65 complexes in nfκb1–/– cells, and p50:cRel complexes in rela–/– cells (Figure 4B; data not shown). However, we note that the H2 κB-probe does not show p65 homodimer complexes in nfκb1–/– cells and appears to impart somewhat lower transcriptional activity than the other κB-sites.
Fig. 4. Compensation on different κB-site sequences in extra-chromosomal plasmids. Reporter plasmids driven by the c-fos core promoter alone or fused to two κB-sites derived from the Igκ the MHC H2 and IFNβ promoter, as indicated in (A), were assayed in response to TNFα stimulation for the indicated time in wild-type (empty bars), nfκb1–/– (horizontally striped bars), rela–/– (vertically striped bars) and nfκb1–/–rela–/– (black bars). EMSA (B) was used to monitor κB-binding activity in TNFα-stimulated NF-κB knockout cells with indicated κB-site containing double-stranded oligonucleotide probes. Induced, specific bands are indicated by arrows, a prominent constitutive protein–DNA complex is indicated by an asterisk.
The above-described analyses indicate that molecular compensation occurs within the RHD protein family and results in functional compensation on extra-chromosomal templates; the 10 bp κB-sites did not reveal much specificity for NF-κB isoforms, although certain sequences may certainly provide a restriction with respect to the entire panel of possible dimers.
We next turned to the regulation of endogenous genes to determine whether they exhibit similar promiscuity. However, compensation evident in single knockouts required the inclusion of fibroblast cell lines that are nfκb1–/–nfκb2–/– and rela–/–crel–/– doubly deficient, as well as single knockout controls derived from nfκb2–/– and crel–/– embryos. For example, to address the function of p65 in wild-type cells, rela–/–crel–/– cells are an informative genotype because the results will not be affected by cRel compensation for the loss of p65. In turn, we can address the question of molecular specificity of RHD proteins in transcriptional activation by determining whether molecular compensation by cRel results in functional compensation on endogenous promoters.
Systematic genetic analysis of a transcription factor family
To undertake such a study, we interbred NF-κB knockout mouse strains and derived 3T3 fibroblast lines that were doubly deficient in p50 and p52 or p65 and cRel. We also derived control lines from nfκb2–/–(Caamano et al., 1998) and crel–/– (Kontgen et al., 1995) embryos. Gel mobility shift assays were then used to detect κB-binding complexes in extracts made from each of these cell lines following TNFα stimulation (data not shown); their molecular composition was determined by employing the previously used panel of antibodies. As summarized in Table I, each mutant cell line showed a characteristic set of TNFα-inducible RHD proteins that gave rise to particular κB-binding activities. Therefore, the systematic panel of mutants allowed us to distinguish between NF-κB-dependent promoters that strictly required a particular RHD protein for activation and those promoters that had a broader requirement for one or the other RHD protein subclass. Specifically, we could distinguish genes that require p50 for TNF-induced activation, those on which either p50 or p52 must function, and those that do not require either protein for NF-κB-dependent activation. Similarly, those same genes could be further classified according to their requirement for p65 or cRel.
Table I. RHD proteins induced by TNFα stimulation in NF-κB knockout cells.
| MEFs genotype |
κB-binding proteins |
κB-binding complexes in EMSA |
|||||
|---|---|---|---|---|---|---|---|
| p50 | p52 | p65 | cRel | ||||
| wt | + | – | + | – | p50:p50a | p50:p65 | p65:p65a |
| nfκb1–/– | – | + | + | – | p52:p65 | p65:p65a | |
| nfκb1–/–nfκb2–/– | – | – | + | – | p65:p65 | ||
| nfκb2–/– | + | – | + | – | p50:p50a | p50:p65 | p65:p65a |
| rela–/– | + | – | – | + | p50:p50 | p50:cRel | cRel:cRel |
| rela–/–crel–/– | + | – | – | – | p50:p50 | ||
| crel–/– | + | – | + | – | p50:p50a | p50:p65 | p65:p65a |
| nfκb1–/–rela–/– | – | – | – | – | – | ||
Fibroblast cell lines derived from embryos of indicated genotypes (column 1) exhibit TNFα-inducible κB-binding activity that consists of indicated RHD proteins (column 2). These proteins form κB-binding complexes indicated in column 3. All data are based on EMSA with κB-site-containing probes and antibodies directed against specific RHD proteins.
aComplexes detected with only a subset of κB-site-containing probes.
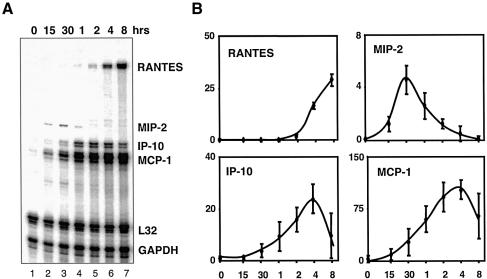
For a quantitative, yet high throughput analysis of endogenous gene expression, we used a multiplex RNase protection assay (RPA) that allows independent monitoring of up to 10 different messenger RNA transcripts (Figure 5A). Two housekeeping genes, GAPDH and L32, allowed for normalization such that consistent expression units can be used to compare different experiments. Multiple data sets from four different wild-type cell lines demonstrated reproducibility, which therefore allows comparison of datasets from different cell lines bearing different mutations. Interestingly, a detailed time course revealed that different genes have widely different kinetics of activation. For example, our panel of genes included several chemokine genes (Figure 5B). After TNFα stimulation, MIP-2 mRNA levels increase as early as 15 min, reaching a peak at 1 h and disappearing by 2 h. On the hand, RANTES transcripts are not detectable until 2 h post-stimulation and continue to rise beyond 8 h. Finally, MCP-1 and IP-10 display an intermediary profile. The dynamic nature of NF-κB activation emphasizes the importance of conducting gene expression assays in detailed time courses.
Fig. 5. Quantitative monitoring of gene expression by multiplex RPA. Multiple double-stranded RNA products indicative of mRNA transcripts derived from indicated NF-κB-dependent and housekeeping genes following TNFα stimulation of wild-type fibroblasts resolved by electrophoresis (A). Three independent experiments with independently derived wild-type 3T3 cell lines were quantitated by PhosphoImager and mRNA abundance was graphed for the indicated chemokine genes in arbitrary units relative to housekeeping genes L32 and GAPDH (B).
Differential NF-κB/Rel protein requirements
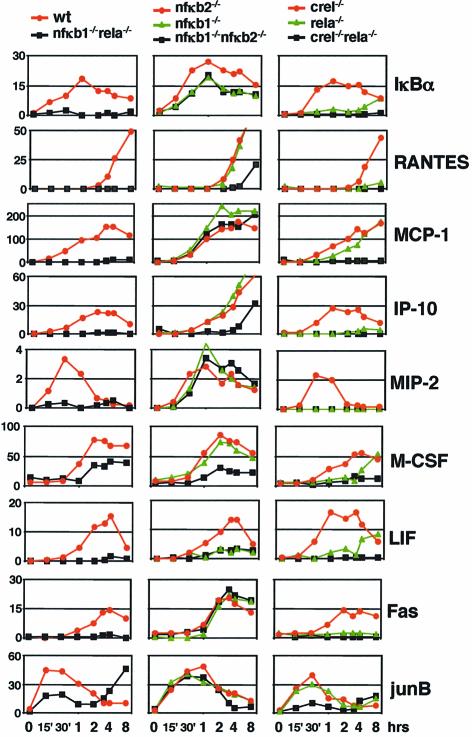
We focused our attention on nine genes that represent diverse classes of NF-κB responsive genes, are induced in fibroblasts by TNFα stimulation, and the mRNA levels of which could be monitored reliably by RPA. These genes were: IκBα; aforementioned chemokine genes RANTES, IP10, MCP-1 and MIP-2; the immune regulatory factor LIF; the growth factor M-CSF; transcription factor junB; and apoptosis-inducing TNF receptor family member Fas. Quantitative RPA results for each gene are summarized in three graphs which group related genotypes (Figure 6). Graphs in the left column demonstrate NF-κB dependence because cells lacking both p50 and p65 have no NF-κB-mediated activity; those in the center column address dependence on p50 and/or p52 proteins, and those in the right column address dependence on p65 and/or cRel proteins. An eight point time course extending to 8 h following the onset of TNFα stimulation revealed diverse kinetics in gene activation. In each case, transcriptional induction proved to be protein synthesis-independent but cycloheximide did affect the transcript levels of some genes at time points following initial activation (A.Hoffmann, unpublished results). TNFα activation of all genes proved to be NF-κB-dependent as determined with cells lacking both p50 and p65 (left column). Two genes, M-CSF and junB, showed residual induction in the absence of NF-κB, but the NF-κB contribution to their activation was found to be reproducible and analyzable with respect to NF-κB protein requirement (see below). Interestingly, junB transcription was found to be highly induced in NF-κB knockouts at the time when these cells undergo widespread TNFα-induced apoptosis, indicating that this gene is subject to additional transcriptional regulation mechanisms independent of NF-κB.
Fig. 6. Transcriptional induction by TNFα of NF-κB target genes in NF-κB knockout cells. Results from a representative RPA experiment are graphed for the indicated genes in three panels. The left panel shows data from wild-type (red line) and nfκb1–/–rela–/– cells (black line), the center panel from nfκb2–/– (red), nfκb1–/– (green) and nfκb1–/–nfκb2–/– (black), and the right panel from crel–/– (red), rela–/– (green), and rela–/–crel–/– (black) cells.
Turning to p50 and p52 NF-κB family members, we expected no activation defects in nfκb2–/– fibroblasts, because our supershift analysis did not reveal any p52 protein in κB-binding complexes in TNFα-stimulated wild-type cells. As shown in the center column graphs, that is in fact the case; however, some genes, notably IP-10, appear to be super-induced at later time points in nfκb2–/– cells. Surprisingly, TNFα-induced gene expression is largely unaffected by p50 deficiency (green line) as well. In fact, only one gene in our collection, LIF, cannot be activated by TNFα stimulation in nfκb1–/– cells. The major reason for such a mild gene expression phenotype appears to be efficient functional compensation by p52, because the activation of several genes, such as RANTES, IP-10 and M-CSF, is defective in cells lacking both p50 and p52. Thus, NF-κB-dependent genes appear to fall into three categories: those requiring p50 for induction in TNFα-stimulated fibroblasts, those on whose promoters either p50 or p52 can efficiently function, and those whose TNFα induction can proceed without either of these two RHD proteins.
Transcript quantitation for these genes in TNFα-induced cells deficient in cRel, p65 or both is shown in the right column of Figure 6. Because cRel was not found in κB-binding complexes in wild-type fibroblasts, it is not surprising that none of the nine genes are significantly attenuated in crel–/– cells (red lines). In fact, most genes appear to be dramatically dependent on the p65 protein (green lines), although in some cases (e.g. IκBα, LIF, M-CSF) we saw significant transcript levels at the latest time points that are probably the cumulative result of residual transcriptional initiation activity. However, the chemokine MCP-1 and the transcription factor junB did not exhibit such a strict requirement for p65 because cRel could also function on these promoters (compare green and black lines). Importantly, we find that no NF-κB-dependent promoter can be activated in the absence of both p65 and cRel. This observation confirms that these two proteins contain the principal activation functions within κB-binding complexes, while p50 and p52 can be considered binding partners that have essential functions in a subset of gene induction events.
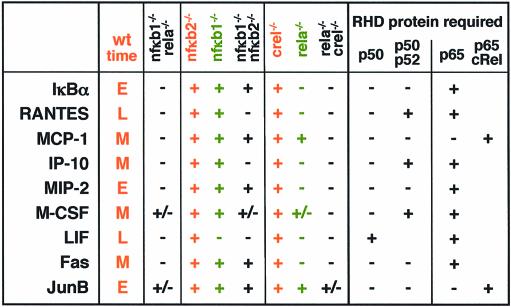
Our panel of knockout cell lines revealed that endogenous genes have remarkably different requirements for RHD proteins when induced by TNFα in fibroblasts. While none could be induced in cells lacking both p50 and p65, or p65 and cRel, the ability to be induced in nfκb1–/–, nfκb1–/–nfκb2–/– or rela–/– cells was gene-specific (Table II, left columns). Therefore, specific RHD protein requirements are determinable for every TNFα-induced gene in fibroblasts, as summarized for the group of nine NF-κB-dependent genes that were part of this study (Table II, right columns). Within this small sample of genes, the pattern of RHD protein requirement does not appear to correlate with the temporal control of gene expression (indicated in the second column) or the known or presumed function of the gene product; the six secreted immune response regulatory proteins represented here (RANTES, MCP-1, IP-10, MIP-2, M-CSF and LIF) cover diverse temporal kinetics as well as diverse combinations of RHD protein requirements.
Table II. Distinct RHD protein requirements for NF-κB-dependent genes.
Transcriptional regulation results are summarized for each of the ten genes (column 1) monitored: temporal regulation (column 2), expression in indicated knockout cells (columns 3–5) and deduced RHD protein requirement (columns 6 and 7).
Discussion
We have presented genetic evidence that mammalian gene activation requires not only specific transcription factors, but specific members of the RHD transcription factor family for the activation of endogenous NF-κB-responsive genes. However, in vitro binding assays with extracts from cells harboring deletions of specific family members reveal DNA binding characteristics that are sufficiently overlapping that functional specificity of family members cannot be reproduced on naked templates in transient transfections. In fact, in the absence of exogenously expressed family members, transiently transfected promoters appear to be remarkably promiscuous with regard to family member-specific requirements, although some κB-site sequences may be more specialized, restricting access to a subset of NF-κB dimers.
Specificity was revealed when taking a genetic approach to a mechanistic question. The specific RHD protein requirement for the activation of an endogenous promoter implies that a particular subset of all available RHD protein dimers (Table I) is capable of functioning on the promoter in question. Thus the RHD protein requirement (Table II) can be translated into the subset of RHD protein dimers that appear capable of mediating the induction of each gene in our study (Table III). For example, LIF induction by TNFα is abolished in both nfκb1–/– as well as rela–/– single knockout cells leading to the conclusion that of all possible RHD protein dimers only the p50:p65 heterodimer is functional on this promoter. In contrast, IP-10 (as well as M-CSF and RANTES) can be induced by dimers containing either p50 or p52, with p65 being a required partner. A representative of a third group is MIP-2; this chemokine (as well as IκBα and Fas) is induced by p65-containing complexes that may not only be heterodimers with p50 or p52, but can also be p65:p65 homodimers. MCP-1 and junB exhibited the least stringent RHD protein requirement with NF-κB/Rel complexes consisting of either p65 or cRel capable of functioning on these promoters. While cRel protein can partner with p50, p65 can function as a homodimer or as a heterodimer with p50 or p52 to activate transcription of these two genes.
Table III. κB-site sequences do not correlate with RHD dimer specificity.
| Gene | Functioning RHD protein dimers | κB-site sequence | Reference | |||
|---|---|---|---|---|---|---|
| MCP-1 | p50:p65 | p52:p65 | p50:cRel | p65:p65 | –2811GGGAATTTCC –2640GGGAATTCC | Ping et al. (1999) |
| JunB | p50:p65 | p52:p65 | p50:cRel | p65:p65 | +2057GGGGCTTTCC | Phinney et al. (1995) |
| MIP-2 | p50:p65 | p52:p65 | p65:p65 | –66GGGAATTTCC | Widmer et al. (1993) | |
| IκBα | p50:p65 | p52:p65 | p65:p65 | –260GGGGAAGTCC –70GGAAATTCCC | Chiao et al. (1994) | |
| Fas | p50:p65 | p52:p65 | p65:p65 | –59GGAATGCCCA | Zheng et al. (2001) | |
| RANTES | p50:p65 | p52:p65 | –87GGGAGTTTCC | Lee et al. (2000) | ||
| IP-10 | p50:p65 | p52:p65 | –169GGGAAATTCC –113GGGACTTCCC | Ohmori et al. (1992) | ||
| M-CSF | p50:p65 | p52:p65 | –378GGAAAGTCCC | Harrington et al. (1991) | ||
| LIF | p50:p65 | –77GGGGATCCCG | Willson et al. (1992) | |||
NF-κB-dependent genes (column 1) are listed in order of increasingly restrictive RHD protein dimer requirement (column 2). Corresponding gene promoters contain conserved κB-sites of indicated sequences (column 3) that were show to be functional in previously published studies (column 4).
Determinants of NF-κB protein requirements
Because individual NF-κB dimers have been shown to have particular binding sequence preferences in vitro (Kunsch et al., 1992), we examined whether RHD protein requirements can be correlated with the sequence of the κB-site. Such a correlation would indicate that DNA–protein interaction affinity, or more precisely Kon and/or koff rates, of RHD protein dimers to specific κB-site sequences determine which RHD protein dimers are functional on a particular promoter. The promoters for most of the genes in this study have been previously investigated by standard transiently transfected reporter assays that led to the identification of the apparent functional transcription factor-binding sites. For LIF, we used human and mouse genomic databases and previous promoter studies to identify a novel κB-binding element. Grouping the NF-κB-dependent genes in our study according their specific RHD protein requirement in Table III failed to reveal a correlation with κB-site sequences. In fact, genes whose TNFα induction can be mediated by the same set of RHD protein dimers, such as RANTES, IP-10 and M-CSF, contain different κB-site sequences that are responsible for this induction. Conversely, promoters containing the same κB-site sequence (GGGAATTTCC), such as MIP-2 and MCP-1, exhibit different RHD protein requirements. Furthermore, focusing on genes that can be activated by p65 homodimers (MCP-1, junB, MIP-2, IκBα and Fas) we cannot discern any common features in the relevant κB-site sequences. In each case, the κB element is embedded in a promoter that is bound by many proteins, such as neighboring transcription factors, co-activators and chromatin components that may interact with NF-κB/Rel dimers bound at the κB-site. The results of our genetic analysis suggest that the ability of RHD protein dimers to function on a particular promoter is not solely determined by DNA–protein interactions at the κB-site. While the regulatory control regions of the genes in this study have previously been characterized, we cannot rule out that NF-κB may function via unrecognized binding sites, whose sequence may not be conserved between mouse and human. Mutations of endogenous sequences via knock-in technology or methodologies that allow faithful expression of transgenic constructs will be needed to confirm and extend our conclusions.
Interactions with DNA is likely to restrict the subset of possible transcription factors that may function at a particular promoter element, and within the family of κB-sites it is known that p65 or cRel homodimers bind better to 9 bp than 10 bp sites. However, our results (based on the highly divergent set of 10 bp κB-sites) point to the importance of protein–protein interactions within promoter–DNA assemblies in determining a specific family member requirement and generating transcriptional specificity in vivo. Transcriptional synergy (Lin et al., 1990) between adjacent transcription factors and within enhanceosomes (Thanos and Maniatis, 1995), as well as specific co-activator requirements (Merika et al., 1998), have been observed in non-chromosomal experimental systems and have long been thought to play a role in generating specificity through combinatorial control. Differential specificity of NF-κB protein family members in interactions with contextual transcriptional factors may thus account for family member-specific requirements for gene activation. Chromatin has also been shown to control NF-κB/Rel accessibility in vivo (Saccani et al., 2001) and may in fact do so in a manner that is specific for a subset of RHD protein dimers. Furthermore, some genes require chromatin re-organization for gene activation (Lomvardas and Thanos, 2002), and this may be dependent on protein–protein interactions specific to a particular family member.
Other groups have taken complementary approaches to studying transcription factor specificity. In particular, Farnham and colleagues used chromatin immunoprecipitation in combination with microarray technology (ChIP–Chip) to examine the role of a single member of the E2F family across a wide spectrum of genes (Weinmann et al., 2002), while Young and colleagues have begun to characterize extensive networks of transcriptional control in budding yeast (Lee et al., 2002). Recently, ChIP experiments have revealed differential NF-κB dimer recruitment to target promoters that may complement the present study (Saccani et al., 2003). While NF-κB occupancy as assayed by ChIP does not indicate the functional activity of the bound dimer without the use of genetic tools, such assays indicate that transcriptional control may involve the dynamic interplay of different dimers to attenuate or sustain transcriptional activity (also A.Hoffmann and D.Baltimore, manuscript in preparation). Perfecting the ChIP–chip technology to allow for a comprehensive characterization of occupancy states of regulatory regions will shed light on promoter architecture. Complementing that approach with transcript measurements in a variety of relevant knockouts may illuminate the operative promoter code, and in particular the question of family member specificity.
Are distinct κB-site sequences functionally equivalent? While our genetic analysis has failed to correlate functional requirement with κB-site sequence, and NF-κB co-crystal structure studies do not reveal extensive NF-κB dimer-specific DNA-binding contacts (Chen and Ghosh, 1999), this does not rule out that the κB-site sequence plays a functional role. In fact, structural comparisons of the p50:p65 dimer bound to different κB-site sequences have revealed significant differences in conformation (Chen and Ghosh, 1999). The particular conformations of DNA-bound NF-κB complexes (probed by protease sensitivity and X-ray crystallography) have indeed between correlated with their ability to transactivate on transfected promoters (Fujita et al., 1992; Chen-Park et al., 2002); however it is unknown whether the conformational state of NF-κB complexes may have similar importance on chromosomal promoters, and whether a particular conformation of an NF-κB dimer may be functional on one promoter but not another. In addition to examining binding affinities, investigating the conformations of NF-κB dimers bound to κB elements and the functional consequence within the context of endogenous regulatory region is likely to bring further understanding to the promoter code.
Multiple protein–DNA complexes may be able to assemble on a given promoter depending on the cell-type and stimulus (Falvo et al., 2000). Because the κB-site sequence is not the sole determinant of the specific RHD dimer requirement, we may find that the specific RHD protein requirement for the activation of a particular gene is also specific to cell-type and stimulus. If combinatorial control by multiple promoter-bound transcription factors generates specific transcription factor requirements, it can form the basis for the inclusion or exclusion of a particular gene in specific gene expression programs in response to multiple activation pathways.
Materials and methods
Cell culture and transfections
Immortalized fibroblast cell lines were generated from E12.5–14.5 embryos according to the 3T3 protocol in 10% bovine calf serum (Aaronson and Todaro, 1968) and maintained in the same manner. Previously published NF-κB luciferase reporter plasmids (Fujita et al., 1992) were transfected together with β-actin–lacZ control plasmid into subconfluent fibroblasts using Ca-PO4 or fugene6 (Roche) methodologies. After 24 h, cells were starved for 24 h with 0.5% serum containing medium and then stimulated with 10 ng/ml TNFα.
DNA-binding assays and western blot
For whole cell western blots, cells were lysed within six well plates using SDS–PAGE sample buffer, and probed with specific antibodies from Santa-Cruz Biotechnology, Inc. For electrophoretic mobility shift assays (EMSAs), previously described methods and κB-site containing probes were used (Fujita et al., 1992; Hoffmann et al., 2002). Supershift assays were done with antibody cocktails specific to indicated RHD proteins composed of rabbit antibodies available at Santa Cruz Biotechnology. The reactivity and specificity of these is demonstrated in the data shown in Figure 3A–D.
RNase protection assays
Total RNA was made from confluent and starved fibroblasts using Tri-Reagent (Molecular Research Center, Inc). RPA was performed with 5 µg RNA using Riboquant probe sets (Pharmingen) according to the manufacturer’s instructions. Data was quantitated using a Molecular Dynamics PhosphorImager. Following local background subtraction, data was normalized using GAPDH and L32 gene data, which allowed results to be compared across experiments. Every experiment was performed at least twice, many three to five times with high reproducibility.
Acknowledgments
Acknowledgements
A.H. thanks A.Beg, W.C.Sha, B.H.Horwitz, S.Gerondakis, R.Ryseck and R.Bravo for knockout mice, L.Anonueovo, B.Kennedy and S.Pease for their maintenance, S.Tronick for antibodies, and G.Ghosh and T.Huxford for critical reading of the manuscript. T.H.L. is an ARCS Foundation fellow and part of the UCLA/Caltech Medical Scientist Training Program.
References
- Aaronson S.A. and Todaro,G.J. (1968) Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J. Cell Physiol., 72, 141–148. [DOI] [PubMed] [Google Scholar]
- Beg A.A., Sha,W.C., Bronson,R.T., Ghosh,S. and Baltimore,D. (1995) Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature, 376, 167–170. [DOI] [PubMed] [Google Scholar]
- Berkowitz B., Huang,D.B., Chen-Park,F.E., Sigler,P.B. and Ghosh,G. (2002) The x-ray crystal structure of the NF-κB p50.p65 heterodimer bound to the interferon β–κB site. J. Biol. Chem., 277, 24694–24700. [DOI] [PubMed] [Google Scholar]
- Bren G.D., Solan,N.J., Miyoshi,H., Pennington,K.N., Pobst,L.J. and Paya,C.V. (2001) Transcription of the RelB gene is regulated by NF-κB. Oncogene, 20, 7722–7733. [DOI] [PubMed] [Google Scholar]
- Caamano J.H., Rizzo,C.A., Durham,S.K., Barton,D.S., Raventos-Suarez,C., Snapper,C.M. and Bravo,R. (1998) Nuclear factor (NF)-κB2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J. Exp. Med., 187, 185–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiao P.J., Miyamoto,S. and Verma,I.M. (1994) Autoregulation of IκBα activity. Proc. Natl Acad. Sci. USA, 91, 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.E. and Ghosh,G. (1999) Regulation of DNA binding by Rel/NF-κB transcription factors: structural views. Oncogene, 18, 6845–6852. [DOI] [PubMed] [Google Scholar]
- Chen Y.Q., Ghosh,S. and Ghosh,G. (1998) A novel DNA recognition mode by the NF-κB p65 homodimer. Nat. Struct. Biol., 5, 67–73. [DOI] [PubMed] [Google Scholar]
- Chen-Park F.E., Huang,D.B., Noro,B., Thanos,D. and Ghosh,G. (2002) The βB DNA sequence from the HIV long terminal repeat functions as an allosteric regulator of HIV transcription. J. Biol. Chem., 277, 24701–24708. [DOI] [PubMed] [Google Scholar]
- Cheng J.D., Ryseck,R.P., Attar,R.M., Dambach,D. and Bravo,R. (1998) Functional redundancy of the nuclear factor κB inhibitors IκBα and IκBβ. J. Exp. Med., 188, 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y. and Kerppola,T.K. (2001) Close encounters of many kinds: Fos–Jun interactions that mediate transcription regulatory specificity. Oncogene, 20, 2438–2452. [DOI] [PubMed] [Google Scholar]
- Escalante C.R., Shen,L., Thanos,D. and Aggarwal,A.K. (2002) Structure of NF-κB p50/p65 heterodimer bound to the PRDII DNA element from the interferon-β promoter. Structure, 10, 383–391. [DOI] [PubMed] [Google Scholar]
- Falvo J.V. et al. (2000) Stimulus-specific assembly of enhancer complexes on the tumor necrosis factor α gene promoter. Mol. Cell. Biol., 20, 2239–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Nolan,G.P., Ghosh,S. and Baltimore,D. (1992) Independent modes of transcriptional activation by the p50 and p65 subunits of NF-κB. Genes Dev., 6, 775–787. [DOI] [PubMed] [Google Scholar]
- Gerondakis S., Grossmann,M., Nakamura,Y., Pohl,T. and Grumont,R. (1999) Genetic approaches in mice to understand Rel/NF-κB and IκB function: transgenics and knockouts. Oncogene, 18, 6888–6895. [DOI] [PubMed] [Google Scholar]
- Ghosh G., van Duyne,G., Ghosh,S. and Sigler,P.B. (1995) Structure of NF-κB p50 homodimer bound to a κB site. Nature, 373, 303–310. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May,M.J. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Grumont R.J. and Gerondakis,S. (1990) Murine c-rel transcription is rapidly induced in T-cells and fibroblasts by mitogenic agents and the phorbol ester 12-O-tetradecanoylphorbol-13-acetate. Cell Growth Differ., 1, 345–350. [PubMed] [Google Scholar]
- Grumont R.J., Richardson,I.B., Gaff,C. and Gerondakis,S. (1993) rel/NF-κB nuclear complexes that bind kB sites in the murine c-rel promoter are required for constitutive c-rel transcription in B-cells. Cell Growth Differ., 4, 731–743. [PubMed] [Google Scholar]
- Harrington M.A., Edenberg,H.J., Saxman,S., Pedigo,L.M., Daub,R. and Broxmeyer,H.E. (1991) Cloning and characterization of the murine promoter for the colony-stimulating factor-1-encoding gene. Gene, 102, 165–170. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Levchenko,A., Scott,M.L. and Baltimore,D. (2002) The IκB–NF-κB signaling module: temporal control and selective gene activation. Science, 298, 1241–1245. [DOI] [PubMed] [Google Scholar]
- Horwitz B.H., Scott,M.L., Cherry,S.R., Bronson,R.T. and Baltimore,D. (1997) Failure of lymphopoiesis after adoptive transfer of NF-κB-deficient fetal liver cells. Immunity, 6, 765–772. [DOI] [PubMed] [Google Scholar]
- Kontgen F., Grumont,R.J., Strasser,A., Metcalf,D., Li,R., Tarlinton,D. and Gerondakis,S. (1995) Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity and interleukin-2 expression. Genes Dev., 9, 1965–1977. [DOI] [PubMed] [Google Scholar]
- Kunsch C., Ruben,S.M. and Rosen,C.A. (1992) Selection of optimal κB/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol. Cell. Biol., 12, 4412–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E.S. et al. (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Lee A.H., Hong,J.H. and Seo,Y.S. (2000) Tumour necrosis factor-α and interferon-γ synergistically activate the RANTES promoter through nuclear factor κB and interferon regulatory factor 1 (IRF-1) transcription factors. Biochem. J., 350, 131–138. [PMC free article] [PubMed] [Google Scholar]
- Lee T.I. et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science, 298, 799–804. [DOI] [PubMed] [Google Scholar]
- Lin Y.S., Carey,M., Ptashne,M. and Green,M.R. (1990) How different eukaryotic transcriptional activators can cooperate promiscuously. Nature, 345, 359–361. [DOI] [PubMed] [Google Scholar]
- Liou H.C., Sha,W.C., Scott,M.L. and Baltimore,D. (1994) Sequential induction of NF-κB/Rel family proteins during B-cell terminal differentiation. Mol. Cell. Biol., 14, 5349–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S. and Thanos,D. (2002) Modifying gene expression programs by altering core promoter chromatin architecture. Cell, 110, 261–271. [DOI] [PubMed] [Google Scholar]
- Mechta-Grigoriou F., Gerald,D. and Yaniv,M. (2001) The mammalian Jun proteins: redundancy and specificity. Oncogene, 20, 2378–2389. [DOI] [PubMed] [Google Scholar]
- Meffert M.K., Chang,J.M., Wiltgen,B.J., Fanselow,M.S. and Baltimore,D. (2003) NF-κB functions in synaptic signaling and behavior. Nat. Neurosci., in press. [DOI] [PubMed] [Google Scholar]
- Merika M., Williams,A.J., Chen,G., Collins,T. and Thanos,D. (1998) Recruitment of CBP/p300 by the IFN-β enhanceosome is required for synergistic activation of transcription. Mol. Cell, 1, 277–287. [DOI] [PubMed] [Google Scholar]
- Muller C.W., Rey,F.A., Sodeoka,M., Verdine,G.L. and Harrison,S.C. (1995) Structure of the NF-κB p50 homodimer bound to DNA. Nature, 373, 311–317. [DOI] [PubMed] [Google Scholar]
- Ohmori Y. and Hamilton,T.A. (1993) Cooperative interaction between interferon (IFN) stimulus response element and κB sequence motifs controls IFNγ- and lipopolysaccharide-stimulated transcription from the murine IP-10 promoter. J. Biol. Chem., 268, 6677–6688. [PubMed] [Google Scholar]
- Pahl H.L. (1999) Activators and target genes of Rel/NF-κB transcription factors. Oncogene, 18, 6853–6866. [DOI] [PubMed] [Google Scholar]
- Phinney D.G., Tseng,S.W. and Ryder,K. (1995) Complex genetic organization of junB: multiple blocks of flanking evolutionarily conserved sequence at the murine and human junB loci. Genomics, 28, 228–234. [DOI] [PubMed] [Google Scholar]
- Ping D., Boekhoudt,G.H., Rogers,E.M. and Boss,J.M. (1999) Nuclear factor-κB p65 mediates the assembly and activation of the TNF-responsive element of the murine monocyte chemoattractant-1 gene. J. Immunol., 162, 727–734. [PubMed] [Google Scholar]
- Ramji D.P. and Foka,P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J., 365, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastinejad F. (2001) Retinoid X receptor and its partners in the nuclear receptor family. Curr. Opin. Struct. Biol., 11, 33–38. [DOI] [PubMed] [Google Scholar]
- Saccani S., Pantano,S. and Natoli,G. (2001) Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med., 193, 1351–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S., Pantano,S. and Natoli,G. (2003) Modulation of NF-κB activity by exchange of dimers. Mol. Cell, 11, 1563–1574. [DOI] [PubMed] [Google Scholar]
- Sanjabi S., Hoffmann,A., Liou,H.C., Baltimore,D. and Smale,S.T. (2000) Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc. Natl Acad. Sci. USA, 97, 12705–12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M.L., Fujita,T., Liou,H.C., Nolan,G.P. and Baltimore,D. (1993) The p65 subunit of NF-κB regulates I κB by two distinct mechanisms. Genes Dev., 7, 1266–1276. [DOI] [PubMed] [Google Scholar]
- Sha W.C., Liou,H.C., Tuomanen,E.I. and Baltimore,D. (1995) Targeted disruption of the p50 subunit of NF-κB leads to multifocal defects in immune responses. Cell, 80, 321–330. [DOI] [PubMed] [Google Scholar]
- Thanos D. and Maniatis,T. (1995) Virus induction of human IFN-β gene expression requires the assembly of an enhanceosome. Cell, 83, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees,J.A. (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol., 3, 11–20. [DOI] [PubMed] [Google Scholar]
- Weinmann A.S., Yan,P.S., Oberley,M.J., Huang,T.H. and Farnham,P.J. (2002) Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev., 16, 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer U., Manogue,K.R., Cerami,A. and Sherry,B. (1993) Genomic cloning and promoter analysis of macrophage inflammatory protein (MIP)-2, MIP-1α, and MIP-1β, members of the chemokine superfamily of proinflammatory cytokines. J. Immunol., 150, 4996–5012. [PubMed] [Google Scholar]
- Willson T.A., Metcalf,D. and Gough,N.M. (1992) Cross-species comparison of the sequence of the leukaemia inhibitory factor gene and its protein. Eur. J. Biochem., 204, 21–30. [DOI] [PubMed] [Google Scholar]
- Wolfe S.A., Nekludova,L. and Pabo,C.O. (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct., 29, 183–212. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Ouaaz,F., Bruzzo,P., Singh,V., Gerondakis,S. and Beg,A.A. (2001) NF-κB RelA (p65) is essential for TNF-α-induced fas expression but dispensable for both TCR-induced expression and activation-induced cell death. J. Immunol., 166, 4949–4957. [DOI] [PubMed] [Google Scholar]