Abstract
The SH2 domain–containing leukocyte protein of 76 kD (SLP-76) is a pivotal element of the signaling machinery controlling T cell receptor (TCR)-mediated activation. Here, we identify 14-3-3ɛ and ζ proteins as SLP-76 binding partners. This interaction was induced by TCR ligation and required phosphorylation of SLP-76 at serine 376. Ribonucleic acid interference and in vitro phosphorylation experiments showed that serine 376 is the target of the hematopoietic progenitor kinase 1 (HPK-1). Interestingly, either S376A mutation or HPK-1 knockdown resulted in increased TCR-induced tyrosine phosphorylation of SLP-76 and phospholipase C-γ1. Moreover, an SLP-76–S376A mutant induced higher interleukin 2 gene transcription than wild-type SLP-76. These data reveal a novel negative feedback loop involving HPK-1–dependent serine phosphorylation of SLP-76 and 14-3-3 protein recruitment, which tunes T cell activation.
TCR sensing of peptide/MHC generates signals that are translated into biological responses as diverse as cell survival, homeostatic proliferation, clonal deletion or expansion, immune memory generation, and tolerance. A molecular model explaining this functional flexibility posits that the intensity and duration of peptide/MHC stimuli impact the qualitative and/or quantitative composition of the TCR-proximal signaling machinery, triggering activation of all or part of a complex array of immediate signaling pathways (1). This results in diversified gene expression profiles, making cells distinctively receptive to auxiliary environmental cues (e.g., cytokines and/or cell–cell interactions) and therefore shapes a particular functional behavior (2, 3).
TCR engagement activates the protein tyrosine kinases Lck, Fyn, and ζ chain–associated protein of 70 kD that initiate the signaling cascade and contribute to the assembly of a “signalosome,” a multiprotein complex including various enzymes, their substrates, and scaffold/adaptor proteins (4). A major platform for the nucleation of this complex is provided by two docking elements, the lipid microdomain-anchored protein linker for activation of T cells (LAT; reference 5) and its cytoplasmic partner SH2 domain–containing leukocyte protein of 76 kD (SLP-76; reference 6), recruited onto LAT via the SH2-mediated binding of the constitutively associated Grb2-related adaptor downstream of Shc (Gads; reference 7). Phosphorylation of tyrosines on LAT and SLP-76 allows recruitment of effectors that channel signals to downstream pathways. For instance, LAT binds phospholipase C (PLC)-γ1 (5), which regulates Ca2+- and diacylglycerol-dependent events (e.g., activation of the NFAT transcription factor and protein kinase C [PKC]), and Grb2, which recruits the Ras-specific activator SOS or the E3-ubiquitin ligase Casitas B lineage lymphoma proto-oncogene (8). Additional SH2 domain binding motifs in the N-terminal region of SLP-76, encompassing phosphorylated Y113, Y128, and Y145, recruit the adaptor Nck, the guanine-nucleotide exchange factor Vav-1 and the inducible T cell kinase, which regulate actin cytoskeleton reorganization and PLC-γ1 activation (9). Furthermore, SLP-76 associates through its C-terminal SH2 domain with the adhesion- and degranulation-promoting adaptor protein, an essential regulator of inside-out integrin signaling, and with the hematopoietic progenitor kinase 1 (HPK-1, also named mitogen-activated protein kinase kinase kinase kinase [MAP4K]1; see below and reference 9).
Assembly of the signalosome relies on networks of cooperative interactions (10, 11), better suited for precise juxtaposition of its components and global stability. However, this protein ensemble remains relatively dynamic. For instance, SLP-76 detaches from plasma membrane–proximal protein complexes a few minutes after TCR stimulation and translocates to a perinuclear compartment (12). Such architectural organization and dynamic behavior likely ensure timely activation of effectors while providing multiple regulatory checkpoints. Several inputs, generated by the TCR as well as by other receptors (e.g., CD28; reference 13), are integrated at these checkpoints that participate in setting thresholds for signal initiation and propagation.
Although we mostly appreciate how the TCR signalosome works in the “forward” mode toward cellular activation, it is less clear when and how counteracting signals that may tune signal kinetics and intensity are elicited. Various TCR-proximal mechanisms may curtail activation, involving specific negative adaptors (e.g., PAG/Cbp, Gab-2, and Dok proteins; reference 14), protein tyrosine phosphatases (PTPs; reference 15), or ubiquitination and degradation of selected components (16). More recently, examples of negative regulation by Ser/Thr protein kinases have been described. For instance, extracellular signal-regulated kinase (ERK)-dependent Thr phosphorylation of LAT interferes with PLC-γ1 recruitment and consequently reduces NFAT transcriptional activity (17). Moreover, HPK-1 inhibits ERKs and NFAT/AP1 transcription factors in T cells (18, 19). Although TCR- dependent tyrosine phosphorylation of HPK-1 and its interaction with the SH2 domain of SLP-76 are involved in optimal activation of this kinase and consequent inhibition of downstream pathways (20), the underlying mechanisms are incompletely understood. Thus, unraveling the opposing mechanisms that shape the generation and life span of the signalosome (e.g., assembly/disassembly) may help us to better understand how the TCR sets in motion T cell fate.
Given the central role of SLP-76 within the TCR-dependent signalosome, we chose this scaffold protein as an “entry point” for identifying new regulators of T cell activation. Thus, we surveyed proteins associated to SLP-76 by using immuno-affinity purification and mass spectrometry (MS). We found that members of the 14-3-3 protein family bind to SLP-76 upon TCR stimulation, although with delayed kinetics compared with other previously described SLP-76 signaling partners. This interaction was induced by the HPK-1–dependent phosphorylation of S376 of SLP-76. Impairing 14-3-3 recruitment by mutating S376 resulted in increased tyrosine phosphorylation of both SLP-76 and PLC-γ1 and improved IL-2 promoter activation.
Thus, our study reveals the mechanism of a novel negative feedback loop initiated from SLP-76 to modulate signal intensity during T cell activation.
RESULTS
Activation-induced association of 14-3-3 proteins with serine-phosphorylated SLP-76
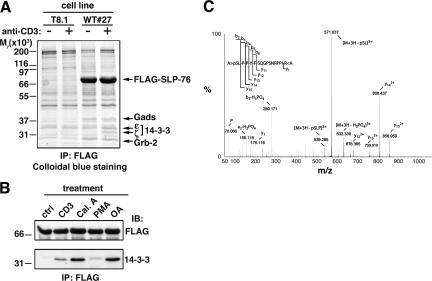
To discover new components of the TCR signalosome nucleated around SLP-76, we used a cell line (WT#27) derived from the murine T cell hybridoma T8.1, which stably expresses a FLAG–SLP-76 construct (21). SLP-76 and associated proteins were isolated by anti-FLAG immuno-affinity purification and fractionated by SDS-PAGE. Proteins absent in anti-FLAG immunoprecipitates from untransfected T8.1 cells or whose intensity increased in WT#27 cells after anti-CD3 stimulation (Fig. 1 A) were identified by matrix-assisted laser desorption and ionization time of flight (MALDI-TOF) peptide mass fingerprinting. In addition to previously described SLP-76 binding partners, such as Gads, Grb-2, and adhesion- and degranulation-promoting adaptor protein (Fig. 1 A and not depicted; reference 9), two members of the 14-3-3 protein family, namely the ζ and ɛ isoforms, were found in FLAG–SLP-76 immunoprecipitates from stimulated cells (Fig. 1 A). This previously unknown interaction was confirmed by different approaches. First, FLAG–SLP-76 immunoprecipitates from anti-CD3–stimulated WT#27 cells were probed with an anti–pan-14-3-3 antibody, which revealed a protein of Mr ≈ 30,000 (Fig. 1 B), the expected size for 14-3-3. Second, human and mouse SLP-76 bound to a glutathione-S-transferase (GST)–14-3-3ζ fusion protein either in pull-down or Far Western assays after anti-CD3 stimulation (not depicted).
Figure 1.
Detection of inducible association of 14-3-3 proteins to SLP-76 and serine 376 phosphorylation. (A) WT#27 cells overexpressing FLAG–SLP-76 and parental T8.1 cells were left unstimulated (−) or activated by anti-CD3 cross-linking (+) for 5 min at 37°C. Lysates were incubated with anti-FLAG affinity resin, and bound proteins were eluted with an excess of FLAG peptide. Eluates were then fractionated by SDS-PAGE, and proteins were detected by colloidal blue staining. Relevant protein bands were excised, digested in gel by trypins, and identified by MALDI-TOF peptide mass fingerprinting. Migration of protein standards is indicated on the left. (B) WT#27 cells were left unstimulated (ctrl), activated at 37°C by CD3 cross-linking as in A, or treated with 1 μM calyculin A (Cal.A), 10 ng/ml PMA, or 1 μM okadaic acid (OA) for 30 min at 37°C. Lysates were immunoprecipitated with anti-FLAG antibodies as described in A and immunoblotted with anti-FLAG (top) or anti–14-3-3 (bottom) antibodies. (C) FLAG–SLP-76 was immunoprecipitated from CD3 cross-linking–stimulated WT#27 cells as described in A. The protein band was excised and digested in gel by trypsin and AspN. The resulting peptide mixture was analyzed by nanoESI MS. The acquired MS/MS dataset was submitted to sequence tag scanning to spot modified peptides. The MS/MS spectrum shows fragment ions of a peptide spanning residues 376–391 of SLP-76, which clearly demonstrates phosphorylation of S376.
In mammals, seven genes encode 14-3-3 protein family members, which are ubiquitously expressed as homodimers or heterodimers. They are implicated in many cellular processes, such as regulation of cell cycle, metabolism, and signaling (22). Hundreds of intracellular proteins, including some signaling proteins in T cells (23–26), have been reported to interact with 14-3-3 isoforms, and in most cases, 14-3-3s associate to their partners by binding to specific sequences containing a phosphorylated Ser or Thr residue (22). This also appears to be the case for SLP-76 because treatment of cells with inhibitors of Ser/Thr phosphatases, i.e., calyculin A and okadaic acid, induced the binding of 14-3-3 proteins to SLP-76 (Fig. 1 B). In contrast, the PKC activator PMA was rather ineffective at stimulating this interaction (Fig. 1 B). These data reveal an interaction between SLP-76 and 14-3-3 proteins induced by TCR engagement and suggest that it is mediated by inducible Ser/Thr phosphorylation of SLP-76.
Phosphorylation of SLP-76 at serine 376 induces the association with 14-3-3 proteins
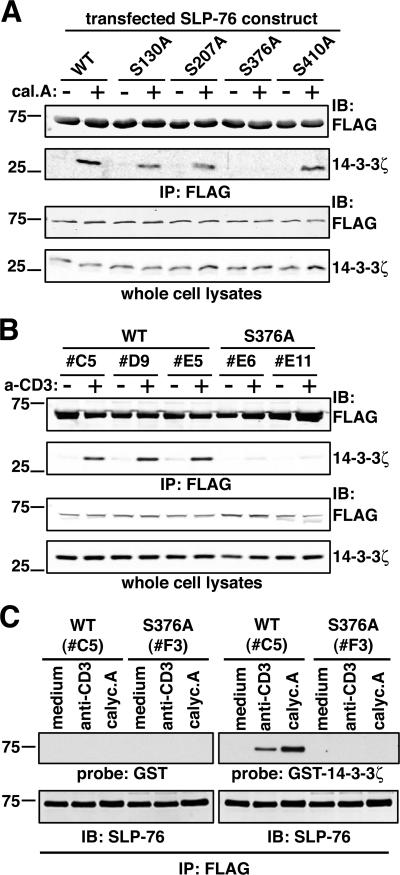
Although three consensus 14-3-3 binding motifs have been described, several unconventional binding motifs have been reported as well (for review see reference 22). Thus, we analyzed FLAG–SLP-76 isolated from resting and stimulated WT#27 cells by nano-electrospray ionization (nanoESI)-tandem MS to identify phosphorylated Ser or Thr residues potentially involved in 14-3-3 binding. These experiments revealed four Ser residues (S130, S207, S376, and S410) that were phosphorylated either constitutively or after anti-CD3 or calyculin A treatment (Fig. 1 C; reference 21). To address the implication of these residues in 14-3-3 binding to SLP-76, we generated FLAG–SLP-76 mutants bearing Ser to Ala mutations at each position. WT or mutant FLAG–SLP-76 constructs were cotransfected together with 14-3-3ζ into COS-7 cells, which were then treated with calyculin A and immunoprecipitated by an anti-FLAG antibody. Fig. 2 A shows that 14-3-3ζ coprecipitated with WT SLP-76 after calyculin A treatment, demonstrating that the interaction could be reconstituted in this heterologous cell system. Moreover, mutation of S130, S207, and S410 only slightly reduced coprecipitation of the two proteins. Importantly, 14-3-3ζ interaction with SLP-76 was undetectable upon mutation of S376, suggesting that its phosphorylation is essential for binding.
Figure 2.
Serine 376 of SLP-76 controls the interaction with 14-3-3 proteins. (A) COS-7 cells were transfected with a myc–14-3-3ζ–encoding plasmid together with FLAG–SLP-76 constructs, either WT or Ser to Ala mutants. After 36 h, cells were harvested and incubated for 10 min at 37°C with medium alone (−) or 1 μM calyculin A (+). Cell lysates were subjected to anti-FLAG immunoprecipitation, and bound proteins were analyzed by anti-FLAG (top row) and anti–14-3-3ζ immunoblotting (second row from top). Protein expression in all samples was assessed by anti-FLAG and anti–14-3-3ζ immunoblotting on whole cell lysates (two bottom rows). (B) J14-derived clones expressing similar amounts of FLAG-SLP-76-WT (clones C5, D9, and E5) or -S376A (clones E6 and E11) were left unstimulated or activated by anti-CD3 cross-linking for 5 min at 37°C. Samples were lysed, immunoprecipitated with anti-FLAG antibodies (top rows), and subjected to anti-FLAG and 14-3-3ζ immunoblotting. Anti-FLAG and anti–14-3-3ζ immunoblots on whole cell lysates (bottom rows) demonstrate comparable expression of transfected SLP-76 constructs and endogenous 14-3-3ζ. (C) J14 cells stably expressing FLAG-SLP-76-WT (clone C5) or -S376A (clone F3) were left unstimulated (medium) or treated with 10 μg/ml anti-CD3 mAb or 1 μM calyculine A (calyc.A) for 5 min at 37°C. Lysates were then immunoprecipitated with an anti-FLAG mAb. After SDS-PAGE and transfer onto nitrocellulose, proteins were denaturated and renaturated, and then probed with either GST-P3 (top left row; reference 45) or a GST–14-3-3ζ fusion protein (top right row). Specific binding was detected by an anti-GST mAb (top rows). Membranes were then stripped and reprobed with an anti–SLP-76 antiserum (M14; bottom rows).
The critical role of S376 in controlling the 14-3-3– SLP-76 interaction was further confirmed in T cell lines. SLP-76–deficient J14 cells (27) were reconstituted by transfecting either WT or S376A FLAG–SLP-76. Several stable clones expressing similar amounts of tagged SLP-76 proteins were selected for further experiments. Fig. 2 B shows that endogenous 14-3-3ζ was coprecipitated from clones expressing SLP-76–WT after anti-CD3 stimulation, whereas only a weak association was observed in unstimulated cells. In contrast, induction of 14-3-3ζ binding was severely impaired in clones expressing SLP-76–S376A, consistent with S376 phosphorylation triggering CD3-dependent 14-3-3 binding to SLP-76.
To assess whether the interaction between 14-3-3 and SLP-76 was direct we used a Far Western assay. SLP-76–WT or SLP-76–S376A was purified by anti-FLAG immunoprecipitation from J14-derived clones, unstimulated or treated with anti-CD3 or calyculin A. As shown in Fig. 2 C, no binding of an irrelevant fusion protein, GST-P3, to membrane-bound proteins was detected (top left), whereas GST–14-3-3ζ specifically bound to SLP-76–WT precipitated from anti-CD3 or calyculin A–treated, but not resting, cells (top right, first three lanes). No binding could be detected on SLP-76–S376A (Fig. 2 C, top right, last three lanes), with protein amounts comparable to SLP-76 WT (bottom). Collectively, these results demonstrate that T cell activation induces a direct association between 14-3-3ζ and SLP-76 that is mediated by a motif encompassing phosphorylated S376.
TCR-induced phosphorylation of serine 376 is delayed compared with tyrosine phosphorylation of SLP-76
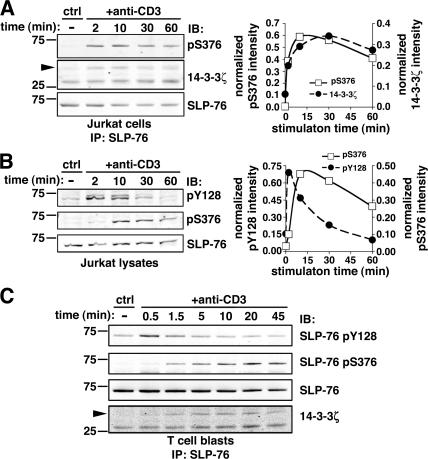
To further characterize S376 phosphorylation, we generated a phospho-specific antiserum against a synthetic peptide spanning residues 371–380 of human SLP-76 and containing phosphorylated S376. Affinity-purified anti–phospho-S376 antibody recognized immunoprecipitated SLP-76 only after cell treatment by anti-CD3 (Fig. 3 A) or calyculin A (not depicted). Using this antibody, we observed that the kinetics of S376 phosphorylation coincided with 14-3-3 binding to SLP-76 (Fig. 3 A), supporting a causal link between these two events. We then compared the kinetics of S376 phosphorylation to the phosphorylation of critical tyrosines in the N-terminal region of SLP-76. Probing this protein with an anti-pY128 mAb demonstrated that in agreement with previous observations (28), Tyr phosphorylation of SLP-76 was rapid and transient, reaching maximal intensity in 2 min or less, decreasing after 10 min, and returning to nearly basal level by 30 min (Fig. 3 B, top). A mAb against pY113 or anti-phosphotyrosine mAb gave similar results (not depicted), suggesting that all tyrosines of SLP-76 are phosphorylated with comparable kinetics. In sharp contrast, S376 phosphorylation was relatively weak at 2 min, peaked at ∼10 min, and decreased slowly thereafter, remaining elevated after 1 h (Fig. 3 B, middle). Phosphorylation of S376 and Y128 upon anti-CD3 stimulation was also detected in human primary CD4+ T cells (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20062066/DC1) and T cell blasts (Fig. 3 C) and followed a kinetics similar to that observed in Jurkat cells. These differential temporal patterns of Tyr and Ser phosphorylation within the same protein suggest a distinct regulatory function of these posttranslational modifications in T cell activation. It is worth noting that S376 phosphorylation correlated with coprecipitation of 14-3-3ζ with SLP-76 in T cell blasts (Fig. 3 C), demonstrating that this interaction is also induced in nontransformed lymphocytes.
Figure 3.
Serine 376 phosphorylation is delayed as compared with tyrosine phosphorylation of SLP-76. (A) Jurkat cells were left unstimulated (ctrl) or incubated with 10 μg/ml anti-CD3 mAbs for the indicated time points. Cells were lysed and immunoprecipitated with an anti–SLP-76 mAb. Samples were then analyzed by Western blotting with anti-pS376 (top left) and anti–14-3-3ζ (middle row, left). Membranes were then stripped and reprobed with an anti–SLP-76 mAb (bottom row, left). Fluorescent immunoblots were acquired and quantified by using an Odyssey scanner. The graph (right) shows the quantification of band intensities in the pS376 (□) and 14-3-3ζ (•) immunoblotting, normalized by the amount of immunoprecipitated SLP-76. (B) Jurkat cells were left unstimulated or stimulated by anti-CD3 mAbs as described in A. SLP-76 phosphorylation was analyzed by immunoblotting with mAb anti-pY128 (top left) and affinity-purified rabbit anti-pS376 antibodies (middle row, left). Membranes were then stripped and reprobed with an anti–SLP-76 mAb (bottom row, left). Quantification of anti-pY128 and anti-pS376 and immunoblots, performed as explained in A, is shown on the right. (C) Human T cell blasts were stimulated with anti-CD3 mAb for the indicated time points. SLP-76 immunoprecipitates were subjected to anti-pY128 (top row), anti-pS376 (second row from top), anti–SLP-76 (third row from top), and anti–14-3-3ζ (bottom row) immunoblotting.
Mutation of serine 376 correlates with increased activation of the IL-2 promoter in T cells
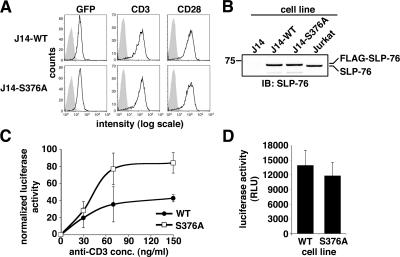
The functional role of S376 phosphorylation was addressed by comparing the ability of SLP-76–WT and SLP-76–S376A to complement T cell activation in SLP-76–deficient J14 cells, which fail to induce IL-2 gene transcription (27 and not depicted). To avoid discrepancies due to clonal variability and protein overexpression, J14 cells were infected with retroviral vectors to express SLP-76–WT or SLP-76–S376A together with GFP expressed from an internal ribosomal entry site. Cells were sorted by FACS to obtain cell populations (named J14-WT or J14-S376A, respectively) expressing comparable amounts of WT and mutant SLP-76, as confirmed by immunoblot assay (Fig. 4, A and B). The protein expression levels of SLP-76 in these cells closely matched that of endogenous SLP-76 in Jurkat cells (Fig. 4 B). J14-WT and J14-S376A cells also expressed comparable levels of both CD3 and CD28 (Fig. 4 A). An IL-2 promoter–dependent luciferase reporter (phIL-2-luc; reference 29) was transiently transfected into these cells, which were then treated with either medium alone or by anti-CD3 and anti-CD28 antibodies for 6 h. After lysis, luciferase activity was measured in protein extracts. Induction of IL-2 promoter–dependent transcription by PMA and Ca2+ ionophore, which bypasses TCR-proximal signaling, was measured to normalize data for transfection efficiency. We found that CD3/CD28-stimulated IL-2 promoter activation was substantially higher in J14-S376A than in J14-WT cells (Fig. 4 C), whereas similar induction of IL-2 promoter activity was observed when cells were stimulated by PMA and Ca2+ ionophore (Fig. 4 D). Similar results were also obtained when the phIL-2-luc reporter was transiently cotransfected in J14 cells together with either SLP-76–WT or SLP-76–S376A expression plasmid (not depicted).
Figure 4.
SLP-76–S376A mutant induces higher IL-2 promoter activation and then WT SLP-76. (A) Cells stably expressing FLAG-SLP-76-WT or -S376A mutant (J14-WT or J14-S376A, respectively) were obtained by retroviral infection of J14 cells (see Materials and methods). GFP+ cells were then enriched by two round of fluorescence-activated cell sorting to obtain stable cell populations expressing comparable levels of GFP (left). Comparable expression of CD3 (middle) and CD28 (right) by these cells was assessed by flow cytometry using PE-conjugated antibodies. (B) J14, J14-WT, J14-S376A, and Jurkat cells were lysed and analyzed by anti–SLP-76 immunoblotting. (C) J14-WT and J14-S376A cells were transfected with the phIL-2 luciferase reporter. 22 h after transfection, cells were incubated for 6 h with or without increasing amounts of anti-CD3 and 10 μg/ml of soluble anti-CD28. Cell lysis and luciferase activity measurement and normalization are described in Materials and methods. Each point represents average and SD of quadruplicate determination. (D) Aliquot of phIL-2 luciferase–transfected J14-WT and J14-S376A cells (see C) were stimulated with PMA and the Ca2+ ionophore A23187 to assess maximal IL-2 promoter activation. Luciferase induction was measured as explained above. Histograms and error bars represent average and SD of raw luciferase activity of quadruplicate determinations. RLU, relative luciferase units.
Collectively, these data indicate that the S376A mutation unleashes TCR-proximal signaling from a repressive action implicating S376 phosphorylation and 14-3-3 binding of SLP-76.
Increased tyrosine phosphorylation of SLP-76 and PLC-γ1 in T cells expressing SLP-76–S376A
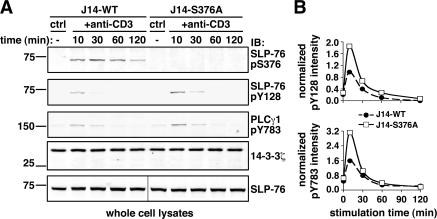
To understand how the S376 mutation affects TCR signal transduction, we looked for potential modifications in the phosphorylation status of critical components of the TCR signalosome. As expected, phosphorylation of S376 was detected only in J14-WT, but not in J14-S376A, cells (Fig. 5 A, top row). Immunoblotting with anti-pY128 mAb revealed that tyrosine phosphorylation of SLP-76 was consistently increased in J14-S376A as compared with J14-WT cells (Fig. 5 A, second row from top, and B, top). Moreover, tyrosine phosphorylation of PLC-γ1 at residue Y783, which is required for the induction of phospholipase activity (30), was found to be increased in J14-S376A cells (Fig. 5 A, third row from top, and B, bottom). These results suggest that phosphorylation of SLP-76 at S376 and recruitment of 14-3-3 modulate TCR signaling by affecting tyrosine phosphorylation of critical effectors, possibly explaining the increased levels of IL-2 gene activation induced by SLP-76–S376A.
Figure 5.
Increased SLP-76 and PLC-γ1 tyrosine phosphorylation in T cells expressing the SLP-76–S376A mutant. (A) J14-WT and J14-S376A cells were left unstimulated or stimulated by anti-CD3 mAb for the indicated time points. Lysates were fractionated on NuPAGE gel and immunoblotted with anti–SLP-76 pS376 (top row) and pY128 antibodies (second row from top), followed by anti–PLC-γ1 pY783 antibodies (third row from top). Equal protein loading in each lane was assessed by 14-3-3ζ immunoblotting (fourth row from top). Aliquots of the same lysates were analyzed on a separate gel to check for equal amount of SLP-76 constructs in all samples (bottom row). (B) Graphs show the comparison of SLP-76 (pY128; top) and PLCγ1 (pY783; bottom) phosphorylation in J14-WT (•) versus J14-S376A cells (□) after normalization for the amount of 14-3-3ζ in each sample.
The serine/threonine kinase HPK-1 phosphorylates serine 376 of SLP-76 and induces the interaction with 14-3-3 proteins
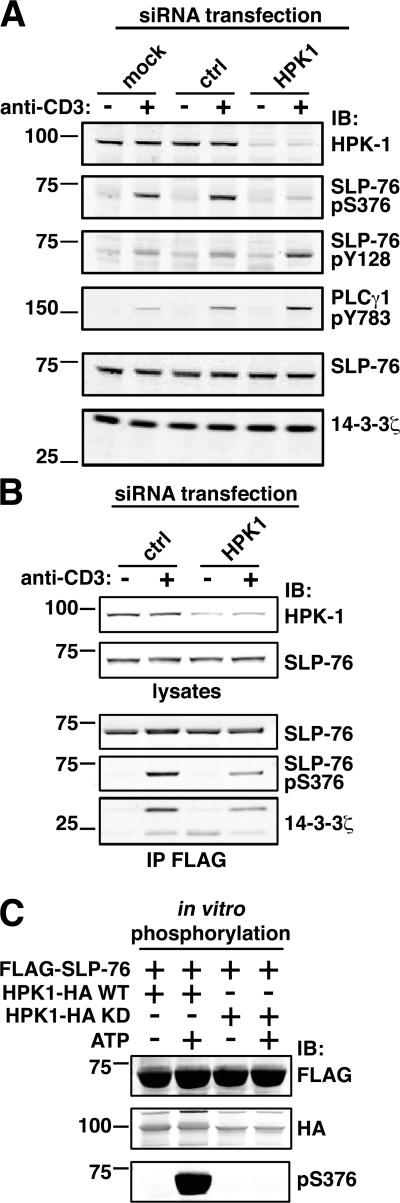
We set out to identify the kinase responsible for S376 phosphorylation. PKCs, protein kinase A (PKA), and the lipid-dependent kinase AKT have been all reported to phosphorylate 14-3-3 binding motifs on different proteins (22). However, using specific pharmacological agonists and antagonists we could exclude a role for these kinases in phosphorylating SLP-76. For instance, raising cellular cAMP levels by forskolin treatment resulted in increased phosphorylation of CREB at S133, a specific PKA substrate, without inducing S376 phosphorylation (Fig. S2 A, available at http://www.jem.org/cgi/content/full/jem.20062066/DC1). Moreover, PMA treatment of Jurkat T cells, which activates classical and novel PKCs, led to robust phosphorylation of ERK kinases but did not increase phosphorylation of S376 (Fig. S2 B). Finally, specific inhibition of anti-CD3–induced PI3K activation by LY294002 virtually abolished AKT activation but did not affect S376 phosphorylation (Fig. S2 C). We then focused on the Ser/Thr protein kinase HPK-1, which binds to the SH2 domain of SLP-76 (20). To address its potential role in SLP-76 phosphorylation, we knocked-down HPK-1 expression by transfection of specific small interfering RNAs (siRNAs) into Jurkat cells. As shown in Fig. 6 A (top row), transfection of a pool of four siRNAs targeting different regions of the human HPK1 transcript resulted in a substantial decrease (∼80%) in HPK-1 protein expression when compared with control cells electroporated with no siRNA or with control siRNAs targeting an unrelated protein. Knock-down of HPK-1 expression was paralleled by a comparable reduction in anti-CD3–induced SLP-76 phosphorylation at S376 (Fig. 6 A, second row from top), implicating HPK-1 in the induction of S376 phosphorylation. In line with the above data obtained with J14-S376A cells, HPK-1 knock-down and decreased S376 phosphorylation of SLP-76 correlated with significantly increased tyrosine phosphorylation of both SLP-76 and PLC-γ1 (Fig. 6 A, third and fourth row from top, respectively). We can exclude that these changes resulted from off-target effects of HPK-1 siRNAs because SLP-76 and 14-3-3 expression was not affected by siRNA transfection (Fig. 6 A, the two bottom rows), and comparable effects on HPK-1 expression and SLP-76 phosphorylation were observed upon independent transfections of three different HPK-1–specific siRNA (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20062066/DC1). Importantly, immunoprecipitation of SLP-76 from J14-WT cells in which HPK-1 expression was reduced by RNA interference (Fig. 6 B, top row) showed not only considerably impaired S376 phosphorylation (Fig. 6 B, fourth row from top), but also reduced 14-3-3ζ binding to SLP-76 (Fig. 6 B, bottom row), thus formally proving that HPK-1 regulates the SLP-76–14-3-3ζ interaction.
Figure 6.
HPK-1 phosphorylates SLP-76 at serine 376. (A) Knockdown of HPK-1 expression in Jurkat T cells was performed by transient transfection of synthetic siRNAs. Cells were electroporated in the presence of medium alone (mock), control siRNAs (ctrl), or HPK-1–specific siRNAs (HPK1). After 72 h, cells were left unstimulated or activated by anti-CD3 mAb, and lysates were fractionated on Nu-PAGE gels. HPK-1 expression was assessed by anti–HPK-1 immunoblotting (top row). Phosphorylation of SLP-76 and PLC-γ1 was then assessed by immunoblotting with anti-pS376 (second row from top), anti-pY128 (third row from top), and anti–PLC-γ1 pY783 (fourth row from top). Equal protein loading in each lane was demonstrated by anti–SLP-76 and anti–14-3-3ζ immunoblotting (two bottom rows). (B) J14-WT cells were transiently transfected with either control siRNAs (ctrl) or HPK-1–specific siRNAs (HPK1) as in A. After 72 h, cells were left unstimulated or activated by anti-CD3 mAb for 10 min, and lysates were immunoprecipitated by anti-FLAG antibodies. Aliquots of the lysate were analyzed by immunoblotting with anti–HPK-1 or anti–SLP-76 antibodies (first and second row from top, respectively). Immunoprecipitates were immunoblotted with anti–SLP-76 (third row), anti-pS376 (fourth row), and anti–14-3-3ζ (bottom row). Quantification of band intensities and normalization by the amount of SLP-76 in each lane indicated that HPK-1 expression, pS376 phosphorylation, and 14-3-3ζ binding to SLP-76 were reduced in this experiment by 68, 58, and 50%, respectively. (C) COS-7 cells were transfected with FLAG–SLP-76 or HPK-1–HA constructs, WT, or a kinase-defective mutant (KD). After 36 h, cells were harvested, lysed, and immunoprecipitated with anti-FLAG (FLAG–SLP-76 transfection) or anti-HA antibodies (HPK-1 WT– or KD-transfected cells). FLAG–SLP-76 was eluted and mixed with bead-bound HPK-1–WT or –KD. Samples were then incubated for 30 min at 37°C in the presence or absence of ATP. Protein mixtures were analyzed by immunoblotting with anti-FLAG (top row), anti-HA (middle row), and anti-pS376 antibodies (bottom row).
The above data did not allow us to distinguish whether HPK-1 phosphorylates S376 directly or by recruiting/activating another kinase. To discriminate between these two possibilities, an in vitro kinase assay was set up with WT or kinase-defective HPK-1–HA and FLAG–SLP-76 constructs isolated separately after transfection of COS-7 cells (see Materials and methods). As shown in Fig. 6 C (bottom row), a robust phosphorylation of S376 was observed when SLP-76 was incubated with WT HPK-1 in the presence, but not in the absence, of ATP. In contrast, no phosphorylation was detected when SLP-76 was mixed with kinase-defective HPK-1. These results demonstrate that HPK1 directly phosphorylates SLP-76 in vitro and strongly suggest that this is also the case in activated T cells.
Collectively, our findings uncover a novel regulatory mechanism that negatively modulates activation-induced transcription in T cells that involves HPK-1–dependent phosphorylation of SLP-76 at S376 and recruitment of 14-3-3 proteins to the TCR-associated signalosome.
DISCUSSION
Current models depicting the TCR signalosome may be incomplete because restricted to tyrosine-phosphorylated components and to prototypic signaling elements shared by many receptors. Thus, novel approaches may be needed to probe for yet unknown signaling elements. Moreover, understanding how TCR signals shape gene activation requires evaluating qualitative and quantitative variations within the signalosome that are likely to occur as a function of stimuli duration and/or intensity. A major hurdle toward these goals is that such a protein complex is of low abundance and highly dynamic, i.e., intrinsically labile. We tried to overcome these difficulties by combining overexpression of epitope-tagged SLP-76 and its tag-dependent isolation, followed by MS identification of associated proteins. The efficacy of this approach was demonstrated by the isolation of known SLP-76 binding partners and the identification of two novel interacting proteins, both members of the 14-3-3 protein family. This finding led us to uncover a novel SLP-76–based negative feedback mechanism that likely serves to fine tune TCR-dependent signals.
Association of 14-3-3 proteins with SLP-76 is mediated by phosphorylation of S376. This residue, which is part of a motif sharing weak homology with the mode I 14-3-3 binding motif (22), lies in the proline-rich region of SLP-76 that mediates association with multiple partners (e.g., Gads, PLC-γ1, and inducible T cell kinase). Ser/Thr phosphorylation of SLP-76 has been reported (31), but neither the specific residues nor the significance of these modifications was identified. Our data (see also reference 21) reveal that four Ser residues of SLP-76 are phosphorylated either constitutively or after cell stimulation, but the role of those residues other than S376 remains to be studied. Different to the immediate induction and rapid decrease of phosphorylation of N-terminal tyrosines involved in the binding of SLP-76 to other key signaling effectors, S376 phosphorylation peaked at 10–15 min and declined slowly, matching the kinetics of 14-3-3 binding. These findings revealed a new mechanism by which SLP-76 recruits signaling partners and a rapid changing pattern of its interactions.
Our results directly implicated HPK-1 in S376 phosphorylation. Previously, this kinase was regarded as MAP4K, a positive upstream regulator of mitogen-activated protein kinases, such as JNK (32, 33). An implication of HPK-1 in TCR-proximal signaling has been suggested (20), but its specific role in this context has remained elusive. We demonstrate here that HPK-1 regulates SLP-76, a major TCR signaling protein, consistent with previous data showing that HPK-1 can inducibly interact with the former (20). We cannot exclude that once recruited to SLP-76, HPK-1 might also phosphorylate other SLP-76 partners and/or act as a MAP4K in T cells.
Data presented herein show that SLP-76 not only orchestrates positive signaling, but in combination with HPK-1 and 14-3-3, also generates a delayed negative signal regulating T cell activation. Both S376A mutation and HPK-1 knock-down correlated with increased TCR signal intensity, as demonstrated by the augmented tyrosine phosphorylation of SLP-76 and PLC-γ1. Reduction of HPK-1 expression by siRNA that increases NFAT activity (18) or overexpression of this kinase that inhibits ERK and AP-1 activation (19) previously suggested that HPK-1 negatively affects TCR-induced transcription. During the revision of this article, Shui et al. (34) have reported a study of HPK-1–deficient mice confirming the role of HPK-1 as a negative regulator of T cell activation and immune response. Similar to our data, HPK-1 deficiency results in increased TCR-induced tyrosine phosphorylation of several signaling proteins, including SLP-76 and PLC-γ1, that correlates with increased calcium mobilization, ERK activation, and enhanced T cell activation. Shui et al. also showed that HPK-1 phosphorylates SLP-76 in vitro and that SLP-76 associates with 14–3-3τ. We have detected 14-3-3ζ, 14-3-3ɛ (this work), and 14-3-3γ (unpublished data) associated with SLP-76 but not 14-3-3τ. The reason for this discrepancy is unclear, but it may be due to differences in the cells used. These results essentially corroborate our model of negative regulation of T cell activation through SLP-76, HPK-1, and 14-3-3. However, our findings that mutation of S376, a specific target of HPK-1 on SLP-76, abolished 14-3-3 binding and resulted in higher TCR-dependent IL-2 promoter induction demonstrate that 14-3-3 recruitment to SLP-76 is one of the main mechanisms by which HPK-1 negatively affects T cell activation. Finally, although Shui et al. proposed that S207 of SLP-76 may be a docking site for 14-3-3, based on conjectures about 14-3-3 consensus binding sites (34), our work indicates that S207 does not play a major role in the 14-3-3–SLP-76 interaction (see Fig. 2). Future studies of knock-in mice expressing SLP-76–S376A would address more precisely T cell abnormalities in the absence of the signal-modulating mechanism describ0ed herein because potential compensatory effects of HPK-1 on other pathways (e.g., JNK) would be avoided.
The 14-3-3ζ and τ isoforms have been implicated in negative regulation of T cell signaling through their interaction with Casitas B lineage lymphoma proto-oncogene, PKCθ, PI3K, or the adaptor 3BP2 (23–26). Hence, 14-3-3 proteins appear to exert a complex control on TCR signaling, relying on multiple mechanisms. The mechanism by which S376 phosphorylation and 14-3-3 recruitment reduces Tyr phosphorylation of SLP-76 and PLC-γ1 (and perhaps other partners) is unknown. 14-3-3 binding may disrupt the interaction of SLP-76 and its partners with an upstream protein tyrosine kinase (e.g., ζ chain–associated protein of 70 kD), thus shortening the duration of tyrosine phosphorylation. Other proteins binding to the proline-rich region of SLP-76 (which contains the 14-3-3 binding site), might be released upon 14-3-3 recruitment. However, Gads association with SLP-76 does not seem to be modified by 14-3-3 binding because the S376A mutation does not affect the Gads–SLP-76 complex (Fig. S4, available at http://www.jem.org/cgi/content/full/jem.20062066/DC1). Nonetheless, 14-3-3 proteins could compete with the binding of PLC-γ1 or inducible T cell kinase. Alternatively, 14-3-3 proteins may recruit/stabilize a PTP in the signalosome or induce dissociation of one or more proteins from the complex, leading to increased availability of phosphorylation sites to PTPs. Another intriguing possibility, not mutually exclusive of the above mechanisms, is suggested by the role of 14-3-3 proteins as regulators of trafficking and subcellular localization of their partners (35). Upon 14-3-3 binding, SLP-76 might be released from the membrane-proximal signalosome, alone or together with some associated proteins, and translocate to a different intracellular site, as suggested by data reporting a relocation of SLP-76 to a perinuclear compartment a few minutes after receptor engagement (12). Hence, the long-lasting phosphorylation of S376 and 14-3-3 binding might also affect potential late functions that SLP-76 would exert after leaving the membrane-proximal signalosome.
An instructive example of signal down-regulation via an adaptor is provided by insulin receptor substrate (IRS)-1, a central effector downstream of insulin receptor. Ser/Thr phosphorylation of IRS-1 counteracts its tyrosine phosphorylation, leading to termination of signaling in a physiological setting or to insulin resistance. Impairment of tyrosine phosphorylation of IRS-1, dissociation of the latter from the receptors and from the signaling complex, or its degradation has been proposed to explain this negative regulation (for review see reference 36). Interestingly, 14-3-3 proteins bind to Ser-phosphorylated IRS-1 and regulate its function (37, 38). It is therefore tempting to speculate that the negative feedback loop by which SLP-76 contributes to TCR signal down-modulation occurs via one of the mechanisms described for IRS-1.
We would like to suggest that negative feedback loops such as the one described herein, acting so early on antigen receptor signals, are not required to stop the signal but rather to shape its intensity and/or duration. Indeed, in a physiological setting, T cells collect inputs from peptide/MHC over long periods of time and tuning mechanisms may be necessary to funnel the signal within a certain range of intensity and duration to orchestrate ordered and dosed gene expression.
Here, we have provided an example of how expressing an epitope-tagged central effector coupled to MS analysis can uncover previously hidden aspects of TCR signaling. Recent advances in sensitivity, mass accuracy, and resolution in MS combined with improvements in the isolation of unstable protein complexes should reveal further intricacies in the molecular mechanisms of T cell activation.
MATERIALS AND METHODS
Antibodies.
The following polyclonal antibodies were used: rabbit antibody against all 14-3-3 isoforms (Upstate Biotechnology) or specific for 14-3-3ζ (Santa Cruz Biotechnology, Inc.); goat anti–HPK-1 (Santa Cruz Biotechnology, Inc.); rabbit antiserum anti–SLP-76 (M14; reference 39); and rabbit antibody against phosphorylated S376 of SLP-76, generated and purified by Eurogentec using the peptide CFPQSA-pS376-LPPY as immunogen. Specific antibodies were purified onto a phosphopeptide affinity column and depleted of antibodies recognizing the unphosphorylated peptide. Phospho-specific antibodies against PLC-γ1 (pY783; Cell Signaling Technology) were used. The following mAbs were used: anti-FLAG M2 and anti-GST (Sigma-Aldrich); anti–SLP-76 (SLP-76/03) and anti–human CD3 (UCHT1; Serotec); anti-HA (F7; Santa Cruz Biotechnology, Inc.); anti-phosphorylated SLP-76 Y113 and Y128 (BD Biosciences); anti-CD28 (CD28.2; Immunotech); and anti–mouse CD3 (145-2C11; reference 40). Phycoerythrin-labeled anti–human CD3 (clone SK7; BD Biosciences) and anti-CD28 (CD28.2; eBiosciences) were used for flow cytometry.
Vectors.
The pEF-FLAG-SLP-76-WT plasmid (41), coding for the FLAG-tagged, WT human SLP-76 cDNA, was provided by G. Koretzky (University of Pennsylvania, Philadelphia, PA). Point mutations were introduced by using the QuikChange kit (Stratagene). The pCDNA3.1 vector coding for MYC-14-3-3ζ was provided by A.S. Shaw (Washington University, St. Louis, MO). The GST-14-3-3ζ coding vector was provided by Y.C. Liu (La Jolla Institute for Allergy and Immunology, San Diego, CA). GST fusion proteins were affinity purified on Gluthatione-Sepharose 4B resin (GE Healthcare). Retroviral vectors were prepared as described previously (42) by first introducing FLAG-SLP-76-WT or FLAG-SLP-76-S376A cDNA in the pDONR221 donor plasmid (Invitrogen), and then transferring it into the Gateway A-pMX-IRES-GFP vector (provided by S. Constantinescu, Ludwig Institute for Cancer Research, Brussels, Belgium; reference 42). The resulting pMX-WT-IRES-GFP and pMX-S376A-IRES-GFP vectors were transduced in J14 cells.
The pMT2-HPK-HA and pMT2-HPK(K46E)-HA vectors (33), coding for HA-tagged WT or kinase-deficient HPK-1 proteins, respectively, were provided by F. Kiefer (Max Planck Institute for Molecular Biomedicine, Münster, Germany).
Cell lines, transfection, and retroviral infection.
Jurkat E6.1 (referred to as Jurkat) and J14 cells (provided by A. Weiss, University of California, San Francisco, San Francisco, CA) were cultured as described previously (27). The murine T cell hybridoma T8.1 and the WT#27 clone overexpressing FLAG-SLP-76-WT were generated and maintained as described previously (21). J14-derived cells stably expressing SLP-76 constructs were obtained by cotransfecting 20 μg pEF-FLAG-SLP-76 plasmid, either -WT or -S376A, and 20 μg pSRα-puromycin vector as described previously (43). J14- derived cell populations stably expressing FLAG-SLP-76-WT or -S376 (named J14-WT and J14-S376A, respectively) were obtained as follows. Viral particles were produced by cotransfection of 293T cells with an encapsidation plasmid (pCMV-MLVgagpol; provided by D. Bohl, Institut Pasteur), the pHCMV-G plasmid coding for VSV-G envelope glycoprotein, and either pMX-WT-IRES-GFP or pMX-S376A-IRES-GFP. Viruses were concentrated from culture supernatants and used to infect J14 cells by two rounds of spinoculation (2,500 rpm for 1 h 30 min at 30°C) in the presence of 8 μg/ml polybrene. GFP+ cells were then enriched by fluorescence- activated cell sorting. COS-7 cells were maintained and transfected as described previously (44). T cell blasts were prepared from human PBMCs as described previously (45).
MS analysis.
To identify SLP-76 associated proteins, anti-FLAG immunoprecipitates from WT#27 cells were eluted with FLAG peptide, fractionated by SDS-PAGE, and stained by colloidal blue (Invitrogen). Relevant bands were excised and digested in gel by trypsin (46), and proteins were identified by MALDI-TOF MS at the proteomic facility of the Institut Pasteur. For the identification of SLP-76 serine phosphorylation sites, immunoprecipitated FLAG–SLP-76 isolated from stimulated WT#27 cells was digested in gel (46) by using a combination of trypsin and AspN in a 1:1 ratio. Mass spectra acquisition by nanoESI-tandem MS and analysis and data filtering by sequence tag scanning were performed as described previously (21).
IL-2 promoter activity assays.
J14-WT and J14-S376A cells were transiently transfected by electroporation at 260 V, 950 μF, with 5 μg of the phIL-2 luciferase reporter plasmid (provided by J.F. Peyron, Université de Nice-Sophia Antipolis, France). Total DNA amount was raised to 30 μg using an empty pEF-Bos vector. 22 h later, cells were left unstimulated or stimulated either with increasing concentrations of plate-bound anti-CD3 (30–150 ng/ml) plus 10 μg/ml of soluble anti-CD28 or a combination of 10 ng/ml PMA (Sigma-Aldrich) and 250 ng/ml of the calcium ionophore A23187 (Calbiochem) in 96-well plates. After 6 h, cells lysis and luciferase activity assays were performed as described previously (29). Percent luciferase activity for anti-CD3/CD28–stimulated samples was calculated as follows: % = ((RLU-MIN)/(MAX-MIN))×100, where RLU is the measured luciferase activity of the sample, MIN is the average RLU for the unstimulated sample, and MAX is the average RLU for sample stimulated with PMA/A23187.
siRNA transfection.
All siRNA duplexes were purchased from Dharmacon Inc. HPK-1 knock-down was obtained by electroporating cells at 300 V, 500 μF essentially as described above in the presence of 400 nM of the MAP4K1 siGENOME SMARTpool. Control cells were electroporated without siRNA or with a pool of unrelated siRNAs (siCONTROL non-targeting siRNA pool). Knock-down efficiency assessment and cell analysis were performed 72 h after transfection.
Immunoprecipitation, immunoblotting, and Far Western assay.
Immunoprecipitations were performed as described previously (29). Protein electrophoresis was performed on standard SDS-PAGE or NuPAGE gels (Invitrogen). Immunoblots were detected by either enhanced chemiluminescence or by near-infrared fluorescence. In the latter case, secondary antibodies labeled with either AlexaFluor 680 (Invitrogen) or IRDye 800 (Rockland Immunochemicals) were used. Fluorescent immunoblot images were acquired and quantified by using an Odyssey scanner and the Odyssey 1.2 software (Li-Cor Biosciences). Far Western assays were performed essentially as described previously (47).
In vitro phosphorylation assay.
COS-7 cells transfected with pEF- FLAG-SLP-76-WT or pMT2-HPK-HA or pMT2-HPK(K46E)-HA were immunoprecipitated with anti-FLAG or anti-HA antibodies, respectively. FLAG-SLP-76-WT was eluted by incubating beads with 0.4 mg/ml FLAG peptide and mixed with either HPK-1 construct in 50 mM Tris-HCl, pH7.4, 10 mM MgCl2, 1 mM EDTA, and 0.1% NP-40 with or without 40 μM ATP. Reaction mixtures were incubated at 30°C for 30 min and analyzed by immunoblotting to detect S376 phosphorylation and protein amounts.
Online supplemental material.
Fig. S1 shows that phosphorylation of SLP-76 in primary CD4+ T cells follows the same kinetics observed in Jurkat cells and human T cell blasts (see Fig. 3). Data in Fig. S2 demonstrate that PKA, PKC, and AKT kinases are not involved in the phosphorylation of S376 of SLP-76. Data shown in Fig. S3 rule out that the effects of HPK-1–specific siRNAs are due to off-target effects. Fig. S4 shows that mutation of S376 of SLP-76 strongly impairs the binding of this protein to 14-3-3ζ (bottom) but does not affect the constitutive interaction between Gads and SLP-76(middle). Figs. S1–S4 are available at http://www.jem.org/cgi/content/full/jem.20062066/DC1.
Acknowledgments
We thank Drs. G. Koretzky, A.S. Shaw, Y.C. Liu, S. Constantinescu, F. Kiefer, A. Weiss, D. Bohl, and J.F. Peyron for sharing reagents and cell lines; Mrs. B. Corre and M.C. Potar for providing purified human T cells; J.C. Rousselle and A. Namane for MALDI-TOF analyses; and B.T. Schurter, G. Langsley, A. Alcover, and R. Weil for critical reading of the manuscript.
This work was supported by grants from the Institut Pasteur, the CNRS, the Association pour la Recherche sur le Cancer, the Ligue Contre le Cancer-Comité Ile-de-France, and the European Union (MUGEN Network of Excellence; contract no. LSGH-CT-2005-005203).
The authors have no conflicting financial interests.
Abbreviations used: ERK, extracellular signal-regulated kinase; Gads, Grb2-related adaptor downstream of Shc; GST, glutathione-S-transferase; HPK-1, hematopoietic progenitor kinase 1; IRS, insulin receptor substrate; LAT, linker for activation of T cells; MALDI-TOF, matrix-assisted laser desorption and ionization time of flight; MAP4K, mitogen-activated protein kinase kinase kinase kinase; MS, mass spectrometry; nanoESI, nano-electrospray ionization; PKA, protein kinase A; PKC, protein kinase C; PLC, phospholipase C; PTP, protein tyrosine phosphatase; siRNA, small interfering RNA; SLP-76, SH2 domain–containing leukocyte protein of 76 kD.
V. Di Bartolo's present address is Lymphocyte Cell Biology Unit, Institut Pasteur, 75724 Paris, Cedex 15, France.
O. Acuto and M. Salek's present address is Sir William Dunn School of Pathology, University of Oxford, OX1 3RE Oxford, UK.
References
- 1.Jun, J.E., and C.C. Goodnow. 2003. Scaffolding of antigen receptors for immunogenic versus tolerogenic signaling. Nat. Immunol. 4:1057–1064. [DOI] [PubMed] [Google Scholar]
- 2.Lanzavecchia, A., and F. Sallusto. 2002. Progressive differentiation and selection of the fittest in the immune response. Nat. Rev. Immunol. 2:982–987. [DOI] [PubMed] [Google Scholar]
- 3.Ansel, K.M., D.U. Lee, and A. Rao. 2003. An epigenetic view of helper T cell differentiation. Nat. Immunol. 4:616–623. [DOI] [PubMed] [Google Scholar]
- 4.Kane, L.P., J. Lin, and A. Weiss. 2000. Signal transduction by the TCR for antigen. Curr. Opin. Immunol. 12:242–249. [DOI] [PubMed] [Google Scholar]
- 5.Zhang, W., J. Sloan-Lancaster, J. Kitchen, R.P. Trible, and L. Samelson. 1998. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 92:83–92. [DOI] [PubMed] [Google Scholar]
- 6.Jackman, J.K., D.G. Motto, Q. Sun, M. Tanemoto, C.W. Turck, G.A. Peltz, G.A. Koretzky, and P.R. Findell. 1995. Molecular cloning of SLP-76, a 76-kDa phosphoprotein associated with Grb2 in T cells. J. Biol. Chem. 270:7029–7032. [DOI] [PubMed] [Google Scholar]
- 7.Liu, S.K., N. Fang, G.A. Koretzky, and C.J. McGlade. 1999. The hematopoietic-specific adaptor protein Gads functions in T-cell signaling via interactions with the SLP-76 and LAT adaptors. Curr. Biol. 9:67–75. [DOI] [PubMed] [Google Scholar]
- 8.Wange, R.L. 2000. LAT, the linker for activation of T cells: a bridge between T cell-specific and general signaling pathways. Sci. STKE. 2000:RE1. [DOI] [PubMed]
- 9.Koretzky, G.A., F. Abtahian, and M.A. Silverman. 2006. SLP76 and SLP65: complex regulation of signalling in lymphocytes and beyond. Nat. Rev. Immunol. 6:67–78. [DOI] [PubMed] [Google Scholar]
- 10.Houtman, J.C., Y. Higashimoto, N. Dimasi, S. Cho, H. Yamaguchi, B. Bowden, C. Regan, E.L. Malchiodi, R. Mariuzza, P. Schuck, et al. 2004. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry. 43:4170–4178. [DOI] [PubMed] [Google Scholar]
- 11.Hartgroves, L.C., J. Lin, H. Langen, T. Zech, A. Weiss, and T. Harder. 2003. Synergistic assembly of linker for activation of T cells signaling protein complexes in T cell plasma membrane domains. J. Biol. Chem. 278:20389–20394. [DOI] [PubMed] [Google Scholar]
- 12.Bunnell, S.C., D.I. Hong, J.R. Kardon, T. Yamazaki, C.J. McGlade, V.A. Barr, and L.E. Samelson. 2002. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 158:1263–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Acuto, O., and F. Michel. 2003. CD28-mediated co-stimulation: a quantitative support for TCR signalling. Nat. Rev. Immunol. 3:939–951. [DOI] [PubMed] [Google Scholar]
- 14.Jordan, M.S., A.L. Singer, and G.A. Koretzky. 2003. Adaptors as central mediators of signal transduction in immune cells. Nat. Immunol. 4:110–116. [DOI] [PubMed] [Google Scholar]
- 15.Cannons, J.L., and P.L. Schwartzberg. 2004. Fine-tuning lymphocyte regulation: what's new with tyrosine kinases and phosphatases? Curr. Opin. Immunol. 16:296–303. [DOI] [PubMed] [Google Scholar]
- 16.Mueller, D.L. 2004. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 5:883–890. [DOI] [PubMed] [Google Scholar]
- 17.Matsuda, S., Y. Miwa, Y. Hirata, A. Minowa, J. Tanaka, E. Nishida, and S. Koyasu. 2004. Negative feedback loop in T-cell activation through MAPK-catalyzed threonine phosphorylation of LAT. EMBO J. 23:2577–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Bras, S., I. Foucault, A. Foussat, C. Brignone, O. Acuto, and M. Deckert. 2004. Recruitment of the actin-binding protein HIP-55 to the immunological synapse regulates T cell receptor signaling and endocytosis. J. Biol. Chem. 279:15550–15560. [DOI] [PubMed] [Google Scholar]
- 19.Liou, J., F. Kiefer, A. Dang, A. Hashimoto, M.H. Cobb, T. Kurosaki, and A. Weiss. 2000. HPK1 is activated by lymphocyte antigen receptors and negatively regulates AP-1. Immunity. 12:399–408. [DOI] [PubMed] [Google Scholar]
- 20.Sauer, K., J. Liou, S.B. Singh, D. Yablonski, A. Weiss, and R.M. Perlmutter. 2001. Hematopoietic progenitor kinase 1 associates physically and functionally with the adaptor proteins B cell linker protein and SLP-76 in lymphocytes. J. Biol. Chem. 276:45207–45216. [DOI] [PubMed] [Google Scholar]
- 21.Salek, M., V. Di Bartolo, D. Cittaro, D. Borsotti, J. Wei, O. Acuto, J. Rappsilber, and W.D. Lehmann. 2005. Sequence tag scanning: a new explorative strategy for recognition of unexpected protein alterations by nanoelectrospray ionization-tandem mass spectrometry. Proteomics. 5:667–674. [DOI] [PubMed] [Google Scholar]
- 22.Aitken, A. 2006. 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16:162–172. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Y.C., C. Elly, H. Yoshida, N. Bonnefoy-Berard, and A. Altman. 1996. Activation-modulated association of 14-3-3 proteins with Cbl in T cells. J. Biol. Chem. 271:14591–14595. [DOI] [PubMed] [Google Scholar]
- 24.Bonnefoy-Berard, N., Y.C. Liu, M. von Willebrand, A. Sung, C. Elly, T. Mustelin, H. Yoshida, K. Ishizaka, and A. Altman. 1995. Inhibition of phosphatidylinositol 3-kinase activity by association with 14-3-3 proteins in T cells. Proc. Natl. Acad. Sci. USA. 92:10142–10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meller, N., Y.C. Liu, T.L. Collins, N. Bonnefoy-Berard, G. Baier, N. Isakov, and A. Altman. 1996. Direct interaction between protein kinase C theta (PKC theta) and 14-3-3 tau in T cells: 14-3-3 overexpression results in inhibition of PKC theta translocation and function. Mol. Cell. Biol. 16:5782–5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foucault, I., Y.C. Liu, A. Bernard, and M. Deckert. 2003. The chaperone protein 14-3-3 interacts with 3BP2/SH3BP2 and regulates its adapter function. J. Biol. Chem. 278:7146–7153. [DOI] [PubMed] [Google Scholar]
- 27.Yablonski, D., M.R. Kuhne, T. Kadlecek, and A. Weiss. 1998. Uncoupling of nonreceptor tyrosine kinases from PLC-γ 1 in an SLP-76-deficient T cell. Science. 281:413–416. [DOI] [PubMed] [Google Scholar]
- 28.Houtman, J.C., R.A. Houghtling, M. Barda-Saad, Y. Toda, and L.E. Samelson. 2005. Early phosphorylation kinetics of proteins involved in proximal TCR-mediated signaling pathways. J. Immunol. 175:2449–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michel, F., G. Attal-Bonnefoy, G. Mangino, S. Mise, and O. Acuto. 2001. CD28 as a molecular amplifier extending TCR ligation and signaling capabilities. Immunity. 15:935–945. [DOI] [PubMed] [Google Scholar]
- 30.Irvin, B.J., B.L. Williams, A.E. Nilson, H.O. Maynor, and R.T. Abraham. 2000. Pleiotropic contributions of phospholipase C-γ1 (PLC-γ1) to T-cell antigen receptor-mediated signaling: reconstitution studies of a PLC-γ1-deficient Jurkat T-cell line. Mol. Cell. Biol. 20:9149–9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clements, J.L. 2003. Known and potential functions for the SLP-76 adapter protein in regulating T-cell activation and development. Immunol. Rev. 191:211–219. [DOI] [PubMed] [Google Scholar]
- 32.Dan, I., N.M. Watanabe, and A. Kusumi. 2001. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11:220–230. [DOI] [PubMed] [Google Scholar]
- 33.Kiefer, F., L.A. Tibbles, M. Anafi, A. Janssen, B.W. Zanke, N. Lassam, T. Pawson, J.R. Woodgett, and N.N. Iscove. 1996. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 15:7013–7025. [PMC free article] [PubMed] [Google Scholar]
- 34.Shui, J.W., J.S. Boomer, J. Han, J. Xu, G.A. Dement, G. Zhou, and T.H. Tan. 2007. Hematopoietic progenitor kinase 1 negatively regulates T cell receptor signaling and T cell-mediated immune responses. Nat. Immunol. 8:84–91. [DOI] [PubMed] [Google Scholar]
- 35.Muslin, A.J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell. Signal. 12:703–709. [DOI] [PubMed] [Google Scholar]
- 36.Zick, Y. 2005. Ser/Thr phosphorylation of IRS proteins: a molecular basis for insulin resistance. Sci. STKE. 2005:pe4. [DOI] [PubMed]
- 37.Ogihara, T., T. Isobe, T. Ichimura, M. Taoka, M. Funaki, H. Sakoda, Y. Onishi, K. Inukai, M. Anai, Y. Fukushima, et al. 1997. 14-3-3 protein binds to insulin receptor substrate-1, one of the binding sites of which is in the phosphotyrosine binding domain. J. Biol. Chem. 272:25267–25274. [DOI] [PubMed] [Google Scholar]
- 38.Craparo, A., R. Freund, and T.A. Gustafson. 1997. 14-3-3 (epsilon) interacts with the insulin-like growth factor I receptor and insulin receptor substrate I in a phosphoserine-dependent manner. J. Biol. Chem. 272:11663–11669. [DOI] [PubMed] [Google Scholar]
- 39.Michel, F., G. Mangino, G. Attal-Bonnefoy, L. Tuosto, A. Alcover, A. Roumier, D. Olive, and O. Acuto. 2000. CD28 utilizes Vav-1 to enhance TCR-proximal signaling and NF-AT activation. J. Immunol. 165:3820–3829. [DOI] [PubMed] [Google Scholar]
- 40.Marquez, M.E., W. Ellmeier, V. Sanchez-Guajardo, A.A. Freitas, O. Acuto, and V. Di Bartolo. 2005. CD8 T cell sensory adaptation dependent on TCR avidity for self-antigens. J. Immunol. 175:7388–7397. [DOI] [PubMed] [Google Scholar]
- 41.Motto, D.G., S.E. Ross, J. Wu, L.R. Hendricks-Taylor, and G.A. Koretzky. 1996. Implication of the GRB2-associated phosphoprotein SLP-76 in T cell receptor–mediated interleukin 2 production. J. Exp. Med. 183:1937–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Royer, Y., C. Menu, X. Liu, and S.N. Constantinescu. 2004. High-throughput gateway bicistronic retroviral vectors for stable expression in mammalian cells: exploring the biologic effects of STAT5 overexpression. DNA Cell Biol. 23:355–365. [DOI] [PubMed] [Google Scholar]
- 43.Mège, D., V. Di Bartolo, V. Germain, L. Tuosto, F. Michel, and O. Acuto. 1996. Mutation of tyrosines 492/493 in the kinase domain of ZAP-70 affects multiple T-cell receptor signaling pathways. J. Biol. Chem. 271:32644–32652. [DOI] [PubMed] [Google Scholar]
- 44.Di Bartolo, V., M. Malissen, E. Dufour, E. Sechet, B. Malissen, and O. Acuto. 2002. Tyrosine 315 determines optimal recruitment of ZAP-70 to the T cell antigen receptor. Eur. J. Immunol. 32:568–575. [DOI] [PubMed] [Google Scholar]
- 45.Blanchet, F., A. Cardona, F.A. Letimier, M.S. Hershfield, and O. Acuto. 2005. CD28 costimulatory signal induces protein arginine methylation in T cells. J. Exp. Med. 202:371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850–858. [DOI] [PubMed] [Google Scholar]
- 47.Liu, Y.-C., Y. Liu, C. Elly, H. Yoshida, S. Lipkowitz, and A. Altman. 1997. Serine phosphorylation of Cbl induced by phorbol ester enhances its association with 14-3-3 proteins in T cells via a novel serine-rich 14-3-3 binding motif. J. Biol. Chem. 272:9979–9985. [DOI] [PubMed] [Google Scholar]