Abstract
A PCR based assay was established to screen for potential changes in the heterogeneity of Vβ TCR expressing CD4+ T cells from rhesus macaques (RM) and sooty mangabeys (SM) prior to and following SIV infection. The rationale for these studies was to determine whether the progressive CD4+ T cell loss in SIV infected disease susceptible RM and the moderate CD4+ T cell loss in disease resistant SM leads to the depletion of select Vβ TCR families of CD4+ T cells. Results show that whereas SIV infection leads to the loss of Vβ TCR heterogeneity in disease susceptible RM, the CD4+ T cells from SM retain their degree of Vβ TCR heterogeneity, suggesting that the mechanism(s) of SIV induced CD4+ T cell loss maybe distinct in these 2 species and contribute to the differences in the clinical outcome.
Keywords: AIDS, SIV, T cell receptor, V beta, CD4+ T cell
INTRODUCTION
One of the hallmarks of human HIV-1 infection is the gradual impairment of CD4+ T cell responses attributed to both a decline in the CD4+ T cell counts and function. Although the CD4+ T cell lineage represents a major target of the virus, it is unclear at present, whether the virus progressively depletes select Vα/Vβ TCR families with specificity for the various viral peptides or both viral and non-virus specific antigen activated CD4+ T cells. Data from several previous studies have suggested that indeed in AIDS patients, T cells expressing particular Vβ gene families are selectively deleted (Bansal et al., 1993; Hodara et al., 1993; Imberti et al., 1991) which could be because virus selectively infects HIV-1 antigen specific CD4+ T cells (Douek et al., 2002) and induces perturbations in the CD4+ T cell repertoire which are associated with progression to AIDS (Gorochov et al., 1998). Data from other studies however suggest that while there is a general decrease in the heterogeneity of Vβ TCR expressing T cells in AIDS patients, the depletion of cells expressing certain Vβ TCR gene families is random (Boldt-Houle, Rinaldo, and Ehrlich, 1993; Boyer et al., 1993; Posnett et al., 1993). Conversely, others showed that HIV infection in fact leads to an expansion instead of depletion of T cells expressing certain Vβ gene families (Dalgleish et al., 1992; Roglic et al., 1997).
SIV infection of rhesus macaques (RM) is a widely utilized non-human primate model of human AIDS. The availability of sequential samples pre- and post- infection makes this model ideal for addressing the issue with regards to the selective depletion of SIV induced repertoire of Vβ TCR expressing CD4+ T cell populations. Unfortunately, once again, while some earlier longitudinal studies reported that the diversity of TCR Vβ repertoire in CD4+ T cell population was maintained in SIV infected rhesus macaques (Chen et al., 1993), some later data suggested that even as early as following acute SIV infection, there is an apparent disruption of the TCR Vβ repertoire in CD4+ T cells with clonal expansion of select Vβ TCR spectratypes (Zhou et al., 1999). Interestingly, certain non-human primate species, such as the sooty mangabeys (SM) are naturally infected with SIV, but do not develop any detectable disease. A number of previous cross sectional studies have documented a lower frequency and absolute number of CD4+ T cells in SIV positive as compared with the SIV negative SM. This sustained depletion of CD4+ T cells in the SM was shown to be due to SIV infection in studies of a cohort of individual SM prior to and post experimental SIV infection (Muthukumar et al., 2005). Once again the question whether such depletion is due to select elimination of select Vβ TCR expressing CD4+ T cells in SM has to date not been addressed. Limited studies of TCR repertoire analysis have to date been performed on PBMC samples from RM (Chen et al., 1993; Zhou et al., 1999) and none in other nonhuman primate species including SM. The studies in RM utilized spectratyping techniques, which we submit is labor intensive and unless the antigenic peptides and the cognate MHC allele are known, such analysis is not very informative. To further our understanding of the SIV mediated CD4+ T cell depletion we developed a relatively different approach for defining changes in TCR repertoire, which allowed us to re-examine this issue with regards to the effect of SIV infection on depletion of select Vβ TCR expressing T cells in disease susceptible RM and for purposes of comparison disease resistant SM. Here we show that while SIV infection in disease susceptible RM indeed leads to a relative reduction of TCR Vβ heterogeneity in the CD4+ T cell population, the CD4+ T cells from the disease resistant SM maintain the extent of TCR Vβ heterogeneity after both natural and experimental SIV infection.
RESULTS AND DISCUSSION
Analysis of TCR variability has historically been a procedure involving spectratyping of each Vα/β subtype individually with a Gaussian distribution expected as representing the normal distribution of each sub-family (Currier et al., 1999). When we previously utilized such techniques to study differences in SIV infection of NHP, the results showed that samples even from uninfected healthy animals showed a considerable degree of skewing within various individual Vβ TCR families for reasons that are unclear at present. Although this analytical approach yields relatively detailed data concerning a full spectrum in each of the sub-families, it is very tedious and extensive. Problems encountered with such an analysis prompted us to seek other assays, which would allow us to reliably and efficiently examine the global instead of subtle sub-family effects of lentivirus infection in NHP on CD4+ T cell repertoire.
We thus developed a screening method, which covers a wide range of TCR Vβ sub-families. We submit that while this assay does not identify changes in specific TCR Vβ sub-families, it does allow for the identification of the relative overall TCR heterogeneity. In this assay, a pool of amplicons is generated first by PCR from cDNA isolated from a single aliquot of cells using a mixture of 10 Vβ specific and 10 Jβ specific primers. To ensure that the Jβ primers were indeed specifically amplifying the V-J regions from both NHP species, we first tested each of the new Jβ primers separately with three randomly selected Vβ primers using cDNA from either RM or SM as a template. Each PCR reaction yielded one specific PCR product of predicted size (range 90 – to 120 nt long; data not shown).
As a next step we performed a cross sectional study which involved screening a set of SIV seronegative and SIV seropositive (i.e. naturally SIV infected) sooty mangabeys for the heterogeneity of the CD4+ T cell TCR Vβ repertoire utilizing the PCR/hybridization based screening method which included the analysis of the V-J fragments generated from 3 aliquots of cells per PBMC sample as described in the methods. We selected our sub-aliquot size at 50,000 cells based on calculations from comparisons with similar studies conducted previously in humans (Naylor et al., 2005). In addition, in our preliminary experiments this number was found to be optimal as increasing the aliquot size decreased the sensitivity of the method and yielding hybridization results approaching 100% identity in most aliquots (Figure 1A). As seen in Figure 1B, no major differences with regards to the extent of TCR Vβ heterogeneity were noted based on the SIV status of the sooty mangabey monkeys from which the PBMC samples were obtained. In each group the majority of the animals showed a relatively high degree of TCR heterogeneity giving the mean values for relative similarity between the aliquots of 0.59 (or 59%) and 0.48 (48%) for the SIV seronegative and SIV seropositive SM, respectively (p>0.2). Interestingly, regardless of the SIV status, in each group there were one or two animals exhibiting a relatively low degree of TCR Vβ heterogeneity (relative similarity above 0.7). This data indicated that the TCR Vβ repertoires in CD4+ T cells from individual SM exhibit a wide spectrum of variability both in SIV naïve and positive animals precluding us from deriving any conclusions regarding the potential effect of SIV infection on the TCR Vβ repertoire from such a cross-sectional study.
Figure 1. Relative TCRβ heterogeneity in sooty mangabeys.
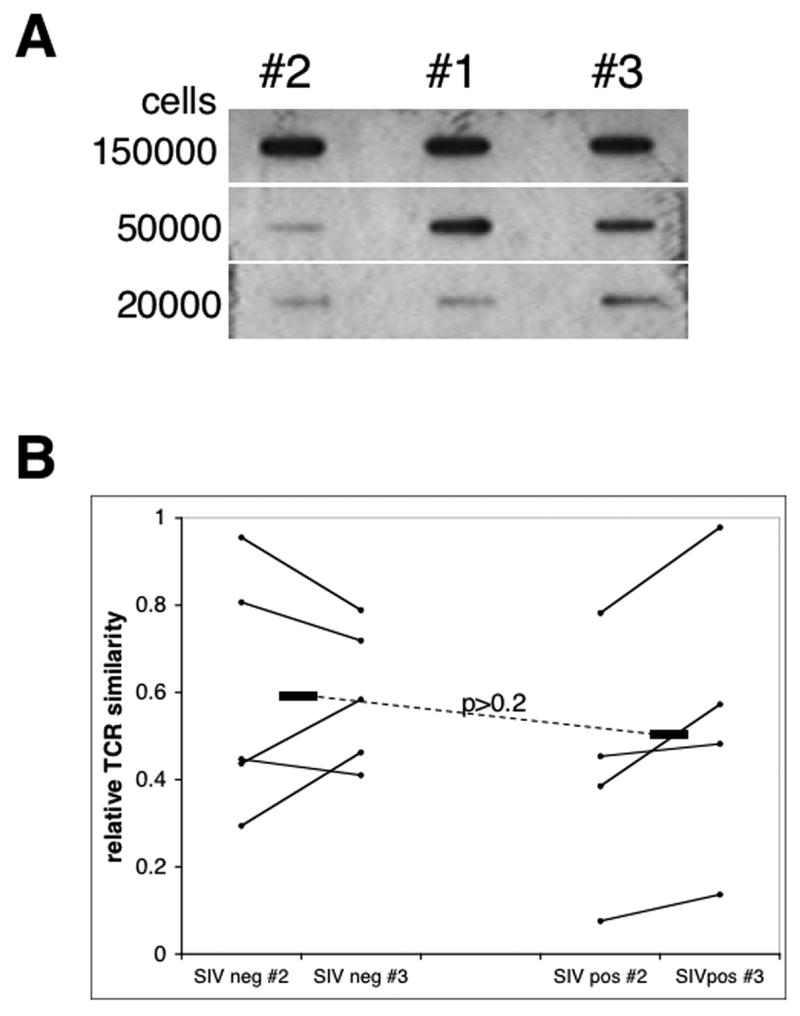
CD4+ T cells were SIV seronegative and seropositive sooty mangabeys and analyzed for the heterogeneity of TCR Vβ region as described in the methods section. A) Optimization of the cell number aliquot. Representative slot blot data with target DNA amplicons and probes generated from aliquots containing variable cell numbers as indicated from one CD4+ T cell sample. Hybridization signal differences increase with a decrease of aliquot size with optimum around 50000cells/aliquot. B) TCR Vβ-Jβ heterogeneity analysis of CD4+ T cells from SIV seronegative (SIVneg; n=4) and SIV seropositive (SIVpos; n=5) sooty mangabeys. Slot blot data were quantitated by densitometry and values from sub-aliquots #2 and #3 were derived as a ratio of the value obtained with the “control” aliquot #1. Horizontal bars indicate means for each animal group with p value shown.
These findings prompted us to carry out a prospective study in which samples of PBMC from 6 RM and 6 SM were obtained sequentially pre and post SIV infection. Since previous data (Figure 1B) indicated that values obtained from the two aliquots (#2 and #3) from each single T cell specimen were within a comparable range, we derived mean values from the two aliquots for each individual sample and compared pre and post infection samples from individual monkeys (Figure 2 B). The data show that CD4+ T cells from the majority of SM (5/6) exhibited the pre-SIV infection relative level of TCR Vβ heterogeneity within the same range as cells from uninfected RM (0.4–0.65 similarity). One sooty mangabey showed a relative low degree of TCR Vβ heterogeneity before SIV infection, which was maintained after the infection. Interestingly, while the SIV infected SM maintained the TCR Vβ heterogeneity at levels similar to the pre-infection level (0.4–0.6 similarity), the SIV infection in RM led to a decrease in TCR heterogenity (0.7–0.9 similarity) which was highly significant (p<0.001). This contraction of TCR Vβ repertoire was detectable relatively early after SIV infection as sequential samples from 3 of the RM post infection showed similar decrease in heterogeneity as early as 5 months p.i. (data not shown). To further corroborate the data obtained by this assay we quantitated the relative abundance of select 5 TCR Vβ mRNA transcripts in CD4+ T cell aliquots from 2 animals from each species pre and post SIV infection using a real-time PCR approach (Fig 3). The data show that while the target TCR sequences post SIV infection in CD4+ T cells from SM were present at similar relative quantity compared to the pre-infection status, in RM there was a relative decrease in abundance of the transcripts in 3 out of 5 TCR Vβ families tested – Vβ 3, 7 and 9 – and changes in two of these were statistically significant. These results therefore validated our hybridization approach as a suitable assay to detect significant overall changes in the TCR Vβ repertoire.
Figure 2. Effect of SIV infection on TCRβ heterogeneity RM and SM.
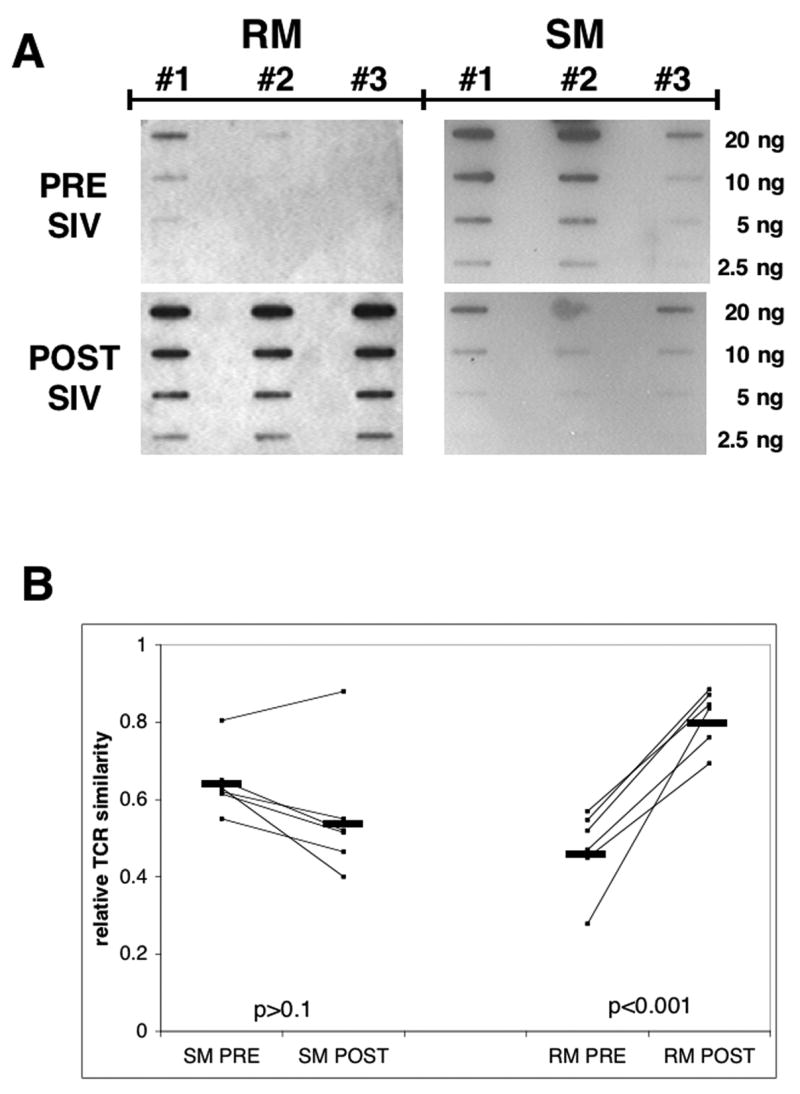
CD4+ T cells were isolated from rhesus macaques (RM) and sooty mangabeys (SM) pre and post SIV infection and analyzed for the heterogeneity of TCR Vβ region as described in the methods section. A) Representative slot blot data pre and post SIV infection from one animal from each species. Serial dilutions of Vβ-Jβ amplicons (20ng – 2.5 ng) from three sub-aliquots (control # 1 sub-aliquot, #2 and #3 sub-aliquots) of 50, 000 CD4+ T cells from a single PBMC sample from each species were hybridized to the probe generated from the control amplicon prepared from sub-aliquot #1. B) TCR Vβ-Jβ analysis of 6 animals from each species pre (RMpre, SMpre) and post (RMpost, SMpost) experimental SIV infection. Slot blot data were quantitated by densitometry and values from sub-aliquots #2 and #3 averaged and expressed as TCR similarity relative to the “control” cell sub-aliquot #1 from each CD4+ T cell sample. Horizontal bars indicate means for each group and p values are shown.
Figure 3. Real-time PCR quantitation of TCR Vbeta.
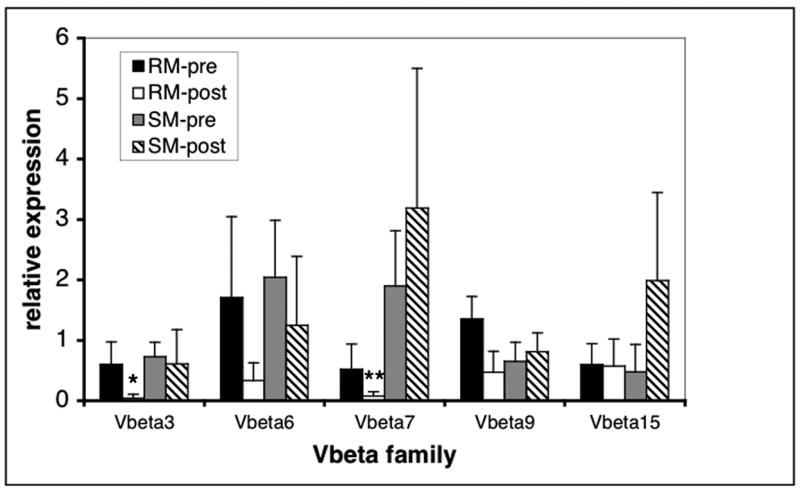
Relative abundance of select TCR Vbeta subfamily-specific transcripts was assssed in cDNA samples from 3 independent aliquots of CD4+ T cells from 2 RM and 2 SM utilizing real time PCR quantitation of TCR Vβ-Cβ target sequence. The target quantitation was then normalized to the GAPDH and the copy numbers of the target gene in each animal are expressed as relative to the copy numbers from the calibrator sample pre-SIV infection. The resulting relative quantitation is expressed as mean+SD. Star indicates p<0.03, double star indicates p<0.005.
Taken together, these data suggest that while SIV infection in disease susceptible NHP species such as the RM leads to a contraction of the TCR Vβ repertoire, similar experimental SIV infection in the disease resistant species – i.e. sooty mangabeys – does not exhibit any detectable effect on TCR diversity. The observed moderate but readily detectable depletion of CD4+ T cells noted consistently in SM after SIV infection is therefore most likely affecting cells expressing a diverse set of TCR Vβ across the board leading to a reduction of total numbers of cells, but not selective TCR Vβ subpopulations. The finding that SIV infection of rhesus macaques leads to a contraction of the TCR Vβ repertoire corroborates some previous experimental data (Zhou et al., 1999). On the other hand, using the TCR spectratyping method, others including our lab have either failed to demonstrate any reduction of TCR repertoire in SIV infected rhesus macaques or obtained inconsistent results (Chen et al., 1993). As outlined above while the method used herein does not provide specific identification of select Vβ families that show deletion in RM after SIV infection, it does allow for higher throughput screening, but does not provide separate data for each Vβ family tested. This clearly should be a subject for further, potentially real-time PCR quantification based studies that will allow for discrimination between each of the Vβ subfamilies in those samples showing disturbed TCR repertoire. We have utilized the V-J region amplification, rather than the V-C utilized previously in the majority of spectratyping studies, which yields shorter products that do not contain any sequences common to all the gene families therefore increasing the sensitivity of the assay. It is recognized the further analysis of subsets of CD4+ cells from not only the peripheral blood but also other tissues such as the gut associated lymphoid tissue will yield more refined data regarding the mechanisms of CD4+ T cell depletion in lentivirus infection and such analysis constitutes a basis for ongoing studies. Taken together we have developed a methodology that can simplify and serve as a valuable tool for relative TCR heterogeneity screening and herein show that SIV disease resistance is associated with the maintenance of TCR repertoire in non-human primate model of AIDS.
MATERIALS AND METHODS
Cells
The PBMC samples for the cross sectional studies were obtained from SIV naïve and naturally SIV infected SM (Cercocebus atys) colony maintained at the Yerkes National Primate Center (YNPRC) of Emory University. The PBMC samples for the prospective study were obtained from 6 adult RM (Macaca mulatta) pre and post infection with SIVmac239 and 6 adult SM pre and post infection with plasma from SM naturally infected with SIVsm. The post infection samples were obtained during the chronic infection stage (mean sampling time 8 months, range 4 – 12 months p.i.) with CD4+ T cell counts >400 and viral loads between 105– 107/ml of plasma. Animals were housed at the YNPRC and maintained according to the guidelines of the Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council and the Health and Human Services guidelines “Guide for the Care and Use of Laboratory Animals.” CD4+ T cells were isolated from fresh or cryopreserved PBMC sing Dynabeads M450 CD4 (Dynal, Lake Success, NY). The purity of the cell population was always > 95.0% as determined by FACS analysis.
TCR variability analysis
The hybridization assay developed for screening of the TCR diversity is a modification of a previously published procedure (Naylor et al., 2005). Total RNA was isolated from 3 aliquots (#1, 2, 3) of 50,000 highly purified CD4+ T cells from a single PBMC sample from a single monkey using the RNeasy kit (Qiagen, Valencia, CA) and cDNA was synthesized using the ProtoScript cDNA synthesis kit (NEB, Beverly, MA). The cDNA from each aliquot was amplified in a PCR reaction containing a mixture of TCR Vβ beta and TCR Jβ primers with 40 cycles of 95°C for 20 s and 60°C for 50 s. The mixture of TCR Vβ primers consisted of primers specific for Vβ 1, 2, 3, 4, 6, 7, 9, 11, 14, and 15 as previously described (Chen et al., 1993). The Jβ region specific primers JB are listed in Table 1. Each of the primers was utilized at 100nM final concentration. The resulting V-J amplification products were purified using the Wizard PCR purification kit (Promega, Madison, WI) and 4 serial two-fold dilutions of each amplification product (20 ng – 2.5 ng) were blotted onto a Hybond-XL membrane (GE Healthcare, Piscataway, NJ). An α32P-dCTP labeled probe was prepared from the amplicon #1 (control) from each sample by PCR as above, hybridized to the membrane with immobilized dilutions of the amplicons from all 3 aliquots (#1,#2 and #3) and hybridization signals were quantitated by densitometry. The TCR diversity ratio was calculated as a ratio of the hybridization signals obtained from each dilution of the two aliquots (#2 and #3) derived from the same CD4+ T cell donor, to the hybridization signal obtained from the same dilution of the “control” aliquot #1. The signal obtained from a given dilution of the positive control ratio was denoted as 1, i.e. 100% homology between the probe and target. Ratios from the aliquots #2 and #3 were averaged and expressed as relative TCR similarity. Those that gave values <1 indicate less homology between the probe and the 2 other different cell aliquots suggesting a diverse content and therefore greater variability/lower similarity of V-J region sequences in these aliquots.
Table 1.
TCR Jβ region primers.
| JB 1.2 | CTGAACCGAAGGTGTAGTC |
| JB 1.3 | CCTCTCCAAAATACACGGTG |
| JB 1.4 | AGAGCTGGGTTCCACTGCCAA |
| JB 1.5 | GTGCCATCTCCAAAATACTGG |
| JB 2.1 | CCCAAAGAACTGCTCACCGT |
| JB 2.2 | CTTCTCCAAAGAACAGCTGC |
| JB 2.3 | GTGCCTGGGCCAAAATACTG |
| JB 2.4 | GCGCCGAAGTACTGAGTGTT |
| JB 2.5 | CGTGCCTGGTCCGAAGTAC |
| JB 2.7 | TGCCCGGCCCGAAGTACTGCT |
Real time PCR quantification of TCR beta cDNA
cDNA samples from 2 RM and 2 SM were prepared from RNA isolated from 3 independent aliquots of CD4+ T cells from each animal ex vivo pre and post-SIV infection and subjected to real-time PCR in an iCycler (BioRad) and SYBR-Green fluorescence quantification utilizing primer pairs consisting of TCRBC primer and appropriate TCR Vβ primer (Chen et al., 1993). As a control, an amplification of the GAPDH fragment was performed using the primers 5′-ACCACCATGGAGAAGGCTGG-3′ and 5′-CAGTTGGTGGTGCAGGAGGC-3′. Parameters of the cycle were 95° for 15″ and 60° for 1′. The target cDNA quantitation in duplicate samples was then performed by first normalizing the threshold cycle number of the target gene to the GAPDH. The copy numbers of the target gene were then expressed relative to the copy numbers from aliquot #1 pre-SIV infection for each animal. The resulting relative quantitation is expressed as mean+SD.
Acknowledgments
This work was supported by the NIH RO1 AI65362 to P.B.
We are grateful to Dr. Guido Silvestri for providing select PBMC samples and to Dr. Jorg Goronzy for his advice in establishing the assay.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bansal AS, Green LM, Khoo SH, Pumphrey RS, Haeney MR, Mandal BK. HIV induces deletion of T cell receptor variable gene product-specific T cells. Clin Exp Immunol. 1993;94(1):17–20. doi: 10.1111/j.1365-2249.1993.tb05970.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt-Houle DM, Rinaldo CR, Jr, Ehrlich GD. Random depletion of T cells that bear specific T cell receptor V beta sequences in AIDS patients. J Leukoc Biol. 1993;54(5):486–91. doi: 10.1002/jlb.54.5.486. [DOI] [PubMed] [Google Scholar]
- Boyer V, Smith LR, Ferre F, Pezzoli P, Trauger RJ, Jensen FC, Carlo DJ. T cell receptor V beta repertoire in HIV-infection individuals: lack of evidence for selective V beta deletion. Clin Exp Immunol. 1993;92(3):437–41. doi: 10.1111/j.1365-2249.1993.tb03417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZW, Kou ZC, Shen L, Reimann KA, Letvin NL. Conserved T-cell receptor repertoire in simian immunodeficiency virus-infected rhesus monkeys. J Immunol. 1993;151(4):2177–87. [PubMed] [Google Scholar]
- Currier JR, Stevenson KS, Kehn PJ, Zheng K, Hirsch VM, Robinson MA. Contributions of CD4+, CD8+, and CD4+CD8+ T cells to skewing within the peripheral T cell receptor beta chain repertoire of healthy macaques. Hum Immunol. 1999;60(3):209–22. doi: 10.1016/s0198-8859(98)00109-8. [DOI] [PubMed] [Google Scholar]
- Dalgleish AG, Wilson S, Gompels M, Ludlam C, Gazzard B, Coates AM, Habeshaw J. T-cell receptor variable gene products and early HIV-1 infection. Lancet. 1992;339(8797):824–8. doi: 10.1016/0140-6736(92)90277-a. [DOI] [PubMed] [Google Scholar]
- Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417(6884):95–8. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- Gorochov G, Neumann AU, Kereveur A, Parizot C, Li T, Katlama C, Karmochkine M, Raguin G, Autran B, Debre P. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat Med. 1998;4(2):215–21. doi: 10.1038/nm0298-215. [DOI] [PubMed] [Google Scholar]
- Hodara VL, Jeddi-Tehrani M, Grunewald J, Andersson R, Scarlatti G, Esin S, Holmberg V, Libonatti O, Wigzell H. HIV infection leads to differential expression of T-cell receptor V beta genes in CD4+ and CD8+ T cells. Aids. 1993;7(5):633–8. doi: 10.1097/00002030-199305000-00004. [DOI] [PubMed] [Google Scholar]
- Imberti L, Sottini A, Bettinardi A, Puoti M, Primi D. Selective depletion in HIV infection of T cells that bear specific T cell receptor V beta sequences. Science. 1991;254(5033):860–2. doi: 10.1126/science.1948066. [DOI] [PubMed] [Google Scholar]
- Muthukumar A, Zhou D, Paiardini M, Barry AP, Cole KS, McClure HM, Staprans SI, Silvestri G, Sodora DL. Timely triggering of homeostatic mechanisms involved in the regulation of T-cell levels in SIVsm-infected sooty mangabeys. Blood. 2005;106(12):3839–45. doi: 10.1182/blood-2005-01-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor K, Li G, Vallejo AN, Lee WW, Koetz K, Bryl E, Witkowski J, Fulbright J, Weyand CM, Goronzy JJ. The influence of age on T cell generation and TCR diversity. J Immunol. 2005;174(11):7446–52. doi: 10.4049/jimmunol.174.11.7446. [DOI] [PubMed] [Google Scholar]
- Posnett DN, Kabak S, Hodtsev AS, Goldberg EA, Asch A. T-cell antigen receptor V beta subsets are not preferentially deleted in AIDS. Aids. 1993;7(5):625–31. doi: 10.1097/00002030-199305000-00003. [DOI] [PubMed] [Google Scholar]
- Roglic M, Macphee RD, Duncan SR, Sattler FR, Theofilopoulos AN. T cell receptor (TCR) BV gene repertoires and clonal expansions of CD4 cells in patients with HIV infections. Clin Exp Immunol. 1997;107(1):21–30. doi: 10.1046/j.1365-2249.1997.d01-886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Kou Z, Ibegbu C, Shen Y, Lee-Parritz D, Shen L, Sehgal PK, McClure HM, Morrison P, Bogle C, Sehgal N, Nahmias AJ, Chen ZW. The disruption of macaque CD4+ T-cell repertoires during the early simian immunodeficiency virus infection. J Med Primatol. 1999;28(4–5):174–80. doi: 10.1111/j.1600-0684.1999.tb00267.x. [DOI] [PubMed] [Google Scholar]


