Abstract
Microsomal triglyceride transfer protein (MTP), an endoplasmic reticulum lipid transfer protein critical for apolipoprotein B (apoB) secretion, regulates CD1d antigen presentation. We identified MTP variant 1 (MTPv1), a novel splice variant of mouse MTP, by polymerase chain reaction and Northern analysis in non–apoB-secreting tissues, including thymocytes and antigen-presenting cells (APCs). Edman degradation of MTPv1 isolated from transfected cells revealed three unique residues; however, recombinant MTP and MTPv1 had an equivalent protein disulfide isomerase association, subcellular localization, triglyceride transfer, phospholipid transfer, response to inhibitors, and ability to support apoB secretion. MTP and MTPv1 efficiently transferred phosphatidylethanolamine to CD1d in vitro. NKT cells fail to develop in fetal thymic organ culture (FTOC) treated with MTP antagonists. MTP-inhibited FTOCs produced negligible numbers of CD1d tetramer–positive cells and exhibited marked defects in IL-4 production upon stimulation with anti-CD3 or α-galactosylceramide–pulsed APCs. CD1d expression on CD4+CD8+ FTOC cells was unaffected by MTP inhibition. Thus, our results demonstrate that MTPv1 in thymocytes is critical to NKT cell development. We hypothesize that, when MTP is inactive, CD1d traffics to the cell surface and presents no lipid or a lipid that is incapable of mediating NKT cell selection and/or is refractory to lysosomal editing.
Microsomal triglyceride transfer protein (MTP) is a mainly ER-resident 97-kD lipid transfer protein that, when complexed with protein disulfide isomerase (PDI), transfers lipids to apolipoprotein B (apoB) for the production of very low density lipoprotein (VLDL) from hepatocytes or chylomicrons from intestinal epithelial cells (IECs) (1–3). MTP transfers phospholipids, triglycerides, and cholesterol to nascent apoB during its translocation into the ER. Kinetic studies of MTP-mediated transfer of various lipid classes revealed that MTP has at least two lipid transfer sites: a fast site that transfers triglycerides and cholesterol esters and a slow site that transfers phospholipids (2, 3). Although apoB-containing lipoproteins are largely composed of triglycerides, drosophila MTP, which cannot transfer triglycerides but can transfer phospholipids, can support apoB secretion (4). This finding demonstrates that the phospholipid transfer function of MTP is sufficient for apoB secretion. MTP is highly abundant in hepatocytes and IECs and has been found, along with modest apoB secretion, in cardiac myocytes, retina, and placenta (5–8). MTP deletion is embryonic lethal in mice, presumably because of insufficient lipid nutrient transfer from the placenta (9, 10). Humans homozygous for mutations in MTP develop abetalipoproteinemia, a disease characterized by malabsorption of lipids and lipophilic vitamins (11).
NKT cells are positively selected by CD1d on CD4+CD8+ thymocytes (12). Negative selection can also occur, as shown by deletion of NKT cells in fetal thymic organ cultures (FTOCs) treated with the potent agonist α-galactosylceramide (α-galcer); negative selection is mediated largely by thymus-resident DCs, but negative selection by CD4+CD8+ cells may also occur (13–17). NKT cells develop in mice that have CD1d expression restricted to T lineage cells, indicating that thymocyte expression of CD1d is sufficient for NKT development (18, 19).
Endogenous lipids, derived from both the ER and the endosomal compartment, likely play a role in NKT cell selection. Phosphatidylinostitol and phosphatidylcholine, presumably of ER origin, have been found associated with CD1d, and at least one phosphatidylinositol-reactive NKT cell hybridoma has been characterized (20–22). The vesicular trafficking protein AP-3 transports CD1d from the cell surface to the endosomal/lysosomal compartment. Lysosomal saposins, generated by cathepsin L–mediated cleavage of prosaposin, and GM2 activator protein can exchange ER-derived lipids bound by CD1d for lysosomal lipids such as iGb3. Loss of AP-3, prosaposin, or cathepsins L or S; decreased production of iGb3 caused by β-hexosaminidase B deficiency; or altered lipid storage caused by loss of Niemann-Pick type C1 all result in impaired NKT cell development (23–29). ER and endosomal lipids differ in their potential to select NKT cells; a tail-deleted form of CD1d, which remains at the cell surface and fails to traffic to endosomes, can activate NKT hybridomas with diverse, but not invariant, TCRs and exclusively supports development of diverse NKT cells (30, 31).
MTP has been shown to regulate CD1d antigen presentation in vitro and in vivo (32, 33). These previous studies found MTP mRNA, protein, and activity in multiple mouse and human tissues, including APCs. Silencing and chemical inhibition of MTP in APCs caused a selective defect in CD1d presentation. Furthermore, purified MTP directly transfers phospholipid to recombinant CD1d in vitro (33). In the current studies, we report that mouse hematopoietic cells express a novel isoform of MTP, which accounts for ubiquitous low level expression of MTP. We further explore the role of MTP in NKT cell development using FTOC and show that MTP lipid transfer activity is required for production of invariant NKT cells.
RESULTS
Mouse MTP is regulated by alternate splicing
Previous work from our laboratory made use of the MTPMx1 mouse to demonstrate the role of MTP in CD1d presentation on hepatocytes and IECs (32). This mouse, which contains loxp sites flanking the promoter and exon 1 of the mttp gene, undergoes Cre-mediated deletion upon injection with the TLR3 ligand, polyinosinic-polycytidylic acid (34). Deletion is highly efficient in the liver, resulting in the loss of MTP-mediated lipidation of apoB and, consequently, loss of VLDL secretion and low serum triglycerides (34). The MTPMx1 mouse has therefore been used as a model for lipoprotein disorders (35, 36).
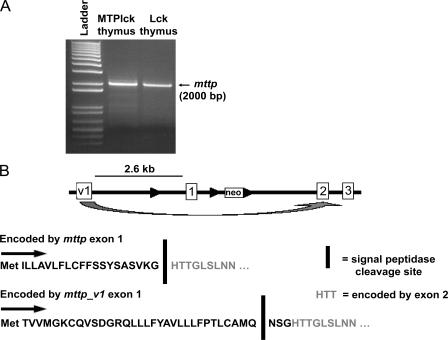
The Mx1 promoter displays low activity in the thymus (37). Therefore, in an attempt to parse out the role of MTP in thymic selection of NKT cells, we intercrossed the MTP-floxed mouse to an Lck-Cre mouse that has Cre expression driven by the proximal Lck promoter. The resulting MTPlck mice had >95% deletion of mttp at the genomic level (unpublished data) but showed no decrease in mttp transcription. We amplified and sequenced full-length cDNAs from wild-type and MTPlck thymi using 5′ and 3′ rapid amplification of cDNA ends (RACE), and identified a novel splice variant of MTP (Fig. 1 A). This alternate transcript of mttp (mttp_v1) makes use of an alternate exon 1 (exon v1) with a predicted start codon 2.6 kb upstream of the previously characterized start site. Based on sequence analysis of 5′ RACE products, exon v1 splices directly in frame with exon 2, skipping exon 1 (Fig. 1 B). Deletion of mttp exon 1 in the MTPMx1 or MTPlck mouse therefore does not affect transcription of mttp_v1.
Figure 1.
Mouse MTP in thymocytes is transcribed from an alternate promoter. (A) 5′ RACE was performed on cDNA from MTPlck thymocytes. mttp from Lck-Cre (Lck) thymocytes is shown as a positive control. The resulting bands were excised, cloned, and sequenced. (B) Diagram of mttp and mttp_v1 coding regions.
We sequenced full-length mttp cDNAs from a variety of other tissues, including DCs (splenic and BM-derived), the DN32 NKT hybridoma, and a fibroblast line (Table I). In each of these tissues, we found the mttp_v1 transcript. As a positive control, mouse hepatocytes contained the previously described mttp transcript.
Table I.
Summary of sequencing of MTP 5′ RACE products
| Tissue | Transcript | Number of clones |
|---|---|---|
| Thymus (wild type) | mttp_v1 | 1 |
| Thymus (MTPlck) | mttp_v1 | 2 |
| L cells | mttp_v1 | 1 |
| DN32 | mttp_v1 | 1 |
| Flt3L splenocytes | mttp_v1 | 1 |
| BM-derived DCs | mttp_v1 | 2 |
| Hepatocytes | mttp | 3 |
| aEyeball | mttp_v1 | NCBI (gi:74200200) |
Recently reported and available from GenBank/EMBL/DDBJ under accession no. 74200200.
mttp and mttp_v1 differ in their abundance and tissue distribution
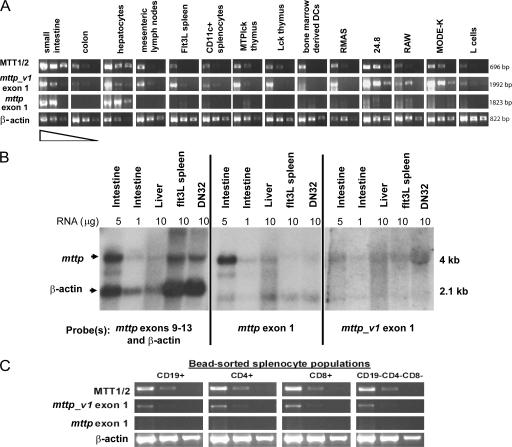
To analyze the tissue distribution of mttp and mttp_v1, we performed semiquantitative RT-PCR on cDNA dilutions from a panel of tissues and cell lines using primers that amplified mttp, mttp_v1, or both (MTT1/2). mttp_v1 was ubiquitously expressed, including the small intestine and hepatocytes, whereas mttp expression was restricted to the intestine, the liver, and the NKT cell hybridoma 24.8 (Fig. 2 A). mttp_v1 expression in intestine and hepatocyte samples may have resulted from contaminating leukocytes in these preparations; however, an IEC line, MODE_K, expressed mttp_v1, suggesting that IECs themselves are capable of transcribing mttp_v1, albeit at substantially lower levels than mttp.
Figure 2.
mttp and mttp_v1 differ in their tissue distribution. (A) RT-PCR was performed using cDNA synthesized from 1 μg of total RNA from the indicated tissues and cell lines. Panels show that RT-PCR products from cDNA used neat, 1:10 and 1:100 dilutions. MTT1/2 primers amplify total MTP transcripts; mttp_v1 exon 1 and mttp exon 1 primers amplify mttp_v1 and mttp, respectively. All RT-PCR for MTP used the same reverse primer (MTT2). RT-PCR for β-actin is shown as a loading control. Flt3L spleen indicates splenocytes from a mouse treated with Flt3L-secreting melanoma to increase numbers of splenic DCs; RMAS represents the mouse thymoma line; 24.8 indicates mouse autoreactive NKT hybridoma; RAW represents the mouse macrophage line; MODE-K indicates the mouse IEC line; and L cells represent the mouse fibroblast line. (B) Northern blot analysis was performed on total RNA from the indicated tissues. Each sample was run in triplicate, and identical blots were hybridized with probes corresponding to total MTP (mttp exons 9–13), mttp (exon 1), mttp_v1 (exon v1), or β-actin. DN32 indicates the mouse invariant NKT hybridoma. Blots were exposed to film for 8 h (left and middle) or 24 h (right). (C) Wild-type splenocytes were sorted using magnetic bead separation. Semiquantitative RT-PCR was performed as in A.
To confirm the RT-PCR data, we performed Northern blot analysis on RNA from the intestine, liver, DCs (Flt3L spleen), and an NKT hybridoma using probes specific for mttp, mttp_v1, or a downstream portion of the MTP gene common to both transcripts (mttp probe exons 9–13). Although MTP transcripts are clearly present in all four tissues, mttp was expressed predominantly in intestine and liver, whereas mttp_v1 accounted for the MTP expression in DCs and NKT cells (Fig. 2 B).
We further analyzed mttp and mttp_v1 expression in sorted splenic populations. mttp_v1 was detected in all populations, but we were unable to detect mttp (Fig. 2 C and not depicted). We therefore conclude that mttp_v1 is the major, and perhaps the only, transcript encoding MTP in mouse hematopoietic cells.
MTP transcription is differentially regulated in the mouse and human
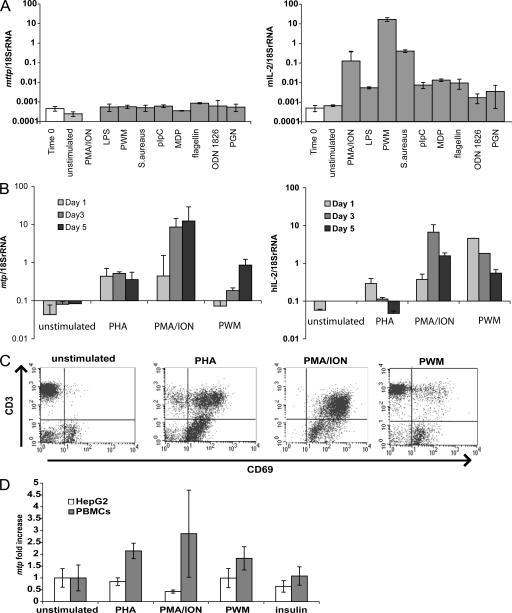
To assess the regulation of mttp_v1 transcription, we stimulated whole-mouse splenocytes with a panel of mitogens and TLR ligands and examined mttp_v1 expression by quantitative real-time PCR (Fig. 3 A). Although IL-2 transcription and CD69 surface expression were up-regulated in each case (Fig. 3 A and not depicted), no changes in mttp_v1 expression were observed.
Figure 3.
Mouse and human MTP exhibit different mechanisms of transcriptional regulation. (A) Wild-type mouse splenocytes were stimulated for 24 h with the indicated compounds. Total RNA was harvested, and 1 μg was used for cDNA synthesis. MTP (left) and IL-2 (right) transcript levels were quantified by real-time PCR and adjusted to 18S rRNA levels. Both mttp and mttp_v1 are amplified in this assay. (B) Human PBMCs were stimulated for 1, 3, or 5 d with the indicated mitogens. MTP (left) and IL-2 (right) transcript levels were quantified by real-time PCR as in A. (C) Human PBMCs stimulated for 3 d with the indicated mitogens were stained with anti-CD3 and anti-CD69 and analyzed by flow cytometry. (D) HepG2 cells and human PBMCs were stimulated for 3 d with the indicated compounds. MTP transcript levels, quantified by real-time PCR as in A, are shown as the fold change compared with unstimulated cells. Values are ±SD.
We next determined whether human tissues also show alternate splicing. Sequencing of mtp from monocyte-derived DCs, PBMCs, or a B cell lymphoma line revealed transcripts corresponding to the previously published sequence from human liver (38), indicating that human MTP is not regulated by alternate splicing (unpublished data). When human PBMCs were stimulated with a panel of mitogens, mtp transcription was up-regulated (Fig. 3 B), which is in contrast to observations in mouse splenocytes (Fig. 3 A). PBMC activation was confirmed by increased IL-2 production (Fig. 3 B) and surface CD69 expression (Fig. 3 C).
HepG2 is a human hepatoma cell line known to express MTP (39). HepG2 cells express 300-fold more mtp transcripts than PBMCs (unpublished data). When PBMCs and HepG2 cells were stimulated in parallel with PHA, PMA, and ionomycin (ION), or pokeweed mitogen, mtp transcription was up-regulated in PBMCs. In contrast, mtp was unchanged or down-regulated in HepG2 cells (Fig. 3 D). Insulin treatment, which is known to down-regulate mtp in HepG2 cells (39), decreased mtp transcription in HepG2 cells but had no effect on mtp transcription in PBMCs (Fig. 3 D).
MTP and MTP variant 1 (MTPv1) proteins are functionally equivalent
The amino acid sequence encoded by mttp_v1 suggested the presence of a signal peptide. To determine the exact signal peptidase cleavage site, we transduced Ld cells with a retrovirus encoding MTPv1 tagged at the C terminus with the FLAG epitope (MTPv1-FLAG). We purified MTPv1-FLAG by immunoprecipitation and observed that it associated with a 55-kD protein, presumably PDI, the “P” subunit of the microsomal triglyceride transfer complex (Fig. 4 A). We excised the 97-kD band corresponding to MTP and determined the N-terminal sequence by Edman degradation. These studies show that signal peptidase cleaves MTPv1 between residues 21 and 22, generating three novel amino acids (asn-ser-gly) in the mature protein that are not present in the original MTP sequence (Fig. 1 B).
Figure 4.
MTP and MTPv1 proteins are functionally equivalent. (A) Lysates from Ld cells transduced with MTP-FLAG or MTPv1-FLAG were immunoprecipitated with M2 resin. FLAG peptide–eluted fractions were subjected to SDS-PAGE and stained with Coomassie blue. (B) Confocal images were taken of Ld cells transduced with MTPv1-FLAG (top) or MTP-FLAG (bottom) and stained with M2 anti-FLAG and either anti-calnexin (right) or anti-PDI (left). Bar, 20 μm. (C) Lysates from Ld cells transduced with MTP or MTPv1 were normalized to contain equal amounts of MTP protein, as determined by immunoblot, and compared with lysates from untransduced Ld cells. Transfer of NBD-triglycerides to unlabeled acceptor vesicles is shown. (D) MTP-FLAG and MTPv1-FLAG were purified from transduced Ld cells, and 0.1 μg of purified protein was used per replicate. Transfer of NBD-PE to unlabeled acceptor vesicles is shown. (E) Ld and COS-7 cells untransduced and transduced with MTP or MTPv1 were transiently transfected with apoB48. Secretion of apoB48 was measured by ELISA of culture supernatants. (F) Ld cells were cultured in media containing vehicle or 13 μM BMS212122 for 5 d, harvested, fixed with glutaraldehyde, and washed before co-culture with 80,000 24.8 NKT cells. IL-2 was measured by ELISA of 24-h culture supernatants. (G) Ld untransduced cells and Ld cells transduced with MTP-FLAG or MTPv1-FLAG were cultured with 20,000 24.8 NKT cells (left) or pulsed with α-galcer for 6 h, washed, and cultured with 20,000 DN32 NKT cells (right). IL-2 was measured by ELISA of 24-h culture supernatants. Values are ±SD.
Although the N terminus of MTP is not part of a previously described functional domain, we investigated whether the addition of asn-ser-gly resulted in a functional difference between MTP and MTPv1. To this end, we generated retroviruses encoding MTP, MTPv1, MTP-FLAG, and MTPv1-FLAG. The FLAG tag was used for immunofluorescence or when purification of MTP was required; the untagged versions of MTP were used when whole cells or whole cell lysates would suffice. No differences in lipid transfer activity were observed between tagged and untagged MTP, either in the current study or in a previously published work (4).
Both forms of mouse MTP associated with PDI. Coomassie blue staining revealed equal amounts of the 55-kD “P” subunit coprecipitating with MTP-FLAG and MTPv1-FLAG (Fig. 4 A), and immunofluorescent staining of Ld cell transductants showed colocalization of MTP-FLAG and MTPv1-FLAG with PDI (Fig. 4 B). Furthermore, both isoforms of MTP were ER localized, as demonstrated by colocalization of MTP-FLAG and MTPv1-FLAG with the ER marker calnexin (Fig. 4 B).
We next addressed the lipid transfer function of MTP using a fluorescence-based assay (40, 41). MTP and MTPv1 were equally efficient at transferring triglyceride, as shown using whole cell lysates from transduced Ld cells (Fig. 4 C) or purified MTP-FLAG and MTPv1-FLAG (not depicted). Both isoforms of purified FLAG-tagged MTP were equally efficient at transferring phospholipid (Fig. 4 D). The triglyceride transfer and phospholipid transfer functions of MTPv1 could be inhibited by several chemical inhibitors of MTP, including BMS212122, BMS200150, and CP346086 (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20062006/DC1; and not depicted). Thus, MTPv1 lipid transfer is sensitive to at least some of the same compounds that block MTP activity. This is of particular importance because BMS212122 and the control compound 9-fluoronyl carboxylic acid (9-FL) were previously used to demonstrate the functional role of MTP in CD1d antigen presentation in APCs (31), and we now show that MTPv1 is the relevant form of MTP in these cells.
The major function of MTP in the liver and intestine is to transfer lipid to apoB. Therefore, we investigated whether MTPv1 could lipidate apoB. Using either Ld cells or COS cells stably transduced with MTP or MTPv1 and transiently transfected with apoB48, we found that both isoforms of MTP supported apoB secretion (Fig. 4 E). Although COS cells produced more apoB than Ld cells, no difference in apoB secretion could be observed between MTP- and MTPv1- transduced cells.
MTP-mediated transfer is the rate-limiting step in apoB secretion; studies show that production of buoyant apoB particles in cell culture and serum lipoprotein levels in vivo are positively correlated with MTP activity (42, 43). In contrast, MTP is not rate limiting for CD1d presentation, and trace amounts of MTP have been shown to be sufficient for maximal CD1d presentation in hematopoietic cells (33). In addition, the low endogenous levels of MTPv1 in commonly used CD1d-transfected hematopoietic and epithelial cell lines (e.g., C1R, HELa, or L cells) are capable of supporting high level CD1d expression. Ld MTP and Ld MTPv1 are fibroblast lines transfected with CD1d and transduced with MTP-FLAG and MTPv1-FLAG. The parent cell line, Ld, expresses MTPv1 (Fig. 2 A) and requires MTPv1 activity for antigen presentation on CD1d (Fig. 4 F). Although BMS212122 inhibition of MTP diminishes CD1d presentation in Ld cells, transduction of Ld cells with MTP or MTPv1 does not enhance CD1d presentation (Fig. 4, F and G). Ld, Ld MTP, and Ld MTPv1 stimulate the autoreactive NKT hybridoma 24.8 and present α-galcer to the NKT hybridoma DN32 with equal efficiency (Fig. 4 G). Overexpression of MTP did not confer any added benefit to CD1d presentation, confirming the hypothesis that MTP is not rate limiting for CD1d expression.
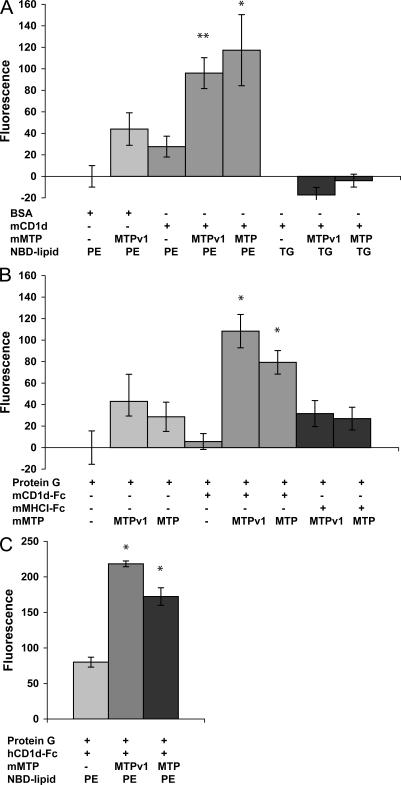
MTP and MTPv1 directly transfer phospholipid to mouse and human CD1d
We previously reported that MTP purified from rat liver could transfer fluorescently labeled phosphatidylethanolamine (PE) to immobilized mouse CD1d in vitro (33). Using the same in vitro lipid transfer assay, we found that MTP-FLAG and MTPv1-FLAG lipidate CD1d with equal efficiency, indicating that the relevant form of MTP in APCs is capable of transferring phospholipid to CD1d (Fig. 5). Mouse CD1d, both from a baculovirus expression system (Fig. 5 A) and as an Fc fusion protein (Fig. 5 B), accepted MTP-transferred phospholipid. When 7-nitro-2,1,3-benzoxadiazol-4-yl (NBD)– labeled triglyceride, which does not fit into the CD1d groove, was used in place of NBD-PE, no transfer of fluorescence to CD1d was observed (Fig. 5 A). As further evidence of the specificity of transfer to CD1d, MTP-FLAG and MTPv1-FLAG did not transfer phospholipid to immobilized MHC class I (Fig. 5 B). These data strongly indicate that MTP and MTPv1 load phospholipid into the CD1d antigen-binding groove. Of note, mouse MTP and MTPv1 also transferred phospholipid to human CD1d (Fig. 4 C). Given the 85% identity between mouse and human MTP and the fact that human NKT cells can recognize mouse CD1d and vice versa, this result is perhaps not surprising (44). Overall, we found that MTP lipidation of CD1d is robust, reproducible, and displays cross-species activity.
Figure 5.
MTP and MTPv1 directly transfer phospholipid to CD1d in vitro. (A) 2 μg BSA or recombinant mouse CD1d, produced in a baculovirus expression system, were coated onto a microtiter plate. Vesicles containing NBD-labeled PE or triglyceride were added with 0.1 μg of purified MTP-FLAG or MTPv1-FLAG, as indicated, for 2.5 h. After washing, isopropanol was added to each well, and the total fluorescence was measured. Values shown are total fluorescence minus background. *, P < 0.05; and **, P < 0.01 versus no MTP. (B) Lipid transfer assays were performed as in A except that microtiter wells were coated with protein G, protein G plus mouse CD1d-Fc, or protein G plus mouse MHC I-Fc. *, P < 0.05 versus transfer to protein G and P < 0.005 versus transfer to MHCI-Fc. (C) Lipid transfer assays were performed as in A, except that microtiter wells were coated with protein G or protein G plus human CD1d-Fc. *, P < 0.002 versus no MTP. Values are ±SD.
MTP activity is critical for NKT cell development
Thymocytes exclusively express the mttp_v1 transcript of MTP; thus, only this isoform is available to lipidate CD1d (Table I and Fig. 2 A). This is of particular importance given that CD1d-bearing thymocytes are responsible for NKT cell selection. The currently available MTP conditional knockout mice delete mttp exon 1 but retain expression of mttp_v1; thus, thymic MTP expression is not diminished in these mice (Fig. 1 A). To address the role of MTP in NKT cell selection, we examined FTOC. NKT cells develop after prolonged culture of fetal thymic lobes (14, 16), and our preliminary experiments showed that embryonic day 16 thymic lobes cultured for 18 d produced the maximal yield of NKT cells, as defined by staining with α-galcer–loaded CD1d tetramer (unpublished data).
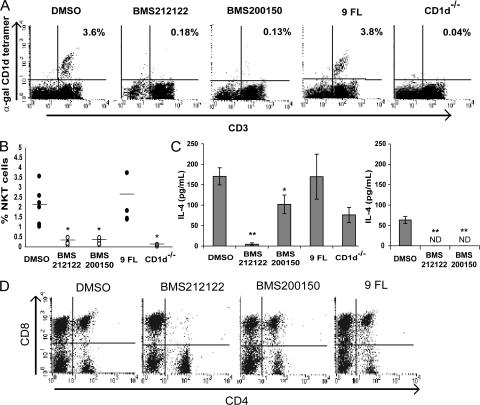
FTOCs treated with either BMS212122 or BMS200150 exhibited a profound defect in NKT cell development (Figs. 6, A and B). Although the percent yield of CD1d tetramer–positive NKT cells that developed in vehicle-treated or 9-FL–treated cultures was somewhat variable, we were unable to detect NKT cells by tetramer staining in either MTP-inhibited culture in six independent replicates (Fig. 6 B). Because noninvariant NKT cells or invariant NKT cells that have down-regulated their TCRs would not be detected by tetramer staining, we also looked for the functional presence of NKT cells. FTOC cells stimulated with plate-bound anti-CD3 produce IL-4, which is largely attributable to NKT cells (16). To verify that IL-4 production in this system is caused by the presence of NKT cells, FTOC cells from CD1d−/− thymic lobes were stimulated with anti-CD3. CD1d−/− FTOC cells produced less than half as much IL-4 as anti-CD3–stimulated wild-type FTOC cells, thus ascribing the production of IL-4 mainly to NKT cells in this assay (Fig. 6 C). Likewise, MTP-inhibited FTOC cells displayed a profound defect in IL-4 production, indicating the lack of NKT cells in these cultures (Fig. 6 C). Finally, MTP-inhibited FTOC cells failed to produce IL-4 in response to α-galcer–pulsed CD1d transfectants, demonstrating a specific loss of α-galcer–reactive NKT cells (Fig. 6 C).
Figure 6.
Inhibition of MTP in FTOC abrogates NKT cell development. (A) Embryonic day 16.5 fetal thymic lobes from wild-type embryos were cultured for 18 d in media containing vehicle (DMSO), 13 μM MTP inhibitor BMS212122, 10 μM MTP inhibitor BMS200150, or 13 μM of the inactive compound 9-FL. FTOC cells were harvested, stained for NKT cells using anti-CD3 and α-galcer–loaded CD1d tetramer, and analyzed by flow cytometry. FTOC cells from CD1d−/− fetal thymic lobes are shown as a negative control. Results are representative of six independent experiments, and percentages indicate CD3+ CD1d tetramer–positive NKT cells. (B) Quantification of NKT cell yields from six independent FTOC experiments, as in A. Horizontal lines represent the means. *, P < 0.02. (C) FTOC cells from wild-type fetal thymic lobes cultured with the indicated compounds were stimulated with plate-bound anti-CD3 (left) or CD1d-transfected RMAS cells pulsed with α-galcer (right). FTOC cells from CD1d−/− fetal thymic lobes are shown as a negative control. IL-4 concentration was measured by ELISA of 48-h culture supernatants. *, P = 0.01 versus DMSO and P = 0.1 versus 9-FL; **, P < 0.01. ND, not detectable. Values are ±SD. (D) Embryonic day 16.5 fetal thymic lobes from CD1d−/− embryos were cultured as in A. FTOC cells were harvested, stained with αCD4 and αCD8, and analyzed by flow cytometry.
To investigate nonspecific effects of the MTP inhibitor compounds in FTOCs, CD4 and CD8 T cell development was also analyzed. Although cultures treated with BMS200150 or 9-FL were indistinguishable from vehicle-treated cultures, BMS212122 affected CD4 and CD8 T cell development such that fewer double-positive and more single-positive T cells were observed (Fig. 6 D and Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20062006/DC1). BMS212122 also caused a modest decrease in surface CD1d expression in some, but not all, experimental replicates (Table II and Fig. S2). The effect of BMS212122 on T cell development was unrelated to the role of MTP on NKT cells in these cultures for the following reasons: the MTP inhibitor BMS200150 did not affect T cell development, and the effect of BMS212122 was observed in both wild-type and CD1d-deficient mice (Table III and Fig. 6 D). Table III shows the average CD4 and CD8 T cell yields from six independent FTOCs. BMS212122 showed a statistically significant alteration in CD4 and CD8 T cell development; BMS200150 and 9-FL did not.
Table II.
MFI of CD1d on FTOC cells
| FTOC culture | CD1d on total FTOC cells |
CD1d on CD4+CD8+ subset |
|---|---|---|
| DMSO | 20.9 ± 5.2 | 36.0 ± 3.7 |
| BMS 212122 | 12.8 ± 2.7 (0.04) | 37.0 ± 13.7 (NS) |
| BMS 200150 | 18.7 ± 4.2 (NS) | 38.1 ± 1.2 (NS) |
| 9-FL | 19.7 ± 2.6 (NS) | 38.1 ± 2.2 (NS) |
MFI of CD1d on total FTOC cells from wild-type fetal thymic lobes cultured with the indicated compounds. Results are averaged from four independent experiments ± SD. The numbers in parentheses indicate p-values versus DMSO controls.
Table III.
Thymic subsets in FTOCs treated with MTP inhibitor compounds
| CD4+CD8+ | CD4+ | CD8+ | |
|---|---|---|---|
| DMSO | 20.4% | 15.1% | 33.8% |
| BMS212122 | 4.8% (0.025) | 29.9% (0.05) | 21.9% (0.13) |
| BMS200150 | 20.2% (0.97) | 19.5% (0.41) | 29.3% (0.37) |
| 9-FL | 18.4% (0.76) | 14.9% (0.98) | 34% (0.97) |
Values are the average percent yield from six independent FTOC cultures. The numbers in parentheses indicate p-values versus DMSO controls.
The lack of NKT cells in MTP-inhibited FTOCs cannot also be attributed to loss of surface CD1d available for selection. Table II shows the median fluorescence intensity (MFI) of total and double-positive (CD4+CD8+) FTOC cells averaged from four independent experiments. The MTP inhibitor BMS200150 caused no alterations in CD1d surface expression on double-positive cells nor any of the other thymic subsets (Fig. S2). Thus, MTP-inhibited FTOCs produce normal numbers of double-positive cells expressing normal surface levels of CD1d; yet, without MTP function, NKT cells fail to develop.
DISCUSSION
In this paper, we report the discovery of mttp_v1, an alternate transcript of mouse mttp, and show that mttp_v1 is the major transcript in hematopoietic cells. The protein MTPv1 contains three novel amino acids but is functionally identical to MTP. Importantly, MTPv1 localizes to the ER and can transfer phospholipid to CD1d in vitro, which is consistent with its role as a chaperone and lipid transfer protein for CD1d. In thymocytes, MTP lipid transfer is required for NKT cell development; MTP inhibition in FTOC results in a complete loss of NKT cells.
Very little MTP protein is required to support CD1d antigen presentation (33). Low level ubiquitous expression of MTP in a wide range of CD1d-positive tissues seems ideal for supporting CD1d antigen presentation, and this is exactly the distribution pattern observed for mttp_v1. On the other hand, MTP is rate limiting for apoB secretion, and production of chylomicrons and VLDL is dependent on a large supply of MTP (42, 43). The discovery that MTP is regulated by transcription of mttp_v1 in virtually all tissues, including hematopoietic cells, whereas transcription of mttp is mainly detected in apoB-secreting tissues, provides a possible mechanism for regulating the dual roles of MTP.
Peroxisome proliferator–activated receptor α binds to a DR-1 site in the mttp promoter and is a known transcriptional activator of mttp in hepatocytes (45). Analysis of the 1,000 base pairs upstream of exon v1 showed neither a DR-1 site nor any other common cis elements, suggesting that different transcription factors regulate mttp versus mttp_v1. The lack of a TATA box in the promoter sequence, the relatively constant low level expression, and the absence of obvious cis-elements imply that mttp_v1 is part of the TATA-boxless housekeeping gene family (46). In contrast, human mtp, which is transcribed from a single promoter, is not expressed in resting PBMCs but is strongly up-regulated in response to mitogenic stimuli. The human hepatoma cell line HepG2 expresses high levels of mtp but down-regulates mtp transcription in response to inflammatory signals such as LPS and IL-1 (39) or PHA and PMA/ION treatment, as shown in this study. Inflamation-induced MTP down-regulation in hepatocytes presumably accounts for the presence of hepatic steatosis in many inflammatory conditions of human liver (39). Thus, tissue-specific regulation of MTP is achieved by alternate promoters in the mouse and by cell-type–specific transcription factors, such as hepatocyte nuclear factor–4, in humans (47).
Hepatocytes and IECs likely produce both MTP transcripts, but until now no group has detected mttp_v1 from these tissues, presumably because of the much higher abundance of mttp (48). The MTPMx1 knockout mouse and an intestinal-specific MTP knockout mouse show severe defects in VLDL and chylomicron secretion, respectively (34, 49). Because these mice still express mttp_v1, it is possible that MTPv1 cannot compensate for the loss of MTP with respect to lipoprotein production. Given that MTPv1 can support apoB secretion as efficiently as MTP when overexpressed, the most likely explanation for the MTPMx1 phenotype is that hepatocytes and IECs do not express trace amounts of MTPv1 protein and that the low level expression of MTPv1 compared with MTP is insufficient to rescue apoB secretion in the MTPMx1 mouse.
MTP and CD1d are highly conserved across species. We previously reported that rat MTP can lipidate mouse CD1d (33); we now show that mouse MTP lipidates mouse and human CD1d. In preliminary assays, bovine MTP is also capable of transferring lipid to mouse CD1d (unpublished data), an important point for future studies because bovine MTP can be obtained commercially and in large quantities.
MTP is presumed to load the first endogenous lipid into the CD1d groove. This initial lipid antigen could be responsible for the selection of diverse NKT cells, as this population of NKT cells is unaffected by alterations in CD1d trafficking to endolysosomes, indicating that ER-derived lipids are sufficient for selection (30, 31). Lysosomal lipid transfer proteins act on CD1d by editing the lipid antigen. We propose that, in the absence of MTP function, the CD1d groove is collapsed or contains a lipid refractory to editing. Thus, without MTP function, saposins and other lipid transfer proteins in the endosomal pathway are disabled in their ability to transfer lipid to CD1d.
CD1d has a remarkable ability to escape ER quality control mechanisms and can be expressed on the cell surface in the absence of β2-microglobulin or calnexin (50, 51). MTP inhibition in primary APCs and hepatocytes causes a modest decrease in surface CD1d, but MTP-inhibited CD1d transfectants show no decrease in surface protein despite marked defects in CD1d antigen presentation (32, 33). The MTP inhibitor BMS200150 had no effect on either the number of double-positive cells or CD1d surface expression on FTOC cells. The profound block in NKT cell development is therefore not caused by a quantitative lack of CD1d molecules but rather by a qualitative difference in the lipid ligand or structural folding of the CD1d groove. Similar observations were made in both AP-3–deficient mice and in mice lacking the lipid storage protein Niemann-Pick type C1; although thymocyte CD1d surface expression was unaffected or increased in these mice, NKT cells failed to develop (24, 27).
MTP is a critical chaperone and lipid transfer protein for CD1d in both mice and humans (32, 33). MTP activity in APCs is required for NKT cell activation, and we have now shown that MTP lipid transfer in thymocytes is required for NKT cell development. NKT cells play a causal role in many diseases, including asthma, autoimmune hepatitis, and inflammatory bowel disease (52–55). Understanding how MTP contributes to NKT cell activation and development may provide insight into possible therapies for these diseases.
MATERIALS AND METHODS
Animals.
Mice with two floxed mttp alleles were bred with mice transgenic for Lck promoter–driven Cre recombinase (Jackson ImmunoResearch Laboratories) to generate MTPlck mice (34). C57BL/6J animals (Jackson ImmunoResearch Laboratories) were used as wild-type controls. All animal experimentation was done in accordance with institutional guidelines and the review board of Harvard Medical School, which granted permission for this study.
Antibodies and reagents.
Cells were stained with the following reagents: FITC-conjugated anti-CD3 (2C11; BD Biosciences), FITC-conjugated anti-CD4 (H129.19; BD Biosciences), PE-conjugated anti-CD8 (ly 3.2; BD Biosciences), PE-conjugated CD1d (1B1; BD Biosciences), PE-conjugated CD1d tetramer loaded with α-galcer (National Institutes of Health [NIH] Tetramer Facility), and PE-conjugated anti-CD69 (H1.2F3; BD Biosciences). Staining was performed in the presence of Via-Probe (BD Biosciences) and analyzed with a flow cytometer (FACSort; Becton Dickinson). Cells were cultured with 5 μg/ml PHA (Sigma-Aldrich), 25 ng/ml PMA (Sigma-Aldrich), 1 μg/ml ION (Sigma-Aldrich), 300 ng/ml LPS (Sigma-Aldrich), 40 μg/ml pokeweed mitogen (Sigma-Aldrich), 0.012% wt/vol heat-killed Staphylococcus aureus (Calbiochem), 100 μg/ml polyinositic-polycytodylic acid (Sigma-Aldrich), 1 μg/ml muramyl dipeptide (InvivoGen), flagellin (5 μg/ml, Invivogen), ODN1826 (0.5 μM, InvivoGen), 10 μg/ml peptidoglycan (Sigma-Aldrich), and 100 nM insulin (Sigma-Aldrich).
RACE PCR.
Total RNA was isolated and quantified from mouse tissues or cell lines using TRIzol (Invitrogen). RNA was quantified, and 0.2–1 μg was used for 5′ or 3′ RACE–ready cDNA synthesis using a RACE cDNA amplification kit (SMART; CLONTECH Laboratories, Inc.). RACE products were amplified using: five cycles of 94°C for 30 s and 72°C for 3 min; five cycles of 94°C for 30 s, 70°C for 30 s, and 72°C for 3 min; and 25 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min. mttp-specific primers were as follows: 5′ RACE, TGACAGCAGCGGCAGTTGTGCGCACCGT; and 3′ RACE, ATGCCTTGCTTCCCGAAGGCATCCCGCT. Amplification products were visualized by agarose gel electrophoresis and ethidium bromide staining. DNA was purified from excised gel slices (QIAGEN), ligated into TOPO TA Cloning vector (Invitrogen), and used to transform bacteria (OneShot TOP10; Invitrogen). Plasmids were reisolated from bacterial cultures (QIAGEN) and sequenced.
Semiquantitative and real-time PCR.
Total RNA was extracted from tissues and cell lines, and 1 μg RNA was used for cDNA synthesis. For semiquantitative RT-PCR, 5 μl of neat, 1:10 or 1:100 diluted cDNA was used per 25-μl PCR. MTP transcripts were amplified using the following primers: mttp exon 1 (forward primer located in exon 1), 5′-ATGATCCTCTTGGCAGTGCTT-3′; mttp_v1 exon 1 (forward primer located in exon v1), 5′-GGTCTCACTGAATCTGGAGAGGAA-3′; MTT1 (forward primer located in exon 9), 5′-GGACTTTTTGGATTTCAAAAGTGAC-3′; and MTT2 (reverse primer located in exon 13), 5′-GGAGAAACGGTCATAATTGTG-3′. Beta-actin was amplified using the following primers: 5′-ATCTGGCACCACACCTTCTACATTGAGCTGCG-3′ and 5′-CGTCATACTCCTGCTTGCTGATCCACATCTGC-3′. For real-time PCR, the following primers were used: mouse MTP forward, 5′-GTGGAGGAATCCTGATGGTGA-3′; mouse MTP reverse, 5′-TGATCTTAGGTGTACTTTTGCCC-3′; mouse IL-2 forward, 5′-GTGCTCCTTGTCAACAGCG-3′; mouse IL-2 reverse, 5′-GGGGAGTTTCAGGTTCCTGTA-3′; human MTP forward, 5′-TGTGGCCTTACTATGGAGGAA-3′; human MTP reverse, 5′-AAGGAGCGTAGGTCTTTGCAG-3′; human IL-2 forward, 5′-TCCTGTCTTGCATTGCACTAAG-3′; human IL-2 reverse, 5′-CATCCTGGTGAGTTTGGGATTC-3′; 18S RNA forward, 5′-GAGGGAGCCTGAGAAACGG-3′; and 18S RNA reverse, 5′-GTCGGGAGTGGGTAATTTGC-3′. Real-time PCR was performed using SYBR green (BioRad Laboratories) and 40 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Final PCR amplification products were subjected to agarose gel electrophoresis and ethidium bromide staining to confirm product size.
Northern blotting.
PCR products corresponding to mttp and mttp_v1 (primers MTT1 and MTT2; 699-bp amplicon), mttp_v1 (primers alt1F and alt1R [5′-GTAGGAAGGAGTAACAGCACGGCA-3′]; 207-bp amplicon), or mttp (primers orig1Fb [5′-GCAGAGGGAGCCAGCATGATCCTCTTGGC-3′] and orig1R [5′-TGAGAGGCCAGTTGTGTGAC-3′]; 96-bp amplicon) were labeled with 50 μCi [32P]dCTP (Redivue nucleotides; GE Healthcare) using a labeling kit (DecaPrime II; Ambion). Northern blotting was performed using Northern Max (Ambion), according to the manufacturer's instructions. In brief, total RNA samples were loaded in triplicate on a formaldehyde agarose gel, electrophoresed, transferred by downward capillary transfer to a nylon membrane (Brightstar; Ambion), and UV cross-linked. Radiolabeled probes were denatured and hybridized to the membranes at 42°C in buffer (UltraHyb; Ambion) for 16 h; washed twice in 2× SSC, 0.1% SDS at room temperature; washed twice in 0.1× SSC, 0.1× SDS at 42°C; and exposed to film (BioMax MS; Kodak).
Cell lines and retroviral constructs.
Cells were maintained in DMEM-10 (2 mM l-glutamine, 10% fetal bovine serum) unless otherwise indicated. MODE-K, a mouse IEC line, has been previously described (56). DN32.D3 (termed DN32 in these studies), a mouse Vα14Jα18 invariant TCR-positive T cell hybridoma, was provided by A. Bendelac (University of Chicago, Chicago, IL [57]). The autoreactive mouse Vα14Jα18 invariant TCR-positive hybridoma 24.8 was provided by S. Behar (Brigham and Women's Hospital, Boston, MA [22, 58]). RMAS is a previously described TAP-deficient mouse T cell lymphoma (59); RAW is an Abelson virus–transformed mouse macrophage cell line (American Type Culture Collection); and HepG2 is a human hepatoma cell line (American Type Culture Collection). COS-7 cells were purchased from the American Type Culture Collection. Ld is a previously described fibroblast cell line transfected with the pSRαCD1d plasmid (60). MTP and MTPv1 sequences were cloned into pMFG and used to transfect the packaging cell line 293GPG, as previously described (61, 62). 293GPG and pMFG were provided by R. Mulligan (Children's Hospital, Boston, MA) and G. Dranoff (Dana Farber Cancer Institute, Boston, MA). MTP-FLAG and MTPv1-FLAG vectors were produced by PCR with a 3′ antisense primer encoding the FLAG sequence (DYKDDDDK), followed by an in-frame termination codon. Stably transduced cell lines were generated by three rounds of infection with transfected 293GPG culture supernatants.
Immunofluorescence.
Ld cells transduced with MTP-FLAG or MTPv1-FLAG were grown at low density on glass coverslips in 24-well plates. Cells were fixed and permeabilized in methanol for 15 min at −20°C. The fixed cells were blocked with PBS containing 1 mM MgCl2, 0.5 mM CaCl2, 3% BSA, and 1% goat serum and incubated with antibodies diluted 1:100 (unless stated otherwise) in the same buffer at room temperature for 1 h. The cells were incubated with mouse anti-FLAG M2 (Sigma-Aldrich) and either rabbit anti-calnexin (StressGen Biotechnologies) or anti-PDI (1:50; StressGen Biotechnologies), followed by treatment with Alexa Fluor 488 (green fluorescence)–conjugated goat anti–rabbit IgG1 and Alexa Fluor 594 (red fluorescence)–conjugated goat anti–mouse IgG1 (Invitrogen). The coverslips were mounted in phosphate-buffered saline containing 10% glycerol and 12% triethyldiamine (Sigma-Aldrich), to prevent fluorescent bleaching, and visualized using a confocal microscope (Radiance 2000; Bio-Rad Laboratories).
FLAG purification and SDS-PAGE.
MTP-FLAG– and MTPv1-FLAG–expressing cells were washed three times in cold PBS, once in buffer K (1 mM Tris, pH 7.4, 1 mM MgCl2, 1 mM EGTA, and complete protease inhibitor cocktail; Roche), and lysed by collecting cells in buffer K with a cell scraper, followed by repeated passage through a 30-gauge needle. Lysates were centrifuged to remove debris, and the salt concentration was increased to 10 mM Tris, pH 7.4, 150 mM NaCl. Lysates were incubated with M2 resin (Sigma-Aldrich) for 4–6 h at 4°C. Proteins were eluted with FLAG peptide (Sigma-Aldrich), and an aliquot of each preparation was subjected to SDS-PAGE, followed by Coomassie blue staining.
Triglyceride and PE transfer assays.
The lipid transfer assay using NBD-labeled triglyceride or NBD-labeled PE embedded in donor phospholipid vesicles (Chylos Inc.) has been previously described (40, 41). PE transfer to CD1d has also been previously described (33). In brief, black, high binding microtiter plates (Costar) were coated overnight at 4°C with 2 μg BSA, 0.0125 mol of recombinant protein G (Pierce Chemical Co.) and/or 0.025 mol/well of recombinant mouse CD1d (NIH Tetramer Facility), mouse CD1d-Fc (DimerX; BD Biosciences), mouse MHC class I–Fc (DimerX; BD Biosciences), or human CD1d-Fc (DimerX; BD Biosciences). Wells were washed three times with PBS. FLAG-tagged MTP or MTPv1, and PE vesicles containing 6:1 PE/NBD-PE were resuspended in transfer buffer (1 mM Tris-HCl, pH 7.4, 0.2 mM EDTA, 15 mM NaCl, 0.1% fatty acid–free BSA; Sigma-Aldrich), added to the wells, and incubated at 37°C for 2.5 h. Wells were then washed three times with PBS, and 100 μl isopropanol was added to each well and incubated for 2 min at room temperature. Fluorescence was read with a fluorescence plate reader using 485-nm excitation and 550-nm emission wavelengths.
ApoB secretion assay.
COS-7 and Ld cells grown in DMEM-10 (2 mM l-glutamine, 10% fetal bovine serum) were stably transduced with retroviruses encoding MTP or MTPv1. The cells were initially dislodged from the plate with trypsin and seeded in six-well plates (400,000 cells/well). Transfection of an apoB48 plasmid was performed using a transfection reagent (FuGENE 6; Roche). At 48 h after transfection, the media was aspirated, and 1 ml of lipid-containing medium (DMEM including 0.4 mM oleic acid complexed with 1.5% BSA and 1 mM glycerol) was added. ApoB concentration was measured by ELISA of 24-h culture supernatants. Protease inhibitors (Sigma-Aldrich) were added to culture supernatants before ELISA.
FTOC.
Embryonic day 16 fetal thymic lobes were harvested from timed pregnant C57BL/6J or CD1d−/− mice and cultured as previously described, with minor modifications (16). Three to six fetal thymic lobes per well were cultured on nitrocellulose filters (Whatman) placed on a sponge (Gelfoam size 4; Upjohn Pharmacia). Lobes were cultured for 18–20 d in 3 ml DMEM containing 20% fetal bovine serum per well on six-well tissue culture plates. 0.1% DMSO, 13 μM BMS212122 dissolved in DMSO (provided by R. Gregg, Bristol-Myers Squibb, Princeton, NJ), 10 μM BMS200150 dissolved in DMSO (provided by R. Gregg), or 13 μM 9-FL dissolved in DMSO (Sigma-Aldrich) was added to the media throughout the culture period. Media was changed every 3–4 d. Cells were harvested by mechanical disruption of the thymic lobes and passage through a 70-μm cell strainer. FTOC cells were resuspended in fresh media containing no MTP inhibitors and cultured in 96-well plates coated with 10 μg/ml anti-CD3 (2C11; BD Biosciences). IL-4 concentration was determined by ELISA of 48-h culture supernatants. A portion of the FTOC cells were used for flow cytometry and/or RNA isolation.
Statistics.
The Student's t test was used to determine significance.
Online supplemental material.
Fig. S1 shows that MTP inhibitors BMS212122 and BMS200150 block MTPv1-mediated phospholipid transfer. Fig. S2 shows a representative FACS plot of CD4 and CD8 thymocyte subsets in MTP inhibitor–treated FTOCs, as well as the CD1d surface expression on each subset. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20062006/DC1.
Acknowledgments
We thank Drs. M. Brenner, G. Dranoff, H. Ploegh, R. Gregg, J. Wetterau, and M. Dougan for advice and reagents.
M.M. Hussain was supported by NIH grants DK46900 and HL64272. R.S. Blumberg was supported by NIH grants DK44319, DK53056, and DK51362 and Harvard Digestive Disease Center grant P30 DK034854.
The authors have no conflicting financial interests.
Abbreviations used: 9-FL, 9-fluoronyl carboxylic acid; α-galcer, α-galactosylceramide; apoB, apolipoprotein B; FTOC, fetal thymic organ culture; IEC, intestinal epithelial cell; ION, ionomycin; MFI, median fluorescence intensity; MTP, microsomal triglyceride transfer protein; MTPv1, MTP variant 1; NBD, 7-nitro-2,1,3-benzoxadiazol-4-yl; PDI, protein disulfide isomerase; PE, phosphatidylethanolamine; RACE, rapid amplification of cDNA ends; VLDL, very low density lipoprotein.
References
- 1.Hussain, M.M., J. Shi, and P. Dreizen. 2003. Microsomal triglyceride transfer protein and its role in apoB-lipoprotein assembly. J. Lipid Res. 44:22–32. [DOI] [PubMed] [Google Scholar]
- 2.Jamil, H., J.K. Dickson Jr., C.H. Chu, M.W. Lago, J.K. Rinehart, S.A. Biller, R.E. Gregg, and J.R. Wetterau. 1995. Microsomal triglyceride transfer protein. Specificity of lipid binding and transport. J. Biol. Chem. 270:6549–6554. [DOI] [PubMed] [Google Scholar]
- 3.Atzel, A., and J.R. Wetterau. 1994. Identification of two classes of lipid molecule binding sites on the microsomal triglyceride transfer protein. Biochemistry. 33:15382–15388. [DOI] [PubMed] [Google Scholar]
- 4.Rava, P., G.K. Ojakian, G.S. Shelness, and M.M. Hussain. 2006. Phospholipid transfer activity of microsomal triacylglycerol transfer protein is sufficient for the assembly and secretion of apolipoprotein B lipoproteins. J. Biol. Chem. 281:11019–11027. [DOI] [PubMed] [Google Scholar]
- 5.Boren, J., M.M. Veniant, and S.G. Young. 1998. Apo B100-containing lipoproteins are secreted by the heart. J. Clin. Invest. 101:1197–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, C.M., J.B. Presley, X. Zhang, N. Dashti, B.H. Chung, N.E. Medeiros, C. Guidry, and C.A. Curcio. 2005. Retina expresses microsomal triglyceride transfer protein: implications for age-related maculopathy. J. Lipid Res. 46:628–640. [DOI] [PubMed] [Google Scholar]
- 7.Madsen, E.M., M.L. Lindegaard, C.B. Andersen, P. Damm, and L.B. Nielsen. 2004. Human placenta secretes apolipoprotein B-100-containing lipoproteins. J. Biol. Chem. 279:55271–55276. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen, L.B., M. Veniant, J. Boren, M. Raabe, J.S. Wong, C. Tam, L. Flynn, T. Vanni-Reyes, M.D. Gunn, I.J. Goldberg, et al. 1998. Genes for apolipoprotein B and microsomal triglyceride transfer protein are expressed in the heart: evidence that the heart has the capacity to synthesize and secrete lipoproteins. Circulation. 98:13–16. [DOI] [PubMed] [Google Scholar]
- 9.Terasawa, Y., S.J. Cases, J.S. Wong, H. Jamil, S. Jothi, M.G. Traber, L. Packer, D.A. Gordon, R.L. Hamilton, and R.V. Farese Jr. 1999. Apolipoprotein B-related gene expression and ultrastructural characteristics of lipoprotein secretion in mouse yolk sac during embryonic development. J. Lipid Res. 40:1967–1977. [PubMed] [Google Scholar]
- 10.Raabe, M., L.M. Flynn, C.H. Zlot, J.S. Wong, M.M. Veniant, R.L. Hamilton, and S.G. Young. 1998. Knockout of the abetalipoproteinemia gene in mice: reduced lipoprotein secretion in heterozygotes and embryonic lethality in homozygotes. Proc. Natl. Acad. Sci. USA. 95:8686–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berriot-Varoqueaux, N., L.P. Aggerbeck, M. Samson-Bouma, and J.R. Wetterau. 2000. The role of the microsomal triglygeride transfer protein in abetalipoproteinemia. Annu. Rev. Nutr. 20:663–697. [DOI] [PubMed] [Google Scholar]
- 12.Bendelac, A. 1995. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J. Exp. Med. 182:2091–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumann, J., P. Pittoni, E. Tonti, H.R. Macdonald, P. Dellabona, and G. Casorati. 2005. Targeted expression of human CD1d in transgenic mice reveals independent roles for thymocytes and thymic APCs in positive and negative selection of Valpha14i NKT cells. J. Immunol. 175:7303–7310. [DOI] [PubMed] [Google Scholar]
- 14.Pellicci, D.G., A.P. Uldrich, K. Kyparissoudis, N.Y. Crowe, A.G. Brooks, K.J. Hammond, S. Sidobre, M. Kronenberg, M.J. Smyth, and D.I. Godfrey. 2003. Intrathymic NKT cell development is blocked by the presence of alpha-galactosylceramide. Eur. J. Immunol. 33:1816–1823. [DOI] [PubMed] [Google Scholar]
- 15.Hayakawa, Y., S.P. Berzins, N.Y. Crowe, D.I. Godfrey, and M.J. Smyth. 2004. Antigen-induced tolerance by intrathymic modulation of self-recognizing inhibitory receptors. Nat. Immunol. 5:590–596. [DOI] [PubMed] [Google Scholar]
- 16.Chun, T., M.J. Page, L. Gapin, J.L. Matsuda, H. Xu, H. Nguyen, H.S. Kang, A.K. Stanic, S. Joyce, W.A. Koltun, et al. 2003. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J. Exp. Med. 197:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gapin, L., J.L. Matsuda, C.D. Surh, and M. Kronenberg. 2001. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2:971–978. [DOI] [PubMed] [Google Scholar]
- 18.Wei, D.G., H. Lee, S.H. Park, L. Beaun, L. Teyton, A. Lehuen, and A. Bendelac. 2005. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J. Exp. Med. 202:239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmer, M.I., A. Colmone, K. Felio, H. Xu, A. Ma, and C.R. Wang. 2006. A cell-type specific CD1d expression program modulates invariant NKT cell development and function. J. Immunol. 176:1421–1430. [DOI] [PubMed] [Google Scholar]
- 20.Park, J.J., S.J. Kang, A.D. De Silva, A.K. Stanic, G. Casorati, D.L. Hachey, P. Cresswell, and S. Joyce. 2004. Lipid-protein interactions: biosynthetic assembly of CD1 with lipids in the endoplasmic reticulum is evolutionarily conserved. Proc. Natl. Acad. Sci. USA. 101:1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giabbai, B., S. Sidobre, M.D. Crispin, Y. Sanchez-Ruiz, A. Bachi, M. Kronenberg, I.A. Wilson, and M. Degano. 2005. Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. J. Immunol. 175:977–984. [DOI] [PubMed] [Google Scholar]
- 22.Gumperz, J.E., C. Roy, A. Makowska, D. Lum, M. Sugita, T. Podrebarac, Y. Koezuka, S.A. Porcelli, S. Cardell, M.B. Brenner, and S.M. Behar. 2000. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 12:211–221. [DOI] [PubMed] [Google Scholar]
- 23.Cernadas, M., M. Sugita, N. van der Wel, X. Cao, J.E. Gumperz, S. Maltsev, G.S. Besra, S.M. Behar, P.J. Peters, and M.B. Brenner. 2003. Lysosomal localization of murine CD1d mediated by AP-3 is necessary for NK T cell development. J. Immunol. 171:4149–4155. [DOI] [PubMed] [Google Scholar]
- 24.Elewaut, D., A.P. Lawton, N.A. Nagarajan, E. Maverakis, A. Khurana, S. Honing, C.A. Benedict, E. Sercarz, O. Bakke, M. Kronenberg, and T.I. Prigozy. 2003. The adaptor protein AP-3 is required for CD1d-mediated antigen presentation of glycosphingolipids and development of Vα14i NKT cells. J. Exp. Med. 198:1133–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou, D., J. Mattner, C. Cantu III, N. Schrantz, N. Yin, Y. Gao, Y. Sagiv, K. Hudspeth, Y.P. Wu, T. Yamashita, et al. 2004. Lysosomal glycosphingolipid recognition by NKT cells. Science. 306:1786–1789. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, D., C. Cantu III, Y. Sagiv, N. Schrantz, A.B. Kulkarni, X. Qi, D.J. Mahuran, C.R. Morales, G.A. Grabowski, K. Benlagha, et al. 2004. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 303:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sagiv, Y., K. Hudspeth, J. Mattner, N. Schrantz, R.K. Stern, D. Zhou, P.B. Savage, L. Teyton, and A. Bendelac. 2006. Cutting edge: impaired glycosphingolipid trafficking and NKT cell development in mice lacking Niemann-Pick type C1 protein. J. Immunol. 177:26–30. [DOI] [PubMed] [Google Scholar]
- 28.Honey, K., K. Benlagha, C. Beers, K. Forbush, L. Teyton, M.J. Kleijmeer, A.Y. Rudensky, and A. Bendelac. 2002. Thymocyte expression of cathepsin L is essential for NKT cell development. Nat. Immunol. 3:1069–1074. [DOI] [PubMed] [Google Scholar]
- 29.Riese, R.J., G.P. Shi, J. Villadangos, D. Stetson, C. Driessen, A.M. Lennon-Dumenil, C.L. Chu, Y. Naumov, S.M. Behar, H. Ploegh, et al. 2001. Regulation of CD1 function and NK1.1(+) T cell selection and maturation by cathepsin S. Immunity. 15:909–919. [DOI] [PubMed] [Google Scholar]
- 30.Chiu, Y.H., S.H. Park, K. Benlagha, C. Forestier, J. Jayawardena-Wolf, P.B. Savage, L. Teyton, and A. Bendelac. 2002. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat. Immunol. 3:55–60. [DOI] [PubMed] [Google Scholar]
- 31.Chiu, Y.H., J. Jayawardena, A. Weiss, D. Lee, S.H. Park, A. Dautry-Varsat, and A. Bendelac. 1999. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J. Exp. Med. 189:103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brozovic, S., T. Nagaishi, M. Yoshida, S. Betz, A. Salas, D. Chen, A. Kaser, J. Glickman, T. Kuo, A. Little, et al. 2004. CD1d function is regulated by microsomal triglyceride transfer protein. Nat. Med. 10:535–539. [DOI] [PubMed] [Google Scholar]
- 33.Dougan, S.K., A. Salas, P. Rava, A. Agyemang, A. Kaser, J. Morrison, A. Khurana, M. Kronenberg, C. Johnson, M. Exley, et al. 2005. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J. Exp. Med. 202:529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raabe, M., M.M. Veniant, M.A. Sullivan, C.H. Zlot, J. Bjorkegren, L.B. Nielsen, J.S. Wong, R.L. Hamilton, and S.G. Young. 1999. Analysis of the role of microsomal triglyceride transfer protein in the liver of tissue-specific knockout mice. J. Clin. Invest. 103:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieu, H.D., S.K. Withycombe, Q. Walker, J.X. Rong, R.L. Walzem, J.S. Wong, R.L. Hamilton, E.A. Fisher, and S.G. Young. 2003. Eliminating atherogenesis in mice by switching off hepatic lipoprotein secretion. Circulation. 107:1315–1321. [DOI] [PubMed] [Google Scholar]
- 36.Marschang, P., and J. Herz. 2003. Mouse models as tools for dissecting disorders of lipoprotein metabolism. Semin. Cell Dev. Biol. 14:25–35. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn, R., F. Schwenk, M. Aguet, and K. Rajewsky. 1995. Inducible gene targeting in mice. Science. 269:1427–1429. [DOI] [PubMed] [Google Scholar]
- 38.Sharp, D., B. Ricci, B. Kienzle, M.C. Lin, and J.R. Wetterau. 1994. Human microsomal triglyceride transfer protein large subunit gene structure. Biochemistry. 33:9057–9061. [DOI] [PubMed] [Google Scholar]
- 39.Navasa, M., D.A. Gordon, N. Hariharan, H. Jamil, J.K. Shigenaga, A. Moser, W. Fiers, A. Pollock, C. Grunfeld, and K.R. Feingold. 1998. Regulation of microsomal triglyceride transfer protein mRNA expression by endotoxin and cytokines. J. Lipid Res. 39:1220–1230. [PubMed] [Google Scholar]
- 40.Rava, P., H. Athar, C. Johnson, and M.M. Hussain. 2005. Transfer of cholesteryl esters and phospholipids as well as net deposition by microsomal triglyceride transfer protein. J. Lipid Res. 46:1779–1785. [DOI] [PubMed] [Google Scholar]
- 41.Athar, H., J. Iqbal, X.C. Jiang, and M.M. Hussain. 2004. A simple, rapid, and sensitive fluorescence assay for microsomal triglyceride transfer protein. J. Lipid Res. 45:764–772. [DOI] [PubMed] [Google Scholar]
- 42.Jamil, H., C.H. Chu, J.K. Dickson Jr., Y. Chen, M. Yan, S.A. Biller, R.E. Gregg, J.R. Wetterau, and D.A. Gordon. 1998. Evidence that microsomal triglyceride transfer protein is limiting in the production of apolipoprotein B-containing lipoproteins in hepatic cells. J. Lipid Res. 39:1448–1454. [PubMed] [Google Scholar]
- 43.Tietge, U.J., A. Bakillah, C. Maugeais, K. Tsukamoto, M. Hussain, and D.J. Rader. 1999. Hepatic overexpression of microsomal triglyceride transfer protein (MTP) results in increased in vivo secretion of VLDL triglycerides and apolipoprotein B. J. Lipid Res. 40:2134–2139. [PubMed] [Google Scholar]
- 44.Brossay, L., and M. Kronenberg. 1999. Highly conserved antigen-presenting function of CD1d molecules. Immunogenetics. 50:146–151. [DOI] [PubMed] [Google Scholar]
- 45.Ameen, C., U. Edvardsson, A. Ljungberg, L. Asp, P. Akerblad, A. Tuneld, S.O. Olofsson, D. Linden, and J. Oscarsson. 2005. Activation of peroxisome proliferator-activated receptor alpha increases the expression and activity of microsomal triglyceride transfer protein in the liver. J. Biol. Chem. 280:1224–1229. [DOI] [PubMed] [Google Scholar]
- 46.Azizkhan, J.C., D.E. Jensen, A.J. Pierce, and M. Wade. 1993. Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit. Rev. Eukaryot. Gene Expr. 3:229–254. [PubMed] [Google Scholar]
- 47.Sheena, V., R. Hertz, J. Nousbeck, I. Berman, J. Magenheim, and J. Bar-Tana. 2005. Transcriptional regulation of human microsomal triglyceride transfer protein by hepatocyte nuclear factor-4alpha. J. Lipid Res. 46:328–341. [DOI] [PubMed] [Google Scholar]
- 48.Nakamuta, M., B.H. Chang, R. Hoogeveen, W.H. Li, and L. Chan. 1996. Mouse microsomal triglyceride transfer protein large subunit: cDNA cloning, tissue-specific expression and chromosomal localization. Genomics. 33:313–316. [DOI] [PubMed] [Google Scholar]
- 49.Xie, Y., E.P. Newberry, S.G. Young, S. Robine, R.L. Hamilton, J.S. Wong, J. Luo, S. Kennedy, and N.O. Davidson. 2006. Compensatory increase in hepatic lipogenesis in mice with conditional intestine-specific Mttp deficiency. J. Biol. Chem. 281:4075–4086. [DOI] [PubMed] [Google Scholar]
- 50.Kang, S.J., and P. Cresswell. 2002. Calnexin, calreticulin, and ERp57 cooperate in disulfide bond formation in human CD1d heavy chain. J. Biol. Chem. 277:44838–44844. [DOI] [PubMed] [Google Scholar]
- 51.Kim, H.S., J. Garcia, M. Exley, K.W. Johnson, S.P. Balk, and R.S. Blumberg. 1999. Biochemical characterization of CD1d expression in the absence of beta2-microglobulin. J. Biol. Chem. 274:9289–9295. [DOI] [PubMed] [Google Scholar]
- 52.Akbari, O., P. Stock, E. Meyer, M. Kronenberg, S. Sidobre, T. Nakayama, M. Taniguchi, M.J. Grusby, R.H. DeKruyff, and D.T. Umetsu. 2003. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat. Med. 9:582–588. [DOI] [PubMed] [Google Scholar]
- 53.Akbari, O., J.L. Faul, E.G. Hoyte, G.J. Berry, J. Wahlstrom, M. Kronenberg, R.H. DeKruyff, and D.T. Umetsu. 2006. CD4+ invariant T-cell-receptor+ natural killer T cells in bronchial asthma. N. Engl. J. Med. 354:1117–1129. [DOI] [PubMed] [Google Scholar]
- 54.Fuss, I.J., F. Heller, M. Boirivant, F. Leon, M. Yoshida, S. Fichtner-Feigl, Z. Yang, M. Exley, A. Kitani, R.S. Blumberg, et al. 2004. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J. Clin. Invest. 113:1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tupin, E., A. Nicoletti, R. Elhage, M. Rudling, H.G. Ljunggren, G.K. Hansson, and G.P. Berne. 2004. CD1d-dependent activation of NKT cells aggravates atherosclerosis. J. Exp. Med. 199:417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van de Wal, Y., N. Corazza, M. Allez, L.F. Mayer, H. Iijima, M. Ryan, S. Cornwall, D. Kaiserlian, R. Hershberg, Y. Koezuka, et al. 2003. Delineation of a CD1d-restricted antigen presentation pathway associated with human and mouse intestinal epithelial cells. Gastroenterology. 124:1420–1431. [DOI] [PubMed] [Google Scholar]
- 57.Bendelac, A., O. Lantz, M.E. Quimby, J.W. Yewdell, J.R. Bennink, and R.R. Brutkiewicz. 1995. CD1 recognition by mouse NK1+ T lymphocytes. Science. 268:863–865. [DOI] [PubMed] [Google Scholar]
- 58.Rauch, J., J. Gumperz, C. Robinson, M. Skold, C. Roy, D.C. Young, M. Lafleur, D.B. Moody, M.B. Brenner, C.E. Costello, and S.M. Behar. 2003. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J. Biol. Chem. 278:47508–47515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Townsend, A., T. Elliott, V. Cerundolo, L. Foster, B. Barber, and A. Tse. 1990. Assembly of MHC class I molecules analyzed in vitro. Cell. 62:285–295. [DOI] [PubMed] [Google Scholar]
- 60.Sugita, M., R.M. Jackman, E. van Donselaar, S.M. Behar, R.A. Rogers, P.J. Peters, M.B. Brenner, and S.A. Porcelli. 1996. Cytoplasmic tail-dependent localization of CD1b antigen-presenting molecules to MIICs. Science. 273:349–352. [DOI] [PubMed] [Google Scholar]
- 61.Cone, R.D., and R.C. Mulligan. 1984. High-efficiency gene transfer into mammalian cells: generation of helper-free recombinant retrovirus with broad mammalian host range. Proc. Natl. Acad. Sci. USA. 81:6349–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ory, D.S., B.A. Neugeboren, and R.C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 93:11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]