Abstract
Pax proteins are DNA-binding transcription factors that regulate embryonic development through the activation and repression of downstream target genes. The Pax2 gene is absolutely required for kidney development and for patterning specific regions of the nervous system such as the eye, ear and hindbrain. The Pax2/5/8 family of proteins contains both transcription activation and repression domains. The activation domain of Pax2 is phosphorylated by the c-Jun N-terminal kinase (JNK) to enhance Pax2-dependent transcription. In this report, we demonstrate that the Groucho/TLE family protein, Grg4, interacts with Pax2 to suppress transactivation. Grg4 is able to specifically inhibit phosphorylation of the Pax2 activation domain, even in the presence of activated JNK. Furthermore, the Grg4 interaction and suppression of phosphorylation depends on Pax2 binding to its target DNA sequence and is independent of histone deacetylation. These data suggest a new model for Groucho mediated suppression of transcription through the specific inhibition of modifications in the activation domain of a transactivator.
Keywords: c-Jun/Groucho/N-terminal kinase/Pax2/transcription repression
Introduction
The Pax family of transcription factors are important developmental regulators in a wide variety of tissues across divergent species. In mouse and man, the Pax2 gene is essential for the differentiation of epithelial cells in the urogenital tract, fusion of the optic fissure and patterning the hindbrain and inner ear (Torres et al., 1995, 1996; Favor et al., 1996; Bouchard et al., 2000). The closely related Pax2, Pax5 and Pax8 genes constitute a subfamily that encodes DNA-binding proteins with both repression and activation domains. The Pax2/5/8 activation domain is located at the carboxy terminus and is rich in serine, threonine and proline residues (Dorfler and Busslinger, 1996; Lechner and Dressler, 1996). In addition, the Pax2/5/8 proteins contain a conserved octapeptide with homology to the engrailed-type repressor sequence. The ability of Pax2 to activate transcription is enhanced by phosphorylation of its activation domain by the c-Jun N-terminal kinase (JNK) (Cai et al., 2002). Furthermore, the phenotypes of a JNK1/JNK2 double null embryos exhibit both optic nerve coloboma and kidney hypoplasia (Weston et al., 2003), which is strikingly similar to a Pax2 partial loss of function. Thus, the activity of Pax2/5/8 proteins may be regulated by external stimuli that mediate JNK signaling in the developing kidney or nervous system. However, at least in the case of Pax5, transactivation activity can also be negatively regulated through interactions with the Groucho/TLE family of co-repressors (Eberhard et al., 2000).
The Groucho/TLE family encodes modular, nuclear proteins that interact with DNA binding factors and promote formation of the repressosome, a complex implicated in short and long term repression of transcription (Courey and Jia, 2001). In yeast, the Groucho related protein Tup1 can repress transcription in vitro, presumably through interactions with subunits of the RNA polymerase II holoenzyme. However, more long term repression by Groucho may involve interactions with histone deacetylases (HDACs) and the chromatin silencing machinery (Chen et al., 1999). Expression of the Groucho proteins Grg1, 3 and 4 is widespread during development and overlaps partially with the Pax2/5/8 family in the neural tube and the kidney (Dehni et al.,1995; Koop et al., 1996; Leon and Lobe, 1997; Muhr et al., 2001). In the nervous system, Groucho-mediated repression, through direct interactions with the Nkx family of homeodomain DNA-binding proteins, is necessary for patterning of the embryonic neural tube (Muhr et al., 2001). This spatially controlled repression, used to create boundaries among sets of neural precursor cells, has been termed the derepression strategy for neural cell specification. In the pituitary gland, Groucho/TLE1 binds to the homeodomain protein Hesx1/Rpx to suppress transcription of candidate target genes such as fgf8 and Prop1 (Dasen et al., 2001). However, deacetylation of histones through potential interactions with HDACs is not the only possible mechanism of Groucho-mediated repression, as experiments with deletion mutants (Fisher et al., 1996), deacetylase inhibitors (Chen et al., 1999; Yao et al., 2001) and anti-HDAC antibodies (Dasen et al., 2001) would suggest. Given the importance of Groucho/TLE interactions in the embryo, characterizing the potential biochemical mechanisms of Groucho-mediated repression will have profound implications for understanding patterning and differentiation during development.
In this report, we examine the mechanism of the Groucho protein Grg4 to negatively regulate Pax2-dependent gene activation. Grg4 can completely suppress Pax2-dependent activation of a reporter gene even in the presence of activated JNK. With increasing amounts of Grg4 expression, phosphorylation of the Pax2 activation domain is blocked. The ability of Grg4 to interact with and block Pax2 phosphorylation requires binding of Pax2 to a DNA target sequence. Thus, the data provide a new mechanism for Groucho-dependent suppression of activation whereby Groucho inhibits phosphorylation of the activation domain of a DNA-bound protein.
Results
Grg4 interacts with the Pax2/DNA complex
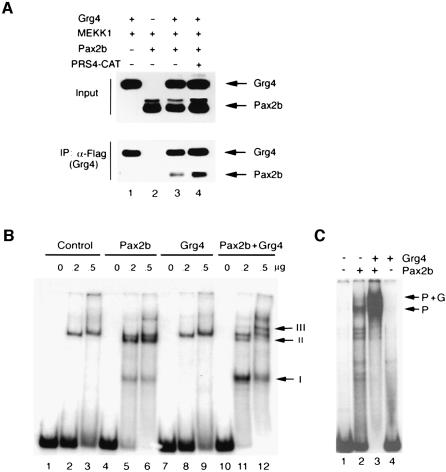
Given the known interaction of the Groucho family protein Grg4 with Pax5 (Eberhard et al., 2000), it was reasonable to postulate that Grg4 may also interact with the related protein Pax2. Within the carboxy terminal 190 amino acids, which contain the activation domain, Pax2 and Pax5 share >67% amino acid identity and >80% amino acid homology. To confirm that Pax2 and Grg4 did interact in cells, whole cell lysates from transfected 293 cells were immunoprecipitated with antibodies against epitope tagged Grg4 and immunoblotted for Pax2 (Figure 1A). Pax2 could be readily detected after immunoprecipitation for Grg4. In order to determine the effects of the Pax2 DNA binding site, PRS4, on the ability to interact with Grg4, we co-transfected the PRS4 containing plasmids prior to immunoprecipitation. The added PRS4 sequence increased the amount of Pax2 that co-precipitated with Grg4 (Figure 1A, lane 4). That Grg4 and Pax2 could interact upon the DNA template was further suggested by electrophoretic mobility shift assay (EMSA) using nuclear lysates from transfected cells (Figure 1B). With the PRS4-labeled sequence as a target, Pax2b transfected 293 cells show two specific shifted complexes (I and II), whereas the Grg4 protein does not exhibit any shifted complexes other than those observed with untransfected 293 cells. However, when Pax2 and Grg4 were present in the lysate, a third higher molecular weight shifted complex was observed (Figure 1B, III). Complex III was only observed when both Pax2 and Grg4 were present and most likely represents an interaction between Pax2, Grg4 and the Pax2 DNA binding site. Since it was difficult to definitively determine the composition of the protein/DNA complex III, we utilized recombinant Pax2 and Grg4 proteins to confirm their ability to form a complex with the PRS4 DNA sequence (Figure 1C). Poly-His tagged Pax2b was expressed in bacteria and purified by nickel affinity chromatography under denaturing conditions and dialyzed stepwise into binding buffer, whereas GST–Grg4 was purified by glutathione affinity under native conditions. Recombinant Pax2b binds PRS4 with a major shifted complex and several smaller complex present (P, Figure 1C). These smaller complexes may be due to breakdown or cleaved products of the recombinant Pax2b, which are evident on Coomassie blue stained gels from the final, purified fraction (data not shown). Addition of recombinant GST–Grg4 results in a slower mobility complex, presumably due to increased molecular mass of the Pax2/Grg4/DNA complex (P + G, Figure 1C). Also, the addition of Grg4 appears to increase the efficiency of DNA binding in vitro, as much of the free probe is now bound in the presence of both proteins. However, recombinant Grg4 does not bind PRS4 in the absence of Pax2 (Figure 1C, lane 4).
Fig. 1. Grg4 physically interacts with Pax2. (A) 293 cells were co-transfected with Grg4 (lanes 1, 3 and 4), MEKK1 (all lanes), Pax2b (lanes 2, 3 and 4) and PRS4 (lane 4) as indicated. One tenth of total lysate was analyzed by SDS–PAGE for verifying protein expression (Input). The remaining lysates were immunoprecipitated with anti-Flag antibody (IP) washed and analyzed by western blotting for Grg4 or Pax2. Co-IP of Pax2 is demonstrated by anti-Pax2 antibody (lanes 3 and 4). Note that Co-IP of Pax2 with Grg4 was enhanced by the presence of PRS4 plasmid (lane 4). (B) Interaction of Grg4 with Pax2 was examined by EMSA using oligonucleotides corresponding to the Pax2 binding site PRS4 and increasing amounts (0, 0.2 and 0.5 µg) of nuclear extracts from control cells (lanes 1, 2 and 3), Pax2b transfected cells (lanes 4, 5 and 6), Grg4 transected cells (lanes 7,8 and 9), and Pax2b and Grg4 co-transfected cells (lanes 10, 11 and 12). The arrows indicate the shifted species. Note the presence of two Pax2 specific gel shifts (I and II). Shift I and II are reduced in intensity and a higher molecular weight species III is evident upon Grg4 co-expression. (C) Recombinant Pax2 and Grg4 interact on the PRS4 DNA template. His-tagged Pax2b and GST–Grg4 were used to bind the PRS4 site as indicated. The predominant Pax2/DNA species is indicated (arrow, P; lane 2) and a higher molecular weight complex is observed upon the addition of Grg4 (arrow, P + G; lane 3). Grg4 alone does not bind the PRS4 sequence (lane 4).
Grg4 suppresses JNK-dependent Pax2 transcription activation
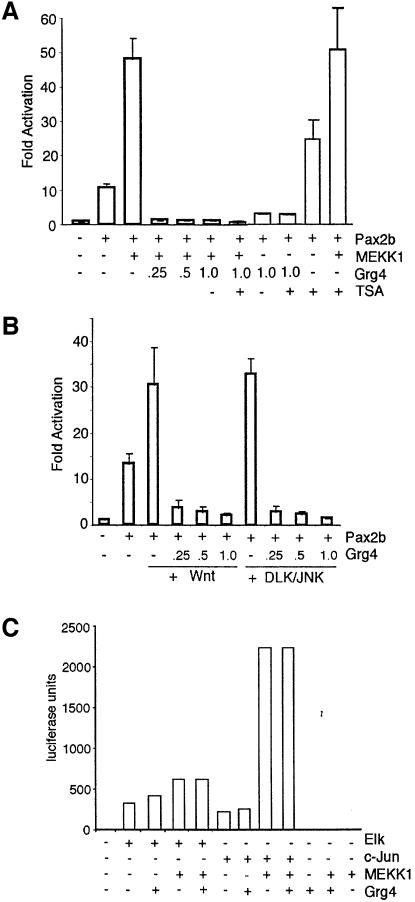
We next examined the ability of Grg4 to affect Pax2 mediated transcription activation. Given that the activation of the JNK pathway results in increased Pax2 phosphorylation and Pax2 dependent transactivation (Cai et al., 2002), cells were transfected with Pax2 and its reporter, PRS4CAT, and activators of the JNK pathway (Figure 2). Three different activators of JNK were utilized. These included the upstream kinase MEKK1, the mixed lineage kinase DLK and the secreted signaling protein Wnt4. While MEKK1 is able to increase Pax2-dependent transactivation from ∼10- to 50-fold, transfection of as little as 250 ng of Grg4 plasmid completely inhibits Pax2-dependent activation in this system (Figure 2A). In fact, Grg4 is better able to suppress Pax2 activation in the presence of MEKK1. Given the ability of Groucho proteins to interact with histone deactylases, we examined the effects of the HDAC inhibitor Trichostatin A (TSA) (Yoshida et al., 1995) on Grg4 suppression of Pax2-mediated activation (Figure 2A). TSA was able to increase Pax2-dependent activation ∼2-fold in the absence of MEKK1. TSA had little effect when MEKK1 was co-transfected with Pax2. Upon co-expression of Grg4 with Pax2, either with or without MEKK1, inhibition of HDACs did not affect the ability of Grg4 to suppress Pax2-dependent activation (Figure 2A). These data suggest that Grg4 suppression of Pax2 activation is not mediated simply by recruitment of HDACs to the chromatin template. Similar results were obtained if Wnt4 or DLK/JNK (Figure 2B) were used to activate JNK and enhance Pax2 dependent transactivation. In all cases examined, Grg4 completely blocked Pax2-mediated activation of the PRS4-CAT reporter construct. In fact, Grg4 inhibited basal levels of Pax2 dependent activation, in the absence of exogenous JNK activators.
Fig. 2. Grg4 inhibits the JNK enhanced transactivation potential of Pax2. (A) Induction of PRS4-CAT reporter plasmid expression in response to transfection with Pax2b, MEKK1 and increasing amounts of Grg4 (µg). MEKK1 (100 ng) induces a 5-fold increase in Pax2 transactivation. Grg4 completely inhibits Pax2 dependent activation in the presence or absence of MEKK1. The HDAC inhibitor TSA does not affect the ability of Grg4 to suppress Pax2-dependent activation, but does increase the basal level of Pax2-dependent activation. (B) Induction of the CAT reporter plasmid expression in response to transfection with Pax2b, Wnt4 or DLK/JNK and increasing amounts of Grg4 are shown. Grg4 suppresses Pax2-dependent activation regardless of how JNK is activated. (C) Grg4 does not affect c-Jun or Elk1 mediated activation. Transactivation assays using a Gal-Luc reporter plasmid and the Gal4 DNA binding domain fused to the activation domain of Elk1 or c-Jun are shown. Activation is expressed as relative light units of the luciferase reporter gene. Note, Grg4 has no inhibitory effect on MEKK1 activation of c-Jun or Elk1. Representative results from duplicate experiments are shown. All experiments except C are averages from at least three independent experiments with error bars corresponding to one standard deviation from the mean. All transfections were normalized to a β-galactosidase internal control vector.
As a control, we examined the effects of Grg4 on the activation of Elk1 or c-Jun dependent transcription using the gal4-dbd fused to the activation domains of Elk1 or c-Jun and a luciferase reporter (Figure 2C). In both cases, co-expression of Grg4 with MEKK1 did not suppress the activation potential of c-Jun or Elk1. Thus, Grg4 did not have a general suppressive effect on transient transfection based activation assays.
Inhibition of Pax2 phosphorylation by Grg4 is DNA dependent
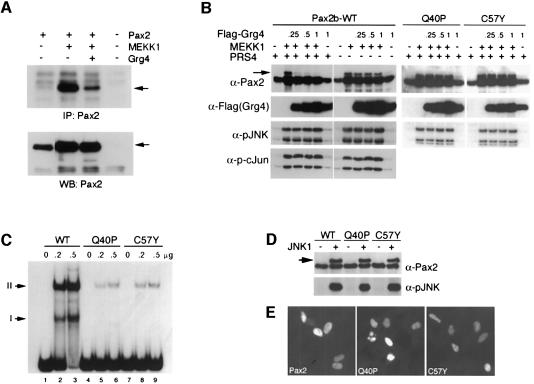
To more closely examine the mechanism of Grg4 mediated suppression, we examined the effect of Grg4 on the status of Pax2 phosphorylation (Figure 3). The level of Pax2 phosphorylation was observed in 293 cells by a 2 h in vivo labeling with 32P after transfection (Figure 3A). As previously published, MEKK1 significantly elevated levels of phosphorylated Pax2 (P-Pax2). Co-expression of Grg4 reduced levels of P-Pax2 on average 5–10-fold under the conditions used for this assay.
Fig. 3. Grg4 inhibits phosphorylation of Pax2 in a DNA-dependent manner. (A) In vivo phospho-labeling of 293 cells after transfection with the indicated plasmids is shown. After 2 h of 32P labeling, cells were washed and lysates were immunoprecipitated for Pax2 with the anti-HA monoclonal antibody, separated on SDS–PAGE, electroblotted and autoradiographed (top panel). The bottom panel shows the same blot probed with a rabbit anti-Pax2 antibody. Note the reduced levels of Pax2 phosphorylation upon co-expression of Grg4. (B) Steady state levels of Pax2 and P-Pax2 in cells co-transfected with the plasmids as indicated. Protein lysates were assayed by western blotting for Pax2, Grg4, P-JNK and P-cJun. Transfections were done in the presence of 1 µg PRS4 containing plasmid (+) or 1 µg of control vector (–), which has deleted the PRS4 sites. Note that increasing amounts of Grg4 reduce the levels of the wild-type P-Pax2 (arrow) but only in the presence of the PRS4 plasmid. The Pax2 DNA binding mutants, Q40P and C57Y, are phosphorylated in response to MEKK1 but do not exhibit reduced P-Pax2 levels with increasing Grg4 even in the presence of PRS4 vector. Grg4 has no effect on P-cJun levels. (C) Single point mutations in the Pax2 paired domain inhibit DNA binding. Increased amounts of nuclear extracts from cells transfected with wild-type (lanes 1–3) or Pax2 paired domain point mutations (Q40P, lane 4–6; C57Y, lane 7–9) were used for EMSAs. Wild-type Pax2 shows two shifted species (I and II) and very little free probe. Only a faint shift (II) is observed with either mutant and the amount of free probe is not reduced significantly. (D) Pax2 DNA binding mutants are efficient substrates for JNK. In vitro kinase reactions using recombinant activated JNK and lysates from cells transfected with either wild-type Pax2 or DNA binding mutants are shown. The arrow points to higher migrating, phosphorylated form of Pax2. (E) The Pax2 DNA binding mutants Q40P and C57Y locate to the nucleus in transfected cells by immunostaining with anti-Pax2 antibodies.
To examine the dose dependence and other parameters of the effects on Pax2 phosphorylation, we used the western blotting assay to measure the relative amounts of P-Pax2 in transfected 293 cells. As previously reported (Cai et al., 2002), phosphorylation of Pax2 by JNK results in an indicative shift in apparent molecular weight. Using the upstream kinase MEKK1, the hyperphosphorylated form of Pax2 is readily observed as an ∼50 kDa species migrating larger than the hypophosphorylated 48 kDa Pax2 isoform (Figure 3B). As the amount of Grg4 expression vector is increased, the amount of phosphorylated Pax2 (P-Pax2) clearly decreases. However, this decrease in P-Pax2 is only observed in the presence of excess PRS4-containing plasmid. By co-transfecting either PRS4-CAT or PRS4 cloned into Bluescript (Stratagene Inc.), Grg4 is able to inhibit P-Pax2 levels. Using vectors alone that have deleted the PRS4 sites, little decrease in P-Pax2 levels is observed. Furthermore, there is no inhibition of JNK activity as phospho-c-Jun levels are unaffected by Grg4. Similar results were obtained using DLK/JNK to phosphorylate Pax2 in the presence or absence of PRS4 (data not shown). These data indicate that Grg4 can inhibit or reduce the steady-state levels of P-Pax2 and that this effect is enhanced by the presence of excess Pax2 DNA binding sites.
If Pax2 must first bind to DNA in order for efficient interaction with Grg4 in vivo, then mutations in the Pax2 paired domain that inhibit DNA binding may not show Grg4 suppression of phosphorylation. While Pax2 point mutants that effect DNA binding have not been well described, the Pax2 and Pax8 paired domain are >95% homologous. Inactivating mutations in the DNA binding domain of Pax8 have been characterized (Macchia et al., 1998). Thus, two DNA binding mutants were generated in Pax2, based on naturally occurring Pax8 mutations mapped in humans (Congdon et al., 2001; Vilain et al., 2001). The glutamine to proline mutation at position 40 (Q40P) or the cysteine to tyrosine mutation at position 57 (C57Y) significantly inhibited the ability of Pax2 to bind to the PRS4 sequence (Figure 3C). As predicted, neither Q40P nor C57Y were able to transactivate the PRS4CAT reporter in the presence or absence of MEKK1 (data not shown). However, both mutant proteins were still phosphorylated with similar efficiency as wild-type Pax2 in an in vitro kinase assay using purified JNK (Figure 3D). Strikingly, in transfected cell lysates, using increasing amounts of Grg4 and the upstream activator MEKK1, Grg4 had no effect on the levels of phosphorylated Q40P or C57Y (Figure 3B). Similar amounts of hyperphosphorylated mutant Pax2 proteins were observed even with as much as 1.0 µg of Grg4 expression plasmid. This was in contrast to the marked inhibition of wild-type Pax2 phosphorylation by Grg4 (Figure 3B). As Grg4 is a nuclear protein, we tested to make sure both Pax2 DNA binding mutants were still able to locate to the cell nucleus (Figure 3E) Taken together, the data suggest that Grg4-mediated inhibition of Pax2 phosphorylation requires binding of Pax2 to its DNA target sites.
Co-localization of Pax2 and Grg4
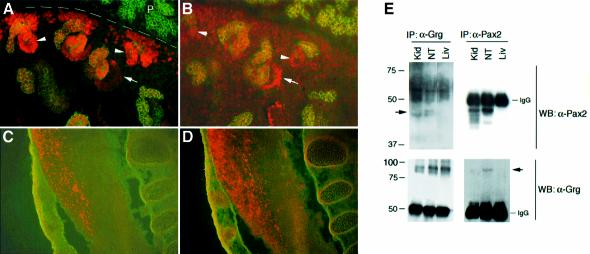
In order to examine the potential Pax2/Grg4 interaction in vivo, we utilized polyclonal antibodies for immunostaining and immunoprecipitation (Figure 4). Grg4 expression is widespread in the embryo, but exhibits local areas with high nuclear protein levels. In the developing kidney at E15.5, mesenchymal cells at the periphery of the tissue are aggregating around the tips of the ureteric bud epithelia and forming polarized vesicles (for review see Dressler, 2002). The polarized vesicles are the precursors of the s-shaped bodies and generate much of the glomerular, proximal tubular and distal tubular epithelia. As described (Dressler and Douglass, 1992; Ryan et al., 1995), Pax2 protein is found in the mesenchymal aggregates, the polarized vesicles and in the part of the s-shaped bodies. Grg4 is found at low levels in much of the mesenchyme at the periphery and within the interstitium. Grg4 levels are slightly higher in the polarized vesicles and are maximal in the proximal portion of the s-shaped body, just at a time when Pax2 begins to be down-regulated. Based on their characteristic position, the cells expressing maximal amounts of Grg4 are the precursors of the glomerular podocytes.
Fig. 4. Co-expression of Pax2 and Grg4 in the embryonic kidney and neural tube. Immunostaining with anti-Pax2 (red, A and C) or anti-Grg4 (red, B and D) and E-cadherin (green, A–D) was performed on E15.5 embryos. Sagital sections through the kidney (A and B) and spinal cord (C and D) are shown. (A) Pax2 localizes to the peripheral mesenchyme and early polarized renal vesicles (arrowhead) and is down-regulated in the proximal part of the s-shaped body (arrow). The dashed line marks the periphery of the developing kidney, with the embryonic pancreas (P) above. (B) In an adjacent kidney section, low levels of Grg4 staining are observed in peripheral and interstitial mesenchyme, with increasing protein levels in polarized renal vesicles (arrowhead) and s-shaped bodies (arrow). (C) In the neural tube, Pax2 expression is seen in many post-mitotic interneurons along the dorsal half of the intermediate zone. (D) Adjacent section to C showing the domain of Grg4 positive cells. (E) Immunoprecipitation of Grg4 and Pax2 from kidney (Kid), neural tube (NT) and liver (Liv) nuclear extracts prepared from E16 tissues. Nuclear extracts were immunoprecipitated with either anti-Grg4 (left panels) or anti-Pax2 (right panels) as indicated. The immunoprecipitates were then blotted for Pax2 (top panels) or Grg4 (bottom panels) as indicated. Pax2 is easily immunoprecipitated from kidney and neural tube and runs just below the IgG heavy chain (top right). Pax2 can also be seen in kidney and neural tube after immunoprecipitation with Grg4 (arrow, top left). Grg4 is immunoprecipitated from all three tissues (bottom left). Grg4 can also be seen co-precipitating with anti-Pax2 in neural tube and faintly in kidney (arrow, bottom right). Note that Grg4 is not detected after immunoprecipitation of liver extracts with anti-Pax2, indicating that co-precipitation is dependent on the presence of Pax2 protein.
In the neural tube Pax2 expression is complex, but is generally restricted to post-mitotic neurons as they populate discrete regions of the spinal chord (Burrill et al., 1997). At E15.5, high levels of Pax2 and Grg4 protein co-localize in the dorsal half at a time when Pax2 positive interneurons have migrated from the ventricular zone (Figure 4C and D). Grg4 expression overlaps well with Pax2 in this region of the neural tube. The same antibodies were used to immunoprecipitate Pax2 or Grg4 from embryonic nuclear lysates made from E16 kidneys, neural tube or liver (Figure 4E). Immunoprecipitation with anti-Grg4 followed by western blotting with anti-Pax2 indicated Pax2 protein co-precipitating with Grg4 in both kidney and neural tube extracts. The reciprocal immunoprecipitation with anti-Pax2 indicated Grg4 co-precipitation in the neural tube and with a very weak signal present in kidney extracts. Even though high levels of Grg4 are present in liver, no Grg4 signal was detected after anti-Pax2 immunoprecipitation of liver extracts indicating that the Grg4 signal observed in neural tube was dependent of Pax2 protein being present. These data indicate that Pax2 and Grg4 can co-localize in both neural tube and kidney, where Pax2 activity is known to be important. Furthermore, at least some fraction of Pax2 and Grg4 can associate in nuclear extracts from these tissues.
Discussion
Previously, we have shown that activation of JNK by upstream kinases or Wnt signaling enhanced Pax2 phosphorylation and transactivation potential (Cai et al., 2002). Grg4 is able to specifically block Pax2 phosphorylation and suppress transactivation, even in the presence of activated JNK. In the absence of exogenous JNK activators, Grg4 suppress Pax2-dependent activation below basal levels. The base level of Pax2 activation in 293 cells may also be due to low levels of P-Pax2, detected after longer exposures (Cai et al., 2002), as low endogenous JNK activity is still present. That phosphorylation of Pax2 by JNK may be an important regulator of activity is further underscored by the phenotype of JNK1/JNK2 double mutant mice (Weston et al., 2003), which exhibit optic nerve coloboma and renal hypoplasia in a manner very reminiscent of a Pax2 hypomorphic allele or Pax2 gene dosage reduction (Sanyanusin et al., 1995; Schimmenti et al., 1995; Torres et al., 1995, 1996; Favor et al., 1996).
The data presented in this report point to a model whereby Grg4 inhibits Pax2 phosphorylation once Pax2 is bound to DNA, or recruits specific phosphatases to deactivate Pax2. This implies that Pax2 is phosphorylated in the nucleus by nuclear kinases, such as activated JNK. This event must be independent of DNA binding, as the data with our Pax2 mutants indicate. Once bound to Pax2, Grg4 could recruit other factors known to associate with the repressosome to block access of JNK to the Pax2 activation domain and thus inhibit Pax2 transactivation. The ability of Grg4 to suppress the activation potential of Pax2 provides a new level of control for regulating genes transiently induced by Pax2, such as GDNF (Brophy et al., 2001) and WT1 (Dehbi et al., 1996). This mechanism could account for rapid short-term repression of Pax2 target genes within the kidney or nervous system once Grg4 is expressed. Such short term repression may be independent of HDACs. While this presumes Pax2 to be an exclusive activator of transcription, there may be additional targets of Pax2 that are normally repressed through these interactions with Groucho. The related gene Pax5 both activates and represses different genes within the same lymphoid precursor lineage (Nutt et al., 1998, 1999), yet it is unclear how Pax5 can distinguish among this set of target genes within the same cell. Our data argue that DNA binding is essential for the Pax2/Groucho interaction, suggesting that the paired domain binding to a DNA target alters or stabilizes the conformation of Pax2 and allows Groucho to recognize protein–protein interaction domains. Given the size and diversity of paired domain DNA binding sequences, whether the Pax2/Groucho interaction depends on the type of DNA target sequence must be considered.
These data suggest that the Pax2/Grg4 interaction can occur on the DNA template and is in fact strengthened when Pax2 is bound to DNA. In our EMSAs with nuclear lysates from Pax2 transfected cells, the identities of the shifted species I and II are not entirely clear, though in the presence of Grg4 the species II disappears more readily while the levels of species I are slightly enhanced. This could indicate preferential binding of Grg4 with the species II complex. Given the differences in motility, species II may represent two molecules of Pax2 bound to the optimal PRS4 sequence and this could favor an interaction with Grg4. Alternatively, the shifted species I and II could be two different conformations of the Pax2 protein or the result of proteolytic processing during preparation. That Pax proteins undergo conformational changes upon binding to DNA has been observed previously with Pax6 using circular dichroism spectroscopy (Epstein et al., 1994). The interaction of Grg4 with Pax5 is complex and involves both the Pax5 octapeptide and the transactivation domain (Eberhard et al., 2000). Thus, the conformation of Pax2 protein may determine how accessible these sites are to Grg4 interaction. We have confirmed the Pax2/Grg4/DNA interaction using recombinant proteins purified from Escherichia coli. These in vitro assays with recombinant proteins also suggest that Grg4 enhances binding of Pax2 to the PRS4 sequence, yet this could not explain the decreased level of transcription activation observed in vitro. To date, no evidence for Groucho modification of Pax DNA binding activity has been observed in vivo.
While the tissue-specific expression of Pax2/5/8 is precisely regulated to ensure correct patterning during development, the amount of Pax activity is also essential, as too little or too much can be detrimental. Reduced activity in heterozygous mice and humans is commonly associated with hypoplasia in the kidney, eye and neural crest (for review see, Chi and Epstein, 2002). Conversely, increased levels of Pax6 activity in the eye, due to multiple gene copy number, also disturbs normal development (Shedl et al., 1996). In the kidney, down-regulation of Pax2 expression is evident as renal epithelial progenitor cells differentiate and exit mitosis. Ectopic or persistent expression of Pax2 is associated with a variety of renal diseases such as Wilms’ tumor (Dressler and Douglass, 1992; Eccles et al., 1992), renal cell carcinoma (Gnarra and Dressler, 1995; Igarashi et al., 2001) and polycystic kidney (Ostrom et al., 2000). The Groucho proteins could play an important role in limiting the activity of the Pax2/5/8 family during terminal differentiation. Thus, understanding the biochemical mechanisms underlying Groucho-mediated inhibition of Pax gene function may provide novel avenues for therapeutic interventions.
Materials and methods
Plasmids
The CMV-Pax-2bHA and PRS4-CAT reported plasmid, which contains five tandem copies of the Pax2 binding site, PRS4, cloned upstream of the herpes simplex virus thymidine kinase promoter, were constructed as described previously (Fickenscher et al., 1993). The CMV-MEKK1 wild-type expression plasmid was a gift from Dr B. Margolis (University of Michigan). The HA-DLK, Flag-JNK, HA-epitope tagged N-terminal of JIP were provided by Dr Lawrence B. Holzman (University of Michigan). The Wnt4 cDNA (gift from A. McMahon) was inserted upstream of the CMV expression plasmid CB6. A partial Grg4 cDNA (gift from C. Lobe) was spliced with a 5′ Grg4 PCR product amplified from an embryonic kidney cDNA library and cloned into pFLAG-CMV2. All expression plasmids were sequenced for verification.
Antibodies
The antibodies used were anti-Pax2 (1:3000; Zymed Labs.), anti-Flag (M2 Monoclonal Antibody; Sigma, 1:2000), anti-HA (16B12; Covance, 1:2000). Anti-Myc (9E10; Covance, 1:2000), anti-Phospho-JNK (Thr183/Tyr185; Cell Signaling Technology, 1:2000) and anti-Phospho-c-Jun (Ser63; Cell Signaling Technology).
Transient transfection
HEK 293 cells were cultured in Dulbecco’s modified Eagle’s medium (glucose concentration, 450 mg/dl) supplemented with heat-inactivated 10% fetal bovine serum and 100 U/ml penicillin and 100 mg/ml streptomycin under humidified 5% CO2/95% air at 37°C. Subconfluent cells were transfected using 2–3 µg of plasmid DNA and 6 µl of Fugene 6 as described by the manufacturer (Roche Molecular Biochemicals). For 5 × 105 of 293 cells, 0.5 µg of CAT reporter plasmid was co-transfected with various doses of effector plasmid and 0.25 µg of pCH110, a β-galactosidase reporter plasmid used to monitor transfection efficiency. Forty hours after transfection, cells were harvested and extracts were prepared and normalized for transfection efficiency. CAT analysis was performed essentially as described previously (Cai et al., 2002). For HDAC inhibition, 100 ng/ml TSA (1 mg/ml stock in ethanol, Sigma) was added 20 h post transfection and the cells harvested after an additional 20 h. Percent acetylation was determined by scintillation counting, and a value of 1.0 was arbitrarily assigned to control parental plasmids. Each experiment was repeated three times and the results presented as average ±1 SD from the mean. Luciferase assays were done with the PathDetect (Stratagene Inc.) signal transduction reporting system according to the manufacturer’s protocols.
Immunoprecipitation, immunostaining and western blotting
For immunoprecipitation, cells were transfected with the expression plasmids as indicated in the legends. After 40 h, lysates were prepared in IP buffer (20 mM Tris–HCl pH 8.0, 100 mM NaCl, 0.5% Triton X-100, 10 mM NaF, 1 mM Na3VO4, 1 mM phenylmethysulfonyl fluoride, and protease inhibitor mixture). Lysates were incubated with desired antibodies for 2 h at 4°C. Antibodies were captured with protein G-Sepharose for 1 h and washed three times with IP-wash buffer (same as IP buffer except 0.1% Triton X-100). Proteins were eluted from protein G-Sepharose by boiling in 2× SDS–PAGE sample buffer and analyzed by SDS–PAGE. Western blots were performed with equal amounts of protein obtained by lysis of transiently transfected cells in PK lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 1 mM Na3VO4, 50 mM NaF, 1% Triton X-100, 10% glycerol and a mixture of protease inhibitors (Roche Molecular Biochemicals, cat 1836170). The lysates were separated by SDS–PAGE and immunoblotted with antibodies indicated in the Figure legends. Horseradish peroxidase-conjugated secondary antibodies were used to detect antigen–antibody complexes, which were visualized with an ECL detection system (Amersham).
Embryonic lysates were made from microdissected tissues by first isolating nuclei with a sucrose step-gradient as described (Phelps and Dressler, 1996) and lysing nuclei in IP buffer. Incubation with primary antibodies against Pax2 or Grg4 were done overnight at 4°C followed by 2 h with protein A/G sepharose and three washes with 1 ml IP buffer.
For immunocytochemistry, embryos were dissected free of extra-embryonic tissue, fixed for 1 h in 4% paraformaldehyde/PBS washed 2 × 30 min in PBS, and perfused overnight in 0.5 M sucrose/PBS at 4°C. Tissues were frozen, sectioned and air dried. Optimal Grg4 staining was observed after a 10 min fixation in methanol at –20°C. Sections were rinsed in PBS/0.1% Tween-20 and incubated with primary antibodies for 2 h. After additional washes, fluorescent-conjugated secondary antibodies (Sigma) were used to visualize Pax2 (Dressler and Douglass, 1992), Grg4 (Yao et al.,1998), and E-cadherin (mouse monoclonal, Transduction labs). Sections were photographed with a Nikon ES800 fluorescent microscope using a digital Spot camera.
In vivo labeling
293 cells were transfected with expression plasmids as indicated in the legends. The cells were cultured in phosphate-free Dulbecco’s modified medium for 2 h after 40 h of transfection, and then labeled with 200 µCi/ml [32P]orthophosphate for 2 h. Lysates were prepared in IP buffer and incubated with anti-HA antibodies at 4°C for 2 h. The immunocomplexes were adsorbed to protein G-Sepharose and were subjected to SDS–PAGE followed by blotting onto nitrocellulose and autoradiography. Quantitation was achieved by using PhosphoImager. The amount of Pax2 that was immunoprecipitated was assessed by re-immunoblotting of the PVDF membranes.
EMSAs
A 25 bp PRS fragment was excised from the PRS4-Cat vector via SPE1 digestion. The fragment was purified and isolated on a 4% Nuseive gel and end labeled with [32P]dCTP via the Klenow fill in reaction. Nuclear extracts from transfected 293 cells were made 40 h post transfection as described (Phelps and Dressler, 1996) and quantified with the Bradford assay. Binding reactions were performed in a total volume of 15 µl for 30 min at room temperature and contained increasing amounts of nuclear extract protein (0, 250 and 500 ng), 100 ng poly (dI-dC), 32P-labeled probe (20 000 d.p.m.). Free DNA and DNA–protein complexes were resolved at room temperature on 4 or 6% neutral polyacrylamide gels in 0.5× TBE at 120 V.
Recombinant proteins were expressed in BL21 E.coli at 30°C after induction with IPTG. After lysis and sonication, GST fusion proteins were purified using glutathione agarose and eluted with 5 mM reduced glutathione. His-tagged proteins were purified under denaturing conditions using 6 M GHCl and nickel affinity chromatography. His-tagged proteins were eluted at low pH and dialyzed stepwise into 4, 2 and 1 M urea in PBS, 0.1% NP-40 before final dialysis into Z-buffer (20% glycerol, 25 mM HEPES pH 7.8, 12.5 mM MgCl2, 1 mM DTT, 0.1 M KCl, 0.1% NP4).
Site-directed mutagenesis
Two different substitutions were made in the CMV-Pax2b plasmid utilizing the Quikchange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) as per the manufacturer’s directions. Specifically, a point mutation was introduced using an HPLC purified set of primers. Sequencing analysis confirmed that the substitutions had been introduced.
Acknowledgments
Acknowledgements
We thank L.Holzman for the DLK and JIP expression plasmids, B.Margolis for the MEKK1 expression plasmid and S. Camper for critical reading of the manuscript. This work was supported by a PILD Grant-in-aid and NIH grants DK54723 and DK54740 to G.R.D. P.D.B. and Y.C. are supported in part by fellowships from the Polycystic Kidney Research Foundation and the American Society of Nephrology Gottschalk award. S.S. is a Scholar of the Fonds de la Recherche en Sante du Quebec.
References
- Bouchard M., Pfeffer,P. and Busslinger,M. (2000) Functional equivalence of the transcription factors Pax2 and Pax5 in mouse development. Development, 127, 3703–3713. [DOI] [PubMed] [Google Scholar]
- Brophy P.D., Ostrom,L., Lang,K.M. and Dressler,G.R. (2001) Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development, 128, 4747–4756. [DOI] [PubMed] [Google Scholar]
- Burrill J.D., Moran,L., Goulding,M.D. and Saueressig,H. (1997) PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development, 124, 4493–4503. [DOI] [PubMed] [Google Scholar]
- Cai Y., Lechner,M.S., Nihalani,D., Prindle,M.J., Holzman,L.B. and Dressler,G.R. (2002) Phosphorylation of Pax2 by the c-Jun N-terminal kinase and enhanced Pax2-dependent transcription activation. J. Biol. Chem., 277, 1217–1222. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the co-repressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi N. and Epstein,J.A. (2002) Getting your Pax straight: Pax proteins in development and disease. Trends Genet., 18, 41–47. [DOI] [PubMed] [Google Scholar]
- Congdon T., Nguyen,L.Q., Nogueira,C.R., Habiby,R.L., Medeiros-Neto,G. and Kopp,P. (2001) A novel mutation (Q40P) in PAX8 associated with congenital hypothyroidism and thyroid hypoplasia: evidence for phenotypic variability in mother and child. J. Clin. Endocrinol. Metab., 86, 3962–3967. [DOI] [PubMed] [Google Scholar]
- Courey A.J. and Jia,S. (2001) Transcriptional repression: the long and the short of it. Genes Dev., 15, 2786–2796. [DOI] [PubMed] [Google Scholar]
- Dasen J.S. et al. (2001) Temporal regulation of a paired-like homeo domain repressor/TLE corepressor complex and a related activator is required for pituitary organogenesis. Genes Dev., 15, 3193–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehbi M., Ghahremani,M., Lechner,M., Dressler,G. and Pelletier,J. (1996) The paired-box transcription factor, PAX2, positively modulates expression of the Wilms’ tumor suppressor gene (WT1). Oncogene, 13, 447–453. [PubMed] [Google Scholar]
- Dehni G., Yanling,L., Husain,J. and Stifani,S. (1995) TLE expression correlates with mouse embryonic segmentation, neurogenesis and epithelial determination. Mech. Dev., 53, 369–381. [DOI] [PubMed] [Google Scholar]
- Dorfler P. and Busslinger,M. (1996) C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO J., 15, 1971–1982. [PMC free article] [PubMed] [Google Scholar]
- Dressler G.R. (2002) Development of the excretory system. In Rossant,J. and Tam,P.T. (eds), Mouse Development: Patterning, Morphogenesis and Organogenesis. Academic Press, San Diego, CA, pp. 395–420. [Google Scholar]
- Dressler G.R. and Douglass,E.C. (1992) Pax-2 is a DNA-binding protein expressed in embryonic kidney and Wilms tumor. Proc. Natl Acad. Sci. USA, 89, 1179–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D., Jimenez,G., Heavey,B. and Busslinger,M. (2000) Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J., 19, 2292–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles M.R., Wallis,L.J., Fidler,A.E., Spur,N.K., Goodfellow,P.J. and Reeve,A.E. (1992) Expression of the Pax2 gene in human fetal kidney and Wilms’ tumor. Cell Growth Differ., 3, 279–289. [PubMed] [Google Scholar]
- Epstein J., Cai,J., Glaser,T., Jepeal,L. and Maas,R. (1994) Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J. Biol. Chem., 269, 8355–8361. [PubMed] [Google Scholar]
- Favor J. et al. (1996) The mouse Pax-21neu mutation is identical to a human PAX2 mutation in a family with renal-coloboma syndrome and results in developmental defects of the brain, ear, eye and kidney. Proc. Natl Acad. Sci. USA, 93, 13870–13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickenscher H.R., Chalepakis,G. and Gruss,P. (1993) Murine Pax-2 protein is a sequence-specific trans-activator with expression in the genital system. DNA Cell Biol., 12, 381–391. [DOI] [PubMed] [Google Scholar]
- Fisher A.L., Ohsako,S. and Caudy,M. (1996) The WRPW motif of the hairy-related basic helix–loop–helix repressor proteins acts as a 4-amino-acid transcription repression and protein–protein interaction domain. Mol. Cell. Biol., 16, 2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnarra J.R. and Dressler,G.R. (1995) Expression of Pax-2 in human renal cell carcinoma and growth inhibition by antisense oligonucleotides. Cancer Res., 55, 4092–4098. [PubMed] [Google Scholar]
- Igarashi T., Ueda,T., Suzuki,H., Tobe,T., Komiya,A., Ichikawa,T. and Ito,H. (2001) Aberrant expression of Pax-2 mRNA in renal cell carcinoma tissue and parenchyma of the affected kidney. Int. J. Urol., 8, 60–64. [DOI] [PubMed] [Google Scholar]
- Koop K.E., MacDonald,L.M. and Lobe,C.G. (1996) Transcripts of Grg4, a murine groucho-related gene, are detected in adjacent tissues to other murine neurogenic gene homologues during embryonic development. Mech. Dev., 59, 73–87. [DOI] [PubMed] [Google Scholar]
- Lechner M.S. and Dressler,G.R. (1996) Mapping of Pax-2 transcription activation domains. J. Biol. Chem., 271, 21088–21093. [DOI] [PubMed] [Google Scholar]
- Leon C. and Lobe,C.G. (1997) Grg3, a murine Groucho-related gene, is expressed in the developing nervous system and in mesenchyme-induced epithelial structures. Dev. Dyn., 208, 11–24. [DOI] [PubMed] [Google Scholar]
- Macchia P.E. et al. (1998) PAX8 mutations associated with congenital hypothyroidism caused by thyroid dysgenesis. Nat. Genet., 19, 83–86. [DOI] [PubMed] [Google Scholar]
- Muhr J., Andersson,E., Persson,M., Jessell,T.M. and Ericson,J. (2001) Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell, 104, 861–873. [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Morrison,A.M., Dorfler,P., Rolink,A. and Busslinger,M. (1998) Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. EMBO J., 17, 2319–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Heavey,B., Rolink,A.G. and Busslinger,M. (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature, 401, 556–562. [DOI] [PubMed] [Google Scholar]
- Ostrom L., Tang,M.J., Gruss,P. and Dressler,G.R. (2000) Reduced Pax2 gene dosage increases apoptosis and slows the progression of renal cystic disease. Dev. Biol., 219, 250–258. [DOI] [PubMed] [Google Scholar]
- Phelps D.E. and Dressler,G.R. (1996) Identification of novel Pax-2 binding sites by chromatin precipitation. J. Biol. Chem., 271, 7978–7985. [DOI] [PubMed] [Google Scholar]
- Ryan G., Steele-Perkins,V., Morris,J., Rauscher,F.J.,III and Dressler,G.R. (1995) Repression of Pax-2 by WT1 during normal kidney development. Development, 121, 867–875. [DOI] [PubMed] [Google Scholar]
- Sanyanusin P., Schimmenti,L.A., McNoe,L.A., Ward,T.A., Pierpont,M.E.M., Sullivan,M.J., Dobyns,W.B. and Eccles,M.R. (1995) Mutation of the Pax2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat. Genet., 9, 358–364. [DOI] [PubMed] [Google Scholar]
- Schedl A., Ross,A., Lee,M., Engelkamp,D., Rashbass,P., van Heyningen,V. and Hastie,N.D. (1996) Influence of PAX6 gene dosage on development: overexpression causes severe eye abnormalities. Cell, 86, 71–82. [DOI] [PubMed] [Google Scholar]
- Schimmenti L.A., Pierpont,M.E., Carpenter,B.L., Kashtan,C.E., Johnson,M.R. and Dobyns,W.B. (1995) Autosomal dominant optic nerve cologomas, vesicouretric reflux and renal anomalies. Am. J. Med. Genet., 59, 204–208. [DOI] [PubMed] [Google Scholar]
- Torres M., Gomez-Pardo,E., Dressler,G.R. and Gruss,P. (1995) Pax-2 controls multiple steps of urogenital development. Development, 121, 4057–4065. [DOI] [PubMed] [Google Scholar]
- Torres M., Gomez-Pardo,E. and Gruss,P. (1996) Pax2 contributes to inner ear patterning and optic nerve trajectory. Development, 122, 3381–3391. [DOI] [PubMed] [Google Scholar]
- Vilain C. et al. (2001) Autosomal dominant transmission of congenital thyroid hypoplasia due to loss-of-function mutation of PAX8. J. Clin. Endocrinol. Metab. 86, 234–238. [DOI] [PubMed] [Google Scholar]
- Weston C.R., Wong,A., Hall,J.P., Goad,M.E., Flavell,R.A. and Davis,R.J. (2003) JNK initiates a cytokine cascade that causes Pax2 expression and closure of the optic fissure. Genes Dev., 17, 1271–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Liu,Y., Husain,J., Lo,R., Palaparti,A., Henderson,J. and Stifani,S. (1998) Combinatorial expression patterns of individual TLE proteins during cell determination and differentiation suggest non-redundant functions for mammalian homologs of Drosophila Groucho. Dev. Growth Differ., 40, 133–146. [DOI] [PubMed] [Google Scholar]
- Yao J., Lai,E. and Stifani,S. (2001) The winged-helix protein brain factor 1 interacts with Groucho and Hes proteins to repress transcription. Mol. Cell. Biol., 21, 1962–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M., Horinouchi,S. and Beppu,T. (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin strucutre and fucntion. BioEssays, 17, 423–430. [DOI] [PubMed] [Google Scholar]