Abstract
Drosophila Sex-lethal (dSXL)-mediated translational repression of male-specific lethal 2 (msl-2) mRNA is essential for X-chromosome dosage compensation. Binding of dSXL to specific sites in both untranslated regions of msl-2 mRNA is necessary for inhibition of translation initiation. We describe the organization of dSXL as a translational regulator and show that the RNA binding and translational repressor functions are contained within the two RRM domains and a C-terminal heptapeptide extension. The repressor function is dormant unless dSXL binds to msl-2 mRNA with its own RRMs, because dSXL tethering via a heterologous RNA-binding peptide does not elicit translational inhibition. We reveal proteins that crosslink to the msl-2 3′ untranslated region (3′UTR) and co-immunoprecipitate with dSXL in a fashion that requires its intact repressor domain and correlates with translational regulation. Translation competition and UV-crosslink experiments show that the 3′UTR msl-2 sequences adjacent to dSXL-binding sites are necessary to recruit titratable co-repressors. Our data support a model where dSXL binding to the 3′UTR of msl-2 mRNA activates the translational repressor domain, thereby enabling it to recruit co-repressors in a specific fashion.
Keywords: dosage compensation/in vitro translation/post-transcriptional control/RNA–protein interactions/RNPs
Introduction
RNA-binding proteins play critical roles in the regulation of nearly every aspect of gene expression. They often display a modular architecture, exert multiple functions and participate in more than one step of the gene expression pathway (Wilkinson and Shyu, 2001). Drosophila Sex-lethal (dSXL) is an RRM (RNA recognition motif)-type RNA-binding protein that plays a central role in sex determination and dosage compensation in the fruitfly (Schutt and Nothiger, 2000). dSXL is expressed only in female flies, where it triggers the cascade of female sexual differentiation by regulating the alternative splicing of transformer pre-mRNA (Sosnowski et al., 1989). In addition, dSXL regulates its own expression by inducing the female-specific splicing of its pre-mRNA (Bell et al., 1991). To control dosage compensation, dSXL acts as both a splicing and translational regulator. dSXL blocks the dosage compensation pathway in female flies by inhibiting the expression of the male-specific lethal-2 (msl-2) gene, which encodes a critical subunit of the dosage compensation complex (Bashaw and Baker, 1995; Akhtar, 2003). Inhibition of msl-2 expression requires, first, dSXL-mediated retention of a small intron in the 5′ untranslated region (5′UTR) of the msl-2 pre-mRNA in the nucleus, and subsequent repression of msl-2 mRNA translation in the cytoplasm (Bashaw and Baker, 1997; Kelley et al., 1997; Gebauer et al., 1998). To inhibit translation, dSXL needs to bind to specific U-rich sequences in both the 5′ and 3′UTRs of msl-2 mRNA (Bashaw and Baker, 1997; Kelley et al., 1997; Gebauer et al., 1999), and cooperates with sequences adjacent to its regulatory 3′UTR binding sites (Gebauer et al., 2003).
The N-terminal domain of dSXL [amino acids (aa) 1–121] is enriched in glycine and asparagine residues, and mediates cooperative RNA-binding interactions between dSXL monomers (Wang and Bell, 1994). It is also necessary for regulation of the alternative splicing of tra and sxl pre-mRNAs (Wang and Bell, 1994; Deshpande et al., 1999; Yanowitz et al., 1999). The requirement of the N-terminal domain for the autoregulatory sxl pre-mRNA splicing can be explained by the recent finding that it interacts with SPF45, a spliceosomal protein necessary for this process (Lallena et al., 2002). In contrast, the translational regulation of msl-2 mRNA tolerates at least a partial deletion of the N-terminal domain of dSXL both in vivo and in vitro (Gebauer et al., 1999; Yanowitz et al., 1999). The N-terminal domain is followed by two RRM-type RNA-binding domains (aa 122–294) that are necessary and sufficient for specific, high-affinity RNA-binding (Wang et al., 1997; Samuels et al., 1998). The isolated single RRMs bind to RNA with lower affinity and poor specificity (Samuels et al., 1998). Both examination of natural dSXL-binding sites and in vitro selection experiments indicate that dSXL recognizes single-stranded poly(U)-rich sequences (Sakashita and Sakamoto, 1994; Singh et al., 1995). NMR and X-ray structure determinations revealed that, in the absence of RNA, the two RRMs of dSXL are flexibly tethered in solution (Lee et al., 1997; Crowder et al., 1999). Interestingly, the relative orientation of the RRMs changes upon RNA binding, closing around the single-stranded RNA molecule (Handa et al., 1999; Kim et al., 2000). The RRMs of dSXL also engage in protein–protein interactions (Deshpande et al., 1996; Sakashita and Sakamoto, 1996; Wang et al., 1997; Samuels et al., 1998; Dong and Bell, 1999). RRM1 interacts with the snRNP component SNF, a protein thought to play a role in Sxl splicing (Samuels et al., 1998), and with SIN, a protein of unknown function (Dong and Bell, 1999). Finally, dSXL can interact with itself in vitro via both RRM1 and RRM2 (Sakashita and Sakamoto, 1996; Wang et al., 1997; Samuels et al., 1998). The function of the C-terminal region of dSXL (aa 295–354) is currently unknown.
Here we identify the domains of dSXL that mediate translational repression. In contrast to its role as a splicing regulator, we show that only about half of the dSXL protein—its two RRM domains and 7 aa C-terminal from RRM2—carries all required activities. Based on tethered function, UV-crosslinking/co-immunoprecipitation and translation competition experiments, we suggest a model for how the translational repressor activity is activated: binding of dSXL to msl-2 mRNA induces the recruitment of translational co-repressors, providing a mechanism to restrict translational repression specifically to the bound mRNA.
Results
The translational repressor domain of dSXL overlaps with its RNA-binding domains
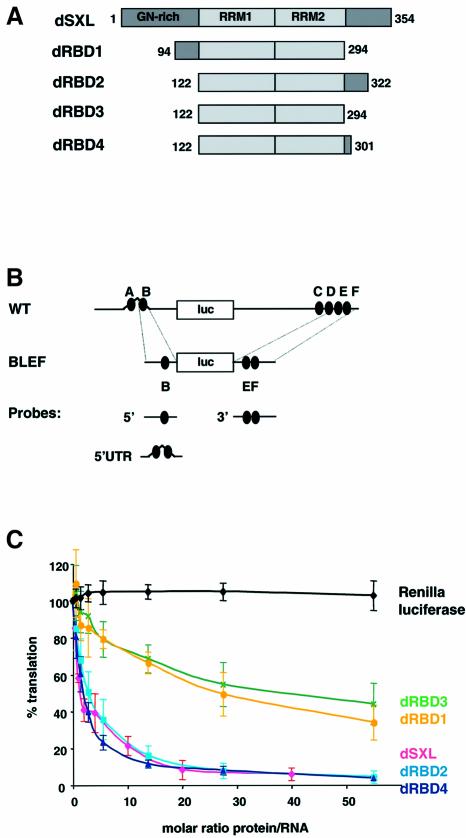
The RRM1 and RRM2 RNA-binding domains of dSXL (354 aa) are located in the middle of the protein (aa 122–294). Previous work showed that a dSXL polypeptide comprising aa 94–322 is fully functional as a translational regulator, whereas deletion of 227 aa from the N-terminus of dSXL yields a non-functional protein (aa 228–354) (Gebauer et al., 1999). To dissect the requirements for translational control, we generated derivatives that comprise the two RRMs (dRBD3, aa 122–294) and sequences extending to the N- (dRBD1, aa 94–294) or C-terminus (dRBD2, aa 122–322, and dRBD4, aa 122–301) of dSXL (Figure 1A). These dSXL variants were analyzed in Drosophila embryo translation extracts using an msl-2 reporter mRNA consisting of the full-length msl-2 5′ and 3′UTRs fused to the open reading frame of firefly luciferase [wild-type (WT) mRNA, Figure 1B; Gebauer et al., 1999]. Renilla luciferase mRNA was cotranslated as an internal control, and full-length dSXL served as a positive control and reference standard.
Fig. 1. Mapping the translational repressor domain of dSXL. (A) Schematic representation and domain organization of dSXL and its deletion derivatives. The amino acid numbers included in each derivative are indicated. (B) Scheme of the RNA constructs used in this study. WT mRNA contains the full-length 5′ (626 nt) and 3′ (1047 nt) UTRs of msl-2 fused to the firefly luciferase open reading frame (Gebauer et al., 1999). The SXL-binding sites are denoted A to F (black ovals). BLEF mRNA contains the minimal msl-2 sequences required for translational repression, which consist of 69 nt in the 5′UTR including site B, and 46 nt in the 3′UTR including sites E and F (Gebauer et al., 2003). Probes used for gel mobility-shift and UV-crosslink assays are also depicted. (C) WT mRNA was translated in Drosophila embryo extracts in the presence of increasing amounts of dSXL (pink line), dRBD1 (yellow line), dRBD2 (light blue line), dRBD3 (green line) or dRBD4 (dark blue line). Renilla luciferase mRNA was co-translated as an internal control (black line). Firefly luciferase values were corrected for Renilla expression and plotted as the percentage of the activity obtained in the absence of recombinant protein against the molar ratio of protein to mRNA.
As described previously (Gebauer et al., 1999), full-length dSXL represses the translation of the msl-2 indicator RNA, while Renilla luciferase values remain unchanged (Figure 1C). Deletion of dSXL polypeptide sequences N-terminal from the RRMs is tolerated without affecting the translational dose–response curve (dRBD2, dRBD4). Whereas dRBD4 also lacks the C-terminal 53 aa and is fully functional, removal of an additional 7 aa from the C-terminus of dRBD4 significantly decreases the ability of dSXL to inhibit translation (compare dRBD4 with dRBD3). UV-crosslink competition and gel mobility-shift assays indicated that the RNA-binding activities of the different deletion mutants are similar (data not shown; see below). In addition, the stabilities of the firefly and renilla mRNAs were equal in the presence of all SXL derivatives tested (data not shown). Thus, the two RRMs followed by the C-terminal 7 aa of dSXL suffice to exert full repressor function (compare dSXL and dRBD4). Furthermore, the two RRMs alone (dRBD3) possess significant, but diminished repressor activity.
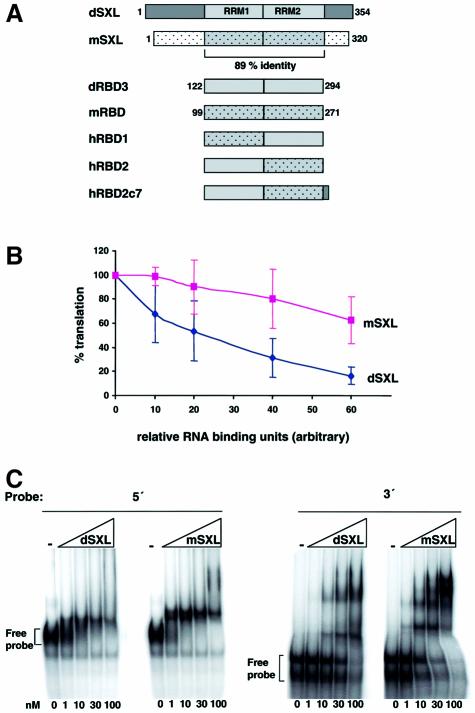
The repressor activity of dRBD3 could either be a consequence of its RNA binding property per se, or it could reflect an additional activity that is required for translational regulation and embedded within the RRMs. To distinguish between these two possibilities, we utilized the SXL homologue from Musca domestica (mSXL). mSXL shares 70% overall identity and 89% identity in the region containing the RNA-binding domains with its Drosophila counterpart (Meise et al., 1998; Figure 2A). When recombinant forms of the Drosophila or Musca proteins are added to a translation reaction, dSXL inhibits translation more efficiently than mSXL (Figure 2B). Importantly, these recombinant proteins bind to both the 5′UTR and the 3′UTR of msl-2 mRNA with comparable affinities (Figure 2C). This result indicates that SXL binding to the mRNA is not sufficient for translational repression. The two RRMs of mSXL, which we refer to as mRBD, contain only 11 non-conservative changes within 172 aa when compared with the RRMs of dSXL (dRBD3). dRBD3 and mRBD display highly similar msl-2 RNA binding (Figure 2E), but the latter totally lacks translational repressor activity (Figure 2D). This shows that the RRMs of dSXL harbour a translational repressor activity that can be distinguished from its RNA binding activity and that is lacking from mRBD.
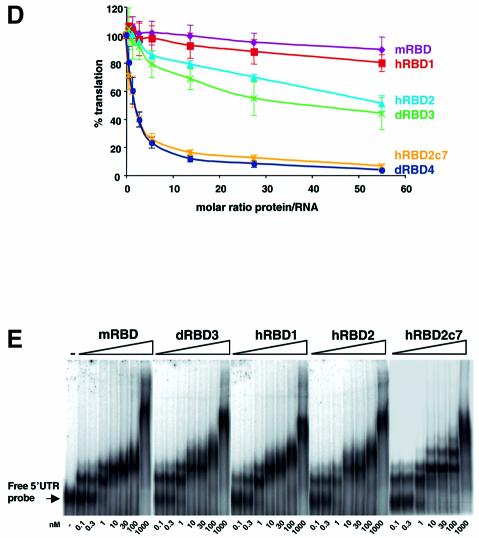
Fig. 2. RRM1 of dSXL contributes to translational repression. (A) Schematic representations of the Drosophila and Musca SXL proteins (dSXL and mSXL, respectively) and their RRMs (dRBD3 and mRBD), as well as of hybrid proteins containing combinations of these (hRBD1, hRBD2 and hRBD2c7). The corresponding amino acid positions of dSXL and mSXL included in the derivatives are indicated. (B) WT and BLEF mRNAs were assayed for repression by dSXL and mSXL, and similar results were obtained for both mRNAs. Shown here is the integration of those results. The translation inhibition curves are normalized for the binding activity of the mSXL and dSXL preparations, as measured by gel mobility-shift assay using the minimal 5′ and 3′ probes (C). (D) Translational repression by the RRM derivatives. Translation reactions were incubated with increasing amounts of dRBD3 (green line), mRBD (purple line), hRBD1 (red line), hRBD2 (light blue line) or hRBD2c7 (orange line) and analysed as described in Figure 1C. dRBD4 (dark blue line) is shown for reference. (E) RNA binding of the RRM derivatives. Gel mobility-shift assays were performed with increasing concentrations of mRBD, dRBD3, hRBD1, hRBD2 and hRBD2c7 and a probe containing 215 nt of the msl-2 5′UTR (positions 189–403).
To further map the translational repressor domain embedded within the RRMs of dSXL, we generated hybrid SXL proteins (hRBD1 and hRBD2) consisting of the combinations of the dSXL and mSXL RRMs (Figure 2A). This approach allowed the clear assignment of the translational repressor function to the RRM1 of dSXL, because hRBD2 controls translation as well as dRBD3, whereas hRBD1 is equally ineffective as mRBD (Figure 2D). In this context, we also wanted to test the contribution of the 7 aa that distinguish dRBD3 from dRBD4 to the repressor activity of hRBD2. As shown in Figure 2D, hRBD2c7 indeed displays the same full repressor activity as dRBD4 (and full-length dSXL). Importantly, all hybrid proteins bind RNA with an affinity comparable to that of mRBD and dRBD3 in gel mobility-shift assays (Figure 2E).
We conclude that the translational repressor domain of dSXL is composed of the two RRMs and the subsequent heptapeptide, and that the repressor function partially overlaps with the RRM1.
Coupling between translational repression and RRM-mediated mRNA binding
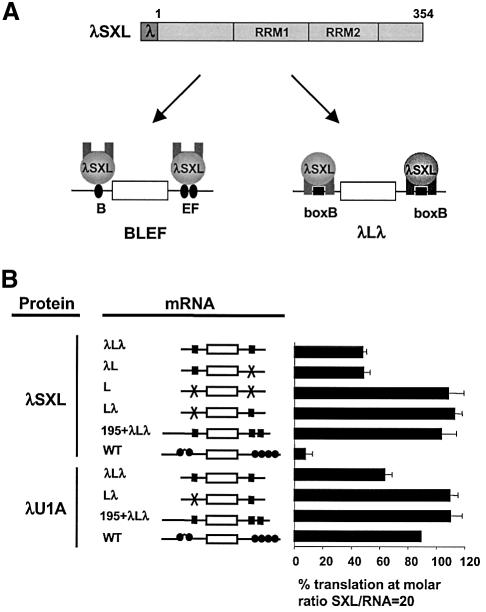
To understand the function of the translational repressor domain, we explored whether translational repression was coupled to RNA binding by the RRMs, or whether the two activities were independent of each other. To address this question, we fused the RNA-binding domain of the λ phage anti-terminator protein, a 22 aa oligomer referred to as the λ peptide, to the N-terminus of dSXL to create the hybrid protein λSXL (Figure 3A). The λ peptide binds a small [19 nucleotide (nt)] RNA stem–loop structure called boxB with high affinity and specificity (Tan and Frankel, 1995). To test whether λSXL is properly folded and fully functional, we used the WT indicator mRNA as a positive control. Importantly, λSXL represses WT mRNA translation with an efficiency equal to dSXL (Figure 3B; see also Figure 1C for inhibition by dSXL). This result shows that the λ peptide does not interfere with dSXL function, and that λSXL acts as a bona fide regulator of translation when it binds to the natural dSXL binding sites of msl-2 mRNA.
Fig. 3. Tethered function analysis of dSXL. (A) Schematic diagram of the experimental approach. The RNA-binding peptide λ was fused to the N-terminus of dSXL to yield λSXL. λSXL binds to mRNAs containing dSXL-binding sites (black ovals) via its RRMs, and to mRNAs harbouring boxB/λ-binding sites (black squares) via the λ peptide. λLλ mRNA is a derivative of BLEF mRNA, in which the dSXL-binding sites are replaced by boxB elements. The open rectangles denote the luciferase open reading frame, and the thin lines represent msl-2 UTR sequences. (B) Translational inhibition of indicated mRNAs by λSXL and λU1A at a 20-fold molar excess of protein to RNA. Consistent results were obtained when the same range of protein concentrations as that used for Figures 1 and 2 was tested (data not shown).
We then replaced the dSXL-binding sites by boxB elements (Figure 3A), using BLEF mRNA as the starting construct. BLEF (see Figure 1B) harbours all msl-2 mRNA sequences required for full regulation by dSXL and λSXL, including the SXL-binding site B in the 5′UTR and sites E and F in the 3′UTR (see Gebauer et al., 2003; data not shown). λLλ mRNA is composed of a λ-binding site (boxB) replacing SXL-binding site B, and an additional boxB replacing SXL-binding sites E and F (Figure 3A). λSXL represses translation of λLλ only 2-fold (Figure 3B, compare λLλ with WT), identical to when a reporter mRNA lacking boxB in the 3′UTR is used (λL). In addition, translation of reporter mRNAs lacking λ-binding sites in their 5′UTRs is not affected by λSXL, regardless of the presence or absence of boxB in the 3′UTR (Lλ and L, respectively). These data show that λSXL represses translation of λLλ mRNA only via its 5′UTR, failing to reflect the genuine mechanism of translational control by dSXL (Bashaw and Baker, 1997; Kelley et al., 1997; Gebauer et al., 1999).
The short distance between the boxB in the 5′UTR and the cap structure in λLλ mRNA suggested that the 2-fold translational repression was achieved by the steric inhibitory mechanism described previously for proteins binding to cap-proximal stem–loop structures (Stripecke and Hentze, 1992; Stripecke et al., 1994). Indeed, increasing the spacing between the cap structure and the boxB by 195 nt abrogates λSXL-mediated repression, even when the E and F sites in the 3′UTR are replaced by two boxB elements (Figure 3B, 195 + λLλ mRNA). Furthermore, replacement of dSXL by a control λ-fusion protein (λU1A) represses λLλ translation to a similar extent as λSXL, and does not affect the translation of an mRNA lacking boxB in the 5′UTR (Figure 3B). Therefore, the 2-fold repression of λLλ by λSXL is indistinguishable from steric inhibition. We conclude that λSXL represses only the translation of WT and BLEF mRNAs, to which it binds with its own RRMs, by the physiological msl-2 mechanism. These data indicate that the translational repressor activity of dSXL is coupled to mRNA binding by the RRMs.
The translational repressor domain of dSXL mediates specific interactions with proteins binding to the 3′UTR of msl-2 mRNA
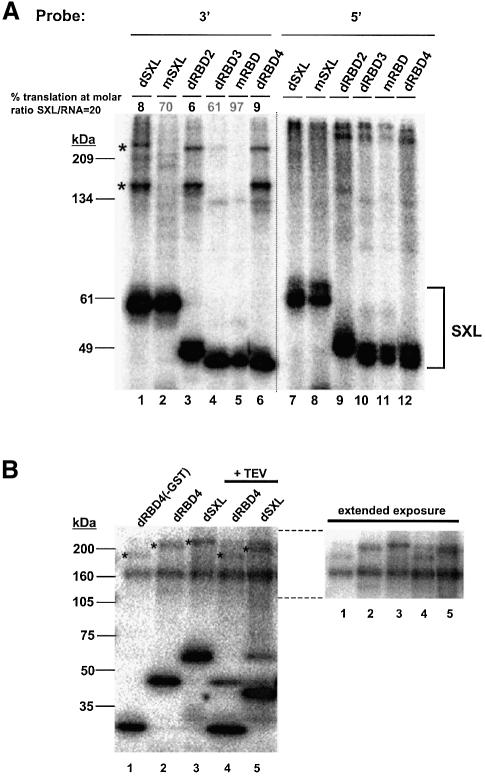
An interesting possibility for the function of the translational repressor domain is that it recruits translational co-repressors upon msl-2 mRNA binding. To identify possible co-repressors, polypeptides present in the Drosophila embryo extract were covalently linked by irradiation with ultraviolet light to an msl-2 3′UTR probe containing the minimal sequences required for translational repression (Figure 1B). These sequences were selected because they provided a function in translational control in addition to SXL binding, suggesting that they could bind co-repressors (Gebauer et al., 2003; see below). Indeed, when the radiolabelled 3′UTR probe is incubated with Drosophila embryo extract under translation conditions, a set of high molecular weight proteins can be detected that specifically crosslink to the RNA in a dSXL-dependent manner (Figure 4, compare lanes 1 and 6). More importantly, these proteins are specifically co-immunoprecipitated with anti-SXL antibody, but not with pre-immune serum (Figure 4, compare lane 8 with lanes 10, 3 and 5), demonstrating that they bind the RNA together with dSXL. These proteins are not detected when the Drosophila extract is omitted from the reaction, indicating that they are not contaminants from the preparation of recombinant dSXL (data not shown). Two of these high molecular weight proteins, of ∼215 and ∼160 kDa, were readily detectable in all of our experiments (high intensity bands marked with an asterisk), while two others were more difficult to detect and less consistently seen (marked with a dot). These results show that high molecular weight Drosophila embryo proteins bind to the 3′UTR of msl-2 RNA in a dSXL-dependent manner.
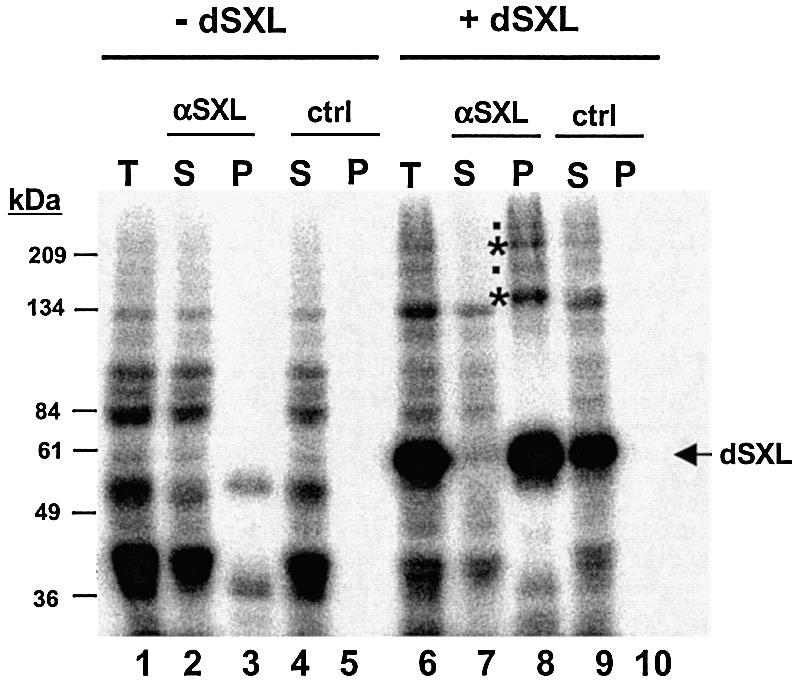
Fig. 4. High molecular weight proteins associate with the msl-2 3′UTR in a dSXL-dependent manner. The 32P-labelled msl-2 3′probe was incubated in a typical translation reaction in the absence (lanes 1–5) or presence (lanes 6–10) of recombinant dSXL. After UV-crosslinking and digestion with RNase T1, complexes were immunoprecipitated with α-SXL antibody or pre-immune serum (ctrl). T, total set of crosslinked proteins before immunoprecipitation; S, supernatant after immunoprecipitation; P, pellet. The position of crosslinked recombinant dSXL is indicated. Asterisks mark proteins that crosslink consistently, bands marked with a dot are not always seen.
To explore further whether the high molecular weight polypeptides were involved in translational regulation, UV-crosslink/co-immunoprecipitation assays were performed with the SXL derivatives that all bind to RNA equally well, but include or lack the translational repressor domain and thus differ in their ability to control translation (Figures 1 and 2). The polypeptides identified in Figure 4 are readily co-immunoprecipitated with dSXL derivatives that harbour the dSXL translation repressor domain (Figure 5A, lanes 1, 3 and 6), but not with SXL variants that lack all or a part of this domain (lanes 2, 4 and 5). It is important to note that all six variant proteins bind to the RNA probe and are readily immunoprecipitated (Figure 5A, lanes 1–6). If the 3′UTR probe is replaced by a probe derived from the 5′UTR of msl-2 (Figure 1B), the high molecular weight polypeptides are not detected even though the six different SXL proteins also bind to this probe (lanes 7–12). These data establish a tight correlation between the presence of the dSXL translational repressor domain, the inclusion of the regulatory 3′UTR msl-2 RNA sequences, and the association of high molecular weight proteins with both the RNA and dSXL in the Drosphila embryo extracts.
Fig. 5. (A) The association of high molecular weight polypeptides with the msl-2 3′UTR correlates tightly with translational repression. 3′ (lanes 1–6) or 5′ (lanes 7–12) RNA probes were incubated in Drosophila translation extracts including recombinant dSXL or SXL derivatives that either repress translation efficiently (dRBD2 and dRBD4) or not (mSXL, dRBD3, mRBD). The ability of each derivative to repress translation is indicated. After UV-crosslinking and immunoprecipitation, proteins present in the pellet were separated in a denaturing 8% acrylamide gel and visualized by autoradiography. Asterisks mark polypeptides of high intensity that specifically co-immunoprecipitate with dSXL (see also Figure 4). (B) The slow mobility band (∼215 kDa) contains SXL. dRBD4 lacking GST (lane 1), or dRBD4 and dSXL (both containing a GST tag, lanes 2 and 3) were incubated in Drosophila translation extracts containing 3′ RNA probe. In lanes 4 and 5, the sample was treated with TEV protease after UV-crosslinking and immunoprecipitation. Proteins were separated in a 6–15% acrylamide gel and visualized by autoradiography. The right panel shows an extended exposure of the upper portion of the gel shown on the left after adjustment of brightness and contrast to reduce background noise.
Close inspection of the high intensity bands with the slowest mobility in Figure 5A, lanes 1, 3 and 6, reveals that the band in lane 1 (where full-length dSXL is used) has a slower mobility than the corresponding bands in lanes 3 and 6 (where smaller SXL variants are used). This suggested the possibility that these bands actually represent denaturation-insensitive complexes containing SXL. To test this possibility, we took advantage of the fact that the dSXL variants are recombinant proteins fused to glutathione S-transferase (GST), with a TEV protease cleavage site in between, which allowed alteration of their sizes. The data shown in lanes 2 and 3 in Figure 5B recapitulate the findings shown in lanes 6 and 1, respectively, in Figure 5A. Figure 5B, lane 1, shows that removal of the GST tag from dRBD4 prior to the crosslinking experiment induces a downshift of the crosslinked band that corresponds to the size of GST. Moreover, when the GST fusion proteins (Figure 5B, lanes 2 and 3) are used for the crosslinking experiments and the GST domain is removed by TEV cleavage just prior to electrophoresis, similar downshifts of both the crosslinked SXL proteins and the high molecular weight bands are observed (lanes 4 and 5). No such changes are seen with the ∼160 kDa band, suggesting that SXL is not part of this crosslinked entity. Interestingly, the size difference between the ∼160 kDa band and the respective higher molecular weight crosslinks correlates with the size of the respective SXL variants. Thus, the two crosslinked bands may represent two different high molecular weight proteins (of which one is bound to SXL), or only one factor of ∼160 kDa that crosslinks with (higher band) and without (lower band) SXL. In the remainder of this paper, we will refer to the two bands as two entities without prejudice to the above two possibilities.
The putative co-repressors assembling on sequences adjacent to the regulatory SXL-binding sites in the 3′UTR of msl-2 mRNA are functionally important, titratable factors
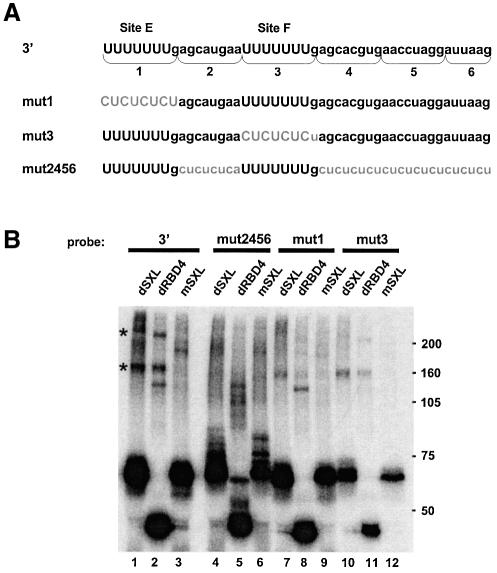
Three independent sets of experiments suggest that the assembly of putative co-repressors on the msl-2 3′UTR sequences is functionally important. First, 5′UTR msl-2 sequences including SXL-binding site B fail to support dSXL-mediated translational repression when placed in the 3′UTR, even though dSXL binds to the 5′UTR sequences with slightly higher affinity than to those in the 3′UTR (Gebauer et al., 2003). Secondly, the putative co-repressors crosslink specifically to 3′UTR, rather than 5′UTR sequences (Figure 5A). Thirdly, the sequences surrounding the SXL-binding sites E and F in the 3′UTR are necessary for translational repression (Gebauer et al., 2003). To test directly whether putative co-repressors binding to the 3′UTR regulatory sequences are functionally important, we replaced the critical sequences flanking the SXL-binding sites by CU repeats (mut2456; Figure 6A) and performed crosslink/co-immunoprecipitation assays similar to those described above. As shown in Figure 6B, the mut2456 probe effectively crosslinks to dSXL, while the binding of the high molecular weight polypeptides is undetectable, indicating that the sequences adjacent to sites E and F are recognized by the putative co-repressors.
Fig. 6. dSXL binding to the msl-2 3′UTR promotes the assembly of high molecular weight polypeptides on sequences adjacent to the regulatory SXL-binding sites. (A) Sequence of the 3′UTR msl-2 segment relevant for translational repression (3′). SXL-binding site E, site F or sequences adjacent to those sites were substituted by CU repeats (mut1, mut3 and mut2456, respectively). (B) Binding of high molecular weight polypeptides to the sequences shown in (A), as determined by UV-crosslink/co-immunoprecipitation assays similar to those described in Figures 4 and 5.
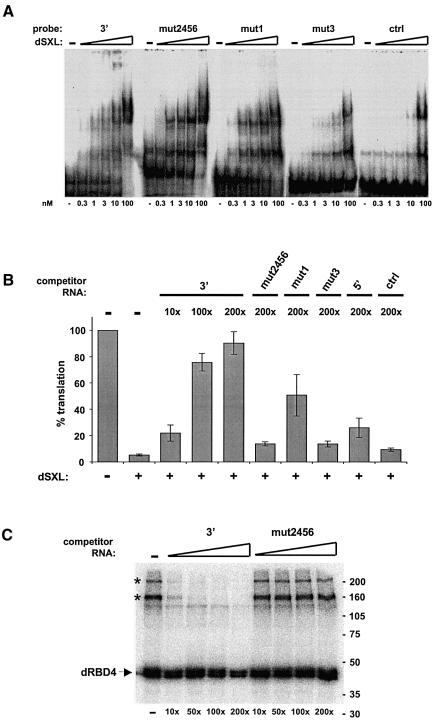
To test whether the recruitment of putative co-repressors correlates with SXL-binding, we took advantage of our previous finding that SXL-binding site F contributes more strongly to translational repression than site E (Gebauer et al., 2003). Substitution of site E by CU repeats (Figure 6A, mut1) resulted in slightly reduced SXL binding, as measured both by crosslinking and gel mobility-shift assays (Figures 6B and 7A, respectively). However, when site F is mutated (Figure 6A, mut3), SXL binding is severely reduced. Both mutants display reduced crosslinking of the putative co-repressors (Figure 6B). These data establish a correlation between dSXL binding and putative co-factor assembly on the 3′UTR of msl-2 mRNA.
Fig. 7. Factors present in the Drosophila embryo extracts are required for translational repression of msl-2 mRNA. (A) Binding of dSXL to the 3′ probe or its derivatives (see Figure 6A), measured by gel mobility-shift assay. A probe containing the NRE of maternal hunchback mRNA was used as a specificity control (ctrl). (B) Functional titration of dSXL co-repressors. Drosophila embryo extracts were pre-incubated with 3′, mut2456, mut1, mut3, 5′ or control (multiple cloning site from pBluescript vector) RNAs, and typical translation reactions were subsequently assembled. The molar amount of competitor RNA was 10-, 100- or 200-fold higher than that of reporter mRNA. The translation assay was performed as described in Figure 1C. For simplicity, only the data obtained at 50-fold molar excess of dSXL over reporter mRNA are shown. Similar results were obtained with WT and BLEF mRNAs. (C) Specific titration of co-repressor crosslinking. Typical translation reactions containing Drosophila embryo extracts, 3′ probe and 50-fold molar excess of dRBD4 over probe were incubated with unlabelled 3′ or mut2456 competitor RNAs. The molar amount of competitor RNA was 10-, 50-, 100- or 200-fold higher than that of the probe. The UV-crosslink/co-immunoprecipitation assays were performed as described in Figure 4.
To test the requirement for co-repressors in dSXL-mediated translational inhibition, we assayed the ability of the different 3′UTR variants to compete for these factors. Drosophila embryo extracts were pre-incubated with WT or derivative msl-2 3′UTR RNAs, as well as with an unrelated RNA, which served as a negative control (Figure 7B). Subsequently, translational inhibition of an msl-2 reporter mRNA by dSXL was tested in the presence of excess dSXL, such that the different competitor RNAs do not de-repress translation on the basis of competition for dSXL. As shown in Figure 7B, translational repression by dSXL is impaired in a dose-dependent manner when the extracts are preincubated with the WT RNA (3′). Reporter mRNA translation is completely de-repressed at the highest concentration used, suggesting that 3′ RNA competes for factor(s) necessary for translational repression. In contrast, little competition is observed when the RNAs lacking SXL-binding site F or the adjacent sequences are used even at the highest concentration (mut3 and mut2456, respectively). Similarly, little competition is seen with the B site (5′ RNA) that binds to dSXL but lacks the flanking sequences of sites E and F, showing that the effect of the 3′ probe cannot be explained by competition for dSXL. In agreement with a weaker contribution of site E to SXL-binding, mut1 RNA competes for translation inhibition more efficiently than mut3. Importantly, the control RNA does not affect translational inhibition at the same concentration. These results indicate that factor(s) assembled on the sequences adjacent to site F are required for dSXL-mediated translational repression. Furthermore, this factor(s) is not dSXL itself, since mut2456 RNA that binds to dSXL with efficiency equal to 3′ RNA (Figure 7A) fails to de-repress translation.
Finally, we tested whether the specific functional competition by the 3′ competitor RNA (compared with the mut2456 RNA) correlates with specific competition for the crosslinked high molecular weight co-repressor(s). The results shown in Figure 7C demonstrate such specific competition for both bands at the same competitor RNA concentrations as those used for the translation assay. We conclude that at least one titratable factor present in the Drosophila embryo extract is required as a co-repressor for translational inhibition by dSXL.
Discussion
We have analysed the architecture and function of dSXL as a translational repressor. In female flies, translational repression of msl-2 mRNA by dSXL represents a critical regulatory step for dosage compensation (Bashaw and Baker, 1997; Kelley et al., 1997). Our results discriminate dSXL’s function as a splicing regulator from its function in translational control. They also suggest that the translational repressor activity of dSXL is dormant, and activated upon binding to the msl-2 mRNA. Finally, we provide functional evidence that dSXL and the msl-2 3′UTR cooperate in the recruitment of proteins that appear to act as translational co-repressors.
Architecture of dSXL as a translational regulator
Modularity represents one of the most common principles of protein architecture. It permits the organization of biological functions into independent domains, and integration of these functions in a single molecule. This work shows that the mRNA binding and translational regulatory functions of dSXL are not organized into independent domains. Rather, both functions reside within aa 122–301 of the protein (Figure 1), a region that was previously recognized as the RNA-binding domain of dSXL (Kanaar et al., 1995; Sakashita and Sakamoto, 1996; Samuels et al., 1998). The N-terminal domain of dSXL contributes an essential function only to alternative splicing (Wang and Bell, 1994; Deshpande et al., 1999; Yanowitz et al., 1999), but is dispensable for translational control of msl-2 mRNA (Figures 1 and 2). The embedding of the translational regulatory function into the RNA-binding domain and a few additional C-terminal amino acids also explains why N-terminal truncations strongly impair dSXL function in sex determination, but not in dosage compensation in vivo (Yanowitz et al., 1999).
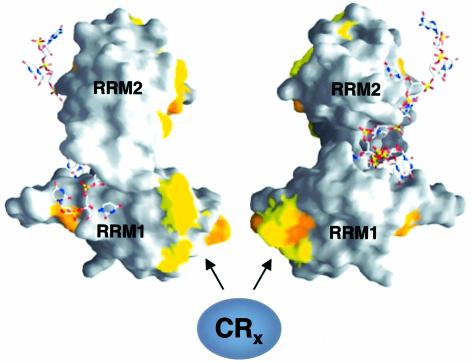
Based on the analysis of hybrid proteins between Drosophila and Musca SXL, we suggest that the region corresponding to RRM1 (aa 122–200) makes an important contribution to translational repression (Figure 2) and represents a critical difference between Drosophila SXL, which efficiently represses translation, and Musca SXL, which does not (Figure 2B). In addition to RRM1, an extension of 7 aa following the RRMs cooperates in translation inhibition. Interestingly, RRM1 has been shown to contact the snRNP component SNF, a protein thought to be involved in the autoregulatory splicing of Sxl pre-mRNA (Samuels et al., 1998). RRM1 also binds SIN, a protein of unknown function (Dong and Bell, 1999). However, both SNF and SIN are much smaller than the crosslinked polypeptides that dSXL recruits to the 3′UTR of msl-2 (Figures 4 and 5). The crystal structure of the dSXL RRMs bound to the RNA reveals that almost all of the residues that differ from those in mSXL are exposed on the accessible outer surface of the protein (yellow and orange patches in Figure 8). None of them contacts the RNA or other parts of the RRMs (Handa et al., 1999). In accordance with these structural data, we find similar RNA-binding affinities of dRBD3 and mRBD in gel mobility-shift assays (Figure 2E). Exposed amino acid residues on the outer surface of the RNA-binding domain of dSXL are ideally placed to mediate the interaction of dSXL with translational co-repressors (Figure 8).
Fig. 8. Molecular surface representation of dSXL RRMs bound to RNA. Two different orientations, vertically rotated by 180°, are shown (Handa et al., 1999). Residues that differ between Drosophila and Musca SXL are highlighted in yellow (conservative changes) and orange (non-conservative changes). A bound RNA oligomer derived from the tra mRNA is depicted using a stick representation. The solvent-exposed surface of RRM1, containing a high proportion of residues that differ from Musca SXL, is suggested to interact with translational co-repressors.
Dormancy and activation of the translational repressor activity of dSXL
Regulatory proteins that bind to specific mRNAs to control their translation employ different modes of action. In all cases, specificity demands that the regulatory protein should affect the translation of the mRNAs to which it binds, but not perturb the general translation machinery in a non-specific way. One solution to this problem is steric control, where the mRNA binding event is both necessary and sufficient for translational repression. In these cases, no translational effector domains per se are required, which could potentially act in trans and interfere with the function of a translation factor while the regulatory protein is not bound to its target mRNA. Steric control has been shown for the iron regulatory proteins (Stripecke et al., 1994; Muckenthaler et al., 1998), and is also evident in the translational inhibition of the λLλ mRNA by the λU1A protein (Figure 3B). msl-2 mRNA regulation by dSXL does not operate by such a steric mechanism (Figures 1–3), and the specificity problem must be solved differently.
The binding of dSXL to its physiological binding sites in the 5′ and 3′UTRs of WT or BLEF mRNA cannot be substituted by tethered dSXL, suggesting that dSXL must bind the RNA with its own RRMs to be active as a translational repressor (Figure 3). This contrasts with other RNA-binding proteins, such as the poly(A)-binding protein (PABP) (Gray et al., 2000) or proteins involved in nonsense-mediated mRNA decay (Lykke-Andersen et al., 2000; Gehring et al., 2003), which retain their respective functions when being tethered to an mRNA. Importantly, the λSXL protein binds to the indicator mRNAs, as evidenced by its steric regulatory function (Figure 3B) and gel mobility-shift assays (data not shown), and it is fully functional as a bona fide msl-2 regulatory protein when binding through its own RRMs (Figure 3B). This excludes trivial misfolding effects as an explanation for the lack of tethered function.
It seems most likely that the dormant translational repressor domain, which is embedded within the RNA-binding region of dSXL, is only activated when the protein binds to msl-2 mRNA, solving the specificity problem. dSXL binding to the EF region of the msl-2 3′UTR induces the binding of additional proteins to this site, which cannot be crosslinked in the absence of dSXL (Figures 4–6). It is possible that two molecules of SXL binding to the adjacent E and F sites, respectively, cooperate in recruiting the co-repressor(s) by creating a binding surface partially lacking from monomeric SXL. The recruitment of these additional proteins has two requirements. First, dSXL but not mSXL can mediate the crosslinking of these factors. This suggests that specific amino acids that are exposed on dSXL (or dSXL molecules binding closely to each other) contribute to the interaction. Secondly, dSXL binding to the 3′UTR probe but not the 5′UTR or mutated 3′UTR probes is required for crosslinking. This suggests that the 3′UTR RNA sequences make an additional specific contribution. Indeed, in addition to SXL-binding sites E and F, sequences adjacent to these sites are essential for recruitment of the additional proteins (Figure 6B). These factors that bind to the 3′UTR regulatory region are functional co-repressors, because only the 3′UTR WT RNA competes for translational inhibition (Figure 7B), while the mut2456 and 5′ RNAs (which bind dSXL but not the co-repressors) fail to do so.
The proposed model for dSXL bears interesting parallels with the function of Pumilio (Pum) and other members of the Puf family of translational regulators (Wickens et al., 2002). Pum inhibits the translation of maternal hunchback mRNA as part of a complex that is sequentially built on the Nanos-response element (NRE) in the 3′UTR of the message, and the translational repressor domain of Pum is embedded within its RNA binding region, the so-called Puf repeat (Wharton et al., 1998). Binding of Pum to the NRE triggers the recruitment of Nanos and this event, in turn, stimulates the binding of Brain Tumor (Sonoda and Wharton, 1999; Sonoda and Wharton, 2001). Hence, the stepwise assembly of co-repressor complexes, which requires multiple specific protein–protein and RNA–protein interactions, emerges as a theme in translational control. The msl-2 regulatory complex inhibits translation initiation by blocking the stable binding of the small ribosomal subunit to the mRNA in a cap-independent fashion (Gebauer et al., 2003). Future work will aim to define the composition of this regulatory complex and reveal its molecular interactions with the machinery that executes early steps in translation initiation.
Materials and methods
DNA constructs
pGEX-dSXL was kindly provided by Dr J.Valcárcel. pGEX-mSXL was obtained by cloning the M.domestica SXL coding sequence (kindly provided by Dr D.Bopp) into the NcoI/EcoRI sites of pGEXcs (Parks et al., 1994). The coding sequences of dRBD1, dRBD2, dRBD3 and dRBD4 were amplified by PCR from pGEX-dSXL and cloned as described for mSXL. Similarly, mRBD was amplified by PCR from pGEX-mSXL and cloned into pGEXcs. To obtain hRBD1, mSXL RRM1 was excised from mRBD by digestion with NcoI and NciI, and dSXL RRM2 was excised from dRBD3 by digestion with NciI and EcoRI. Both fragments were ligated into the NcoI and EcoRI sites of pGEXcs. hRBD2 was cloned in a similar manner, by excising dSXL RRM1 from dRBD3 and mSXL RRM2 from mRBD. Similarly, hRBD2c7 was cloned by excising dSXL RRM1 from dRBD3 and ligating it to RRM2 from mRBD amplified by PCR using primers containing the sequence for additional seven dSXL amino acids. The λSXL plasmid was obtained by insertion of NcoI/NcoI fragment from pSGλ4E73–102 (De Gregorio et al., 2001), containing the λ peptide sequence, into the NcoI site of pGEX-dSXL. To obtain λU1A, U1A was excised from U1AΔB12His plasmid (a gift from Dr I.Mattaj) by digestion with NcoI/BamHI and ligated to pGEXcs NcoI/EcoRI. The λ peptide was inserted in the same manner as for λSXL.
The WT mLm plasmid (WT) containing the firefly luciferase open reading frame fused to the 5′ and 3′UTRs of msl-2 has been described previously (Gebauer et al., 1999). BoxB-containing plasmids were obtained using the min construct as the parent vector. This construct includes the luciferase coding region flanked by nt 1–354 of msl-2 5′UTR and 841–954 of the msl-2 3′UTR (Gebauer et al., 2003). λLλ was obtained in two steps. First, a segment generated by hybridization of complementary oligonucleotides including nt 270–341 of the msl-2 5′UTR containing a boxB sequence in place of the U16 at positions 317–332 was cloned into the SacI/SmaI sites of min to yield plasmid 5′boxB. In the second step, a DNA segment containing a boxB followed by nt 916–924 of the msl-2 3′UTR was cloned into the HpaI/NotI sites of 5′boxB plasmid. λL plasmid was derived from 5′boxB and contained no boxB sequence, but 67 nt of unrelated sequence in the 3′UTR. Plasmid Lλ was obtained by replacing the 5′UTR sequences of the min plasmid with a DNA segment containing nt 270–341 of the msl-2 5′UTR with positions 316–333 substituted by a non-functional mutated boxB sequence. The 3′UTR of Lλ was cloned as described for λLλ. Plasmid L was obtained as described for Lλ using the corresponding DNA fragments containing mutated boxB sequences.
To generate 195 + λLλ, a fragment containing nt 1–195 of the msl-2 5′UTR was cloned into the SacI site of 5′boxB plasmid. The 3′UTR of the resulting construct was then substituted by a fragment containing nt 909–954 of the msl-2 3′UTR, with the two U7 stretches replaced by two boxB sequences. The regulatory sequences flanking site F in msl-2 3′UTR are preserved in this construct.
To obtain the 3′ plasmid, a fragment generated by hybridization of complementary oligonucleotides including nt 909–954 of the msl-2 3′UTR was cloned into HincII/NotI sites of pBluescript vector. mut1, mut3 and mut2456 plasmids were obtained in an analogous manner.
Protein expression and purification
Recombinant proteins were expressed in Escherichia coli as N-terminal GST-tagged fusions, and were purified as described previously (Valcárcel et al., 1993), with some modifications. Briefly, 3 h after induction with 1 mM IPTG, cells were pelleted and resuspended in buffer X [20 mM Tris–HCl pH 7.5, 1 M NaCl, 0.2 mM EDTA, 1 mM dithiothreitol (DTT), protease inhibitor cocktail Complete™ (Roche)]. Cells were lysed and debris was removed by centrifugation at 12 000 g for 20 min. Glutathione–agarose beads were incubated with the supernatant on a rocking platform for 15 min at 4°C. After washing with 40 vol. of buffer X, bound proteins were eluted with 10× 200 µl of elution buffer (50 mM glutathione, 100 mM HEPES pH 8.0, 1 mM DTT). Fractions containing the protein were dialysed against Dignam buffer D (20 mM HEPES pH 8.0, 20% glycerol, 0.2 mM EDTA, 1 mM DTT, 0.01% NP-40).
In vitro transcription and translation
mRNAs were synthesized as described previously (Gray and Hentze, 1994). All mRNAs contained a 5′ 7mGpppG cap and a poly(A) tail of 73 residues. Competitor RNAs were synthesized using the T7-MEGAshortscript™ transcription kit (Ambion). Probes were synthesized in the presence of radiolabelled UTP and ATP. In vitro translation reactions in Drosophila embryo extracts were performed in a final volume of 12.5 µl as described previously (Gebauer et al., 1999). The template mRNA was added at a final concentration of 3.2 ng/µl. Renilla luciferase mRNA was included as an internal control at a concentration of 0.8 ng/µl. Increasing amounts of recombinant dSXL or its derivatives were added at a volume of 1 µl. Translation reactions were incubated for 90 min at 25°C, and the translation efficiency was determined by measuring the luciferase activity using the Dual Luciferase Assay System (Promega). The firefly luciferase values were corrected for renilla expression.
For translational inhibition competition experiments, Drosophila embryo extracts were incubated with competitor RNAs for 30 min at 25°C prior to assembling the translation reactions.
Gel mobility-shift assays
In vitro transcribed 32P-labelled RNA containing 215 nt (positions 189–403), 70 nt (positions 270–339) of the msl-2 5′UTR or 46 nt (positions 909–954) of the msl-2 3′UTR was incubated with increasing amounts of recombinant protein as described previously (Valcárcel et al., 1993). The formation of RNA-protein complexes was analysed by non-denaturing polyacrylamide gel electrophoresis.
UV-crosslink and immunoprecipitation
32P-labelled msl-2 5′UTR RNA (5′, positions 270–339), 3′UTR RNA (3′, positions 909–954) or its derivative RNA (mut1, mut3 or mut2456) was incubated in a typical translation reaction containing 50 ng of dSXL or its derivatives. After 30 min of incubation at 25°C, reactions were crosslinked at 254 nm and 600 mJ on ice, and digested with RNase T1. Proteins were immunoprecipitated with SXL antiserum (a kind gift from Dr J.Valcárcel) or pre-immune serum bound to protein A–Sepharose, using the NET-gel buffer system described by Sambrook et al. (1989). Unless otherwise stated, labelled proteins were separated by 8% SDS–PAGE and visualized using a phosphoimager.
TEV cleavage
For TEV cleavage after co-immunoprecipitation, samples were incubated with 1 µg of TEV protease in a buffer containing 50 mM Tris–HCl pH 8.0, 0.5 mM EDTA and 1 mM DTT for 2 h at 30°C. For TEV cleavage of dRBD4 prior to the crosslinking experiment, 6870 µg of dRBD4 were incubated with 70 µg of TEV protease at 4°C for 16 h. To remove the GST and TEV proteins, dRBD4(–GST) was purified by FPLC using a Mono-S column.
Acknowledgments
Acknowledgements
We are grateful to Drs Daniel Bopp (Zürich), Juan Valcárcel (Barcelona) and Iain Mattaj (Heidelberg) for generously providing reagents. We thank Philipp Selenko for help with the GRASP, and Asifa Akhtar, Kevin Czaplinski, Tobias Walther, Kent Duncan and members of the Hentze lab for helpful discussions and comments on the manuscript. This work was supported by a predoctoral fellowship of the Louis-Jeantet Foundation to M.G., and by a grant from the Deutsche Forschungsgemeinschaft (He 1442/7–1) to MWH.
References
- Akhtar A. (2003) Dosage compensation: an intertwined world of RNA and chromatin remodelling. Curr. Opin. Genet. Dev., 13, 161–169. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J. and Baker,B.S. (1995) The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development, 121, 3245–3258. [DOI] [PubMed] [Google Scholar]
- Bashaw G.J. and Baker,B.S. (1997) The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell, 89, 789–798. [DOI] [PubMed] [Google Scholar]
- Bell L.R., Horabin,J.I., Schedl,P. and Cline,T.W. (1991) Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell, 65, 229–239. [DOI] [PubMed] [Google Scholar]
- Crowder S.M., Kanaar,R., Rio,D.C. and Alber,T. (1999) Absence of interdomain contacts in the crystal structure of the RNA recognition motifs of Sex-lethal. Proc. Natl Acad. Sci. USA, 96, 4892–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gregorio E., Baron,J., Preiss,T. and Hentze,M.W. (2001) Tethered-function analysis reveals that elF4E can recruit ribosomes independent of its binding to the cap structure. RNA, 7, 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G., Samuels,M.E. and Schedl,P.D. (1996) Sex-lethal interacts with splicing factors in vitro and in vivo. Mol. Cell Biol., 16, 5036–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande G., Calhoun,G., Yanowitz,J.L. and Schedl,P.D. (1999) Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell, 99, 271–281. [DOI] [PubMed] [Google Scholar]
- Dong Z. and Bell,L.R. (1999) SIN, a novel Drosophila protein that associates with the RNA binding protein sex-lethal. Gene, 237, 421–428. [DOI] [PubMed] [Google Scholar]
- Gebauer F., Merendino,L., Hentze,M.W. and Valcárcel,J. (1998) The Drosophila splicing regulator sex-lethal directly inhibits translation of male-specific-lethal 2 mRNA. RNA, 4, 142–150. [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Corona,D.F., Preiss,T., Becker,P.B. and Hentze,M.W. (1999) Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J., 18, 6146–6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F., Grskovic,M. and Hentze,M.W. (2003) Drosophila Sex-Lethal Inhibits the stable association of the 40S ribosomal subunit with msl-2 mRNA. Mol. Cell, 11, 1397–1404. [DOI] [PubMed] [Google Scholar]
- Gehring N.H., Neu-Yilik,G., Schell,T., Hentze,M.W. and Kulozik,A.E. (2003) Y14 and hUpf3b form an NMD-activating complex. Mol. Cell, 11, 939–949. [DOI] [PubMed] [Google Scholar]
- Gray N.K. and Hentze,M.W. (1994) Regulation of protein synthesis by mRNA structure. Mol. Biol. Rep., 19, 195–200. [DOI] [PubMed] [Google Scholar]
- Gray N.K., Coller,J.M., Dickson,K.S. and Wickens,M. (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J., 19, 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa N., Nureki,O., Kurimoto,K., Kim,I., Sakamoto,H., Shimura,Y., Muto,Y. and Yokoyama,S. (1999) Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature, 398, 579–585. [DOI] [PubMed] [Google Scholar]
- Kanaar R., Lee,A.L., Rudner,D.Z., Wemmer,D.E. and Rio,D.C. (1995) Interaction of the sex-lethal RNA binding domains with RNA. EMBO J., 14, 4530–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R.L., Wang,J., Bell,L. and Kuroda,M.I. (1997) Sex lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature, 387, 195–199. [DOI] [PubMed] [Google Scholar]
- Kim I. et al. (2000) Interactions of a didomain fragment of the Drosophila sex-lethal protein with single-stranded uridine-rich oligoribonucleotides derived from the transformer and Sex-lethal messenger RNA precursors: NMR with residue-selective [5-2H]uridine substitutions. J. Biomol. NMR, 17, 153–165. [DOI] [PubMed] [Google Scholar]
- Lallena M.J., Chalmers,K.J., Llamazares,S., Lamond,A.I. and Valcárcel,J. (2002) Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell, 109, 285–296. [DOI] [PubMed] [Google Scholar]
- Lee A.L., Volkman,B.F., Robertson,S.A., Rudner,D.Z., Barbash,D.A., Cline,T.W., Kanaar,R., Rio,D.C. and Wemmer,D.E. (1997) Chemical shift mapping of the RNA-binding interface of the multiple-RBD protein sex-lethal. Biochemistry, 36, 14306–14317. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Shu,M.D. and Steitz,J.A. (2000) Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell, 103, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Meise M., Hilfiker-Kleiner,D., Dubendorfer,A., Brunner,C., Nothiger,R. and Bopp,D. (1998) Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development, 125, 1487–1494. [DOI] [PubMed] [Google Scholar]
- Muckenthaler M., Gray,N.K. and Hentze,M.W. (1998) IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell, 2, 383–388. [DOI] [PubMed] [Google Scholar]
- Parks T.D., Leuther,K.K., Howard,E.D., Johnston,S.A. and Dougherty,W.G. (1994) Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase. Anal. Biochem., 216, 413–417. [DOI] [PubMed] [Google Scholar]
- Sakashita E. and Sakamoto,H. (1994) Characterization of RNA binding specificity of the Drosophila sex-lethal protein by in vitro ligand selection. Nucleic Acids Res., 22, 4082–4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita E. and Sakamoto,H. (1996) Protein–RNA and protein–protein interactions of the Drosophila sex-lethal mediated by its RNA-binding domains. J. Biochem., 120, 1028–1033. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbour, NY. [Google Scholar]
- Samuels M., Deshpande,G. and Schedl,P. (1998) Activities of the Sex-lethal protein in RNA binding and protein:protein interactions. Nucleic Acids Res., 26, 2625–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt C. and Nothiger,R. (2000) Structure, function and evolution of sex-determining systems in Dipteran insects. Development, 127, 667–677. [DOI] [PubMed] [Google Scholar]
- Singh R., Valcárcel,J. and Green,M.R. (1995) Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science, 268, 1173–1176. [DOI] [PubMed] [Google Scholar]
- Sonoda J. and Wharton,R.P. (1999) Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev., 13, 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J. and Wharton,R.P. (2001) Drosophila Brain Tumor is a translational repressor. Genes Dev., 15, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski B.A., Belote,J.M. and McKeown,M. (1989) Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell, 58, 449–459. [DOI] [PubMed] [Google Scholar]
- Stripecke R. and Hentze,M.W. (1992) Bacteriophage and spliceosomal proteins function as position-dependent cis/trans repressors of mRNA translation in vitro. Nucleic Acids Res., 20, 5555–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stripecke R., Oliveira,C.C., McCarthy,J.E. and Hentze,M.W. (1994) Proteins binding to 5′ untranslated region sites: a general mechanism for translational regulation of mRNAs in human and yeast cells. Mol. Cell. Biol., 14, 5898–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R. and Frankel,A.D. (1995) Structural variety of arginine-rich RNA-binding peptides. Proc. Natl Acad. Sci. USA, 92, 5282–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J., Singh,R., Zamore,P.D. and Green,M.R. (1993) The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature, 362, 171–175. [DOI] [PubMed] [Google Scholar]
- Wang J. and Bell,L.R. (1994) The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev., 8, 2072–2085. [DOI] [PubMed] [Google Scholar]
- Wang J., Dong,Z. and Bell,L.R. (1997) Sex-lethal interactions with protein and RNA. Roles of glycine-rich and RNA binding domains. J. Biol. Chem., 272, 22227–22235. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Sonoda,J., Lee,T., Patterson,M. and Murata,Y. (1998) The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell, 1, 863–872. [DOI] [PubMed] [Google Scholar]
- Wickens M., Bernstein,D.S., Kimble,J. and Parker,R. (2002) A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet., 18, 150–157. [DOI] [PubMed] [Google Scholar]
- Wilkinson M.F. and Shyu,A.B. (2001) Multifunctional regulatory proteins that control gene expression in both the nucleus and the cytoplasm. BioEssays, 23, 775–787. [DOI] [PubMed] [Google Scholar]
- Yanowitz J.L., Deshpande,G., Calhoun,G. and Schedl,P.D. (1999) An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of sex-lethal. Mol. Cell. Biol., 19, 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]