Abstract
The 3;8 chromosomal translocation, t(3;8)(p14.2;q24.1), was described in a family with classical features of hereditary renal cell carcinoma. Previous studies demonstrated that the 3p14.2 breakpoint interrupts the fragile histidine triad gene (FHIT) in its 5′ noncoding region. However, evidence that FHIT is causally related to renal or other malignancies is controversial. We now show that the 8q24.1 breakpoint region encodes a 664-aa multiple membrane spanning protein, TRC8, with similarity to the hereditary basal cell carcinoma/segment polarity gene, patched. This similarity involves two regions of patched, the putative sterol-sensing domain and the second extracellular loop that participates in the binding of sonic hedgehog. In the 3;8 translocation, TRC8 is fused to FHIT and is disrupted within the sterol-sensing domain. In contrast, the FHIT coding region is maintained and expressed. In a series of sporadic renal carcinomas, an acquired TRC8 mutation was identified. By analogy to patched, TRC8 might function as a signaling receptor and other pathway members, to be defined, are mutation candidates in malignant diseases involving the kidney and thyroid.
The 3;8 chromosomal translocation, t(3;8)(p14.2;q24.1), was described in a family with classical features of hereditary renal cell carcinoma (RCC), i.e., autosomal dominant inheritance, and early onset and bilateral disease (1). The translocation and RCC segregated concordantly, and a follow-up analysis reported the occurrence of thyroid cancer in two translocation carriers with RCC (2). Frequent 3p loss of heterozygosity in sporadic RCC led to the initial assumption that a critical tumor suppressor gene (TSG) would be located at 3p14. However, identification of the von Hippel-Lindau (VHL) gene at 3p25 (3), which frequently is mutated in RCC, provided an alternative explanation for at least some observed 3p loss of heterozygosity. Also, van den Berg and Buys (4) subsequently have reported that region 3p21 may be a target involved in the malignant progression of renal tumors.
Within 3p14, Ohta et al. (5) identified the fragile histidine triad gene (FHIT), which was interrupted in its 5′ untranslated region by the 3;8 translocation. The human gene, like its yeast homologue, encodes diadenosine-5′,5′′′-P1,P3-triphosphate hydrolase activity (6), an unprecedented TSG function. Although several reports have described FHIT alterations in diverse carcinomas by using nested reverse transcriptase–PCR (RT-PCR) (5, 7–10), other results have been contradictory (11–17). In fact, most FHIT abnormalities occur in the presence of wild-type transcripts and result from low-abundance splicing alterations, similar to those seen for TSG101 (13, 18). We described a series of 3p14 homozygous deletions, primarily in cervical and colorectal carcinoma cell lines, which coincided with FRA3B, the most inducible fragile region in the genome (14). Interestingly, p53 alterations appeared to be a prerequisite. The proximity of FHIT exon 5 with FRA3B suggested that its loss might be primarily related to genomic instability in contrast to negative selection during tumor development (14).
These results made FHIT an unlikely, or at least suspect, causative gene in the hereditary t(3;8) family and led us to continue a search for alternatives. We noted that FHIT, in one parotid adenoma, underwent fusion with the high mobility group protein gene (HMGIC), the causative gene in a variety of benign tumors (19). In fact, other HMGIC translocations with unrelated genes indicated that FHIT could be a bystander in this fusion. However, it suggested that the 3;8 translocation might fuse FHIT to an alternative candidate gene on chromosome 8. By using 5′ rapid amplification of cDNA ends (RACE) (20), we were able to identify a gene, TRC8, which was related to the Drosophila segment polarity gene patched. In addition to being the receptor for sonic hedgehog (21, 22), patched is responsible for both hereditary and sporadic basal cell carcinomas as well as medulloblastomas (23–25). Together with the identification of a TRC8 mutation in a sporadic renal carcinoma, our results indicate that TRC8 may define an additional pathway of mutations leading to the development of renal and thyroid cancer.
MATERIALS AND METHODS
Tumors, Cell Lines, and Genomic Clones.
Tumor cell lines were obtained from the American Type Culture Collection, and somatic cell hybrids were generated by us previously (26). The hybrids TL12–8 and 3;8/4–1 contain the der(3) and der(8) chromosomes, respectively, from the t(3;8) lymphoblastoid cell line TL9944 (without either normal 3 or 8 chromosomes). The human lymphoblastoid line AG4103 served as a normal control. Isolation of DNA and RNA was performed by using standard methods. The HD-7 genomic phage clone carrying the 3;8 translocation breakpoint from the der(8) chromosome was isolated from a library prepared from the TL9944 cell line in λ FIXII (Stratagene). A chromosome 3 probe (λ4040; ref. 27), which maps just distal to the 3;8 breakpoint and detects the rearrangement, was used for screening this library.
RACE.
5′ RACE was performed essentially as described (28). First-strand cDNA synthesis was primed by using a FHIT exon 4-specific primer R1 (TCAGAAGACTGCTACCTCTTCG), followed by dCTP tailing with terminal deoxynucleotidyl transferase. Primary amplification used the AAP 5′ RACE primer together with a nested FHIT exon 4-specific primer R2 (TCAGTGGCAGGATGCACAG). Second-round PCR used primer AUAP with a second nested FHIT exon 4-specific primer R3 (GGTCTAAGCAGGCAGGTATTC). Products were cloned into a T-vector (pBluescript II KS), analyzed by hybridization with additional internal FHIT oligonucleotides F4 (TGGAAGGGAGAGAAAGAG) and R4 (GGTATTCCTAGGATAC), and sequenced.
Chromosomal Localization and RT-PCR Analysis.
PCR mapping used TRC8-specific primers R-M (GCCCTGCCTTTACATCATCGAC) and F-O (AGATCTGGAGCACGATGCAGAAC). PCRs were performed under touch-down annealing conditions with Perkin–Elmer Buffer II and Promega AmpliTaq DNA polymerase. Touch-down annealing temperatures started at 70°C and ended at 60°C (ΔT of −0.5°C per cycle) for 20 cycles, followed by 15 cycles at 60°C. Products generated from 10 to 40 ng of template were separated on a 2.0% agarose gel. cDNA synthesis used random hexamer primers along with Superscript II (Life Technologies, Gaithersburg, MD). Subsequent PCRs were performed as above, except touch-down annealing temperatures were adjusted to 65°C–55°C. The EMR primer, specific for the 3′ portion of TRC8, was TCTTGTTAGCCAAAAGACTCG, whereas the F1 primer specific for FHIT exon 1 was TCCCTCTGCCTTTCATTCC.
DNA Sequence Analysis.
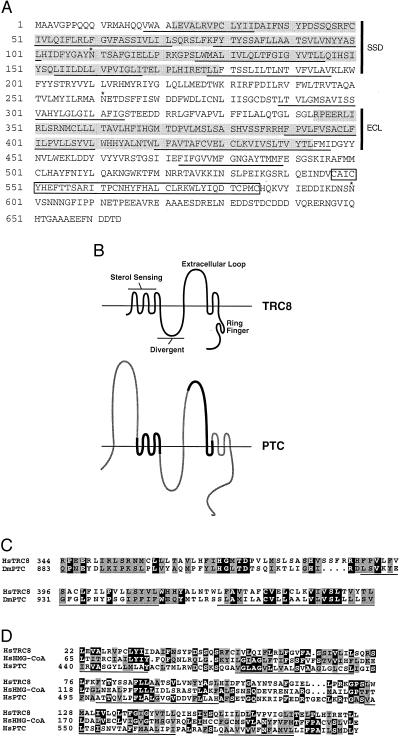
Sequencing was performed on an ABI377 through the Colorado Cancer Center DNA Sequencing Core. Analysis for transmembrane segments (TMs) was performed by using five prediction programs: PHD_htm at EMBL (http://www.embl-heidelberg.de/predictprotein/), TMpred at ISREC (http://ulrec3.unil.ch/software/TMPRED_form.html), SOSUI at Tokyo University (http://www.tuat.ac.jp/∼mitaku/adv_sosui/), DAS at Stockholm University (http://www.biokemi.su.se/∼server/DAS/), and PSORT at Osaka University (http://psort.nibb.ac.jp/). All 10 TMs that are underlined in Fig. 1A were predicted by at least four of the five programs, although in most cases the programs did not agree on the precise boundaries of the segment.
Figure 1.
(A) Predicted amino acid sequence of TRC8. The sequence begins with the first methionine present in the isolated cDNAs. The 3;8 translocation breakpoint occurs between amino acids 60 and 61 (arrowhead). Predicted TMs are underlined, and three potential N-linked glycosylation sites are indicated by ∗. Two regions showing similarity to patched [the SSD and the extracellular loop (ECL)] are shaded. The RING-H2 motif is boxed. (B) Schematic of the predicted structure for TRC8 compared with patched (PTC). The horizontal line represents the lipid bilayer that is crossed by 10 putative TMs of TRC8 and 12 in patched. The divergent loop refers to a region of patched that is nonconserved among patched homologues (45). The diagram of patched shows regions that are similar or different from TRC8 by bold black or thin shaded lines, respectively. The N-terminal extracellular loop of patched is absent from TRC8. The predicted intracellular loop 3′ of the SSD is not conserved between either of the two known murine patched genes (Ptch1 and Ptch2) or in TRC8. (C) Amino acid sequence homology between TRC8 and Drosophila patched. The Dm Ptc sequence (GenBank accession no. M28999) was aligned with TRC8 by gapped blast. Identical amino acids are indicated by white letters on black, and similar amino acids (positive scores in a PAM250 matrix) are shaded. Two TRC8 TMs within this homology region are underlined. (D) Alignment within the putative SSD. Human sequences for HMG-CoA reductase (Swissprot accession no. P04035) and Patched (GenBank accession no. U59464) were aligned with TRC8 by gapped blast. Identities and similarities are indicated as in C; TMs within the putative SSD of TRC8 are underlined.
Single-Stranded Conformational Polymorphism Analysis (SSCP).
SSCP was performed by the method of Spritz et al. (29). Nine primer sets were designed to amplify the entire coding region in segments averaging 325 bp and that also would span any intron-exon boundaries. These primer sets were: set 1 (set1F AGTTGCCCGCCTTAGCC and set1R CCAAAGACACATACTCGACCC), set 2 (set2F CATAACTCTTAGTGGGGAAACATTC and set2R TGTAACGTATCCAATTCCAAATG), set 3 (set3F TGGCACTTATCGTTCTACAGC and set3R TCTTGTTAGCCAAAAGACTCG), set 4 (set4F AGTGTTTGTCCTGGCAGTG and set4R ACAGTTAGTGTAGAATCGCACCC), set 5 (set5F TGGCAAATGAAACTGATTCC and set5R CATGGATAAAATGCAGGACTG), set 6 (set6F AAGACCAGAAGAGAGACTTATTCG and set6R TGCTGTAACTGCAAACAACC), set 7 (set7F TCTTTGGCATCACTATGCAC and set7R CTTCACAGCAGTCCTACGATTC), set 8 (set8F CCAAAAATGGCTGGAAGAC and set8R TGTCAGATTCAGCAGCAGC), and set 9 (set9F CCACCCAATGAAACTCCAG and set9R AGTAGCACATCACAGTAAACGG). In addition, three primer sets were designed to amplify the 5′ untranslated region including: set P1 (setP1F TCCCAGGCAGCTCTGAAC and setP1R ACCATCTTGACCTCGCCC), set P2 (setP2F GTTCGCTTGACTGACGGC and setP2R ATGAGCCGCTGCCACAC), and set P3 (setP3F CACCGAAACCCAGAGACC and setP3R CCAAAGACACATACTCGACCC). These primers were used to amplify genomic DNA (10 ng) under 65°C to 55°C touch-down conditions, as described above. Because of the high GC content present in a 300-bp segment near the amino terminus of TRC8, PCRs for primer sets 1, P2, and P3 contained 2.5 M betaine (30). Reaction products were mixed 50:50 with denaturing dyes (95% formamide, 10 mM NaOH, 20 mM EDTA, 0.02% bromophenol blue, and 0.02% xylene cyanol) and heated to 95°C for 5 min immediately before loading. Samples were separated at 8 W for 16 hr on 0.5× MDE (FMC) gels containing 0.6× Tris-borate buffer and 10% glycerol. Bands were visualized by silver staining.
RESULTS
Identification and Sequence Analysis of TRC8.
To test for a possible fusion gene, RNA was isolated from TL9944 lymphoblastoid cells carrying the 3;8 translocation and subjected to RACE. The t(3;8) breakpoint interrupts FHIT between exons 3 and 4. Therefore, 5′ RACE products were generated by using nested primers within exon 4. Although the cloned amplification products were identified by hybridization to an internal FHIT exon 4 oligonucleotide (R4), nearly 80% of these clones were negative for a FHIT exon 3 oligonucleotide (F4) suggesting they might represent a gene fusion. Twelve such clones were further examined by DNA sequencing, and nine were found to contain an identical novel sequence spliced exactly to the 5′ end of FHIT exon 4. Mapping studies confirmed that the sequences were derived from chromosome 8 (see below). We refer to this gene as TRC8 for translocation in renal cancer from chromosome 8.
The coding region of TRC8 was determined from multiple cDNA clones and PCR products isolated from a human fetal brain library (Stratagene) plus IMAGE clone 331H8 identified from dbEST. Additional genomic sequences extending approximately 2.1 kb upstream of the first in-frame methionine were obtained from phage HD7, which contains both chromosome 3 and 8 material spanning the 3;8 breakpoint. The cDNA sequence contains a predicted 1,992-bp ORF encoding 664 amino acids (Fig. 1A). The upstream cDNA and genomic sequences are GC-rich, indicative of a CpG island. The 5′ most cDNA clone extended to position −286 bp, and additional RACE experiments failed to extend this further. Using a promoter prediction program (http://www-hgc.lbl.gov/projects/promoter.html), four transcriptional start sites were located at −622, −55, −36, and −22 bp of the first methionine. The −22 site corresponded precisely to the two longest RACE products. Thus, it appears that multiple transcription start sites are used.
The ORF is predicted to encode a 664-aa protein of 76 kDa with 10 membrane-spanning segments (Fig. 1B). TRC8 contains two regions of similarity with patched, the receptor for sonic hedgehog (SHH) (21, 22). The region from amino acids 344–443 shows the strongest match with 60% similarity and 23% identity (Fig. 1C). The corresponding segment of patched spans amino acids 883–979 and represents most of the second predicted extracellular domain involved in the binding of SHH (21). A second region of patched similarity involves amino acids 22–179 and encodes a putative sterol-sensing domain (SSD), Fig. 1D. Such domains have been identified in hydroxymethyl glutaryl (HMG)-CoA reductase and the sterol regulatory element binding protein (SREBP) cleavage activating protein (SCAP) (31). Patched contains a putative SSD, of unknown function, from amino acids 440 to 600 (32, 33). This region is 53% similar/17% identical to the SSD of HMG-CoA reductase (34); the corresponding region from TRC8 shows 63% similarity/17% identity. We surmise, based on the multiple TMs and regions of patched similarity, that TRC8 encodes a membrane-bound receptor.
In addition, a perfect match with a ring-finger motif of the RING-H2 subtype (35) was found between amino acids 547 and 585. The RING-H2 motif (CX2CX9–27CXHX2HX2CX6–17CX2C) differs from the standard RING finger by replacement of the fourth cysteine with a histidine. Functionally, RING-H2 motifs have been suggested to be protein–protein or protein–lipid interaction domains (35). We also observed that TRC8 is highly conserved among mammals. This finding was evident from two murine expressed sequence tags (dbEST clones mu78h12 and vl43c01) found to be 93% and 89% identical, respectively, at the nucleotide level over 971 bp.
Expression of TRC8.
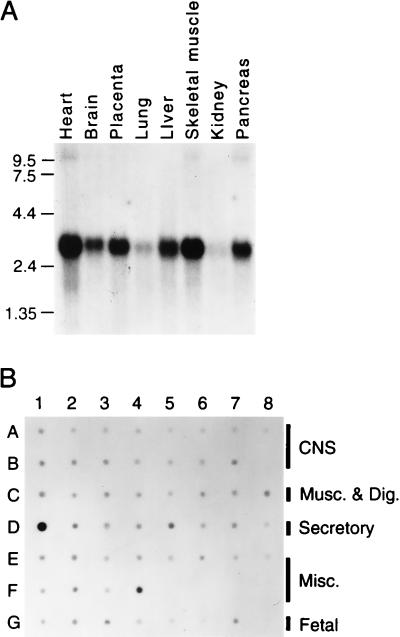
Hybridization of TRC8 to a Northern blot (CLONTECH) prepared from adult human tissues and placenta identified a message of approximately 3.0 kb (Fig. 2A). Although our longest cDNA clones total 2.5 kb, use of the −622-bp promoter, as discussed above, would result in a 2.9-kb message, close to the observed size. Although expression in the lung and kidney appeared reduced, hybridization with a control glyceraldehyde-3-phosphate dehydrogenase probe (data not shown) indicated that there was less RNA present in these samples. A human RNA dot blot (RNA Master blot, CLONTECH) revealed TRC8 message in all tissues examined (Fig. 2B) with the highest levels in testis (D1), placenta (F4), and adrenal (D5). TRC8 is expressed in both fetal (G3) and adult kidney (E1) and in adult thyroid (D6), the suspected target organs for TRC8 aberrations in the 3;8 translocation family.
Figure 2.
Analysis of TRC8 expression by Northern (A) and dot blot (B). (A) Gel resolved polyadenylated RNA (2 μg) from adult human tissues (CLONTECH) was hybridized under recommended conditions with a 1.5-kb 3′ TRC8 cDNA encompassing most of the TMs and the ring finger (bp 83–1623). A second, largely nonoverlapping probe (bp 1446–2212) yielded essentially the same pattern. The filter was exposed for 18 hr at −80°C. (B) A CLONTECH human RNA master dot blot was hybridized with the same probe as in A under recommended conditions and exposed for 15 hr. Final wash conditions were 0.1× standard saline citrate, 0.5% SDS at 55°C for 20 min. Signals were collected on a Molecular Dynamics PhosphorImager. Blank positions included B8, F5-F8, and G8. Central nervous system tissues (A1-A8 and B1-B7) included (in order) whole brain, amygdala, caudate nucleus, cerebellum, cerebral cortex, frontal lobe, hippocampus, medulla oblongata, occipital lobe, putamen, substantia nigra, temporal lobe, thalamus, sub-thalamic nucleus, and spinal cord. Musculature and digestive tissues (C1-C8) included heart, aorta, skeletal muscle, colon, bladder, uterus, prostate, and stomach. Secretory tissues (D1-D8) included testis, ovary, pancreas, pituitary, adrenal, thyroid, salivary, and mammary glands. Miscellaneous tissues (E1-E8 and F1-F4) included kidney, liver, small intestine, spleen, thymus, peripheral leukocytes, lymph node, bone marrow, appendix, lung, trachea, and placenta. Fetal tissues (G1-G7) included brain, heart, kidney, liver, spleen, thymus, and lung. All control spots (yeast and Escherichia coli RNAs, human Cot1, and total human DNAs) were blank (not shown).
Mapping of TRC8.
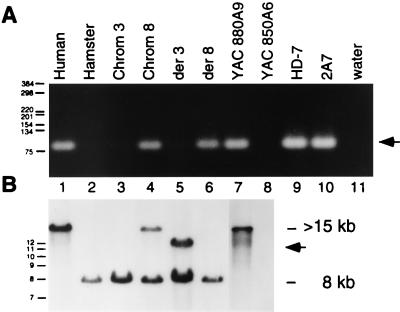
TRC8 sequences were localized to the immediate region of the breakpoint on chromosome 8 by both PCR and Southern blot analysis of hybrids, yeast artificial chromosome (YACs), and phage clones (Fig. 3). Primers derived from the 5′ coding portion of TRC8 yielded the expected product in the chromosome 8-only hybrid (Fig. 3A, lane 4) but not in a chromosome 3-only hybrid (lane 3). The same product was also present on the der(8) but not the der(3) chromosome from the 3;8 translocation. Similarly, the 8q24 YAC 880A9 was positive as was the lambda clone, HD7, which contained both chromosome 8 and 3 material from the breakpoint junction. Southern blot analysis (Fig. 3B) demonstrated that the remaining 3′ portion of TRC8 was contained on the der(3) chromosome (lane 5). Notably, the probe hybridized to an altered band (arrow) in the der(3) hybrid consistent with the t(3;8) rearrangement. Together, these data indicate that TRC8 is localized to 8q24 and is interrupted by the 3;8 translocation and that its 5′ to 3′ orientation is centromere to telomere.
Figure 3.
(A) Localization of 5′ TRC8 sequences to chromosome 8q. Primers R-M and F-O amplify an 82-bp fragment specific for the 5′ portion of TRC8. Templates in lanes 1–11 included, respectively, AG4103 (normal human), CHO glyA− (hamster), UCTP-2A3 (chromosome 3 only hybrid), 706-B6, clone17 (chromosome 8 only hybrid), TL12–8 [t(3;8) der(3) hybrid], 3;8/4–1 [t(3;8) der(8) hybrid], YAC 880A9 (chromosome 8-specific YAC spanning 3;8 breakpoint), YAC 850A6 (chromosome 3-specific YAC spanning 3;8 breakpoint), HD-7 (genomic phage clone carrying the 3;8 breakpoint region from chromosome 8), 2A7 (longest 5′RACE clone), water control. Molecular size standards are indicated in bp. (B) The same hybrid and YAC DNAs used in A were digested with EcoRI and Southern-blotted. The filter was hybridized with a 1.4-kb TRC8 cDNA fragment that derives from the 3′ end. The normal human TRC8 fragment is >15 kb, which is reduced to ≈12 kb by the translocation (arrow). The cross-hybridizing fragment in hamster DNA (lanes 2–6) is 8 kb.
Both Reciprocal Products are Expressed in t(3;8) Lymphoblastoid Cells.
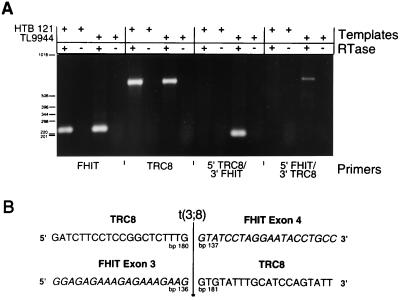
To determine whether both reciprocal products were expressed, we performed RT-PCR analysis on RNA isolated from TL9944 lymphoblastoid cells carrying the 3;8 translocation. Primers that flanked the breakpoint and were specific for the 5′ and 3′ portions of either TRC8 or FHIT were used. As can be seen in Fig. 4, primers specific for wild-type FHIT and TRC8 generated bands of the expected size from both t(3;8) and control RNAs. In contrast, the 5′ TRC8 primer together with the 3′ FHIT primer produced a product only from the (3;8) translocation cell line. Similarly, the reciprocal 5′ FHIT/3′ TRC8 product also was observed only in the translocation line. No products were detected in the absence of RT. Sequence analysis (Fig. 4B) confirmed that these unique RT-PCR products contained fusions of TRC8 and FHIT sequences, as expected. Thus, although FHIT is interrupted in its 5′ untranslated region, its complete coding sequence is contained in the product from the der(8) chromosome. In contrast, TRC8 is interrupted within the predicted SSD.
Figure 4.
(A) RT-PCR analysis of fusion product expression. RNAs isolated from the t(3;8) lymphoblastoid cell line TL9944 (36) and from a control breast carcinoma cell line (HTB121) were treated with or without RT, as indicated (+ or −) and analyzed for expression of FHIT and TRC8 by PCR. Four primers specific for 5′ and 3′ portions of each gene, F1 and R1 for FHIT and R-M and EMR for TRC8, were used in combination to detect both wild-type and putative chimeric transcripts. The FHIT primer pair generated a product of the expected size (231 bp) as did the TRC8 primer pair (651 bp). Reciprocal chimeric products were amplified by using R-M plus R1 for 5′ TRC8/3′ FHIT and F1 plus EMR for 5′ FHIT/3′ TRC8. Predicted sizes of the chimeric products are 188 and 694 bp, respectively. (B) Sequences of 3;8 chimeric transcripts. The RT-PCR-amplified cDNAs in lanes 11 and 15, corresponding to the reciprocal chimeric transcripts, were purified and sequenced on both strands. Forty bp surrounding the boundary between FHIT exons 3 and 4 are shown with FHIT sequences italicized. The precise position of the fusion on both TRC8 and FHIT transcripts is indicated.
A Tumor-Specific Mutation in Sporadic RCCs.
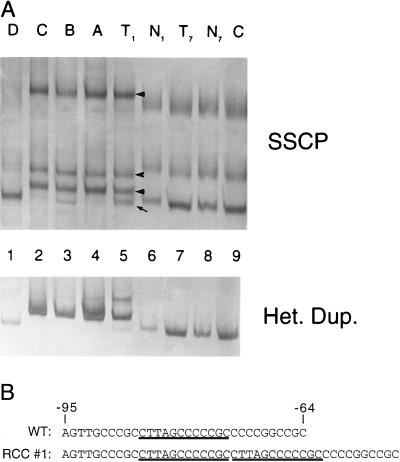
An SSCP analysis was performed by using 12 primer pairs covering the coding sequence and the 5′ untranslated region in 32 renal carcinomas. A duplication of 12 nucleotides in the 5′ untranslated region was identified in one tumor that was absent in matched normal DNA and is thus tumor specific (Fig. 5A, lane 5). This mutation was verified by multiple separate PCR amplifications, SSCP analyses and sequencing, as well as by the use of an alternative primer set, thus eliminating the possibility of a PCR artifact. The duplication (Fig. 5B) was found to occur in a consistent RNA stem loop structure predicted by the GCG program mfold in both energetically optimal and all suboptimal folds.
Figure 5.
(A) Detection of a tumor-specific somatic mutation by SSCP and heteroduplex analysis. DNA samples were PCR-amplified by using primers flanking the first coding exon of TRC8. The products were denatured, separated on a nondenaturing MDE gel and detected by silver staining. Samples included matched tumor and normal DNAs from patients 1 and 7 (lanes 5–8, respectively) and an unrelated normal control (AG4103, lane 9). A separate SSCP gel was used to isolate four individual SSCP bands from RCC no. 1 (lane 5, marked by an arrow or arrowheads). The excised bands A-D corresponded to the indicated bands in lane 5 from top to bottom. These last templates were reamplified and analyzed by SSCP to determine whether they contained mutant or wild-type sequences. Comparison to lane 5 suggested that bands A and C contained primarily mutant DNA, band B was a mixture of mutant and wild-type, and band D was wild-type only. Results for SSCP (Upper) and heteroduplex (Lower) are shown. (B) RCC no. 1 contains a 12-bp duplication in the 5′ untranslated region. Purified PCR products shown in A (lanes 1–4) were sequenced. The mutation consisted of a 12-bp direct duplication (underlined) at bp position −73, which was present in the tumor sample but not in the corresponding normal DNA.
DISCUSSION
The relationship between the 3;8 translocation and kidney cancer has been a long-term project of this laboratory (27, 36–38) and the source of considerable interest among investigators seeking to identify a 3p TSG. Although the 3p14 FHIT gene was postulated to be a tumor suppressor (5, 39, 40), this role was questioned (41) based on its biochemical function as a di-adenosine hydrolase (6) combined with the lack of substantial mutations in tumors and the fact that most reported PCR alterations resulted from low-abundance splicing variations (11, 13–15, 17). Moreover, there is little support for the involvement of FHIT in renal cancers (16, 42). Similarly, the reintroduction of FHIT into tumorigenic cell lines was inconsistent in suppressing tumors, including the fact that a hydrolase “dead” mutant appeared active (39). Otterson et al. (43) introduced FHIT into six carcinoma cell lines and observed no effects on proliferation, morphology, cell-cycle kinetics, or tumorigenesis. Also, there was no correlation between the presence of truncated RT-PCR products and FHIT protein. We identified a series of 3p14 deletions, many not involving FHIT exons, which overlapped FRA3B in various carcinoma cell lines (14). However, spontaneous deletions also were observed in nontumor backgrounds, and we proposed that the deletions were the result of genomic instability. Although another 3p14 gene might exist, our sequence data totaling 160 kb from FRA3B (14) (plus GenBank updates AF023460 and AF023461) together with 135 kb of nonoverlapping sequence from Inoue et al. (44) did not identify any additional definitive genes.
The observation that FHIT was involved in a translocation-derived fusion with HMGIC (19), the causative gene in a variety of benign tumors, led us to consider that the same event might have occurred in the 3;8 translocation. By using RACE, we were able to identify an additional gene, TRC8, from the 8q24 breakpoint. TRC8 encodes a predicted 664-aa, multitransmembrane protein with similarity to patched. This similarity includes the second extracellular domain of patched, which is involved in binding sonic hedgehog, as well as its putative SSD. In addition, the first 480 amino acids of TRC8 and amino acids 440-1100 of patched share an organizational similarity (Fig. 1B). This similarity begins with the common SSD, followed by the divergent region that is nonconserved among patched homologues (45), and finally by the conserved second extracellular loop. TRC8 lacks the first extracellular loop of patched and likewise shows no similarity after the second extracellular loop. Therefore, although TRC8 has similarity to patched and is predicted to be a plasma membrane protein by psort (46), it is not the type of direct homologue as is the Patched 2 gene, for instance (45).
TRC8 may be the critical gene in the 3;8 translocation based on the following: (i) its similarity to patched, which in turn is responsible for the hereditary basal cell carcinoma syndrome (23, 25), (ii) the preservation and expression of FHIT coding sequences in 3;8 translocation containing cells (in contrast to the disruption of TRC8 coding sequences), and (iii) its demonstrated mutation in a sporadic renal carcinoma. How TRC8 may function in human tumorigenesis is presently unknown. In human basal cell carcinomas, PATCHED appears to function as a recessive TSG (reviewed in ref. 47). However, medulloblastomas (or perhaps the precursor cell proliferations) occurring in mice heterozygous for a ptc mutation might develop without the requirement for loss of the remaining allele (24, 47). In tumors from the 3;8 translocation family, we do not know whether the wild-type allele was lost. A screen of TRC8 in sporadic renal carcinomas by SSCP demonstrated a mutation in one tumor (Fig. 5A). In that sample, very little of the wild-type heteroduplex product can be seen. This rearrangement resulted in an insertion of 12 bp in the tumor, which was not present in the corresponding normal DNA of that patient. This insertion occurs in a consistently predicted stem-loop structure in the 5′ untranslated region. The consequence of this insertion conceivably affects either transcription or translation. Although the frequency of TRC8 mutations in spontaneous tumors appears low, it is possible this finding is reminiscent of the mutation frequencies observed in BRCA1 and BRCA2 (48, 49). We believe the most exciting aspect of our findings is that TRC8 appears to define an additional mutation pathway in renal and thyroid cancers. Whether it functions in some analogous way to patched or is involved in other developmental processes remains to be determined.
Acknowledgments
We thank Dr. C. Korch and C. Dessev for their assistance with nucleotide sequencing through the auspices of the University of Colorado Cancer Center DNA Sequencing Core (supported by P30 CA 46934 from the National Institutes of Health) and A. Yokomizo for isolation of tumor DNAs. We also thank Dr. R. S. Brown and the members of the hereditary kidney cancer family. This investigation was supported by Grants CA58187 and DAMD17–94J-4391 from the National Institutes of Health and Department of Defense.
ABBREVIATIONS
- RCC
renal cell carcinoma
- TSG
tumor suppressor gene
- FHIT, fragile histidine triad gene
RACE, rapid amplification of cDNA ends
- TRC8
translocation in renal carcinoma, chromosome 8 gene
- TM
transmembrane segment
- SSD
sterol-sensing domain
- RT-PCR
reverse transcriptase–PCR
- SSCP
single-stranded conformational polymorphism analysis
- HMG
hydroxymethylglutaryl
- YAC
yeast artificial chromosome
Footnotes
References
- 1.Cohen A J, Li F P, Berg S, Marchetto D J, Tsai S, Jacobs S C, Brown R S. N Engl J Med. 1979;301:592–595. doi: 10.1056/NEJM197909133011107. [DOI] [PubMed] [Google Scholar]
- 2.Li F P, Decker H J, Zbar B, Stanton V P, Jr, Kovacs G, Seizinger B R, Aburatani H, Sandberg A A, Berg S, Hosoe S. Ann Intern Med. 1993;118:106–111. doi: 10.7326/0003-4819-118-2-199301150-00005. [DOI] [PubMed] [Google Scholar]
- 3.Latif F, Tory K, Gnarra J, Yao M, Duh F M, Orcutt M L, Stackhouse T, Kuzmin I, Modi W, Geil L, et al. Science. 1993;260:1317–1320. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 4.van den Berg A, Buys C H. Genes Chromosomes Cancer. 1997;19:59–76. doi: 10.1002/(sici)1098-2264(199706)19:2<59::aid-gcc1>3.3.co;2-o. [DOI] [PubMed] [Google Scholar]
- 5.Ohta M, Inoue H, Cotticelli M G, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T, et al. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- 6.Barnes L D, Garrison P N, Siprashvili Z, Guranowski A, Robinson A K, Ingram S W, Croce C M, Ohta M, Huebner K. Biochemistry. 1996;35:11529–11535. doi: 10.1021/bi961415t. [DOI] [PubMed] [Google Scholar]
- 7.Sozzi G, Veronese M L, Negrini M, Baffa R, Cotticelli M G, Inoue H, Pilotti S, De Gregorio L, Pastorino U, Pierotti M A, et al. Cell. 1996;85:17–26. doi: 10.1016/s0092-8674(00)81078-8. [DOI] [PubMed] [Google Scholar]
- 8.Virgilio L, Shuster M, Gollin S M, Veronese M L, Ohta M, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1996;93:9770–9775. doi: 10.1073/pnas.93.18.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Negrini M, Monaco C, Vorechovsky I, Ohta M, Druck T, Baffa R, Huebner K, Croce C M. Cancer Res. 1996;56:3173–3179. [PubMed] [Google Scholar]
- 10.Sozzi G, Alder H, Tornielli S, Corletto V, Baffa R, Veronese M L, Negrini M, Pilotti S, Pierotti M A, Huebner K, Croce C M. Cancer Res. 1996;56:2472–2474. [PubMed] [Google Scholar]
- 11.Thiagalingam S, Lisitsyn N A, Hamaguchi M, Wigler M H, Willson J K V, Markowitz S D, Leach F S, Kinzler K W, Vogelstein B. Cancer Res. 1996;56:2936–2939. [PubMed] [Google Scholar]
- 12.Fong K M, Biesterveld E J, Virmani A, Wistuba I, Sekido Y, Bader S A, Ahmadian M, Ong S T, Rassool F V, Zimmerman P V, et al. Cancer Res. 1997;57:2256–2267. [PubMed] [Google Scholar]
- 13.Gayther S A, Barski P, Batley S J, Limin L, de Foy K A F, Cohen S N, Ponder B A J, Caldas C. Oncogene. 1997;15:2119–2126. doi: 10.1038/sj.onc.1201591. [DOI] [PubMed] [Google Scholar]
- 14.Boldog F, Gemmill R M, West J, Robinson M, Robinson L, Li E, Roche J, Todd S, Waggoner B, Lundstrom R, et al. Hum Mol Genet. 1997;6:193–203. doi: 10.1093/hmg/6.2.193. [DOI] [PubMed] [Google Scholar]
- 15.Panagopoulos I, Thelin S, Mertens F, Mitelman F, Aman P. Genes Chromosomes Cancer. 1997;19:215–219. doi: 10.1002/(sici)1098-2264(199708)19:4<215::aid-gcc2>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 16.van den Berg A, Draaijers T G, Kok K, Timmer T, van der Veen A Y, Veldhuis P M, de Leij L, Gerhartz C D, Naylor S L, Smith D I, Buys C H. Genes Chromosomes Cancer. 1997;19:220–227. doi: 10.1002/(sici)1098-2264(199708)19:4<220::aid-gcc3>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 17.Latil A, Bièche I, Fournier G, Cussenot O, Pesche S, Lidereau R. Oncogene. 1998;16:1863–1868. doi: 10.1038/sj.onc.1201703. [DOI] [PubMed] [Google Scholar]
- 18.Muller C Y, O’Boyle J D, Fong K M, Wistuba I I, Biesterveld E, Ahmadian M, Miller D S, Gazdar A F, Minna J D. J Natl Cancer Inst. 1998;90:433–439. doi: 10.1093/jnci/90.6.433. [DOI] [PubMed] [Google Scholar]
- 19.Geurts J M, Schoenmakers E F, Roijer E, Stenman G, Van de Ven W J. Cancer Res. 1997;57:13–17. [PubMed] [Google Scholar]
- 20.Frohman M A. PCR Methods Appl. 1994;4:S40–S58. doi: 10.1101/gr.4.1.s40. [DOI] [PubMed] [Google Scholar]
- 21.Marigo V, Davey R A, Zuo Y, Cunningham J M, Tabin C J. Nature (London) 1996;384:176–179. doi: 10.1038/384176a0. [DOI] [PubMed] [Google Scholar]
- 22.Stone D M, Hynes M, Armanini M, Swanson T A, Gu Q, Johnson R L, Scott M P, Pennica D, Goddard A, Phillips H, et al. Nature (London) 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 23.Hahn H, Wicking C, Zaphiropoulous P G, Gailani M R, Shanley S, Chidambaram A, Vorechovsky I. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 24.Goodrich L V, Milenkovic L, Higgins K M, Scott M P. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 25.Johnson R L, Rothman A L, Xie J, Goodrich L V, Bare J W, Bonifas J M, Quinn A G, Myers R M, Cox D R, Epstein E H, Jr, Scott M P. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- 26.Drabkin H A, Smith D, Jones C, Jonsen M, Sage M, Gold S, Glover T, Dobrovic A, Bradley W E, Gemmill R. Cancer Cells. 1989;7:63–68. [Google Scholar]
- 27.Boldog F L, Gemmill R M, Wilke C M, Glover T W, Nilsson A S, Chandrasekharappa S C, Brown R S, Li F P, Drabkin H A. Proc Natl Acad Sci USA. 1993;90:8509–8513. doi: 10.1073/pnas.90.18.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frohman M A. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 29.Spritz R A, Holmes S A, Ramesar R, Greenberg J, Curtis D, Beighton P. Am J Hum Genet. 1992;51:1058–1065. [PMC free article] [PubMed] [Google Scholar]
- 30.Henke W, Herdel K, Jung K, Schnorr D, Loening S A. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hua X, Nohturfft A, Goldstein J L, Brown M S. Cell. 1996;87:415–426. doi: 10.1016/s0092-8674(00)81362-8. [DOI] [PubMed] [Google Scholar]
- 32.Loftus S K, Morris J A, Carstea E D, Gu J Z, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld M A, Tagle D A, et al. Science. 1997;277:232–235. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- 33.Carstea E D, Morris J A, Coleman K G, Loftus S K, Zhang D, Cummings C, Gu J, Rosenfeld M A, Pavan W J, Krizman D B, et al. Science. 1997;277:228–231. [Google Scholar]
- 34.Chin D J, Gil G, Russell D W, Liscum L, Luskey K L, Basu S K, Okayama H, Berg P, Goldstein J L, Brown M S. Nature (London) 1984;308:613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- 35.Freemont P S. Ann NY Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 36.Gemmill R M, Coyle-Morris J, Ware-Uribe L, Pearson N, Hecht F, Brown R S, Li F P, Drabkin H A. Genomics. 1989;4:28–35. doi: 10.1016/0888-7543(89)90310-8. [DOI] [PubMed] [Google Scholar]
- 37.Boldog F L, Waggoner B, Glover T W, Chumakov I, Le Paslier D, Cohen D, Gemmill R M, Drabkin H A. Genes Chromosomes Cancer. 1994;11:216–221. doi: 10.1002/gcc.2870110403. [DOI] [PubMed] [Google Scholar]
- 38.Drabkin H A, Bradley C, Hart I, Bleskan J, Li F P, Patterson D. Proc Natl Acad Sci USA. 1985;82:6980–6984. doi: 10.1073/pnas.82.20.6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siprashvili Z, Sozzi G, Barnes L D, McCue P, Robinson A K, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L, et al. Proc Natl Acad Sci USA. 1997;94:13771–13776. doi: 10.1073/pnas.94.25.13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huebner K, Hadaczek P, Siprashvili Z, Druck T, Croce C M. Biochim Biophys Acta. 1997;1332:M65–M70. doi: 10.1016/s0304-419x(97)00009-7. [DOI] [PubMed] [Google Scholar]
- 41.Le Beau M M, Drabkin H, Glover T W, Gemmil R, Rassool F V, McKeithan T W, Smith D I. Genes Chromosomes Cancer. 1998;21:281–289. doi: 10.1002/(sici)1098-2264(199804)21:4<281::aid-gcc1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 42.Bugert P, Wilhelm M, Kovacs G. Genes Chromosomes Cancer. 1997;20:9–15. [PubMed] [Google Scholar]
- 43.Otterson G A, Xiao G H, Geradts J, Jin F, Chen W D, Niklinska W, Kaye F J, Yeung R S. J Natl Cancer Inst. 1998;90:426–432. doi: 10.1093/jnci/90.6.426. [DOI] [PubMed] [Google Scholar]
- 44.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce C M. Proc Natl Acad Sci USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Motoyama J, Takabatake T, Takeshima K, Hui C. Nat Genet. 1998;18:104–106. doi: 10.1038/ng0298-104. [DOI] [PubMed] [Google Scholar]
- 46.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ingham P W. Curr Opin Genet Dev. 1998;8:88–94. doi: 10.1016/s0959-437x(98)80067-1. [DOI] [PubMed] [Google Scholar]
- 48.Futreal P A, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett L M, Haugen-Strano A, Swensen J, Miki Y. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 49.Lancaster J M, Wooster R, Mangion J, Phelan C M, Cochran C, Gumbs C, Seal S, Barfoot R, Collins N, Bignell G, et al. Nat Genet. 1996;13:238–240. doi: 10.1038/ng0696-238. [DOI] [PubMed] [Google Scholar]