Abstract
The yeast nucleoporin Nup2p is associated primarily with the nuclear basket of nuclear pore complexes and is required for efficient importin-α:β-mediated nuclear protein import as well as efficient nuclear export of Kap60p/importin-α. Residues 1–51 of Nup2p bind tightly to Kap60p and are required for Nup2p function in vivo. We have determined the 2.6 Å resolution crystal structure of a complex between this region of Nup2p and the armadillo repeat domain of Kap60p. Nup2p binds along the inner concave groove of Kap60p, but its interaction interface is different from that employed for nuclear localization signal (NLS) recognition although there is some overlap between them. Nup2p binds Kap60p more strongly than NLSs and accelerates release of NLSs from Kap60p. Nup2p itself is released from Kap60p by Cse1p:RanGTP only in the presence of the importin-β binding (IBB) domain of Kap60p. These data indicate that Nup2p increases the overall rate of nuclear trafficking by coordinating nuclear import termination and importin recycling as a concerted process.
Keywords: importin-α/nuclear trafficking/nucleoporin/recycling
Introduction
Eukaryotic cells transport proteins and RNAs into and out of the nucleus through nuclear pore complexes (NPCs), which are constructed from a large number of different proteins collectively termed nucleoporins (Nups) (Weis, 2003). NPCs have a central disc with 8-fold rotational symmetry embedded in the nuclear envelope together with extensions that form cytoplasmic filaments and nuclear baskets. In Saccharomyces cerevisiae, NPCs are constructed from ∼30 different Nups with a combined mass of 50 MDa (Rout et al., 2000). Many Nups are symmetrically distributed about the central axis of the NPC, although a subset is localized exclusively to the cytoplasmic or nuclear faces and so may be crucial for regulating initiation and termination of trafficking.
Nuclear transport is a signal-mediated active process that depends on concerted interactions between nucleoporins, soluble transport factors and their cargo macromolecules. Transport factors (such as karyopherins) recognize cargo in one compartment, translocate it through NPCs, and then release it in the other compartment before being recycled to participate in further rounds of transport (reviewed by Weis, 2003). The classical nuclear localization signal (NLS) is characterized by one or two short stretches of basic residues (Dingwall and Laskey, 1998). In the cytoplasm, NLS-containing proteins bind the Kap60p/importin-α adapter, which, in turn, binds Kap95p/importin-β via its importin-β binding (IBB) domain. This cargo:carrier complex is then docked to and translocated through NPCs in a process mediated by interactions between Kap95p/importin-β and Nups that contain phenylalanine–glycine (FG) sequence repeats (Rexach and Blobel, 1995; Bayliss et al., 2000). Once in the nucleus, RanGTP dissociates the cargo:carrier complex and the importins are recycled to the cytoplasm. Kap95p/importin-β returns bound to RanGTP, whereas recycling of Kap60p/importin-α is mediated by CAS/Cse1p complexed with RanGTP (Kutay et al., 1997; Hood and Silver, 1998; Solsbacher et al., 1998). In the cytoplasm, the Ran GTPase is activated by RanGAP, dissociating the importin-β:RanGTP and importin-α:CAS:RanGTP complexes, freeing the importins for another cycle. Finally, the RanGDP generated in the cytoplasm is recycled by NTF2 to the nucleus where it is recharged with GTP by RCC1.
Crystal structures of several key players in the classical NLS-import cycle have been obtained (Conti and Izaurralde, 2001). Kap95p/importin-β is built from 19 α-helical HEAT repeats, whereas Kap60p/importin-α has an N-terminal importin-β binding (IBB) domain, a central armadillo (Arm) repeat domain, and a short C-terminal extension. Arm repeats are constructed from three α-helices and pack into a right-handed superhelix to produce a gently curved elongated molecule (Conti et al., 1998). Both HEAT and Arm repeats provide a structural platform that mediates protein–protein interactions along its surface. The classical NLS sits on the concave inner surface of Kap60p/importin-α, in a groove lined with conserved Trp and Asn residues. There are two NLS binding sites: monopartite NLSs bind primarily to the major site formed by Arm repeats 2–4, whereas bipartite NLSs bind to both this site and a second one, formed by Arm repeats 7–8 (Conti et al., 1998; Conti and Kuriyan, 2000; Fontes et al., 2000).
Translocation through NPCs appears to be a reversible facilitated diffusive process and the disassembly/assembly of cargo:carrier complexes in the appropriate compartments not only imparts directionality, but also allows accumulation of cargo against a concentration gradient (Gorlich et al., 2003; Weis, 2003). Import cargoes dissociate from the carriers in the nucleus after which the carriers are recycled to the cytoplasm. The precise way in which cargo is released in the nucleus has not been established unequivocally. The spontaneous dissociation of a classical NLS from the Kap60p:Kap95p heterodimer is quite slow (Gilchrist et al., 2002). This probably prevents premature release of cargo during the transit across the NPC, but necessitates a mechanism by which cargo release is accelerated in the nucleus (Gilchrist et al., 2002). Possible acceleration mechanisms include competition with the auto-inhibitory region of the IBB domain of Kap60p/importin-α (Kobe, 1999; Catimel et al., 2001; Harreman et al., 2003a), competition with the CAS:RanGTP complex (Kutay et al., 1997), and the action of peripheral Nups such as Nup2p (Solsbacher et al., 2000; Gilchrist et al., 2002).
Nup2p has a multi-domain structure based on an N-terminal Kap60p binding and NPC targeting domain, a central domain containing 16 tandem FxFG sequence repeats that bind Kap95p, and a C-terminal Ran-binding domain (Loeb et al., 1993). Nup2p has little regular secondary structure (Denning et al., 2002). It is localized primarily to the nucleoplasmic face of NPCs (Solsbacher et al., 2000; Dilworth et al., 2001) and is proposed to form part of the nuclear basket, although in some conditions it can also shuttle between nucleus and cytoplasm (Dilworth et al., 2001). The precise mechanism underlying Nup2p function has not been established, although Gilchrist et al. (2002) have shown that it accelerates the release of the Cbp80p NLS from Kap60p:Kap95p. Lindsay et al. (2002) have suggested that Nup2p may be analogous to vertebrate Npap60, which is proposed to bind the importin:substrate complex and accompany it during import through NPCs. Although Nup2p is not essential in normal genetic backgrounds, yeast lacking Nup2p exhibit defects in Kap60p/Kap95p-mediated nuclear protein import and defects in Cse1p-mediated recycling of Kap60p (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000). Because of its multi-domain structure, Nup2p could mediate a variety of protein–protein interactions, raising the possibility that disassembly and assembly of transport complexes, which are often considered in isolation, could occur in a cooperative fashion in vivo.
Here we show that residues 1–50 of Nup2p are required for function in vivo and use X-ray crystallography to define the interaction interface between Nup2p and Kap60p. Nup2p and NLSs have different yet partially overlapping binding sites on Kap60p and the affinity of Nup2p for Kap60p is higher than that of NLSs. Moreover, we present evidence that Nup2p residues 1–51 increase the off-rate of NLS-cargoes from Kap60p and that Nup2p is itself removed from Kap60p by the concerted action of Cse1p:RanGTP and the IBB domain. The unique structural features of the Nup2p:Kap60p interaction, together with these binding and kinetic data, provide insight into the cooperative mechanism by which Nup2p together with the IBB domain, Cse1p and RanGTP orchestrates the efficient release of NLS-cargo in the nucleus concomitant with assembly of the export complexes (Kap95p:RanGTP and Kap60p:Cse1p:RanGTP) required for recycling.
Results
The N-terminus of Nup2p is required for function in vivo
Hood et al. (2000) showed deletion of residues 1–175 inhibited Nup2p targeting to the nuclear envelope as well as binding to Kap60p and suggested a role for the Nup2p N-terminus in providing an initial NPC docking site along the Cse1p:Ran-mediated Kap60p export pathway. Subsequent work suggested that the first 50 residues of Nup2p were critical for Kap60p binding (Denning et al., 2001). To assess the functional importance of these residues, we created a plasmid in which Δ50nup2p (Nup2p residues 51–720) was fused at its C-terminus to GFP. Deletion of residues 1–50 (Δ50nup2p) did not affect the expression of Nup2p (data not shown). The truncated protein was still targeted to the nuclear rim although there was some increased signal in the cytoplasm (Figure 1A). To test the functional importance of Nup2p residues 1–50 in vivo, the Δ50nup2p plasmid, a control wild-type Nup2p plasmid, or vector alone were transformed into Δnup2 srp1-31 double mutant cells (where Nup2p is absolutely essential for viability; Booth et al., 1999) and the SRP1 maintenance plasmid removed by plasmid shuffle (Figure 1B). Although expression of full-length Nup2p complemented Δnup2 srp1-31 cells, expression of Δ50nup2p did not, consistent with residues 1–50 of Nup2p being functionally necessary in vivo. Expression of a construct that expressed only residues 1–51 (1–51nup2p) did not complement the Δnup2 srp1-31 mutant (Figure 1B) and, although a fusion of these residues to GFP was localized to the nucleus, it did not localize specifically to the nuclear rim (data not shown). These data demonstrate that residues 1–50 of Nup2p are necessary, but not sufficient for Nup2p function in vivo.
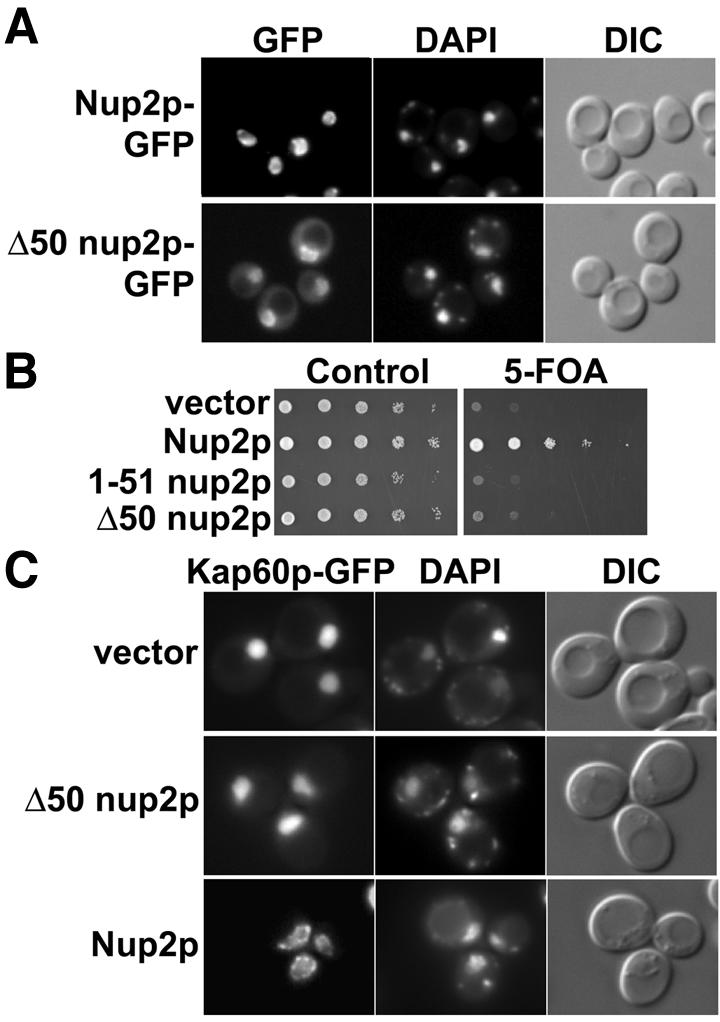
Fig. 1. In vivo functional analysis of NUP2. (A) Nup2p and Δ50Nup2p expressed as C-terminal GFP fusions under the control of NUP2 promoter in Δnup2 yeast cells show nuclear envelope localization as visualized by GFP fluorescence. Corresponding DIC and DAPI images are shown. (B) Δnup2 srp1-31 yeast cells maintained by a plasmid encoding Kap60p and expressing either full-length Nup2p, 1–51 Nup2p or Δ50Nup2p were spotted onto control plates lacking uracil or 5-FOA plates. 5-FOA eliminates the URA3 maintenance plasmid encoding Kap60p. (C) The Nup2p N-terminus promotes docking of Kap60p to the nuclear envelope and is required for efficient recycling of Kap60p to the cytoplasm. Kap60p–GFP was integrated at the endogenous SRP1 locus of Δnup2 yeast cells. The cells were then transformed with plasmids encoding either full-length Nup2p, Δ50Nup2p, or vector alone, and Kap60p–GFP was visualized by GFP fluorescence. Corresponding DIC and DAPI images are shown.
We exploited the observation that Kap60p accumulates within the nucleus of cells that lack Nup2p (Booth et al., 1999; Hood et al., 2000; Solsbacher et al., 2000) to probe whether residues 1–50 of Nup2p are required for Nup2p-dependent recycling of Kap60p to the cytoplasm. In wild-type cells, integrated Kap60p–GFP was localized to the nuclear rim whereas, in Δnup2 cells, it accumulated within the nucleus. The Δnup2 cells were subsequently transformed with plasmids encoding full-length Nup2p, Δ50nup2p, or vector alone, co-stained with DAPI to mark the position of the nucleus, and visualized (Figure 1C). Integrated Kap60p–GFP was concentrated at the nuclear rim in cells expressing Nup2p, as described (Booth et al., 1999; Solsbacher et al., 2000), but was localized throughout the nucleus of Δnup2 cells expressing Δ50nup2p (or vector alone), consistent with residues 1–50 of Nup2p being essential for efficient recycling of Kap60p to the cytoplasm. Binding assays indicated that Δ50nup2p does not bind Kap60p (Denning et al., 2001) and that residues 1–51 of Nup2p retained the ability to bind tightly to Kap60p (Table I and see below), indicating that the effects seen in vivo were due, at least in part, to decreased interaction between Nup2p and Kap60p.
Table I. Dissociation constants determined by microtitre plate binding assay.
| Kap60p construct | Binding partner | KD (nM) |
|---|---|---|
| 1–542, wild-type | GST-Nup2p (1–720), wild-type | 4.4 ± 1.1 |
| 88–530, wild-type | GST-Nup2p (1–720), wild-type | 0.07 ± 0.02 |
| 88–530, wild-type | GST-Nup2p (1–720), R38A/R39A | 3.0 ± 0.4 |
| 88–530, wild-type | GST-Nup2p (1–720), R47A/R48A | 0.13 ± 0.02 |
| 88–530, wild-type | GST-Nup2p (1–720), R38A/R39A/R47A/R48A | 240 ± 60 |
| 1–542, wild-type | GST-Nup2p (1–51), wild-type | 30 ± 5 |
| 88–530, wild-type | GST-Nup2p (1–51), wild-type | 2.1 ± 0.3 |
| 88–530, Y397D | GST-Nup2p (1–51), wild-type | 4.4 ± 0.5 |
| 88–542, wild-type | GST-Nup2p (1–51), wild-type | 2.4 ± 0.4 |
| 88–542, wild-type | GST-Nup2p (1–174), wild-type | 3.5 ± 0.4 |
| 88–542, wild-type | GST-Nup2p (36–51), wild-type | 20 ± 1 |
| BFP-81–542, wild-type | GST-Nup2p (1–51), wild-type | 2.2 ± 0.4 |
| 1–542, wild-type | GST-SV40 NLS | 1000 ± 400 |
| 88–542, wild-type | GST-SV40 NLS | 22 ± 3 |
| BFP-81–542, wild-type | SV40 NLS-GFP | 12 ± 2 |
| 1–542, wild-type | GST-nucleoplasmin NLS | 450 ± 100 |
| 88–542, wild-type | GST-nucleoplasmin NLS | 34 ± 4 |
| BFP-81–542, wild-type | nucleoplasmin NLS-GFP | 9 ± 1 |
| 1–542, wild-type | GST | not detectable |
| 88–542, wild-type | GST | not detectable |
| 88–530, wild-type | GST | not detectable |
| 88–530, Y397D | GST | not detectable |
| BFP-81–542, wild-type | GST | not detectable |
| BFP-81–542, wild-type | GFP | not detectable |
Data represent the best-fit value ± standard error as analyzed by nonlinear regression assuming one site binding. Each assay was performed in duplicate.
Crystal structure of Nup2p residues 1–51 complexed with Kap60Δ
We used X-ray crystallography to address the structural basis of the interaction between Kap60p and Nup2p. The IBB domain of Kap60p is not required for Nup2p binding (Booth et al., 1999; Hood et al., 2000). A truncated construct of Kap60p (residues 88–530; Kap60Δ) bound tightly to full-length Nup2p (KD 0.07 ± 0.02 nM) and a Nup2p N-terminal fragment (residues 1–51; Nup2N) retained nM affinity (KD 2.1 ± 0.3 nM) for Kap60Δ (Table I). The Kap60Δ mutant Y397D that has improved solubility (Conti and Kuriyan, 2000) had comparable affinity for Nup2N (KD 4.4 ± 0.5 nM) and was used for crystallization. We obtained P21212 crystals of the Kap60Δ:Nup2N complex that diffracted to 2.6 Å resolution and solved their structure by molecular replacement. The final model was refined to a free R-factor of 25.7% (R-factor 21.6%) and contained residues 88–526 of Kap60p, residues 36–51 of Nup2p and 158 waters (Table II). The two Kap60Δ:Nup2N complexes in the asymmetric unit were essentially identical, indicating that crystal packing interactions had not significantly altered their conformation. Although the two Kap60Δ chains in the asymmetric unit formed a symmetrical dimer with roughly one third of the N-terminal portion of each contributing to the dimer interface, each Nup2N chain interacted almost exclusively with a single Kap60Δ. Gel filtration indicated that the Kap60Δ:Nup2N complex existed primarily as a monomer in solution (data not shown), indicating that the dimer in the crystals was probably not important physiologically. The Kap60Δ chains in the complex were similar to those in the Kap60Δ:SV40-NLS or Kap60Δ:nucleoplasmin-NLS complexes (Conti et al., 1998; Conti and Kuriyan, 2000): the overall Cα r.m.s. deviations were 0.67 Å and 1.23 Å, respectively, indicating that the binding of Nup2N was not accompanied by a major conformational change.
Table II. Crystallographic statistics.
| Data collection statistics | |
| Space group | P21212 |
| Unit cell dimensions (Å) | a = 129.81, b = 140.08, c = 63.99 |
| Resolution range (Å)a | 20–2.6 (2.74–2.60) |
| Mosaicity | 0.75 |
| Total observationsa | 76810 (10875) |
| Unique reflectionsa | 29670 (4308) |
| Completeness (%)a | 97.5 (98.3) |
| Rmerge (%)a | 8.6 (53.1) |
|
I/σa |
10.0 (1.8) |
| Refinement statistics | |
| Number of reflections (working, test) | 33802/1806 |
| Rcryst/Rfree (%) | 21.6/25.7 |
| Total number of non-H atoms | 7189 |
| Number of water molecules | 158 |
| R.m.s. deviation from ideal bond length (Å) | 0.012 |
| R.m.s. deviation from ideal bond angles (degree) | 1.831 |
| Ramachandran plot (%) | |
| Core region | 92.6 |
| Allowed region | 7.0 |
| Generously allowed region | 0.4 |
| Disallowed region | 0.0 |
aParentheses refer to final resolution shell.
The interface between Kap60Δ and the Nup2p N-terminus
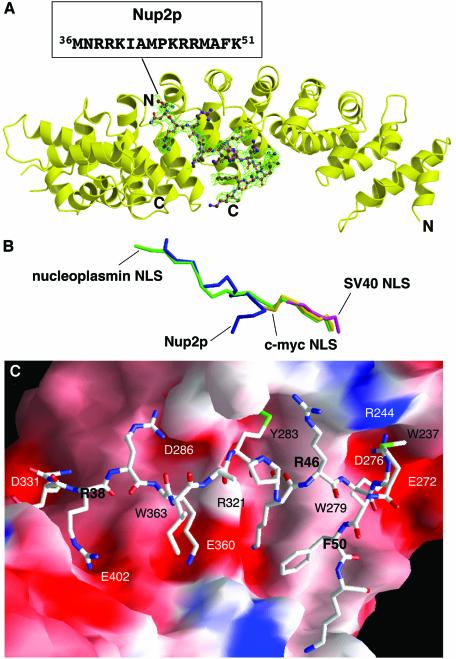
The interaction interface between Nup2p and Kap60Δ was different to that seen with NLSs or the IBB domain (Kobe, 1999; Conti and Kuriyan, 2000). The extensive region of Nup2p visible in the omit map (Figure 2A) contained an elongated region (residues 36–45) that ran along the inner groove of Kap60Δ, together with a type I β-turn (residues 46–50), which reversed the direction of the chain towards the Kap60Δ C-terminus. Nup2p packed against the third helices of Arm repeats 4–8 and was intimately attached at two distinct sites. The first was located between the major and minor NLS binding sites and involved primarily interactions between residues 43–51 of Nup2p and Arm repeats 4–6, whereas the second site corresponded to the minor NLS binding site and involved primarily interactions between Nup2p residues 36–40 and surface pockets in Arm repeats 7 and 8. The Nup2p chains were well defined, with average B-factors of 47 and 56 Å2 reducing to generally 30–50 Å2 at the binding sites. The buried interfacial area was 2393 Å2 and involved a combination of H-bonds, electrostatic interactions and stereospecific hydrophobic interactions. The path of the Nup2p main chain on Kap60Δ partially overlapped with that of the N-terminal half of the bipartite nucleoplasmin NLS, but its overlap with the monopartite c-myc or SV40 NLS was less severe (Figure 2B).
Fig. 2. Structure of residues 1–51 of Nup2p bound to Kap60Δ. (A) Overview of the Kap60Δ:Nup2N complex. The Fo – Fc omit map corresponding to the Nup2p fragment contoured at 2.5 σ and the refined model of Nup2p (residues 36–51) are superimposed. (B) Overlay of Nup2p with NLS peptides. The coordinates of Kap60Δ bound to Nup2N (blue) and the SV40 (purple), c-myc (yellow), and nucleoplasmin (green) NLSs (Conti et al., 1998; Conti and Kuriyan, 2000) were superimposed, then removed to show the relative positions of each ligand. Orientation as in (A). (A and B) were prepared with Bobscript (Esnouf, 1997) and Raster3D (Merritt and Bacon, 1997). (C) Molecular surface of Kap60Δ coloured by electrostatic potential shaded from –13 kT/e (red) to +13 kT/e (blue) (calculated with Nup2N removed using GRASP; Nicholls et al., 1991) shows acidic pockets and a nonpolar surface that recognize Nup2p. Nup2p residues are bold.
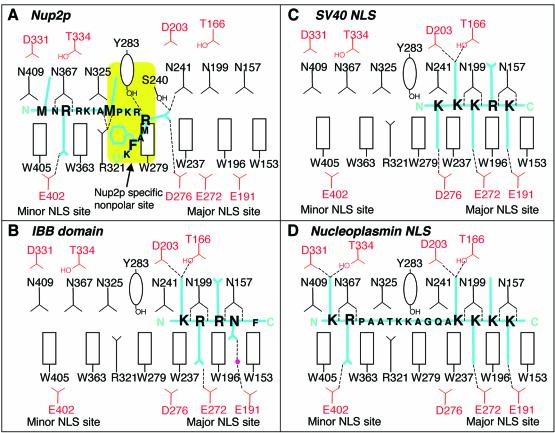
The Nup2p residues contributing to the first binding site included an IBB-like sequence, 45KRR47, together with flanking hydrophobic residues. However, the Nup2p KRR sequence bound in a novel way (Figures 2C and 3) that was quite different from that observed for the IBB domain (Kobe, 1999). Thus, the side chain of Nup2p Met43 was inserted into a hydrophobic pocket around Kap60Δ Tyr283 and the interaction was further stabilized by a H-bond between the OH of Tyr283 and the Nup2p backbone. The side chain of Kap60Δ Arg321 formed H-bonds with the main chain carbonyls of Nup2p Ala42 and Met43, holding Met43 in an appropriate position and orientation for insertion of its hydrophobic side chain into the nonpolar pocket. The side chain of Nup2p Arg47 extended into the P1 acidic pocket in the major NLS binding site of Kap60Δ (Conti et al., 1998; Conti and Kuriyan, 2000) where its guanidinium group formed H-bonds with Ser240, Asn241 and Asp276. There is an electrostatically neutral region on the surface of Kap60Δ between the major and minor NLS binding sites (Figure 2C), and this hydrophobic patch (centred on Trp279) made interactions with the aliphatic regions of the side chains of Nup2p Lys45, Arg46 and Arg47 as well as with Phe50. The side chains of Nup2p Met43, Arg47 and Phe50 were well defined in the electron density map and so were probably key determinants of binding specificity and affinity.
Fig. 3. Schematic comparison of the interactions of Kap60Δ with (A) Nup2p, (B) IBB domain (based on mouse importin α; Kobe, 1999), (C) monopartite NLS (Conti et al., 1998), and (D) bipartite NLS (Conti and Kuriyan, 2000).
At the second Nup2p binding site, Nup2p Met36 and Arg38 fit into the two pockets in the minor NLS binding site in much the same way as observed for a bipartite NLS (Conti and Kuriyan, 2000). The side chain of Arg38 that occupied the P2′ pocket had well defined density and formed a H-bond with Kap60Δ Glu402 as well as a cation-π interaction with the ring of Trp405. The side chains of Kap60Δ Asn367 and Trp363 formed H-bonds with the main chain amide and carbonyl of Nup2p Arg38, stabilizing the interactions at this second site. The side chains of Nup2p Arg39 and Lys40 did not have strong electron density, but did point towards acidic potentials on the Kap60Δ surface around Asp286 and Glu360 (Figure 2C), and so probably make additional contributions to binding.
To evaluate the contribution of different Nup2p residues to Kap60p binding, we constructed full-length Nup2p mutants in which either of the two Arg clusters (Arg38/39 and Arg46/47) or all four Args were mutated to Ala, and measured binding to Kap60Δ (Table I). Full-length Nup2p bound (KD 0.07 ± 0.02 nM) ∼30-fold more strongly than Nup2N, indicating that residues C-terminal to Lys51 also contribute to the interaction. This additional contribution probably derived from the FxFG repeats and/or the Ran binding domain, because residues 1–174 had an affinity for Kap60Δ (KD 3.5 ± 0.4 nM) comparable to Nup2N (KD 2.4 ± 0.4 nM). In this context it is interesting to note that in the Kap60Δ:Nup2N structure, many hydrophobic Nup2p side chains (Ile41, Ala42, Pro44, Met48 and Ala49) are exposed to solvent and so part of the rest of Nup2p may pack over them, reducing the entropic penalty associated with binding Kap60Δ. R38/39A mutations in Nup2p weakened Kap60Δ binding by ∼40-fold (KD 3.0 ± 0.4 nM) whereas the R46/47A mutations were less effective (KD 0.13 ± 0.02 nM). When combined, however, the mutations were synergistic and the affinity of the R38/39/46/47A mutant for Kap60Δ was reduced by three orders of magnitude (KD 240 ± 60 nM). These data show that both Arg clusters are important for the interaction and support the idea that the N-terminal 51 residues of Nup2p represent the primary Kap60p binding site. Although Nup2p residues 1–35 did not have clear electron density, they probably made an additional, minor, contribution to binding because residues 36–51 alone had 10-fold weaker affinity (KD 20 ± 1 nM) for Kap60Δ.
The interaction between Kap60Δ and Nup2p is different from the interaction with either classical NLSs or the IBB domain
In Kap60:NLS structures, clusters of basic residues bind in the major NLS binding site in an extended conformation (Conti et al., 1998; Conti and Kuriyan, 2000; see Figures 2B and 3). This contrasts with the Kap60Δ: Nup2N interaction, where the downstream KRR sequence in Nup2p interacts in a novel way, leaving most of the major NLS binding site accessible (Figure 3). Site-directed mutagenesis confirmed the differences between the binding sites of Nup2p and NLSs on Kap60Δ. Thus, whereas the D203K/N157A mutation in the primary NLS binding site of Kap60Δ abolished binding to the SV40 NLS, Nup2N binding was retained (Figure 4A). Three factors probably contributed to these differences. First, Nup2p residues 36–38 are specifically anchored at the minor NLS site; secondly, the linker sequence between the upstream and downstream basic clusters of Nup2p is shorter than in bipartite NLSs; and thirdly, the hydrophobic residues flanking the KRR sequence allow Nup2p residues 46–50 to form a β-hairpin and pack against a nonpolar patch on Kap60Δ.
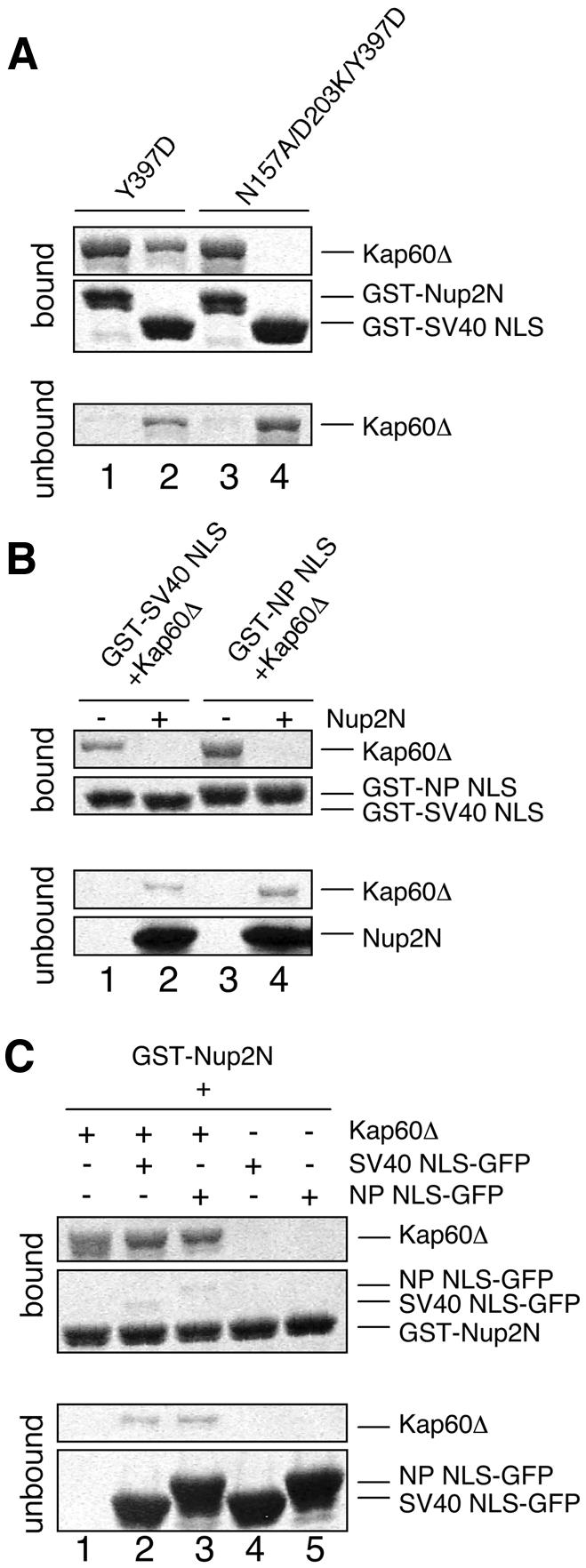
Fig. 4. Nup2p–Kap60p interactions are different from NLS interactions and Nup2p competes with NLSs. (A) N157A/D203K mutations in the major NLS binding site of Kap60Δ abolish SV40 NLS binding but not Nup2N binding. GST–Nup2N (10 µg; lanes 1 and 3) or GST–SV40 NLS (10 µg; lanes 2 and 4) was treated with 15 µg of Kap60Δ. (B) Nup2N competes both monopartite and bipartite NLSs from Kap60Δ. Beads containing 4.5 µg GST–SV40 NLS or GST–nucleoplasmin (NP) NLS were treated with Kap60Δ (10 µg) which remained mainly bound after washing with binding buffer but was subsequently removed by 30 µM His/S–Nup2N. (C) NLS-cargoes do not effectively compete Nup2N from Kap60Δ and only bind weakly in the presence of Nup2N. GST––Nup2N (4.5 µg) was treated with Kap60Δ (10 µg) ± 30 µM NLS–GFP (lanes 1–3) or 30 µM NLS–GFP alone (lanes 4 and 5).
Nup2N accelerates dissociation of NLSs from Kap60Δ
As illustrated in Figure 3, although the Nup2p binding site on Kap60p is different from both NLS binding sites, there is a degree of overlap. Nup2p would clash with monopartite NLSs in the P1 pocket of the major NLS binding site and so could destabilize interaction with neighbouring pockets, particularly the P2 site crucial for NLS recognition (Hodel et al., 2001). Although the bipartite nucleoplasmin NLS does not use the P1 site, it would clash with Nup2p at the minor NLS site and also at the intervening region. Furthermore, Nup2N bound more strongly to Kap60Δ (KD ∼2.4 nM) than the SV40 or nucleoplasmin (NP) NLS (KD ∼10–30 nM; Table I). Consequently, the Nup2p residues 1–51 should compete NLSs from Kap60p. This prediction was confirmed by an equilibrium cross-competition assay (Figure 4B and C), consistent with a previous study using Nup2p residues 1–174 (Solsbacher et al., 2000).
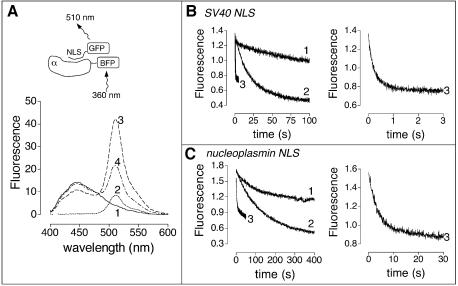
Equilibrium competition could result from either active displacement or passive competition, which can be discriminated by examining the influence of Nup2p on the rate of dissociation of NLSs from Kap60p. Gilchrist et al. (2002) have shown that full-length Nup2p increases the off-rate of Cbp80p-NLS from Kap95p:Kap60p:NLS and Kap60p:NLS, supporting an active displacement mechanism. We used a fluorescence resonance energy transfer (FRET)-based assay to show that Nup2p residues 1–51 are sufficient to accelerate the release of both monopartite (SV40) and bipartite (NP) NLSs from Kap60Δ without the need for Kap95p or RanGTP. Blue fluorescent protein (BFP, donor) was fused to the N-terminus of Kap60Δ and GFP (acceptor) was fused to the C-terminus of each NLS. As expected from the close proximity of the NLS C-terminus and the Kap60Δ N-terminus in the complex (Conti and Kuryan, 2000), a powerful BFP–GFP FRET signal at 510 nm was produced when NLS–GFPs were added to BFP–Kap60Δ (Figure 5A, line 3). This signal decreased on addition of Nup2N (Figure 5A, line 4), consistent with its replacing the NLSs. No FRET decrease was seen when 1 mg/ml BSA was added and no FRET signal was seen when GFP alone was added to BFP-Kap60Δ (data not shown). BFP–Kap60Δ had normal affinity for Nup2N and NLS-GFP fusions (Table I), confirming that the fusions did not interfere with these interactions. Figure 5B shows the dissociation kinetics of the BFP–Kap60Δ:SV40-NLS–GFP complex. The spontaneous dissociation rate (koff of 0.07/s) was estimated from the decrease in the complex’s FRET signal in a 20-fold molar excess of Kap60Δ (Figure 5B) to prevent rebinding and was not increased at higher Kap60Δ concentrations (data not shown). Adding Nup2N to the complex dramatically increased the off-rate (kobs ∼3.6/s; Figure 5B; Table III) of the SV40 NLS. Similarly, Nup2N increased the off-rate of the NP NLS (kobs ∼0.35/s compared with ∼0.007/s for spontaneous dissociation; Figure 5C; Table III. The slower rates for NP were probably due to its two binding regions acting co-operatively.) Thus, Nup2N displaces both monopartite and bipartite NLSs by an active mechanism, probably by forming a transient trimeric Nup2N:Kap60Δ:NLS complex. Moreover, this active displacement does not require Kap95p or the IBB domain of Kap60p and, based on the crystal structure, does not appear to involve a conformational change in Kap60p.
Fig. 5. Nup2N accelerates NLS dissociation. (A) Emission profiles of 0.2 µM BFP–Kap60Δ (line 1), 0.18 µM SV40 NLS–GFP (line 2), 0.2 µM BFP–Kap60Δ and 0.18 µM SV40 NLS–GFP (line 3), 0.2 µM BFP–Kap60Δ, 0.18 µM SV40 NLS–GFP and 2 µM Nup2N (line 4). Excitation was at 360 nm (BFP excitation peak). (B) Nup2N increases the off-rate of SV40 NLS. 0.2 µM BFP–Kap60Δ and 0.18 µM SV40 NLS–GFP alone (line 1), or with 4 µM Kap60Δ (line 2), or 2 µM Nup2N (line 3) added. The line 3 trace is expanded on the right. (C) Nup2N increases the off-rate of nucleoplasmin (NP) NLS. 0.2 µM BFP–Kap60Δ and 0.18 µM NP NLS–GFP alone (line 1), or with 4 µM Kap60Δ (line 2), or 2 µM Nup2N (line 3) added. The line 3 trace is expanded on the right.
Table III. Rates of NLS dissociation from Kap60Δ.
| NLS | Spontaneous dissociation (s–1) | Nup2N-accelerated release (s–1) |
|---|---|---|
| SV40 | 0.068 ± 0.001 | 3.57 ± 0.03 |
| Nucleoplasmin | 0.007 ± 0.002 | 0.35 ± 0.01 |
Data represent mean ± standard error based on four measurements.
Competition between Nup2p and the IBB domain
The crystal structure also suggested that competition could occur between Nup2p and the IBB domain. In the crystal structure of mouse importin-α, residues 49KRRNV53 (mouse numbering) of the IBB domain fit into the P2–P6 pockets, whereas residues further upstream in the IBB domain also appear to interact weakly with the P1 site and minor NLS binding site (Kobe, 1999). The auto-inhibitory KRRNV sequence and Nup2p are positioned close to one another on the surface of Kap60p, and so the IBB domain could obstruct Nup2p binding. Consistent with this hypothesis, Nup2N bound more strongly to Kap60Δ (KD 2.4 ± 0.4 nM) than to Kap60p (KD 30 ± 5 nM; Figure 6A; Table I). Similarly, the affinity of full-length Nup2p for Kap60p (KD 4.4 ± 1.1 nM) was less than that for Kap60Δ (KD 0.07 ± 0.02 nM). Thus, the IBB domain reduced Nup2p affinity for Kap60p. However, the affinity of Nup2p for full-length Kap60p remained higher than that of NLSs (Table I), and so Nup2p appears less sensitive to competition from the IBB domain than NLSs. This is consistent with the observation that the affinity of the NLSs for Kap60Δ is lower than that of Nup2p, and also with the major NLS binding site being the primary binding site for NLS-cargoes and the IBB domain, but not for Nup2p.
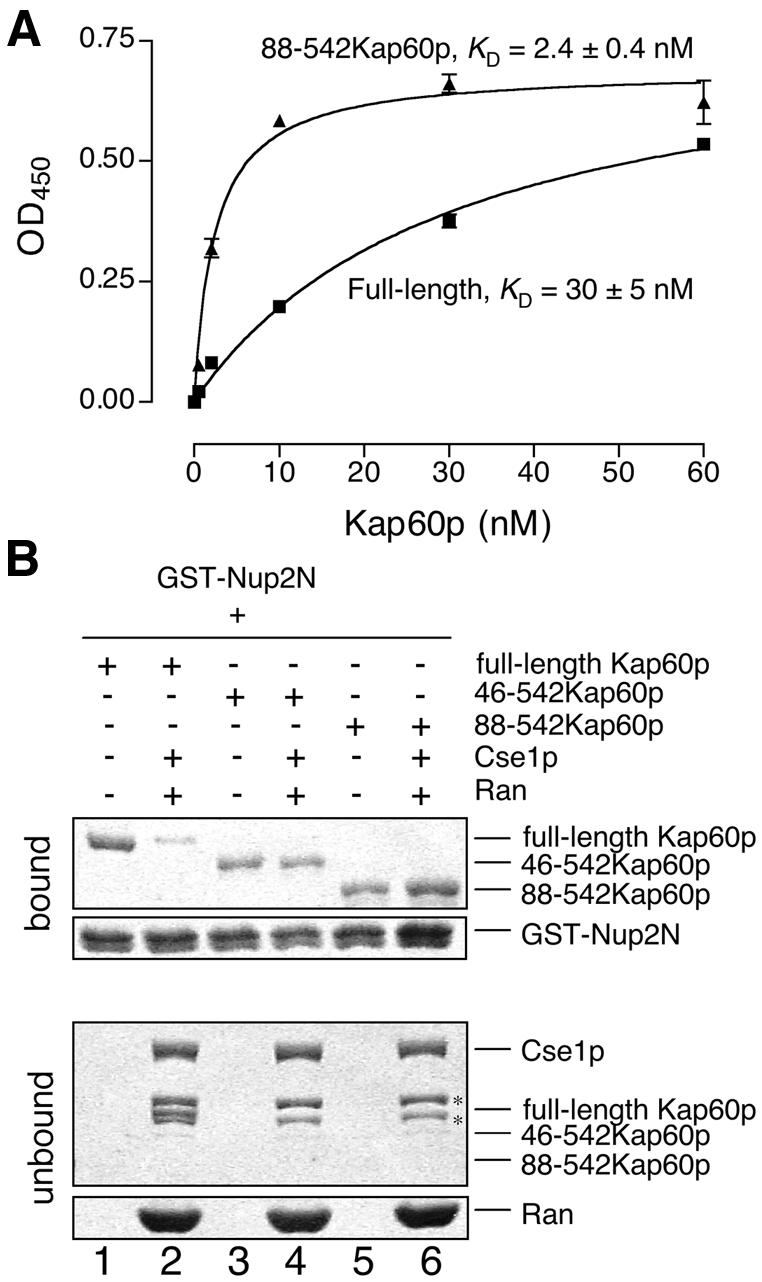
Fig. 6. Competition with the IBB domain and Cse1p/RanGTP. (A) Kap60p (filled squares) binds more weakly to Nup2N than Kap60Δ (filled triangles). Each data point was performed in duplicate and error bars represent SEM. (B) Cse1p:RanGTP competes Kap60p from Nup2N when the IBB domain is present. GST–Nup2N (10 µg) was treated with 3 µM Kap60p ± Cse1p (15 µg):Ran (30 µg) in binding buffer supplemented with 5 mM Mg(OAc)2. Two bands marked with * in the unbound fractions are copurifying bacterial proteins present in Cse1p. Full-length Kap60p can be seen between these two bands in lane 2.
Cse1p:RanGTP and the IBB domain cooperate to remove the Nup2p N-terminus from Kap60p
The 1–10 nM affinity of Nup2N or Nup2p for Kap60p indicated that, if Nup2p is anchored to the nuclear basket of NPCs, the dissociation of Kap60p from Nup2p has to be facilitated by some other factor to enable Kap60p to be recycled efficiently to the cytoplasm. Cse1p:RanGTP dissociates Kap60p from Nup2p residues 1–174 (Booth et al., 1999; Solsbacher et al., 2000) and Figure 6B (lanes 1 and 2) confirmed that Cse1p:RanGTP also dramatically weakened the Kap60p:Nup2N interaction. Thus, although the IBB domain alone weakens Nup2p interactions with Kap60p (Figure 6A), efficient dissociation of Kap60p from Nup2p probably also requires Cse1p:RanGTP. However, the CAS/Cse1p binding site on human importin-α maps to the convex face of Arm repeats 9 and 10 (Herold et al., 1998). The crystal structure of the Nup2N:Kap60Δ complex suggests that Nup2N binding may not compete directly with Cse1:RanGTP binding and raises the question of how Cse1p:RanGTP forms a complex with Kap60p. Cse1p:RanGTP binding to Kap60p dissociates both NLSs (Kutay et al., 1997; Gilchrist et al., 2002) and Nup2N. Moreover, Nup2N binds to Arm repeats 4–8, suggesting that Cse1p:RanGTP binds simultaneously to a site located near the C-terminus of Kap60p and to residues N-terminal to the IBB auto-inhibitory sequence, attaching the IBB domain along the entire concave groove of Kap60p. In this model, the Kap60p N-terminus could remove Nup2p sterically, concomitant with the completion of the formation of the export complex (Kap60p:Cse1p:RanGTP). This would release the export complex from the nuclear basket and would also ensure that Cse1p:RanGTP binds only to cargo-free Kap60p. To test this hypothesis, we engineered N-terminal deletion mutants of Kap60p and found that deletion of residues 1–45 of the IBB domain or the entire IBB domain made Nup2N binding insensitive to Cse1p:RanGTP (Figure 6B). Thus, Nup2p is dissociated from the Arm repeat domain of Kap60p by the concerted action of Cse1p:RanGTP and the IBB domain and at least part of this activity resides within Kap60p residues 1–45, which are N-terminal to the auto-inhibitory 54KRRN57 sequence.
Discussion
We have shown that residues 1–51 of Nup2p are important for function in vivo and have determined the crystal structure of these residues in complex with the Arm repeat domain of Kap60p at 2.6 Å resolution. The crystal structure shows that Nup2p residues 36–51 bind intimately to Kap60p in a distinctly different way to that observed with either monopartite and bipartite NLSs or the IBB domain. However, although the Nup2p N-terminus, NLSs and the IBB domain bind differently, their binding sites are close to each other and partially overlap. Binding studies show that Nup2p weakens NLS binding to Kap60Δ, consistent with the partial overlap of the respective binding sites and also consistent with the affinity of Nup2p to Kap60p being higher than that of NLSs. Moreover, kinetic data indicate that Nup2p actively displaces NLSs by accelerating their release (Gilchrist et al., 2002; Figure 5), thereby facilitating the release of import substrate. However, for Kap60p to be recycled to the cytoplasm, it must first bind to its export factor, Cse1p:RanGTP. Our binding studies indicate that Nup2p must be removed from Kap60p for this binding to occur and, furthermore, it requires the IBB domain of Kap60p. Our structural and biochemical data, combined with the subcellular location of Nup2p and its multidomain structure, are consistent with a model in which Nup2p orchestrates both the disassembly of the cargo:carrier import complex and the subsequent recycling of Kap60p to the cytoplasm.
Because in addition to its high-affinity Kap60p binding sequence, Nup2p has FxFG repeats that can bind Kap95p, it could provide a higher affinity binding site for incoming Kap95p:Kap60p:NLS-cargo complex than other FG-Nups. The crystal structure and the binding data both suggest that, unlike vertebrate Npap60/Nup50 (Lindsay et al., 2002), Nup2p binding would dissociate the NLS:Kap60p complex and so Nup2p is unlikely to accompany the cargo:carrier complex during its import through NPCs. The Nup2p binding site is where the NLS:Kap60p interaction appears to be relatively weak. Thus Nup2p binding would be an effective initial step of NLS dissociation. Although Nup2p binding alone might be sufficient to displace cargo, competition from the binding of IBB domain to the NLS binding site (Kobe, 1999) and Cse1p:RanGTP binding would probably make NLS dissociation more efficient. Nup2p could assist the IBB domain to displace an NLS both by weakening the interaction between the NLS and Kap60p and also by the Nup2p Ran-binding domain facilitating the transfer of RanGTP to Kap95p, thereby facilitating the release of the IBB domain and allowing it to bind to the major NLS binding site. The binding of Nup2p to RanGTP is weak with a µM KD (Denning et al., 2001), and so would have a high off-rate, ideal for the transfer of RanGTP to other proteins retained in the immediate vicinity through their binding to Nup2p.
After the import complex has been dissociated and the NLS-cargo delivered to the nucleus, Kap95p and Kap60p are recycled to the cytoplasm. Kap95p does this complexed with RanGTP, but Kap60p recycling is mediated by Cse1p:RanGTP (Hood and Silver, 1998; Solsbacher et al., 1998). The FxFG repeats and Ran-binding domain of Nup2p could facilitate recruitment of Cse1p and RanGTP to the Nup2p:Kap60p complex. The observations that Nup2p binds along the inner groove of Kap60p and that the N-terminal half of the IBB domain is required for Cse1p:RanGTP to displace Nup2p from Kap60p, together with the importance of C-terminal residues of importin-α in CAS binding (Herold et al., 1998), suggest that the export complex might assemble in such a way that Cse1p:RanGTP binds both the C- and N-termini of Kap60p, ensuring that the IBB domain binds along the entire inner groove of its Arm repeat domain. In this case, Cse1p would act as an allosteric regulator to strengthen the autoinhibitory function of the IBB domain. This would ensure that Cse1p binds only cargo-free Kap60p, preventing rebinding of NLS-cargo to Kap60p as well as preventing the IBB binding to Kap95p during recycling. Consistent with this idea, recent work shows that overexpression of Cse1p can suppress the phenotype of an IBB mutant (Harreman et al., 2003b). However, a full understanding of the molecular basis of export complex assembly requires its atomic structure, which represents an important future challenge.
It has been suggested that Nups may be arranged in a sequence of increasing affinity for importins from the cytoplasmic side to the nuclear side of the NPCs (Ben-Efraim and Gerace, 2001), although other work has argued against such a gradient being necessary (Nachury and Weis, 1999) and has proposed that maintaining a gradient of RanGTP between the nuclear and cytoplasmic compartments is sufficient for at least nuclear protein import (reviewed by Gorlich et al., 2003; Weis, 2003). The asymmetric distribution of Nup2p could contribute to the importin affinity gradient and so might bias their stochastic movement within the NPC towards the nucleus. However, it is thermodynamically impossible to drive an accumulation of cargo against a concentration gradient by an affinity gradient alone (Gorlich et al., 2003). To drive active transport, it is important that docking to a high affinity site such as Nup2p is coupled to Ran-dependent disassembly of cargo:carrier complexes. Therefore, the free energy change that defines the distribution of material between cytoplasm and nucleus (the active component of transport that enables accumulation against a concentration gradient) is almost certainly derived from the difference in RanGTP concentration between nucleus and cytoplasm (Gorlich et al., 2003; Weis, 2003). However, a higher affinity of Nup2p for the import complex could still be used to target the complex to the correct site for disassembly/recycling without contributing directly to the transport itself. In this way, Nup2p would act like a catalyst, increasing the overall rate of transport by facilitating a key step (analogous to lowering the activation energy), but not changing the equilibrium between cytoplasm and nucleus and so the overall free energy change.
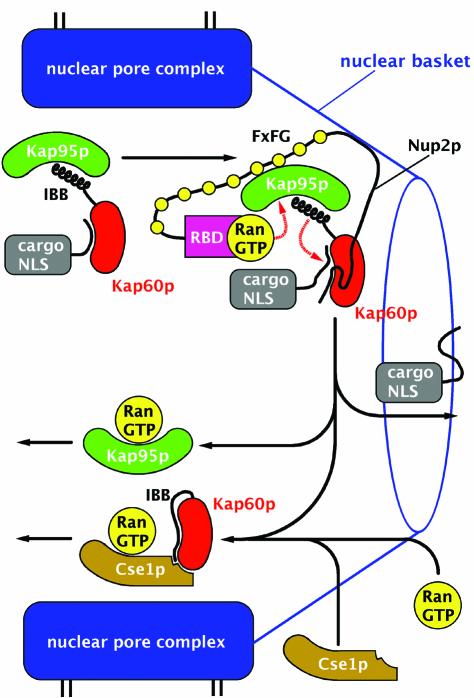
Our structural and binding data suggest a model for how Nup2p orchestrates both dissociation of the Kap60p:cargo complex and subsequent recycling of Kap60p to the cytoplasm (Figure 7). Nup2p provides a high affinity binding site for incoming Kap95p:Kap60p:NLS-cargo complex and the flexibility of Nup2p (Denning et al., 2002) could facilitate rapid capture. The Nup2p N-terminus would displace the NLS, and, by recruiting RanGTP and Cse1p, accelerate dissociation of the NLS-cargo by facilitating assembly of both the Kap95p:RanGTP and Kap60p:Cse1p:RanGTP complexes. By orchestrating both import complex disassembly and Kap60p/Kap95p recycling, Nup2p coordinates these steps and ensures that they proceed as a concerted process. In this way, Nup2p serves to increase the efficiency of the process and so increase the rate of nuclear trafficking, ensuring Kap95p and Kap60p are only recycled after the NLS-cargo has been released. Thus Nup2p provides a series of checkpoints in the nuclear transport cycle. Because Nup2p functions primarily to improve efficiency, Nup2p nulls are not lethal but impair cellular function sufficiently to show synthetic lethality with mutants in other components of the nuclear trafficking machinery (Loeb et al., 1993).
Fig. 7. Proposed mechanism by which Nup2p orchestrates dissociation of the import complex and recycling of Kap95p and Kap60p. Docking of the Kap60p:Kap95p:NLS-cargo complex to Nup2p at the nuclear basket triggers cooperative binding/dissociation interactions leading to release of the NLS-cargo into the nucleoplasm concomitant with release of export complexes from nuclear basket. The Nup2p N-terminus displaces NLS-cargo from Kap60p. By transferring RanGTP to Kap95p and recruiting Cse1p and RanGTP to Kap60p, Nup2p further facilitates NLS-cargo release as well as facilitating formation of export complexes. RanGTP binding to Kap95p dissociates it from the FxFG repeats of Nup2p and the IBB domain of Kap60p. Cse1p and RanGTP then bind to Kap60p so that Cse1p:RanGTP stabilizes the interaction between the IBB domain and the Arm repeat domain of Kap60p, ensuring both NLS and Nup2p dissociation. Thus, Nup2p orchestrates both the termination of nuclear protein import and the initiation of the nuclear export of cargo-free importins through interactions involving the Kap60p IBB domain, RanGTP and Cse1p.
In summary, our data indicate that a primary function of Nup2p is to facilitate the disassembly of import complexes and the assembly of export complexes by acting as a coordinating scaffold. The crystal structure of the Kap60Δ:Nup2N complex provides structural insight into how the Nup2p N-terminus accelerates NLS-cargo release, and also suggests how Nup2p couples termination of NLS-cargo import and recycling of Kap60p and Kap95p.
Materials and methods
In vivo functional analysis
The in vivo function of Nup2p mutants was assessed using a plasmid shuffle technique (Boeke et al., 1987). Plasmids encoding Nup2p (pAC1342, CEN, LEU), Δ50nup2p (pAC1268, CEN, LEU), or 1–51nup2p (pAC1215, CEN, LEU, GFP-tag) expressed from the NUP2 promoter or a control vector (pRS315; Sikorski and Hieter, 1989) were each transformed into Δnup2 srp1-31 cells (ACY789) containing a maintenance plasmid encoding Srp1p/Kap60p (pAC876, CEN, URA). Single transformants were grown in liquid culture for 3 days to saturation, serially diluted 1:10, and spotted on control plates lacking uracil and test plates containing fluororotic acid (5-FOA). 5-FOA eliminates the URA3 maintenance plasmid encoding wild-type Kap60p. Plates were incubated at 25°C for 5 days. The intracellular localization of Nup2p and Δ50nup2p was determined using GFP fusion proteins. The Nup2p-GFP plasmid (pAC1395) and Δ50nup2p-GFP plasmid (pAC1394) were created by amplifying the promoter and coding regions of pAC1298 (Nup2p, CEN, URA) and pAC1268 (Δ50nup2p, CEN, LEU), respectively, and cloned as PstI/XhoI fragment into pAC45 (2µ, URA). The 1–51nup2p-GFP plasmid (pAC1215) was created by cloning the first 51 amino acids of Nup2p into pNOPGFP21 using the BamHI/XhoI sites. To assess the effect of Nup2p on the intracellular localization of Kap60p, Kap60p-GFP was integrated at the endogenous SRP1 locus of Δnup2 cells using a standard integration strategy, creating the strain ACY712. These cells were transformed with plasmids encoding Nup2p (pAC1342, CEN, LEU), Δ50nup2p (pAC1268), or vector control (pRS315). The localization of GFP-tagged proteins was monitored using a GFP-optimized filter on an Olympus BX60 epifluorescence microscope equipped with a Photometrics Quantix digital camera. The position of the nucleus was confirmed by incubating cultures with 1 µg/ml DAPI for 30 min, which, in live cells, stains both nuclear and mitochondrial DNA.
Expression and purification of the Kap60Δ:Nup2N complex
His/S-tagged Nup2p residues 1–51 (Nup2N) were expressed from pET30a (Novagen). Non-tagged Arm repeat domain of Kap60p (residues 88–530, Kap60Δ) was cloned into pET30a and the Y397D point mutation introduced using Quikchange PCR (Stratagene). For crystallization, Nup2N and Kap60Δ were expressed separately in BL21-Gold(DE3) (Stratagene) and, after lysis and clarification, crude bacterial extracts were mixed in buffer A (20 mM Tris-HCl pH 7.5, 0.3 M NaCl, 1 mM PMSF, 3 mM β-mercaptoethanol), with Kap60Δ in excess, and applied to Ni-NTA (Qiagen). The resin was washed extensively with buffer A, followed by buffer A containing 25 mM imidazole and then exchanged into 20 mM Tris–HCl pH 7.5, 50 mM NaCl, 2 mM CaCl2, 3 mM β-mercaptoethanol and incubated with 7 U/ml enterokinase (Novagen) overnight. The Nup2N:Kap60Δ complex released from the resin was applied to enterokinase capture resin (Novagen) and the flow-through fraction concentrated to 39 mg/ml in 10 mM Tris–HCl pH 8.0, 100 mM NaCl, 10% glycerol.
Crystallization, data collection and structure determination
Crystals of the Nup2N:Kap60Δ complex were obtained at 18°C by streak seeding hanging drops containing 2 mg/ml protein, 50 mM HEPES pH 6.8, 0.15 M NaCl, 24% PEG3350 and 2% PEG400. The plate-shaped crystals had P21212 symmetry with a = 129.8 Å, b = 140.1 Å and c = 63.9 Å. A 200×200×400 µm crystal was cryo-protected in 50 mM HEPES pH 6.8, 0.15 M NaCl, 27% PEG3350 and 12% PEG400 and flash-frozen in liquid nitrogen. Mosaic spread was reduced by freeze-thaw annealing and a 2.6 Å dataset collected on beamline 14.2 at Daresbury (UK) using a MAR CCD detector and 0.978 Å wavelength radiation. Molecular replacement solutions were found using CNS (Brunger et al., 1998) for two independent Kap60Δ molecules in the asymmetric unit using a chain from the c-myc NLS complex (Conti and Kuriyan, 2000) as search model. The structure was initially refined using CNS. After rigid body refinement, simulated annealing, positional refinement using conjugate gradient minimization with NCS restraints enforced throughout the molecule, alternating with local rebuilding, the free R-factor was 32.7% (R-factor 29.6%) and unambiguous difference density appeared for the Nup2p fragment along the central groove of Kap60Δ on both chains and a model was built for residues 36–51. The model was then refined using REFMAC5 (CCP4, 1994) and strong NCS restraints based on individual Arm repeats. After iterative cycles of refinement and rebuilding and the addition of 158 waters, the final R-factor was 21.6% (R-free 25.7%).
Cloning, expression and purification of proteins for biochemical assays
GST-Nup2p (residues 36–51), GST-SV40 NLS (SPKKKRKVE), GST-NP NLS (KRPAATKKAGQAKKKKL) were constructed by ligating complementary oligoDNAs into BamHI/XhoI sites of pGEX-4T-1. GST-Nup2N and GST-Nup2p (residues 1–174) were constructed by cloning PCR-amplified fragments into BamHI/XhoI sites of pGEX-20T and pGEX-4T-1, respectively. The GST-Nup2p expression vector was as described (Hood et al., 2000). The coding sequences of Kap60p residues 1–542, residues 46–542, residues 81–542, residues 88–542, or residues 88–530 (either wild type or Y397D) were cloned into BamHI/XhoI sites of pET30a. BFP cDNA was PCR-amplified from pEBFP-C1 (Clontech) and ligated into the BamH1 site of His/S-Kap60p(81–542). D203K/N157A mutations were introduced into Kap60p (residues 88–530, Y397D), and R38/39A or R46/47A or R38/39/46/47A mutations were introduced into GST-Nup2p (full-length) using Quikchange PCR. C-terminal His-tagged Cse1p and N-terminal His-tagged Ran were constructed by cloning CSE1 or GSP1 into pET30a or pET15b respectively. SV40 NLS–GFP and NP NLS–GFP were constructed by cloning the NLS sequence, a 6-residue linker (ASGLVP), and red-shifted S65T/V163A-GFP into the EcoRI/XhoI sites of pET28a. All constructs were verified by sequencing. Recombinant proteins were expressed in BL21-Gold(DE3) or BL21(DE3)RIL cells (Stratagene). GST-fusions were purified over glutathione Sepharose 4B (Amersham Pharmacia) and His-tagged proteins purified over Ni-NTA, following manufacturers’ protocols. Ran was loaded with GTP as described (Bayliss et al., 2000).
Equilibrium binding assays
Pulldowns were performed in binding buffer (PBS, 0.1% Tween-20, 2 mM DTT, 0.2 mM PMSF) unless indicated otherwise. GST fusion protein was immobilized on 10 µl of packed glutathione Sepharose 4B beads and incubated with the other proteins in a total volume of 50 µl for 1–2.5 h at 4°C. Beads were then collected by centrifugation and the supernatant saved. Beads were washed twice with 1 ml of binding buffer and bound proteins eluted with SDS-sample buffer. Bound and unbound fractions were analyzed by SDS–PAGE on 4–20% gradient gels and stained with Coomassie. Solid phase binding assays were carried out on microtitre plates (Bayliss et al., 2002) except that binding reactions were carried out overnight at 4°C. Binding data were analyzed with GraphPad Prism (Biosoft) using nonlinear regression assuming one-site binding.
NLS dissociation kinetics
NLS dissociation kinetics were measured at 25°C in PBS with an Applied Photophysics SX18 stopped-flow spectrophotometer. BFP was excited at 360 nm, with GFP emission monitored using a 475 nm filter. Protein concentrations are those after the mixing in the stopped-flow cell. Data were fitted to double exponentials by nonlinear regression using GraphPad Prism with one of the exponentials being a slow photobleaching term and the other representing NLS dissociation. Photobleaching was determined by fitting single exponential to the decay curve after mixing fluorophores with PBS alone, and these parameters were constrained during double exponential fitting to NLS dissociation time course. Steady-state fluorescence spectra were recorded using a Perkin Elmer LS50B spectrofluorometer at 25°C in PBS. Excitation was at 360 nm, and slit widths were 6 nm for excitation and 10 nm for emission. Samples were incubated for 1 h before measurement.
Accession number
The coordinates of the structure have been deposited in the Protein Data Bank (accession code 1un0).
Acknowledgments
Acknowledgements
We are most grateful to our many friends and colleagues, especially R.Bayliss, L.Clayton, P.Evans, A.Fersht, R.Grant, P.Jemth, C.Johnson, A.Leslie, T.Littlewood, M.Rexach and A.Weeds for their many comments, criticisms and assistance. We thank Pam Silver for NUP2 and CSE1 clones. Y.M. was supported by Uehara Memorial Foundation postdoctoral fellowship. This work was supported in part by research grant P0386/2001 from the Human Frontiers Science Program (A.H.C. and M.S.) and by NIH GM16252-05 (A.H.C.)
References
- Bayliss R., Littlewood,T. and Stewart,M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood,T., Strawn,L.A., Wente,S.R. and Stewart,M. (2002) GLFG and FxFG nucleoporins bind to overlapping sites on importin-β. J. Biol. Chem., 277, 50597–50606. [DOI] [PubMed] [Google Scholar]
- Ben-Efraim I. and Gerace,L. (2001) Gradient of increasing affinity of importin β for nucleoporins along the pathway of nuclear import. J. Cell Biol., 152, 411–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., Trueheart,J., Natsoulis,G. and Fink,G.R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol., 154, 164–175. [DOI] [PubMed] [Google Scholar]
- Booth J.W., Belanger,K.D., Sannella,M.I. and Davis,L.I. (1999) The yeast nucleoporin Nup2p is involved in nuclear export of importin α/Srp1p. J. Biol. Chem., 274, 32360–32367. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 50, 905–921. [DOI] [PubMed] [Google Scholar]
- Catimel B., Teh,T., Fontes,M.R., Jennings,I.G., Jans,D.A., Howlett,G.J., Nice,E.C. and Kobe,B. (2001) Biophysical characterization of interactions involving importin-α during nuclear import. J. Biol. Chem., 276, 34189–34198. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Conti E. and Kuriyan,J. (2000) Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin α. Struct. Fold. Des., 8, 329–338. [DOI] [PubMed] [Google Scholar]
- Conti E. and Izaurralde,E. (2001) Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol., 13, 310–319. [DOI] [PubMed] [Google Scholar]
- Conti E., Uy,M., Leighton,L., Blobel,G. and Kuriyan,J. (1998) Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell, 94, 193–204. [DOI] [PubMed] [Google Scholar]
- Denning D., Mykytka,B., Allen,N.P., Huang,L., Al,B. and Rexach,M. (2001) The nucleoporin Nup60p functions as a Gsp1p–GTP-sensitive tether for Nup2p at the nuclear pore complex. J. Cell Biol., 154, 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D.P., Uversky,V., Patel,S.S., Fink,A.L. and Rexach,M. (2002) The Saccharomyces cerevisiae nucleoporin Nup2p is a natively unfolded protein. J. Biol. Chem., 277, 33447–33455. [DOI] [PubMed] [Google Scholar]
- Dilworth D.J., Suprapto,A., Padovan,J.C., Chait,B.T., Wozniak,R.W., Rout,M.P. and Aitchison,J.D. (2001) Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J. Cell Biol., 153, 1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C. and Laskey,R.A. (1998) Nuclear import: a tale of two sites. Curr. Biol., 8, R922–R924. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced colouring capabilities. J. Mol. Graph. Model., 15, 132–136. [DOI] [PubMed] [Google Scholar]
- Fontes M.R., Teh,T. and Kobe,B. (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J. Mol. Biol., 297, 1183–1194. [DOI] [PubMed] [Google Scholar]
- Gilchrist D., Mykytka,B. and Rexach,M. (2002) Accelerating the rate of disassembly of karyopherin:cargo complexes. J. Biol. Chem., 277, 18161–18172. [DOI] [PubMed] [Google Scholar]
- Gorlich D., Seewald,M.J. and Ribbeck,K. (2003) Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J., 22, 1088–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harreman M.T., Hodel,M.R., Fanara,P., Hodel,A.E. and Corbett,A.H. (2003a) The auto-inhibitory function of importin α is essential in vivo. J. Biol. Chem., 278, 5854–5863. [DOI] [PubMed] [Google Scholar]
- Harreman M.T., Cohen,P.E., Hodel,M.R., Truscott,G.J., Corbett,A.H. and Hodel,A.E. (2003b) Characterization of the auto-inhibitory sequence within the N-terminal domain of importin-α. J. Biol. Chem., 278, 21361–21369. [DOI] [PubMed] [Google Scholar]
- Herold A., Truant,R., Wiegand,H. and Cullen,B.R. (1998) Determination of the functional domain organization of the importin α nuclear import factor. J. Cell Biol., 143, 309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodel M.R., Corbett,A.H. and Hodel,A.E. (2001) Dissection of a nuclear localization signal. J. Biol. Chem., 276, 1317–1325. [DOI] [PubMed] [Google Scholar]
- Hood J.K. and Silver,P.A. (1998) Cse1p is required for export of Srp1p/importin-α from the nucleus in Saccharomyces cerevisiae. J. Biol. Chem., 273, 35142–35146. [DOI] [PubMed] [Google Scholar]
- Hood J.K., Casolari,J.M. and Silver,P.A. (2000) Nup2p is located on the nuclear side of the nuclear pore complex and coordinates Srp1p/importin-α export. J. Cell Sci., 113, 1471–1480. [DOI] [PubMed] [Google Scholar]
- Kobe B. (1999) Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol., 6, 388–397. [DOI] [PubMed] [Google Scholar]
- Kutay U., Bischoff,F.R., Kostka,S., Kraft,R. and Gorlich,D. (1997) Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell, 90, 1061–1071. [DOI] [PubMed] [Google Scholar]
- Lindsay M.E., Plafker,K., Smith,A.E., Clurman,B.E. and Macara,I.G. (2002) Npap60/Nup50 is a tri-stable switch that stimulates importin-α:β-mediated nuclear protein import. Cell, 110, 349–360. [DOI] [PubMed] [Google Scholar]
- Loeb J.D., Davis,L.I. and Fink,G.R. (1993) NUP2, a novel yeast nucleoporin, has functional overlap with other proteins of the nuclear pore complex. Mol. Biol. Cell, 4, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Nachury M.V. and Weis,K. (1999) The direction of transport through the nuclear pore can be inverted. Proc. Natl Acad. Sci. USA, 96, 9622–9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Rexach M. and Blobel,G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell, 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J., Maurer,P., Bischoff,F.R. and Schlenstedt,G. (1998) Cse1p is involved in export of yeast importin-α from the nucleus. Mol. Cell. Biol., 18, 6805–6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solsbacher J., Maurer,P., Vogel,F. and Schlenstedt,G. (2000) Nup2p, a yeast nucleoporin, functions in bidirectional transport of importinα. Mol. Cell Biol., 20, 8468–8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. (2003) Regulating Access to the Genome. Nucleocytoplasmic Transport throughout the Cell Cycle. Cell, 112, 441–451. [DOI] [PubMed] [Google Scholar]