Abstract
Several modes of cell death are now recognized, including necrosis, apoptosis, and autophagy. Oftentimes the distinctions between these various modes may not be apparent, although the precise mode may be physiologically important. Accordingly, it is often desirable to be able to classify the mode of cell death. Apoptosis was originally defined by structural alterations in cells observable by transmitted light and electron microscopy. Today, a wide variety of imaging and cytochemical techniques are available for the investigation of apoptosis. This review will highlight many of these methods, and provide a critique on the advantages and disadvantages associated with them for the specific identification of apoptotic cells in culture and tissues.
Keywords: Apoptosis, Cell death, Cytochemistry, Morphology, Microscopy, Immunostaining
Introduction
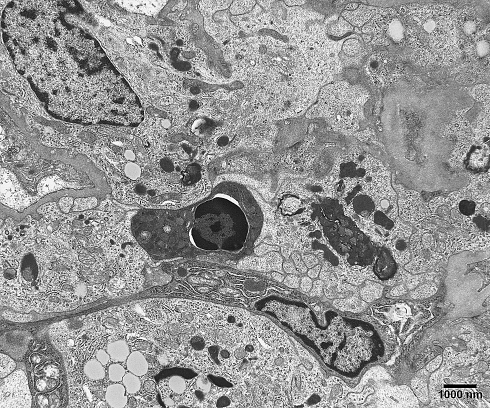
The mode of cell death referred to as apoptosis, and now defined by a variety of techniques, including biochemistry, flow cytometry, and imaging was originally defined based solely upon morphological criteria. In 1972, Kerr et al. (1972) introduced the neologism “apoptosis” to describe a type of cell death which had previously been referred to as “shrinkage necrosis”. Recognizing the importance of its kinetic mechanism in controlled cell deletion, either occurring spontaneously or in response to a stimulus, they offered the term apoptosis (from Greek, meaning “to fall away from”, as in leaves from a tree; thus, the falling away of cells from a tissue). They observed apoptotic cells in a wide variety of tissues, including during development and neoplastic transformation. Although they could detect apoptotic cells in many instances by light microscopy, it was their observations by transmission electron microscopy that established the characteristic ultrastructural features now considered the hallmark of apoptosis. These features include (1) cytoplasmic and nuclear condensation (pyknosis); (2) nuclear fragmentation (karyorrhexis); (3) normal morphological appearance of cytoplasmic organelles; and (4) an intact plasma membrane (Kerr et al. 1972; Wyllie et al. 1980; Galluzzi et al. 2007). Frequently, the pyknotic nucleus assumes the appearance of a half-moon or crescent shape, a feature most indicative of an apoptotic cell (Fig. 1). Following nuclear fragmentation, the cell disaggregates into a number of membrane-bound apoptotic bodies, which are engulfed via phagocytosis by neighboring epithelial cells or macrophages.
Fig. 1.
Transmission electron microscopic image of an apoptotic cell in a human kidney biopsy. Note the pyknotic, shrunken nucleus and the very condensed cytoplasm
In more recent years, evaluation of cells and tissues for apoptosis has evolved towards staining for light microscopic and flow cytometric analysis. As the biochemical and cell signaling events involved in the apoptotic cascade have been revealed, new tools for the analysis of apoptosis have emerged. Many of these tools are in the form of antibodies raised against proteins specific for the apoptotic pathway, or against neoepitopes on proteins resulting from action of an activated enzyme. While these new probes may specifically target aspects of the apoptotic pathway, they do not address the ultrastructural changes upon which the term apoptosis was originally defined.
This review will focus on some morphological and cytochemical techniques used to demonstrate the presence of apoptotic activity in tissue and cultured cells. For further information on this topic, the reader is referred to the earlier excellent reviews by Willingham (1999), Barrett et al. (2001) and Watanbe et al. (2002). However, before we begin to describe these specific detection methods, we need to examine more closely the different classifications of cell death, and then delve into the mechanisms responsible for initiating the apoptotic cascade.
Classification of cell death
Cell death is now known to be perpetrated through a variety of mechanisms. According to Galluzzi et al. (2007), cell death can be classified into four different types, based upon morphological characteristics: apoptosis (Type 1), autophagy (Type 2), necrosis (oncosis, Type 3), and mitotic catastrophe. The morphological changes accompanying apoptosis have been described in detail above. Whereas apoptosis is manifested by volume reduction of the nucleus and cytoplasm (cell shrinkage), necrosis (the mode of cell death with which apoptosis is most often confused) is evinced by cytoplasmic swelling, rupture of the plasma membrane, swelling of cytoplasmic organelles (particularly mitochondria), and some condensation of nuclear chromatin (Galluzzi et al. 2007). Autophagy is distinguished by the accumulation of cytoplasmic vacuoles and membranes, and mitotic catastrophe by multinucleation. Clearly, cellular morphological characteristics must be taken into consideration when determining the mode of cell death. This is of utmost importance when employing the TUNEL assay, since this technique stains both apoptotic and necrotic cells, which as just described display widely different morphological characteristics. This issue will be addressed in detail below.
Pathways of apoptosis
Cell death through apoptosis is known to occur through two primary pathways, an extrinsic pathway involving death receptors, and an intrinsic pathway via members of the Bcl-2 family (Adams and Cory 1998). The extrinsic pathway uses the classical death receptors such as Fas (CD95/APO1), TNF Receptor1, and TRAIL Receptors. Engagement of these receptors triggers a now well-defined process of recruitment of proteases known as caspases (cysteine-dependent aspartate-cleaving proteases). In the case of Fas, oligomerization by its ligand causes recruitment of the adaptor protein FADD (Fas-associated via death domain) through interactions between the death domains (DD) found in each protein (Krueger et al. 2003). FADD then recruits caspase-8 through their common death effector domains (DED). Homodimerization of caspase-8 results in its activation through sequential cleavage at critical aspartate residues. Fully active caspase-8 then cleaves downstream targets such as the Bcl-2 family member Bid to form truncated Bid or tBid (Li et al. 1998). tBid localizes to the outer mitochondrial membrane where it promotes oligomerization of another Bcl-2 member, Bax, which results in breakdown of the mitochondria and release of cytochrome c and Smac/Diablo (Verhagen et al. 2000). Cytochrome c engages a complex containing Apaf-1 and caspase-9 that in the presence of ATP forms a large molecular weight complex of active caspase-9 known as the apoptosome (Cain et al. 2000). The apoptosome can then activate the effector caspases, such as caspase-3 and caspase-6 that ultimately lead to breakdown of cellular components. Among these is cleavage of an inhibitor of caspase-activated DNAse (ICAD) that allows the DNAse to degrade DNA. This process is fundamental to various assays of DNA breaks that are described in this review.
In parallel with the extrinsic death pathway, the intrinsic pathway ultimately engages the same mitochondria-initiated downstream death pathways. In this case the cascade is initiated by any of a variety of sensors that detect cellular stress. The initiators in this case include a panel of Bcl-2 family members that contain only the BH3 domain (so-called BH3-only proteins) (Strasser 2001). These pro-apoptotic Bcl-2 family members are located throughout the cell somewhat as sentinels that detect cellular perturbations. Upon their activation they induce aggregation and activation of Bax or Bak, which then feeds into the mitochondrial death pathway described above. Activation of the intrinsic death pathway may occur in the form of DNA damage by UV irradiation that activates p53 and then two BH3-only proteins, Puma and Noxa. Glucocorticoids similarly activate Puma and Bim that are critical to induction of cell death by steroids. In fact most chemotherapeutic agents ultimately engage a BH3-only protein in the process of killing cells. As a consequence of these pathways, several death assays measure mitochondrial integrity or membrane potential as described in this review. Additional assays might assess cleavage of caspases or their substrates.
Given the localization of many Bcl-2 family members at the mitochondria, considerable interest is now developing over the concept that these proteins may function as well in some normal metabolic pathways. An early and somewhat overlooked study from the Korsmeyer laboratory showed that increased expression of Bcl-2 in T lymphocytes decreased their production of the growth cytokine IL-2 and resulted in somewhat reduced proliferative capacity following stimulation of the T cell antigen receptor (TCR) (Linette et al. 1996). Bcl-2-deficient T cells manifested the opposite phenotype. More recent studies suggest that this may function via regulation of calcium signaling (White et al. 2005). Other studies suggest that BAD may regulate glucokinase activity in mitochondria and that BAD-deficient β-islet cells are less responsive with insulin secretion to glucose levels (Wikstrom et al. 2007). In a similar manner, caspase-8 has been shown to be critical to growth of various cell types including T lymphocytes (Chun et al. 2002; Salmena et al. 2003). Thus, analysis of activation of “death” molecules will, in the future, need to consider these potential overlapping functions with normal cell metabolism.
Detection of apoptosis by electron microscopy
It was the early electron microscopic investigations of Kerr et al. (1972) and Wyllie et al. (1980) that led to their formulation of the concept of apoptosis as a wide-spread form of cell death in a variety of tissues and physiological situations. Electron microscopy is still considered the “gold standard” for the identification of apoptotic cells. However, likely due to the costs associated with performing electron microscopy, the specialized equipment and technical expertise required, and the limited tissue area represented, this technique is no longer routinely used as an apoptosis assay. In many instances, the degree of apoptosis detected, for example following treatment with a specific reagent, is an experimental endpoint. This can easily be accomplished with light microscopy or flow cytometry-based methodologies, but is exceedingly laborious by transmission electron microscopy. However, in instances where quantitation is unimportant, and the mere presence of specifically identifiable apoptotic morphology is the goal, electron microscopy is indispensable. For instance, Abbate et al. (2007) recently employed this approach as a means to characterize the nature of cell death in the myocardium after an ischemic event. They investigated the ultrastructural appearance of cardiomyocytes by transmission electron microscopy in failing rabbit hearts 16 days following coronary artery ligation. They described cardiomyocytes with severe nuclear derangements, peripheral coarse chromatin condensation, and prevalent mitochondrial and myofibrillar alterations. Although these ultrastructural characteristics are not typical of apoptotic cells, the cells were deemed to be apoptotic based upon an intact cardiomyocyte plasma membrane. These results would suggest that the “typical” ultrastructural features ascribed to apoptotic cells may in some instances be cell-type specific. However, further experimentation will be necessary to confirm this interpretation.
Scanning electron microscopy has also been used to examine apoptotic cells, with their characteristic rounded-up phenotype well represented by this imaging modality (Wyllie et al. 1980; Germain et al. 2007).
Detection of apoptosis by cytochemical optical microscopy
A wide variety of probes are commercially available today for the detection of apoptosis (see Table 1). Many of these are in the form of antibodies raised against proteins specific for the apoptotic pathway, or against neoepitopes on proteins resulting from action of an activated enzyme. These will be discussed later, however, since we will begin with ostensibly the most common cytochemical method for the determination of apoptosis, the TUNEL assay.
Table 1.
Commercial sources for apoptosis reagents
| Reagent | Commercial source |
|---|---|
| TUNEL assay kit | MBL International, Woburn MA Millipore Corporation, Billerica, MA Invitrogen Corporation, Carlsbad, CA Promega, Madison, WI Roche Applied Science, Indianapolis, IN |
| Apo stain (F7-26) (anti-ssDNA) | Axxora, LLC, San Diego, CA |
| Anti-cleaved caspase 3 (ASP 175) | Cell Signaling Technology, Inc., Danvers, MA |
| Anti-cleaved cytokeratin 18 | Chemicon, Temecula, CA |
| Annexin V-FITC | BD Biosciences Pharmingen, San Diego, CA Roche Applied Science, Indianapolis, IN Invitrogen Corporation, Carlsbad, CA |
| Lectins specific for α-d-mannose and β-d-galactose | Lectinotest Laboratory, Lviv, Ukraine EY laboratories, San Mateo, CA Calbiochem, San Diego, CA Sigma-Aldrich, St Louis, MO |
| Anti-cytochrome c oxidase subunit 1 | Invitrogen Corporation, Carlsbad, CA BD Biosciences Pharmingen, San Diego, CA |
| Anti-apoptosis-inducing factor | Santa Cruz Biotechnology, Santa Cruz, CA Syntem, Nimes, France |
This list is not meant to be exhaustive, but to serve as a starting point to search for reagents. Apologies to those companies not listed
The TUNEL assay
The presence of apoptotic cells has often been inferred from a positive reaction to the TUNEL assay [terminal deoxynucleotidyl transferase (TdT)-mediated dUTP in situ nick end labeling]. However, the lack of specificity of the TUNEL assay for apoptotic cells has been well documented (Sloop et al. 1999; Jung et al. 2000; Walker and Quirke 2001; Watanabe et al. 2002; Groos et al. 2003; Takemura and Fujiwara 2006). TUNEL will detect free 3′-OH terminals of both single- and double-strand DNA breaks, thus potentially labeling both apoptotic and necrotic derived DNA strand breaks. Moreover, the TUNEL assay has also been reported to label mitotic cells (Ito et al. 2006), cells in the process of gene transcription (Kockx et al. 1998), cells undergoing DNA repair (Kanoh et al. 1999), and cells with DNA breaks caused by the microtomy sectioning procedure (Sloop et al. 1999). The effects of tissue fixation and processing may also alter TUNEL reactivity (Negoescu et al. 1996; Labat-Moleur et al. 1998; Tateyama et al. 1998). In a rat model, we found that exposure to asbestos and cigarette smoke resulted in strong TUNEL positivity in bronchiolar epithelial cells from paraffin-embedded sections stained by immunoperoxidase (Jung et al. 2000). However, when bronchioles from the same animals were examined by transmission electron microscopy, only necrotic epithelial cells were observed, indicating that the TUNEL-positive cells detected by light microscopy were likely necrotic, not apoptotic. The TUNEL-positive nuclei did not display the characteristic shrunken appearance expected for an apoptotic cell, again emphasizing the need to corroborate staining results with morphological observations. Similar conclusions were reached by Negoescu et al. (1996) and Labat-Moleur et al. (1998) in their efforts to improve tissue preparation and staining procedures for the TUNEL method.
The TUNEL assay maintains its popularity as a marker of apoptotic cells; it is available in a kit form, is easy to use, and has been available for a number of years, becoming entrenched in the literature. However, given the concerns raised above with its specificity for apoptotic cells, we are of the opinion that other methods should be employed concomitantly with this assay in order to make a determination that the mode of cell death observed is the result of an apoptotic mechanism.
Anti-single-stranded DNA antibody (Apostain)
Frankfurt et al. (1994, 1996) and Frankfurt and Krishan (2001) have pioneered the application of a monoclonal antibody raised against single-stranded DNA (ssDNA) as a more specific cytochemical probe for apoptotic cells than the TUNEL assay. They have shown that the combination of a commercially available ssDNA antibody with formamide-induced denaturation of condensed chromatin, results in specific labeling of apoptotic cells in culture and in formalin-fixed, paraffin embedded tissues (Frankfurt and Krishan 2001). Ito et al. (2006) expanded upon these observations by adapting the formamide DNA denaturation followed by anti-ssDNA antibody staining technique for the electron microscopic level. They show very convincingly by immunoelectron microscopy that the TUNEL assay results in staining of condensed chromatin in mitotic and necrotic cells in addition to apoptotic cells, whereas the formamide-ssDNA technique stained only the condensed chromatin from apoptotic cells.
We have also used this Apostain method for the specific detection of apoptotic pulmonary epithelial (Buder-Hofmann et al. 2001) or rat pleural mesothelial cells (Shukla et al. 2003) after exposure to crocidolite asbestos in culture, and in human lung adenocarcinoma cells (A-549) following treatment with the chemotherapeutic agents paclitaxel and vinorelbine (Jung et al. 2004). Moreover, we recently combined staining with the ssDNA antibody with a second specific marker of apoptosis in a novel dual staining method on tissue sections, as described below. In our hands, in agreement with studies described above, we find the formamide condensed DNA denaturation/ssDNA technique to be a reliable indicator of apoptotic cells in a variety of cell cultures and tissues.
Apoptosis-related neoepitopes
Antibodies raised against caspase-cleaved proteins are now commercially available. The enzymatic cleavage of proteins during the apoptotic cascade results in the formation of “neoepitopes” on these proteins which can be used as antigens for the production of caspase-mediated apoptotic reagents. Although several such antibodies are currently available, those raised against neoepitopes present on cleaved cytokeratin 18 and caspase 3 have been most frequently used for the detection of apoptosis. Willingham and colleagues (Barrett et al. 2001) demonstrated the utility of using an anti-cleaved cytokeratin 18 antibody for the specific detection of apoptotic cells in culture and in archived paraffin embedded tissues. Specifically stained cells showed the shrunken condensed morphology expected of an apoptotic cell. Naturally, this antibody can only be employed for cells and tissues that express cytokeratin 18.
Likewise, Gown and Willingham (2002) used a commercially available antibody raised against cleaved caspase 3 to specifically label apoptotic cells in culture and paraffin embedded tissues. Given the ubiquitous presence of caspase 3 in most cells, and the formation of the neoepitope following activation of the apoptotic cascade, this reagent should prove to be a most useful and specific label for cells undergoing caspase-mediated apoptotic cell death.
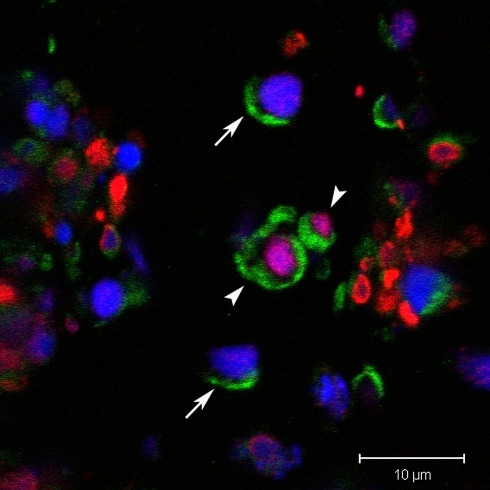
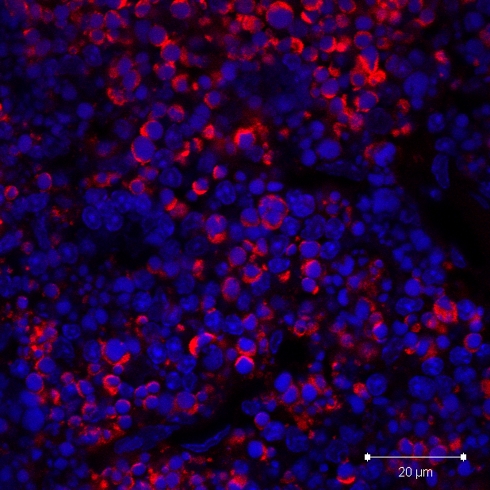
Recently, we combined immunostaining for anti-cleaved caspase 3 with the formamide denaturation/ssDNA method for the detection of two specific apoptosis probes in the same tissue section using confocal scanning laser microscopy (Taatjes et al. 2007). In dual stained cells in paraffin sections from infarcted mouse ventricle, the anti-cleaved caspase 3 antibody labeled the cytoplasm, whereas the anti-ssDNA antibody stained the cell nucleus. Cells were observed stained singly with either of the two probes, suggesting that cells in different phases of the apoptotic cascade may be identified by their staining properties with these antibodies. We include sections from the thymus of dexamethasone-treated mice as a positive control (Figs. 2, 3) for the apoptosis assays (Taatjes et al. 2007), since steroid hormone treatment has been shown by transmission electron microscopy to induce thymic apoptosis (Wyllie et al. 1980).
Fig. 2.
Paraffin section of thymus from mouse treated with dexamethasone and stained with anti-cleaved caspase 3 antibody (green), and anti-ssDNA antibody (red). Confocal microscopy demonstrates doubly stained thymic cells (arrowheads). Note that the cleaved caspase 3 is localized to the cytoplasm, whereas the ssDNA is localized to the nucleus. Some cells only stained by cleaved caspase 3 antibody are indicated by solid arrows. The nucleus is labeled with DAPI (blue)
Fig. 3.
Paraffin section of thymus from mouse treated with dexamethasone and stained with anti-cleaved caspase 3 antibody (red). Confocal microscopy image shows intensely stained cells in the cortex, with neighboring unstained cells. Nuclei have been stained with DAPI (blue). This represents a good positive control for the anti-cleaved caspase 3 antibody technique
Annexin V
Some markers for apoptotic cells require the use of live cells. For instance, early in the apoptotic cascade of events, phosphatidyl serine residues flip from the internal to external side of the cells’ plasma membrane. Accordingly, annexin V, which binds specifically to phosphatidyl serine residues has been used in this manner as a specific probe for apoptosis (Koopman et al. 1994). However, as pointed out by Willingham (1999), this assay does not seem to label all apoptotic cells, and moreover will stain necrotic cells once the plasma membrane is compromised.
Other probes for apoptosis
Bilyy et al. (2004, 2005) and Bilyy and Stoika (2003) have championed the examination of plasma membrane glycoconjugates as indicators of apoptosis. They have detected an augmentation in the expression of α-d-mannose- and β-d-galactose-containing glycoproteins in the plasma membrane in response to apoptotic signals. These glycoconjugates can be visualized using a battery of lectins (carbohydrate-binding proteins of nonimmune origin), in conjunction with fluorescence- or peroxidase-conjugated probes for light microscopy. Unlike most of the markers for apoptosis which bind to cytoplasmic or nuclear constituents, these plasma membrane-specific probes offer the means to assess activity occurring at the cell surface during the apoptotic cascade (similar to annexin V binding to phosphatidylserine). Given the enormous diversity in carbohydrate structure, and the role played by carbohydrates in a variety of cell–cell interactions, it is feasible that they may offer more insight into cellular mechanisms of apoptosis in the future.
An early event in the apoptotic cascade is the release of cytochrome c from the outer mitochondrial membrane. Accordingly, the intracellular localization of cytochrome c could serve as an indicator for apoptosis. Jones and colleagues (Jiang et al. 1999) used this approach to show that in a control (untreated) osteosarcoma cell line, immunostaining for cytochrome c (as assessed by confocal microscopy) was observed in a punctuate cytoplasmic pattern, coincident with mitochondrial staining by MitoTracker Red. When cells were treated with staurosporine to induce apoptosis, the immunolabel for cytochrome c was found to be diffusely present throughout the cytoplasm, consistent with its release from the mitochondria. We have also used immunolabeling for cytochrome c as a marker for mitochondria in intact cells (Fung et al. 1998), and its further application as a potential marker for apoptotic cells dependent upon its intracellular staining pattern should be further explored.
Apoptosis-inducing factor (AIF) is a mitochondrial membrane protein involved in caspase-independent cell death (Daugas et al. 2000). Immunostaining for AIF revealing a translocation from the mitochondrion to the nucleus has been used as a marker for caspase-independent apoptosis (Daugas et al. 2000; Susin et al. 2000). However, Daugas et al. (2000) have shown that the nuclear translocation of AIF also occurs during a necrotic response, so other methods should be employed in conjunction with AIF immunostaining to differentiate between these two modes of cell death. Indeed, we have used immunostaining of AIF translocation from the cytoplasm (mitochondrion) to the nucleus, together with cell morphological assessment by phase contrast microscopy, as an indicator of apoptosis in lung C10 cells in response to asbestos exposure (Yuan et al. 2004).
Detection of apoptosis by video optical microscopy
Morphological assessment of cultured cells by video microscopy has been successfully used to follow the cellular changes induced by apoptosis. Cells treated with apoptosis-inducing agents are imaged with a light microscope, typically operating in phase contrast or differential interference contrast mode, and time-lapse recording is made with a ccd camera connected to a video recorder (Collins et al. 1997; Willingham 1999). A major advantage of this type of temporal microscopy is the ability to document cellular morphological changes associated with apoptosis with the time course of the event. Differences in cellular changes observed in necrosis versus apoptosis can be well documented, and the slow course of apoptosis is well suited to time-lapse recording (Collins et al. 1997). Using phase contrast microscopy in conjunction with video recording has produced dramatic footage of cells exposed to apoptotic agents rounding up and displaying intense surface blebbing (Collins et al. 1997). Of course, the full apoptotic cascade may require up to 24 h in cultured cells, so movies can also be made by merging files from single ccd camera images and exporting them in a movie-type format. Thorburn and colleagues (Morgan et al. 2002) used a combination of fluorescence and phase contrast microscopy to image apoptosis in live HeLa cells. A Hamamatsu digital camera was used to acquire combination images every 30 min over a 24 h period. The merged image series were exported in Quicktime format, and a representative movie sequence can be seen in the online supplemental material referenced in the manuscript (Morgan et al. 2002). This type of analysis is particularly useful for determining the temporal sequence of morphological events associated with the introduction of an apoptosis-inducing agent to cell culture media.
Detection of apoptosis by laser scanning cytometry
Laser scanning cytometry (LSC) is a method which combines the efficiency and precision of flow cytometry with the morphological characterization ability of a microscope-based imaging system (Kamentsky and Kamentsky 1991; Kamentsky et al. 1997; Taatjes et al. 2001; Pozarowski et al. 2006). The ability to examine the morphological appearance of cells following flow cytometry-like analysis overcomes the limitations posed by flow cytometry itself in the analysis of apoptosis; that is, apoptotic cells appear in the subdiploid peak of a flow cytometric histogram, together with apoptotic bodies and fluorescence debris, and cannot be directly visualized. Darzynkiewicz and colleagues have pioneered the use of LSC for analysis of apoptosis, and the reader is referred to their excellent review for further details (Bedner et al. 1999). In particular, they have used LSC in combination with a variety of apoptosis markers, including annexin-V, markers of mitochondrial transmembrane potential (rhodamine 123 and DiOC6), nuclear markers (DAPI, 7-ADD, PI), and markers of DNA strand breaks. Importantly, in all of these investigations, they emphasized the combination of fluorescence staining and morphological determination with LSC as being of utmost importance for the identification of apoptotic cells.
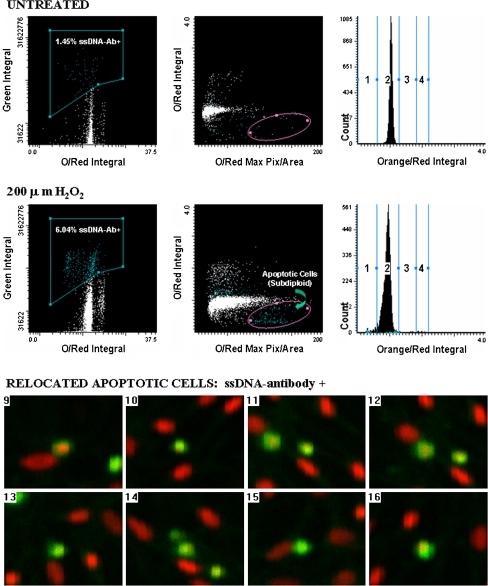
We have also used LSC for the cytometric and morphological characterization of apoptotic cultured mouse lung epithelial cells (C10 cell line) (Taatjes et al. 2001). Cells were exposed to hydrogen peroxide for 24 h to induce apoptosis, followed by staining with the anti-ssDNA antibody and an Oregon green-conjugated secondary antibody to stain apoptotic cells. All cells were then stained with propidium iodide and analyzed by LSC. Scattergrams from the analysis (Fig. 4) showed that hydrogen peroxide induced an increase in apoptotic cells, evinced by an increase in the population of cells depicted in the subdiploid area. Cells located in this subdiploid portion were then visually confirmed through relocation using the automated stage and microscope of the LSC. These cells were found to be labeled with the anti-ssDNA antibody, and showed the condensed morphological characteristics of apoptotic cells (Fig. 4).
Fig. 4.
Laser scanning cytometry determination of apoptosis in lung epithelial cells using anti-ssDNA antibody. Upper panel scattergrams showing percentage of ssDNA-positive cells (green integral vs. orange/red integral), the apoptotic fraction (orange/red integral vs. orange/red maximum pixel), and a DNA histogram for untreated C10 cells. Middle panel scattergrams showing percentage of ssDNA-positive cells (green integral vs. orange/red integral), the apoptotic fraction (orange/red integral vs. orange/red maximum pixel), and a DNA histogram for C10 cells treated with 200 μM H2O2 for 24 h. Note the increased number of cells ssDNA-positive, and the enhanced subdiploid DNA fraction in cells treated with H2O2 as compared with sham controls. Bottom panel cell relocation feature of LSC demonstrated for eight cells. Cells within the subdiploid fraction (elliptical region highlighted on scattergram in middle panel) were relocated and visually confirmed as apoptotic by morphological appearance and positive staining with the ssDNA antibody. Reprinted with permission from BioTechniques (Taatjes et al. 2001)
Detection of apoptosis by atomic force microscopy (AFM)
The atomic force microscope is a member of the scanning probe microscope family (Binnig et al. 1986). This instrument utilizes a tactile sensing system, in which a probe (typically composed of silicon nitride) is attached to the end of a cantilever and raster scanned in x- and y-directions across a specimen through the activity of a piezoelectric tube to which the cantilever assembly is mounted (Montigny et al. 2006). A laser beam is directed onto the surface of the cantilever, and the reflected beam is directed up to a four quadrant photodiode, which converts laser movement on the surface into a measurable electrical voltage change. This voltage change is then translated via computer interface into height and amplitude information about the specimen. The complexity of the system belies the simplicity of specimen preparation; fixed or unfixed cells grown on glass coverslips can be readily imaged by AFM without further processing, or importantly introduction of artifacts. Although AFM initially found its greatest utility in the imaging of isolated molecules and their dynamic interactions (Hansma and Ho 1994; Taatjes et al. 1997; Rand et al. 2003), more recently whole cell studies have become more prevalent (Madl et al. 2006). With this development, and since AFM typically images surface phenomena, it was a natural extension to use this technology to image cells undergoing apoptosis. Hessler et al. (2005) used AFM to image and measure the cellular volume decrease occurring during apoptosis. They treated the human epidermoid carcinoma cell line KB with staurosporine to induce apoptosis, and determined that after a 3-h exposure, the total cell height decreased 32%, the total cell volume was decreased by 50%, and the cell diameter changed by 15%. Given the temporal experimental capabilities of AFM, it seems as if this would be an ideal modality, in combination with other imaging methods, to investigate the cellular dynamics accompanying the apoptotic cascade, as elegantly shown by Hessler et al.
An example of the importance of delineating apoptosis: cell death in the heart
In this brief review, we have attempted to highlight some of the morphological and cytochemical techniques currently in use to classify cell death as an apoptotic response. There are a wide ranging variety of methods to choose from, rendering the choice a matter of the experimental system employed and personal preference. However, the determination of cell death as apoptotic, or through another mode should not be viewed simply as a matter of semantics. For instance, an example from clinical cardiovascular disease will illustrate the importance of the precise determination of the mode of cell death. For several decades the conceptual basis underlying treatment of acute coronary syndromes has been that limitation of myocardial oxygen requirements by diminution of afterload and heart rate coupled with augmentation of myocardial oxygen supply through restoration of myocardial perfusion are the pivotal components (Maroko et al. 1972; Shell and Sobel 1974; Sobel and Braunwald 1980). The potential value of “tissue protective” interventions including favorable modification of myocardial metabolism has long been recognized as well (Maroko et al. 1972).
Several different types of cell death have been characterized in the heart including coagulation necrosis, the typical manifestation of ischemia sufficient to render myocardium nonviable, contraction band necrosis typical of ischemia followed by reperfusion and presumably mediated in part by calcium overload, autophagy typical of cells insulted by deprivation of nutrients that consequently consume their own components, and apoptosis. The extent to which this final modality, programmed cell death or apoptosis, contributes to overall cardiomyocyte cell death is far from a trivial matter (Sobel et al. 2007). If apoptosis plays a substantial role as a contributor to total cardiomyocyte cell death and if it can be modified or prevented despite the presence of ischemia, the potential, favorable impact of tissue protective interventions is likely to be considerable. Conversely, if apoptosis plays a small role in overall cardiomyocyte cell death consequent to ischemia, an anti-apoptotic strategy would likely be ineffective. Unfortunately, the implication of apoptosis in cardiomyocyte cell death in studies in laboratory animals has been based virtually exclusively on results with the TUNEL assay (see above for issues of nonspecificity of this assay). As noted by Takemura and Fujiwara (2006), there is no unequivocal evidence that apoptosis occurs in zones of infarction.
As described above, we have recently sought to quantify apoptosis in myocardium and have developed a novel dual staining method for confocal microscopic identification of apoptotic cells (Taatjes et al. 2007). Our initial findings indicate that despite massive necrosis induced by ischemia in a transgenic strain of mice deficient in plasminogen activator inhibitor type-1 (PAI-1) hardly any cells were apoptotic as judged from positive reactions to antibodies against both cleaved caspase 3 and ssDNA. Others have observed an absence of apoptosis as judged from electron microscopic criteria (Ohno et al. 1998) despite TUNEL positivity in hearts of animals subject to transitory ischemia followed by reperfusion for several hours.
In view of these considerations, it appears important to determine definitively whether the presence of ssDNA coupled with the presence of caspace 3 cleavage product does, indeed, map isomorphically with the cells that exhibit electron microscopic criteria of apoptosis. Furthermore, it will be important to determine the extent to which the positivity of each of these criteria of apoptosis occurs in hearts of experimental animals subjected to ischemia for selected intervals followed by reperfusion for selected intervals as well as in those from animals subjected to myocardial ischemia that is persistent and to define the time course of the evolution of apoptosis, if present, under both conditions. It is, of course, possible that apparently scanty apoptosis at any given time could be of considerable biological importance since apoptotic cells are rapidly destroyed or removed by macrophages. Thus, if a given percentage of cells, though a small one, were exhibiting apoptosis repetitively, the cumulative amount could be substantial. Accordingly, both the temporal and the spatial evolutions of apoptosis associated with myocardial ischemia and infarction require elucidation. This is particularly important because apoptosis in the heart may be secondary to necrosis rather than a primary mode of cell death, a distinction that needs to be clarified because of the disparate therapeutic implications of the two possibilities. Only when armed with information needed to characterize apoptosis in the heart more definitively will it be possible to delineate the potentially beneficial effects of tissue protective interventions designed to retard or prevent apoptotic cell death in cardiomyocytes subjected to ischemia.
Concluding remarks
Determining the mechanisms of cell death is an area of intense research interest in a wide range of fields including cancer biology, pathology, and toxicology. Certainly, the role of apoptosis as a mode of “programmed cell death” during embryonic development has been unequivocally established (Penaloza et al. 2006). We hope that this brief review has highlighted a variety of imaging modalities suitable for differentiating amongst the different modes of cell death. What should be apparent is that rarely will one method be sufficient for such a determination. Each imaging method has its own strengths with respect to the morphological information it can provide. Such morphological information, combined with biochemical or immunohistochemical assessment can provide a strong indication of the specific mode of cell death observed. The key, in our estimation, is a multiparameter observational approach.
Accordingly, the take-home message for any morphological or cytochemical investigation of apoptosis is to employ more than one detection method and run multiple controls.
Acknowledgments
We would like to thank Dr. Brooke Mossman for her many years collaborating on apoptosis, in the lung.
References
- Abbate A, De Falco M, Morales C, Gelpi R, Prisco M, De Luca A, Palleiro J, Fedele V, Feroce F, Baldi F, Vetrovec GW, Baldi A (2007) Electron microscopy characterization of cardiomyocyte apoptosis in ischemic heart disease. Int J Cardiol 114:118–120 [DOI] [PubMed]
- Adams JM, Cory S (1998) The Bcl-2 protein family: arbiters of cell survival. Science 281:1322–1326 [DOI] [PubMed]
- Barrett KL, Willingham JM, Garvin AJ, Willingham MC (2001) Advances in cytochemical methods for detection of apoptosis. J Histochem Cytochem 49:821–832 [DOI] [PubMed]
- Bedner E, Li X, Gorczyca W, Melamed MR, Darzynkiewicz Z (1999) Analysis of apoptosis by laser scanning cytometry. Cytometry 35:181–195 [DOI] [PubMed]
- Bilyy RO, Stoika RS (2003) Lectinocytochemical detection of apoptotic murine leukemia L1210 cells. Cytometry A 56A:89–95 [DOI] [PubMed]
- Bilyy RO, Antonyuk VO, Stoika RS (2004) Cytochemical study of role of α-d-mannose- and β-d-galactose-containing glycoproteins in apoptosis. J Mol Histol 35:829–838 [DOI] [PubMed]
- Bilyy R, Kit Y, Hellman U, Tryndyak V, Kaminskyy V, Stoika R (2005) In vivo expression and characteristics of novel α-d-mannose-rich glycoprotein markers of apoptotic cells. Cell Biol Int 29:920–928 [DOI] [PubMed]
- Binnig G, Quate CF, Gerber C (1986) Atomic force microscope. Phy Rev Lett 56:930–933 [DOI] [PubMed]
- Buder-Hoffman S, Palmer C, Vacek P, Taatjes D, Mossman B (2001) Different accumulation of activated extracellular signal-regulated kinases (ERK1/2) and role in cell-cycle alterations by epidermal growth factor, hydrogen peroxide, or asbestos in pulmonary epithelial cells. Am J Respir Cell Mol Biol 24:405–413 [DOI] [PubMed]
- Cain K, Bratton SB, Langlais C, Walker G, Brown DG, Sun XM, Cohen GM (2000) Apaf-1 oligomerizes into biologically active approximately 700-kDa and inactive approximately 1.4-MDa apoptosome complexes. J Biol Chem 275:6067–6070 [DOI] [PubMed]
- Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, Skoda-Smith S, Atkinson TP, Straus SE, Lenardo MJ (2002) Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature 419:395–399 [DOI] [PubMed]
- Collins JA, Schandl CA, Young KK, Vesely J, Willingham MC (1997) Major DNA fragmentation is a late event in apoptosis. J Histochem Cytochem 45:923–934 [DOI] [PubMed]
- Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prevost M-C, Leber B, Andrews D, Penninger J, Kroemer G (2000) Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J 14:729–739 [PubMed]
- Frankfurt OS (1994) Detection of apoptosis in leukemic and breast cancer cells with monoclonal antibody to single-stranded DNA. Anticancer Res 14:1861–1870 [PubMed]
- Frankfurt OS, Krishan A (2001) Identification of apoptotic cells by formamide-induced DNA denaturation in condensed chromatin. J Histochem Cytochem 49:369–378 [DOI] [PubMed]
- Frankfurt OS, Robb JA, Sugarbaker EV, Villa L (1996) Monoclonal antibody to single-stranded DNA is a specific and sensitive cellular marker of apoptosis. Exp Cell Res 226:387–397 [DOI] [PubMed]
- Fung H, Kow YW, van Houten B, Taatjes DJ, Hatahet Z, Yanssen YMW, Vacek P, Faux SP, Mossman BT (1998) Asbestos increases mammalian AP-endonuclease gene expression, protein levels, and enzyme activity in mesothelial cells. Cancer Res 58:189–194 [PubMed]
- Galluzzi L, Maiuri MC, Vitale I, Zischka H, Castedo M, Zitvogel L, Kroemer G (2007) Cell death modalities: classifications and pathophysiological implications. Cell Death Differ 14:1237–1243 [DOI] [PubMed]
- Germain F, Fernandez E, De La Villa P (2007) Morphological signs of apoptosis in axotomized ganglion cells of the rabbit retina. Neuroscience 144:898–910 [DOI] [PubMed]
- Gown AM, Willingham MC (2002) Improved detection of apoptotic cells in archival paraffin sections: immunohistochemistry using antibodies to cleaved caspase 3. J Histochem Cytochem 50:449–454 [DOI] [PubMed]
- Groos S, Reale E, Luciano L (2003) General suitability of techniques for in situ detection of apoptosis in small intestinal epithelium. Anat Rec 272:503–513 [DOI] [PubMed]
- Hansma H, Ho J (1994) Biomolecular imaging with the atomic force microscope. Annu Rev Biophys Biomol Struct 23:115–139 [DOI] [PubMed]
- Hessler JA, Budor A, Putchakayala K, Mecke A, Rieger D, Banaszak Holl MM, Orr BG, Bielinska A, Beals J, Baker J Jr (2005) Atomic force microscopy study of early morphological changes during apoptosis. Langmuir 21:9280–9286 [DOI] [PubMed]
- Ito Y, Shibata M-A, Kusakabe K, Otsuki Y (2006) Method of specific detection of apoptosis using formamide-induced DNA denaturation assay. J Histochem Cytochem 54:683–692 [DOI] [PubMed]
- Jiang S, Cai J, Wallace DC, Jones DP (1999) Cytochrome c-mediated apoptosis in cells lacking mitochondrial DNA. Signaling pathway involving release and caspase 3 activation is conserved. J Biol Chem 274:29905–29911 [DOI] [PubMed]
- Jung M, Davis WP, Taatjes DJ, Churg A, Mossman BT (2000) Asbestos and cigarette smoke cause increased DNA strand breaks and necrosis in bronchiolar epithelial cells in vivo. Free Radic Biol Med 28:1295–1299 [DOI] [PubMed]
- Jung M, Grunberg S, Timblin C, Buder-Hoffman S, Vacek P, Taatjes DJ, Mossman BT (2004) Paclitaxel and vinorelbine cause synergistic increases in apoptosis but not in microtubular disruption in human lung adenocarcinoma cells (A-549). Histochem Cell Biol 121:115–121 [DOI] [PubMed]
- Kamentsky LA, Kamentsky LD (1991) Microscope-based multiparameter laser scanning cytometer yielding data comparable to flow cytometry data. Cytometry 12:381–387 [DOI] [PubMed]
- Kamentsky LA, Burger DE, Gershman RJ, Kamentsky LD, Luther E (1997) Slide-based laser scanning cytometry. Acta Cytol 41:123–143 [DOI] [PubMed]
- Kanoh M, Takemura G, Misao J, Hayakawa Y, Aoyama T, Nishigaki K, Noda T, Fujiwara T, Fukuda K, Minatoguchi S, Fujiwara H (1999) Significance of myocytes with positive DNA end-labeling (TUNEL) in hearts with dilated cardiomyopathy. Circulation 99:2757–2764 [DOI] [PubMed]
- Kerr JF, Wyllie AH, Currie AR (1972) Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer 26:239–257 [DOI] [PMC free article] [PubMed]
- Kockx MM, Muhring J, Knaapen MW, De Meyer GR (1998) RNA synthesis and splicing interferes with DNA in situ end labeling techniques used to detect apoptosis. Am J Pathol 152:885–888 [PMC free article] [PubMed]
- Koopman G, Reutelingsperger CP, Kuijten GA, Keehnen RM, Pasla ST, van Oers MH (1994) Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84:1415–1420 [PubMed]
- Krueger A, Fas SC, Baumann S, Krammer PH (2003) The role of CD95 in the regulation of peripheral T-cell apoptosis. Immunol Rev 193:58–69 [DOI] [PubMed]
- Labat-Moleur F, Guillermet C, Lorimier P, Robert C, Lantuejoul S, Brambilla E, Negoescu A (1998) TUNEL apoptotic cell detection in tissue sections: critical evaluation and improvement. J Histochem Cytochem 46:327–334 [DOI] [PubMed]
- Li H, Zhu H, Xu CJ, Yuan J (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94:491–501 [DOI] [PubMed]
- Linette GP, Li Y, Roth K, Korsmeyer SJ (1996) Cross talk between cell death and cell cycle progression: BCL-2 regulates NFAT-mediated activation. Proc Natl Acad Sci USA 93:9545–9552 [DOI] [PMC free article] [PubMed]
- Madl J, Rhode S, Stangl H, Stockinger H, Hinterdorfer P, Schutz GJ, Kada G (2006) A combined optical and atomic force microscope for live cell investigations. Ultramicroscopy 106:645–651 [DOI] [PubMed]
- Maroko PR, Libby P, Sobel BE, Bloor CM, Sybers HD, Shell WE, Covell Jw, Braunwald E (1972) Effect of glucose–insulin–potassium infusion on myocardial infarction following experimental coronary artery occlusion. Circulation 45:1160–1172 [DOI] [PubMed]
- Montigny WJ, Quinn AS, Wu X-X, Bovill EG, Rand JH, Taatjes DJ (2006) Atomic force microscopy in the study of macromolecular interactions in hemostasis and thrombosis: utility for investigation of the antiphospholipid syndrome. In: Jena BJ, Horber JKH (eds) Force microscopy: applications in biology and medicine, Wiley, Hoboken, pp 267–286
- Morgan M, Thorburn J, Pandolfi PP, Thorburn A (2002) Nuclear and cytoplasmic shuttling of TRADD induces apoptosis via different mechanisms. J Cell Biol 157:975–984 [DOI] [PMC free article] [PubMed]
- Negoescu A, Lorimier P, Labat-Moleur F, Drouet C, Robert C, Guillermet C, Brambilla C, Brambilla E (1996) In situ apoptotic cell labeling by the TUNEL method: improvement and evaluation on cell preparation. J Histochem Cytochem 44:959–968 [DOI] [PubMed]
- Ohno M, Takemura G, Ohno A, Misao J, Hayakawa Y, Minatoguchi S, Fujiwara T, Fujiwara H (1998) Apoptotic myocytes in infarct area in rabbit hearts may be oncotic myocytes with DNA fragmentation: analysis by immunogold electron microscopy combined with in situ nick end-labeling. Circulation 98:1422–1430 [DOI] [PubMed]
- Penaloza C, Lin L, Lockshin RA, Zakeri Z (2006) Cell death in development: shaping the embryo. Histochem Cell Biol 126:149–158 [DOI] [PubMed]
- Pozarowski P, Holden E, Darzynkiewicz Z (2006) Laser scanning cytometry: principles and applications. In: Taatjes DJ, Mossman BT (eds) Methods in molecular biology. Cell imaging techniques: methods and protocols, vol 319. Humana Press, Totowa, pp 165–192 [DOI] [PMC free article] [PubMed]
- Rand JH, Wu X-X, Quinn AS, Chen PP, McCrae KR, Bovill EG, Taatjes DJ (2003) Human monoclonal antiphospholipid antibodies disrupt the annexin A5 anticoagulant shield on phospholipid bilayers. Evidence from atomic force microscopy and functional assay. Am J Pathol 163:1193–1200 [DOI] [PMC free article] [PubMed]
- Salmena L, Lemmers B, Hakem A, Matysiak-Zablocki E, Murakami K, Au PY, Berry DM, Tamblyn L, Shehabeldin A, Migon E, Wakeham A, Bouchard D, Yeh WC, McGlade JC, Ohashi PS, Hakem R (2003) Essential role for caspase 8 in T-cell homeostasis and T-cell-mediated immunity. Genes Dev 17:883–895 [DOI] [PMC free article] [PubMed]
- Shell WE, Sobel BE (1974) Protection of jeopardized ischemic myocardium by reduction of ventricular afterload. N Engl J Med 291:481–486 [DOI] [PubMed]
- Shukla A, Jung M, Stern M, Fukagawa NK, Taatjes DJ, Sawyer D, van Houten B, Mossman BT (2003) Asbestos induces mitochondrial DNA damage and dysfunction linked to the development of apoptosis. Am J Physiol Lung Cell Mol Physiol 285:L1018–L1025 [DOI] [PubMed]
- Sloop GD, Roa JC, Delgado AG, Balart JT, Hines MO, Hill JM (1999) Histologic sectioning produces TUNEL reactivity. A potential cause of false-positive staining. Arch Pathol Lab Med 123:529–532 [DOI] [PubMed]
- Sobel BE, Braunwald E (1980) Management of acute myocardial infarction. In: Braunwald E (ed) Heart disease. WB Saunders, Philadelphia, pp 1353–1386
- Sobel BE, Tarikuz Zaman AKM, Budd RC, Schneider DJ, Taatjes DJ (2007) Attenuation of apoptosis and the eye of the beholder. Coron Artery Dis (in press) [DOI] [PubMed]
- Strasser A (2001) BH3-only members of the Bcl-2 family are critical inducers of apoptosis and preclude autoimmunity. Abstract 003 Keystone Symposium on Molecular Mechanisms of Apoptosis, 16–22 January 2001, Keystone, CO
- Susin SA, Daugas E, Ravagnan L, Samejima K, Zamzami N, Loeffler M, Costantini P, Ferri KF, Irinopoulou T, Prevost M-C, Brothers G, Mak TW, Penninger J, Earnshaw WC, Kroemer G (2000) Two distinct pathways leading to nuclear apoptosis. J Exp Med 192:571–579 [DOI] [PMC free article] [PubMed]
- Taatjes DJ, Quinn AS, Jenny RJ, Hale P, Bovill EG, McDonagh J (1997) Tertiary structure of the hepatic cell protein fibrinogen in fluid revealed by atomic force microscopy. Cell Biol Int 21:715–726 [DOI] [PubMed]
- Taatjes DJ, Palmer CJ, Pantano C, Buder-Hoffmann S, Cummins A, Mossman BT (2001) Laser-based microscopic approaches: application to cell signaling in environmental lung disease. Biotechniques 31:880–894 [DOI] [PubMed]
- Taatjes DJ, Wadsworth MP, Tarikuz Zaman AKM, Schneider DJ, Sobel BE (2007) A novel dual staining method for identification of apoptotic cells reveals a modest apoptotic response in infarcted mouse myocardium. Histochem Cell Biol 128:275–283 [DOI] [PubMed]
- Takemura G, Fujiwara H (2006) Morphological aspects of apoptosis in heart diseases. J Cell Mol Med 10:56–75 [DOI] [PMC free article] [PubMed]
- Tateyama H, Tada T, Hattori H, Murase T, Li WX, Eimoto T (1998) Effects of prefixation and fixation times on apoptosis detection by in situ end-labelling of fragmented DNA. Arch Pathol Lab Med 51:252–253 [PubMed]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, Moritz RL, Simpson RJ, Vaux DL (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102:43–53 [DOI] [PubMed]
- Walker JA, Quirke P (2001) Viewing apoptosis through a “TUNEL”. J Pathol 195:275–276 [DOI] [PubMed]
- Watanabe M, Hitomi M, van der Wee K, Rothenberg F, Fisher SA, Zucker R, Svoboda KKH, Goldsmith EC, Heiskanen KM, Nieminen A-L (2002) The pros and cons of apoptosis assays for use in the study of cells, tissues, and organs. Microsc Microanal 8:375–391 [DOI] [PubMed]
- White C, Li C, Yang J, Petrenko NB, Madesh M, Thompson CB, Foskett JK (2005) The endoplasmic reticulum gateway to apoptosis by Bcl-X(L) modulation of the InsP3R. Nat Cell Biol 7:1021–1028 [DOI] [PMC free article] [PubMed]
- Wikstrom JD, Katzman SM, Mohamed H, Twig G, Graf SA, Heart E, Molina AJ, Corkey BE, de Vargas LM, Danial NN, Collins S, Shirihai OS (2007) Beta-cell mitochondria exhibit membrane potential heterogeneity that can be altered by stimulatory or toxic fuel levels. Diabetes 56:2569–2578 [DOI] [PubMed]
- Willingham MC (1999) Cytochemical methods for the detection of apoptosis. J Histochem Cytochem 47:1101–1109 [DOI] [PubMed]
- Wyllie AH, Kerr JFR, Currie AR (1980) Cell death: the significance of apoptosis. Int Rev Cytol 68:251–306 [DOI] [PubMed]
- Yuan Z, Taatjes DJ, Mossman BT, Heintz NH (2004) The duration of nuclear extracellular signal-regulated kinase 1 and 2 signaling during cell cycle reentry distinguishes proliferation from apoptosis in response to asbestos. Cancer Res 64:6530–6536 [DOI] [PubMed]