Abstract
In this study, we describe the ultrastructural changes associated with acid activation of Helicobacter pylori vacuolating cytotoxin (VacA). Purified VacA molecules imaged by deep-etch electron microscopy form ∼30-nm hexagonal “flowers,” each composed of an ∼15-nm central ring surrounded by six ∼6-nm globular “petals.” Upon exposure to acidic pH, these oligomeric flowers dissociate into collections of up to 12 teardrop-shaped subunits, each measuring ∼6 × 14 nm. Correspondingly, glycerol density gradient centrifugation shows that at neutral pH VacA sediments at ∼22 S, whereas at acidic pH it dissociates and sediments at ∼5 S. Immunoblot and EM analysis of the 5-S material demonstrates that it represents ∼90-kD monomers with 6 × 14–nm “teardrop” morphology. These data indicate that the intact VacA oligomer consists of 12 ∼90-kD subunits assembled into two interlocked six-membered arrays, overlap of which gives rise to the flower-like appearance. Support for this interpretation comes from EM identification of small numbers of relatively “flat” oligomers composed of six teardrop-shaped subunits, interpreted to be halves of the complete flower. These flat forms adsorb to mica in two different orientations, corresponding to hexameric surfaces that are either exposed or sandwiched inside the dodecamer, respectively. This view of VacA structure differs from a previous model in which the flowers were interpreted to be single layers of six monomers and the flat forms were thought to be proteolysed flowers. Since acidification has been shown to potentiate the cytotoxic effects of VacA, the present results suggest that physical disassembly of the VacA oligomer is an important feature of its activation.
H elicobacter pylori is the cause of chronic superficial gastritis in humans, and infection with this organism predisposes the host to the development of peptic ulcer disease and gastric adenocarcinoma (Telford et al., 1994b ; Blaser, 1996; Cover and Blaser, 1996). Many H. pylori strains secrete a cytotoxin (VacA) that induces prominent vacuolation and degeneration of cultured eukaryotic cells (Papini et al., 1994; Cover, 1996). Direct introduction of VacA into the stomachs of mice causes epithelial injury (Telford et al., 1994a ), and VacA activity is thought to be important in the pathogenesis of peptic ulcer disease in humans (Figura et al., 1989; Atherton et al., 1995; Marchetti et al., 1995).
Purified H. pylori VacA migrates as an ∼90-kD protein under denaturing conditions but as an ∼1,000-kD complex under nondenaturing conditions (Cover and Blaser, 1992; Manetti et al., 1995). Previous deep-etch electron microscopic work demonstrated that the VacA complex is a “flower”-like oligomeric structure with six or seven visible “petals,” suggesting that it may be constructed similarly to A-B type bacterial toxins (Lupetti et al., 1996). Additionally, previous tissue culture studies demonstrated that the specific activity of VacA increases by more than an order of magnitude after exposure to acidic pH (de Bernard et al., 1995). To analyze further the oligomeric structure of VacA and to determine what happens to its EM structure when activated by low pH, the present study was undertaken.
Materials and Methods
Preparation of H. pylori VacA
H. pylori strain 60190 (American Type Culture Collection 49503; Rockville, MD), a well-characterized cytotoxin-producing strain with a type s1a/m1 vacA genotype (Cover and Blaser, 1992; Cover et al., 1994; Atherton et al., 1995), was cultured in ambient air containing 6% CO2 for 48 h at 37°C in sulfite-free Brucella broth (Hawrylik et al., 1994) containing 0.5% charcoal. After centrifugation of the culture at 10,000 g for 15 min, supernatant proteins were precipitated with a 50% saturated solution of ammonium sulfate and resuspended in 60 mM Tris, pH 7.5, containing 100 mM NaCl. After passage through a 0.2-μm filter, supernatant proteins were fractionated by gel filtration chromatography in 60 mM Tris, pH 7.5, containing 100 mM NaCl, using either a Superose 12 HR 16/50 or a Superose 6 HR 16/50 column (Pharmacia Biotech, Piscataway, NJ) (Cover and Blaser, 1992). The presence of VacA in samples was detected by Western blotting with rabbit anti-VacA serum, as described previously (Cover and Blaser, 1992). In addition to purifying VacA from H. pylori 60190, VacA proteins also were purified from H. pylori strains with type s2/m2 and s1a/m2 vacA genotypes (86-338 and 95-54, respectively) (Atherton et al., 1995).
Isolation and Characterization of VacA by Glycerol Density Gradient Centrifugation
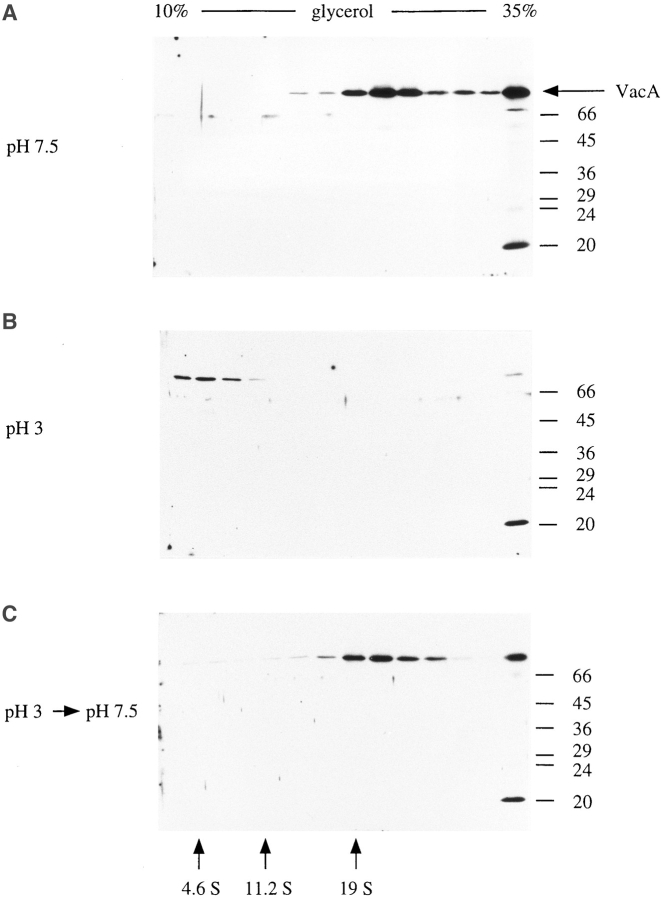
As an alternate approach for purifying VacA, ammonium sulfate–precipitated supernatant proteins were fractionated by centrifugation through glycerol density gradients. 1-ml aliquots of dialyzed samples (containing about 10 mg total protein) were layered on 37-ml 10–35% glycerol gradients in 60 mM Tris, pH 7.5, containing 100 mM NaCl and centrifuged at 28,000 rpm in an SW28 rotor (Beckman Instruments, Palo Alto, CA) for 20 h at 4°C (Martin and Ames, 1961). Gradients were fractionated from the top using an Auto Densi-Flow IIC apparatus (Labconco Corp., Kansas City, MO) and analyzed for VacA content by immunoblotting, as above. Standards were fractionated in parallel gradients and included BSA (4.6 S), catalase (11.2 S), and thyroglobulin (19 S).
For analytical purposes, 5-ml 10–35% glycerol gradients were prepared in either 60 mM Tris, pH 7.5, containing 100 mM NaCl, or 100 mM glycine, pH 3.0, containing 100 mM NaCl. 100-μl aliquots of samples (either dialyzed ammonium sulfate–precipitated proteins from broth culture supernatant or VacA that had been partially purified by gel filtration chromatography as described above) were layered on the gradients and centrifuged for 7.5 h at 40,000 rpm in an SW50.1 rotor (Beckman Instruments). For acidification, samples were typically brought to pH 3 by drop-wise addition of 0.1 N HCl while stirring and then incubated at 30°C for 10 min. Subsequent reneutralization was accomplished by drop-wise addition of Tris base until the sample was brought back to pH 7.5. VacA oligomers also dissociated when layered directly on a pH 3 glycerol gradient (i.e. without prior acidification).
Electron Microscopic Imaging of VacA
In preparation for EM, VacA samples were adsorbed to a suspension of finely ground mica flakes, followed by freeze-drying and platinum replication according to established procedures (Heuser, 1983, 1989). Briefly, this involved adding two drops of a suspension of finely ground mica to 0.5 ml of a solution containing 10–30 μg purified VacA and allowing the toxin to adsorb to the mica for 30 s. The mica flakes were then pelleted by gentle centrifugation, washed twice with a solution of 30 mM Hepes, pH 7.2, containing 70 mM KCl and 5 mM MgCl2, and layered onto a thin slice of aldehyde-fixed lung for support during freezing. This was accomplished by dropping the samples onto a liquid helium–cooled copper block in a homemade “quick-freeze” device.
Quick-frozen mica flakes were then freeze-fractured in a Balzer's freeze etch machine and “deep-etched” for 4 min at −100°C, thereby exposing molecules adsorbed to the surfaces of the mica flakes. These were then rotary-replicated with ∼2 nm of platinum evaporated from an angle of 11° above the horizontal, and finally “backed” or supported with a ∼10-nm film of pure carbon. The replicas were cleaned of mica by floating them overnight in concentrated hydrofluoric acid. The replicas were then washed in water and finally picked up on 75-mesh formvar-coated microscope grids.
Replicas were viewed in a transmission electron microscope (JEOL U.S.A., Inc., Peabody, MA) operated at 100 kV and were photographed in stereo using +10° of tilt with a eucentric side-entry goniometer stage. Finally, representative molecules were chosen by 4× video imaging of micrographs initially taken at 68,000×, and their video images were transferred to a computer. Using Adobe Photoshop, they were enlarged to 300,000× for production of the photomontages shown here.
Cell Culture Methodology
Vacuolating cytotoxin activity was assessed by incubating HeLa cells for 24 h with serial dilutions of samples in a microtiter assay, as described previously (Cover et al., 1991). In each experiment, Eagle's modified minimal essential medium containing 10% FBS was supplemented with 5 mM ammonium chloride at the time of cytotoxin addition. Vacuolation was quantified by a neutral red uptake assay (Cover et al., 1991) or directly visualized in living cells with video-enhanced time-lapse light microscopy (Heuser et al., 1993).
Results
Deep-etch Imaging of the Vacuolating Cytotoxin
In initial experiments, we extracted high–molecular weight proteins produced by H. pylori strain 60190 by ammonium sulfate precipitation of broth culture supernatants and isolated the largest molecules by gel filtration chromatography on a Superose 12 HR 16/50 column. More than 90% of the vacuolating activity of the culture supernatant eluted in the void volume of such columns (Cover and Blaser, 1992). When this material was adsorbed to mica flakes and examined by deep-etch EM (Fig. 1), it was found to contain a mixture of three molecular forms, including spheres, barrel shapes, and the striking flower shapes identified as VacA in a previous deep-etch EM study (Lupetti et al., 1996).
Figure 1.
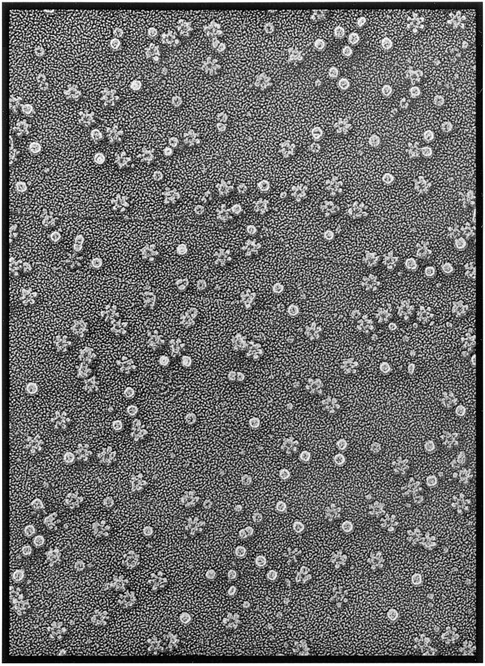
Deep-etch survey view of the high–molecular mass proteins present in broth culture supernatant from H. pylori 60190 as they appear after adsorption to mica and freeze-drying. Three different types of macromolecules are visible, corresponding to urease (spheres), HspB (barrels), and VacA (flowers). The panel represents a field 0.7 μm in width.
Closer examination of the flower shapes in such preparations (10 examples of which are presented in the top two rows of Fig. 2) demonstrated that they are composed of prominent central rings ∼15 nm in diameter, surrounded generally by six globular petals measuring 5–6 nm in diameter, yielding an overall diameter of ∼30 nm for the entire flower. VacA preparations purified further by glycerol density gradient centrifugation yielded identical images of six-sided flowers (not shown). As a control, we examined the high–molecular weight components of culture supernatant from an isogenic mutant strain of H. pylori (60190-v1) that fails to produce VacA (Cover et al., 1994). Flower-shaped complexes were not found in this preparation, which confirmed that the flowers represented oligomeric VacA.
Figure 2.
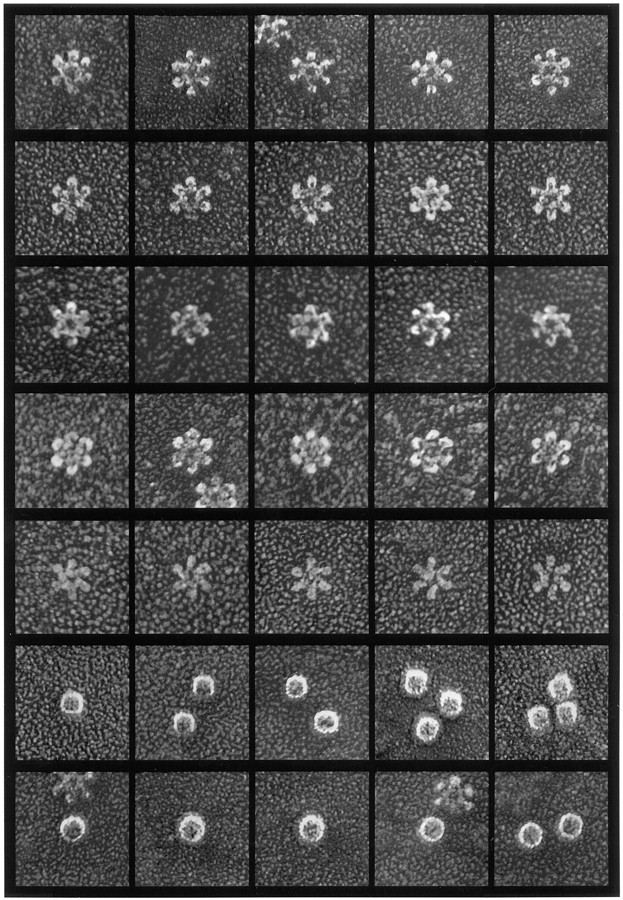
Rotary replicas of purified and freeze-dried H. pylori macromolecules. First and second rows: Typical VacA flowers purified from broth culture supernatants of tox+ H. pylori strain 60190 (type s1a/m1 vacA genotype). Third row: VacA flowers purified from strain 60190 and digested with trypsin for 5 h before adsorption to mica. (See Fig. 5 c for immunoblot analysis of the proteolytic breakdown pattern of this particular preparation.) Fourth row: VacA flowers purified from H. pylori strains 95-54 and 86-338 (type s1a/m2 and s2/m2 vacA genotypes, respectively). Fifth row: Flat forms of VacA found in low abundance among the more typical flowers shown in first through third rows. Sixth row: HspB molecules from culture supernatant of H. pylori 60190-v1 (an isogenic tox [−] strain). Seventh row: Urease molecules from culture supernatant of H. pylori 60190-v1. Magnification 300,00×. In this and all subsequent EM figures, each panel represents a field 75 × 75 nm.
By displaying predominantly six-sided flowers, the VacA preparations generated in this study differed from those generated in the previous deep-etch study of VacA, in which approximately half of the flower-shaped complexes appeared to be seven sided (Lupetti et al., 1996; cf., Fig. 3, top row). A possible reason for this difference will be discussed after presenting further data below. As was seen in the previous deep-etch study of VacA, a second distinct form of VacA oligomer was also observed (Fig. 2, fifth row). It appeared relatively “flat” and lacked the prominent central ring described above. These flat forms generally represented <10% of the total number of flower-like molecules present but turned out to be important indicators of the underlying construction of VacA, as will be elaborated below. The petals comprising such flat forms were elongated ellipsoids measuring about 6 × 14 nm. These radiated from the center of the complex with a distinct clockwise cant or skew. This same chirality was also observed in the earlier deep-etch study, although flat forms were also more commonly seven sided in that study (Lupetti et al., 1996; cf., Fig. 3, bottom row).
Figure 3.
Rotary replicas of VacA molecules drawn from a previous study (Lupetti et al., 1996) in which seven-sided forms predominated. The top row displays whole flowers, and the bottom row displays flat forms. Both are composed of seven radial subunits but are otherwise structurally identical to the six-sided forms shown in Fig. 2.
Although essentially all clinical isolates of H. pylori contain a vacA homologue, individual isolates differ markedly in the quantity and activity of VacA produced, and several different families of vacA alleles have been described (Atherton et al., 1995). To determine whether H. pylori isolates with type m2 vacA alleles produce flower-shaped VacA complexes similar to those shown in Fig. 2, culture supernatants from two isolates with type m2 VacA alleles (strains 95-54 and 86-338) were prepared for EM as above. Each of these strains produced immunoreactive VacA macromolecules that were similar in size to VacA from strain 60190, based on immunoblot analysis of gel filtration chromatography fractions (not shown). Correspondingly, deep-etch EM of VacA preparations from these strains yielded images of six-sided flower-shaped molecules (Fig. 2, fourth row). Interestingly, however, flat forms of VacA were not detected in these preparations.
Two additional macromolecules produced by H. pylori are HspB (a GroEL heat shock protein homologue) and urease. These have estimated molecular masses of 750 and 550–650 kD, respectively (Hu and Mobley, 1990; Austin et al., 1991, 1992). Western blot assays using anti-HspB and antiurease sera indicated that these proteins were present along with VacA in H. pylori broth culture supernatants. HspB and urease eluted from a Superose 6 size exclusion column in fractions corresponding to a slightly smaller size than VacA (not shown). Correspondingly, HspB appeared in deep-etch EMs as 14 × 16–nm barrel-like molecules, and urease appeared as spherical entities 17–18 nm in diameter (Fig. 2, sixth and seventh rows).
Unidirectional Shadow-Castings of H. pylori Macromolecules
To further analyze the three-dimensional structure of the various forms of VacA, urease, and HspB, high–molecular mass proteins were isolated from broth culture supernatant and imaged by “shadow-casting” according to classical techniques (Williams and Wyckoff, 1946), with platinum deposited from 15° above the horizontal in the absence of sample rotation to create shadows roughly 3.5× longer than an object is tall (Fig. 4). Based on the observed shadow lengths, HspB barrels stood ∼12 nm above the mica, and urease stood ∼15 nm above the mica (Fig. 4, fourth and fifth rows). These measurements compared favorably with the 17-nm diameter of rotary-replicated urease spheres since platinum coating should add 2–3 nm to the apparent diameter of all molecules (Heuser, 1989).
Figure 4.

Unidirectional shadow-castings of macromolecules produced by H. pylori 60190. First and second rows: flower forms of VacA, including the rare seven-membered examples found in current preparations. Third row: Flat forms of VacA. Fourth row: HspB molecules. Fifth row: Urease molecules.
Shadow-castings highlighted important differences between the two different forms of VacA oligomers described above. The petals of the more typical VacA flowers cast ∼23-nm shadows, and their central rings cast ∼40-nm shadows, indicating that these domains were raised ∼7 and ∼12 nm above the mica, respectively (Fig. 4, first and second rows). In contrast, the flat forms of VacA cast shadows that were never greater than 13 nm in length (Fig. 4, third row), indicating that no part of these structures was raised above the mica by more than 3.5 nm. Thus, the latter form was indeed much flatter than the flower form. The implications of this roughly twofold difference in molecular thickness will figure centrally in the interpretation of VacA structure presented below.
Relationship between the Structure of VacA and Its State of Proteolysis
VacA secreted by H. pylori 60190 migrates under denaturing conditions as an ∼90-kD band (Cover and Blaser, 1992), but during prolonged storage it undergoes limited proteolytic degradation into ∼34- and ∼58-kD components (Telford et al., 1994a ; Garner and Cover, 1996). To determine whether VacA cleavage into ∼34 and ∼58-kD fragments was associated with an increased abundance of the flat VacA forms, as claimed in the previous deep-etch study of VacA (Lupetti et al., 1996), we compared the ultrastructure of VacA in preparations containing only intact ∼90-kD bands with that of preparations consisting entirely of ∼34- and ∼58-kD VacA cleavage products (the latter resulting from storage of VacA at 4°C for 2 mo before adsorption to mica, or from treatment with exogenous trypsin before adsorption to mica) (Fig. 5). In our hands, neither treatment altered the proportion of intact versus flat VacA oligomers (Fig. 2, third row). Thus, we could not confirm the previous claim that the proportion of flat forms was increased by proteolysis (Lupetti et al., 1996). Instead, we developed the impression that flat forms tended to occur in groups near the fractured surfaces of our replicas, suggesting that they may be artifactual freeze-cleavage products of intact flowers. Furthermore, when proteolysed VacA was fractionated on a Superose 6 HR 16/50 gel filtration column, subsequent SDS-PAGE and immunoblotting indicated that the ∼34- and ∼58-kD fragments remained together in the same high–molecular mass fractions that normally contained the intact ∼90-kD VacA species. This demonstrates that the ∼34- and ∼58-kD domains of VacA remain associated within the VacA flower, even after proteolysis, and rules out the possibility that flat VacA forms are oligomers that have lost one or the other of these proteolytic fragments.
Figure 5.
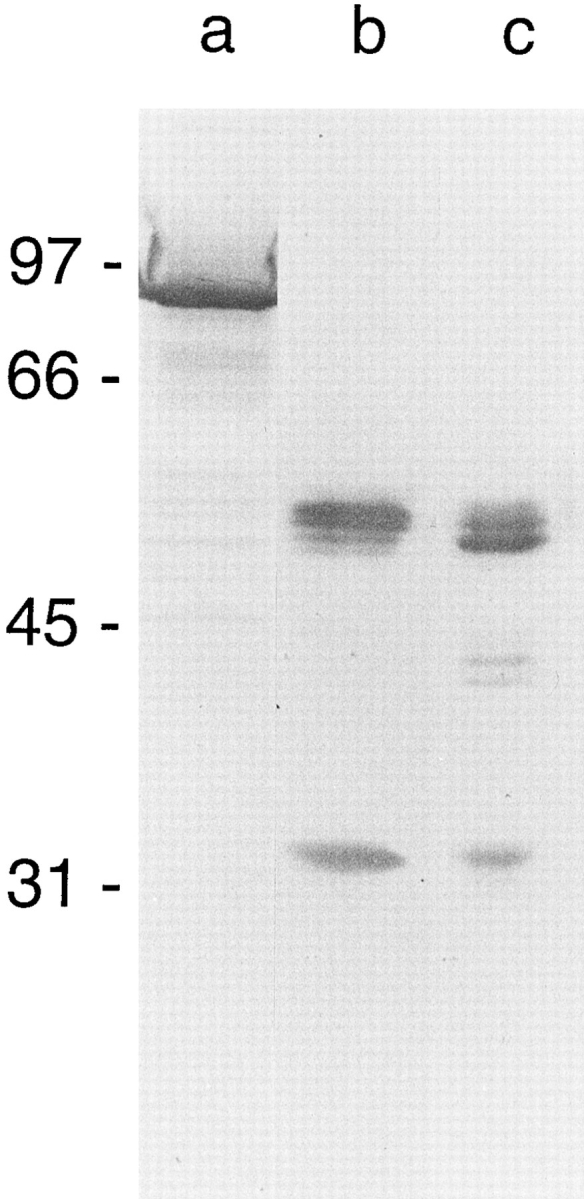
Immunoblot analysis of intact and proteolysed VacA preparations electrophoresed on a 10% acrylamide gel, transferred to nitrocellulose, and reacted with a 1:10,000 dilution of rabbit anti-VacA serum; the antigens were resolved as described previously (Cover and Blaser, 1992). Lane a, intact ∼90-kD VacA from H. pylori 60190; lane b, VacA proteolytic fragments (34 and 58 kD) arising after prolonged storage of purified VacA; lane c, purified VacA treated with trypsin for 5 h at 37°C.
Effect of Acidification on VacA Structure
The vacuolating activity of VacA has been shown to increase substantially after exposure of the toxin to acidic pH (de Bernard et al., 1995). We confirmed this result in the present study (not shown). To determine whether acidification of VacA alters its ultrastructure, VacA was adsorbed onto mica chips at neutral pH in the usual way, and then the mica was washed for 60 s in a 100 mM glycine buffer at pH 3.5. This procedure yielded not the usual flowers, but instead relatively discrete clusters of up to 12 separate petals (Fig. 6, first through third rows). When not too widely separated from each other, the petals in these complexes radiated from the center with a distinct clockwise cant or chirality, identical to that seen previously in the flat VacA forms. However, the prominent central rings that characterize intact VacA flowers were nowhere to be seen. Importantly, the petals generated by this acid- induced dissociation appeared to be identical in size and shape to the individual petals comprising the flat forms of VacA described above. The only difference was that they occurred in groups of up to 12 petals rather than in groups of only six (compare Fig. 6, first row, with Fig. 2, fifth row).
Figure 6.

Acid-induced dissociation of VacA demonstrated in rotary replicas. First through third rows: intact VacA adsorbed to mica at neutral pH as in Fig. 2 and then treated with pH 3.0 glycine buffer. This caused the preadsorbed flowers to “burst” into astral arrays of up to twelve petals. Fourth and fifth rows: VacA acidified to pH 3 before adsorption to mica. This caused nearly complete splaying of the flowers (fourth row) or complete dissociation (fifth row). Note that the individual 6 × 14–nm petals (the presumed VacA monomers) appear slightly curved or bilobed when viewed in isolation (arrow).
A second approach by which we observed acid-induced dissociation of VacA was to bring a solution of VacA oligomers to pH 3 by the slow addition of HCl, incubate it at 30°C for 10 min, and finally adsorb the products of this reaction to mica at pH 3 before EM imaging. This yielded very few remnants of intact VacA oligomers, i.e., there were only rare examples of groups of 6 × 14–nm petals clinging together with any sense of organization (Fig. 6, fourth row). More frequently observed were free 6 × 14– nm petals lying helter-skelter on the mica surface in no apparent relation to each other (Fig. 6, fifth row), or with only an occasional tendency to dimerize (Fig. 6, lower right panel).
Velocity sedimentation on glycerol density gradients also indicated that acidification of VacA induced dissociation of the oligomeric form. Thus, when VacA was sedimented on a glycerol gradient at pH 7.5, a ∼22-S peak was obtained, whereas when VacA was acidified to pH 3 for 10 min at 30°C and applied to a pH 3.0 gradient, nearly 100% of it sedimented in fractions corresponding to ∼5 S (Fig. 7). Immunoblotting the ∼5-S fractions recovered from such pH 3.0 gradients indicated that VacA was still in its intact ∼90-kD form. Hence, VacA dissociation did not seem to be dependent upon nor in any way associated with VacA proteolysis. Deep-etch EM of the purified ∼5-S fractions yielded images of fully dissociated 6 × 14–nm petals identical to those shown in Fig. 6 (not shown). These 5-S fractions induced prominent cell vacuolation when added to tissue culture medium overlying cultured HeLa cells (not shown). Thus, dissociated VacA subunits were clearly not denatured by prolonged exposure to pH 3. However, whether the 5-S VacA monomer is the active vacuolating species or whether it must repolymerize into an oligomeric form when added to tissue culture media (at neutral pH) remains to be determined.
Figure 7.
Glycerol gradient sedimentation of VacA at neutral and acidic pH. VacA-containing samples were centrifuged through 10–35% glycerol gradients, as described in the Materials and Methods. Gradients (5 ml) were fractionated from the top, and aliquots of each fraction were resolved on 10% SDS–polyacrylamide gels. VacA was detected by immunoblotting with rabbit anti-VacA serum and enhanced chemiluminescence reagents. Gradients and samples were at pH 7.5 (A), pH 3 (B), and pH 7.5 after acidification and reneutralization of the sample (C). The position of standards sedimented in a parallel gradient is shown (bovine serum albumin [4.6 S], catalase [11.2 S], and thyroglobulin [19 S]). The peak of VacA immunoreactivity corresponds to ∼22 S at pH 7.5 and to ∼5 S at pH 3.0. High– molecular mass aggregates of VacA were detected in the bottom pellet fractions of each of the gradients.
Reassembly of the VacA Oligomer
To discriminate between these two possibilities, we next sought to determine whether VacA monomers produced by acidification could reassemble into oligomers upon reneutralization to pH 7. A VacA-containing preparation was acidified to pH 3 by the addition of dilute HCl in the presence of constant stirring and incubated at 30°C for 10 min, and then aliquots were either maintained at pH 3 or neutralized by the addition of Tris base. On glycerol gradients at pH 3, the continuously acidified VacA aliquot yielded a ∼5-S monomeric peak, as seen above. In contrast, when sedimented through a pH 7.5 gradient, the acidified and reneutralized aliquot again yielded a ∼22-S peak, indicating that the VacA monomers had reannealed (Fig. 7). This experiment was repeated after a preparation of VacA had remained at pH 3 for 7.5 h, with identical results (not shown).
Deep-etch EM imaging of the reannealed ∼22-S species revealed that they were slightly less orderly in construction than the starting material but had in large part reacquired a flower-like appearance with prominent central rings surrounded by relatively globular petals (Fig. 8). Residual disorder was manifest by an increased frequency of bifurcated or “ectopic” petals (Fig. 8, arrowheads). This gave the immediate visual impression of a much greater than normal frequency of seven-sided flowers, approaching that seen in the earlier deep-etch study (cf. Fig. 3). However, after all of the examples of petal “mismatch” were discounted, the actual increase in the proportion of oligomers with seven petals rather than six (Fig. 8) was more modest (∼10–30% increase).
Figure 8.
Rotary replica of reannealed VacA oligomers after acid-induced dissociation followed by reneutralization. Top row: Examples of complexes that reannealed into six-sided shapes like the original flowers (cf. Fig. 2). Bottom row: Examples of complexes that assumed a seven-sided configuration like those seen in the starting material of an earlier study (Lupetti et al., 1996; cf. Fig. 3). Arrowheads indicate examples of incorrect realignment of petals that typify VacA reannealings.
Three-dimensional Analysis Reveals a Second Type of Flat VacA Oligomer
Careful inspection of many replicas revealed that although >95% of flat forms displayed a clockwise chirality (as shown in Fig. 2), occasional flat forms could be found that exhibited the opposite (counterclockwise) chirality (Fig. 9). Importantly, central rings like those found in the typical VacA flowers were clearly visible in these counterclockwise forms (Fig. 9), whereas central rings were absent from the more common clockwise forms (Fig. 2). Indeed, counterclockwise forms looked so much like the usual flowers that they were not initially recognized as being flatter until the replicas were examined carefully in three dimensions. After presenting our model for VacA structure in the Discussion, we will explain why we interpret these two different types of flat forms as being views of the top and bottom faces of VacA hexamers. The presence of a visible ring in only one of the two views is an indication that the top and bottom surfaces of VacA hexamers are not identical.
Figure 9.
Stereo images of two different views of flat VacA molecules. To correspond with Fig. 10, the newly recognized counterclockwise flat forms with visible central rings are labeled as top halves, while the more common clockwise flat forms are labeled as bottom halves. The VacA flowers themselves are labeled as wholes.
Discussion
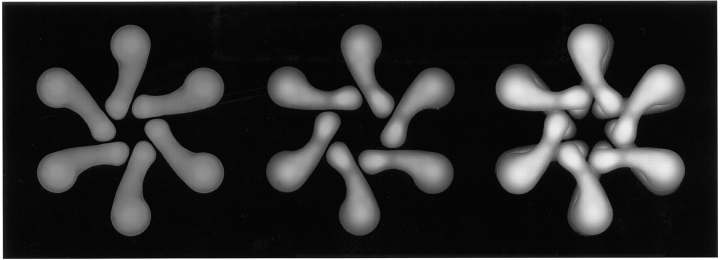
Based on the ultrastructural changes in VacA that are provoked by acidification, we propose a model in which the intact VacA flower is composed of 12 ∼90-kD subunits arranged as a symmetrical pair of six-membered arrays (Fig. 10). In this model, the flat VacA forms are thought to represent the individual six-membered arrays or halves of the complete VacA dodecamer. One critical feature of the model presented in Fig. 10 is that the six-membered flat arrays are predicted to have two distinguishable faces, one relatively featureless and the other displaying a reduced version of the central ring seen in the complete VacA flower. These two views were distinguishable in three- dimensional views such as in Fig. 9. A second feature of the model is that the dodecamer is predicted to be formed with “P2” symmetry, i.e., with its two component hexamers oriented face-to-face. This conclusion was reached because all VacA flowers looked the same in deep-etch replicas, regardless of their orientation on the mica. An important consequence of this P2 symmetry, when combined with the cant of individual VacA petals, is that the opposed hexamers would lock together to form the particularly close-knit complex shown (Fig. 10, right panel). Finally, a third prediction of the model proposed here is that the prominent central ring of the VacA flower would result from two factors: (a) from overlap of central parts of the petals in its two apposed hexamers, and (b) from outward orientation of central rings in each hexamer, as is seen in the rare counterclockwise flat forms shown in Fig. 9. It is important to note, however, that if factor b were true, it would raise the question of why intact VacA flowers do not display the counterclockwise chirality seen in this sort of flat form. We believe that this is most likely due to the vagaries of platinum replication. The intact flowers stand nearly twice as tall above the mica as the flat forms (Fig. 4) and thus receive commensurately heavier coats of platinum. Fig. 11 illustrates how a heavier deposition of platinum could obscure the chirality of the upper hexamer.
Figure 10.
A structural model of VacA based on the three different deep-etch EM views described in this report (Fig. 9). Right: The flower view, interpreted to be a dodecamer composed of two hexameric flat forms interlocked face-to-face. Center: The newly recognized hexameric flat form with counterclockwise chirality and a prominent central ring. The visible surface of this flat form corresponds to the outermost visible face of the VacA flower shown at right. Left: The more common flat form with clockwise chirality and no central ring, interpreted to be the opposite view of a VacA hexamer. This visible surface would be sandwiched in the center of the dodecamer and not normally accessible to view.
Figure 11.
Schematic view of how the deposition of platinum on intact VacA flowers would obscure their chirality. The progressive increase in highlighting is intended to represent increasing amounts of platinum deposition. This would accentuate the central ring but would largely obscure the cant of the subunits.
The model of VacA proposed in Fig. 10 differs significantly from that proposed in a previous deep-etch EM study (Lupetti et al., 1996). There, the flower was considered to be a single-layered complex composed of six or seven ∼90-kD subunits, each divided into distinct ∼34- and ∼58-kD domains, one set of which was thought to give rise to the central ring and the other to the flat form of VacA. This interpretation was based on an experiment in which a preparation of proteolytically nicked VacA yielded an unusually high proportion of flat forms, which were designated “processed” forms, implying that they were derived from flowers that had released the group of subunits that formed the central ring (Lupetti et al., 1996). The present work disagrees with that interpretation because it shows no correlation between the degree of VacA proteolysis and the relative abundance of flat forms. Despite considerable effort to confirm the earlier interpretation, including a thorough examination of VacA preparations that had undergone proteolysis during prolonged storage at 4°C as well as preparations that had been intentionally proteolysed with exogenous trypsin, no increase in the relative abundance of flat forms was ever observed. Furthermore, it became apparent in the present study that the component petals of the flat forms (especially in the clockwise views) were identical to the individual petals observed when VacA is dissociated into its component ∼90-kD monomers by acidification. This observation alone argues strongly that the flat VacA forms represent six intact ∼90-kD subunits arranged in an astral array and thus represent one-half of the complete VacA flower.
We were unable to separate flat forms from intact flowers, either by gel filtration chromatography or by density gradient centrifugation. Coupled with our finding that most flat forms occurred in clusters near the freeze-fractured surface of our replicas (see also Fig. 1 in Lupetti et al., 1996), we conclude that most of the observed flat forms must be artifacts of the EM preparation, resulting from hemisection of intact dodecameric VacA flowers during freeze-fracturing. This would explain why >95% of the observed flat forms—and all of the ones that were clustered—exhibited a clockwise chirality. Thus, regardless of which side of an intact VacA flower had adsorbed to mica, its cleavage during freeze-fracture would invariably leave behind its bottom hexamer (Fig. 10, left image).
The fact that all of the petals in acid-dissociated 12-mers display the clockwise chirality seen in most flat forms (Fig. 6), despite Fig. 10's prediction that VacA is assembled with P2 symmetry (i.e., with the upper hexamer oriented opposite from the lower), suggests that as the oligomers fall apart in acid, the petals of the upper hexamer fall down between the petals of the lower hexamer and rotate to adopt the orientation of the lower ones. An alternate explanation, which we believe is less likely, is that individual VacA monomers or petals actually have an intrinsic polarity that always makes them tend to adsorb to mica with the clockwise orientation that is seen in most flat forms. If this were true, it would suggest that VacA typically became “warped” during adsorption to mica and hence, its observed chirality might be an artifact. However, the identification of clear-cut examples of counterclockwise flat forms (Fig. 9), which are most easily interpreted as views of the opposite sides of the component hexamers, provides strong evidence that the VacA complex does indeed possess the intrinsic chirality shown in Fig. 10.
An additional finding in the present study was that the dissociated ∼90-kD VacA monomers generated by acidification were capable of reannealing into flower-like structures upon return to neutral pH. (This, of course, demonstrated that they were not completely denatured by the exposure to acidic pH.) However, the reannealed VacA oligomers appeared to display an increased proportion of seven-sided forms. Some of these obviously resulted from a mismatch of petals between the two reannealing hexamers, as shown by the arrowheads in Fig. 8; but many were as perfectly assembled as the seven-sided complexes shown in Fig. 3. These demonstrated that VacA has a clear tendency to oligomerize into 14-mers as well as 12-mers under reneutralizing conditions. As such, this raises the possibility that the preponderance of seven-sided VacA molecules observed in the earlier deep-etch study of Lupetti et al. (1996) may have resulted from inadvertent exposure to acidic pH during the purification (Manetti et al., 1995) or EM processing.
The present results demonstrate that VacA behaves somewhat like the cyclic pentameric complexes of cholera toxin and Escherichia coli heat-labile enterotoxin B-rings (CtxB and EtxB), which also disassemble when exposed to acid pH (Ruddock et al., 1995) and spontaneously reassemble at neutral pH (Hardy et al., 1988). Curiously, even though the amino acid sequences of CtxB and EtxB are ∼80% identical to each other (Domenighini et al., 1995), CtxB disassembles at around pH 3.9, whereas EtxB requires pH 2 to disassemble (Ruddock et al., 1995). Within the ∼7-nm pentameric B-rings of these enterotoxins, adjacent subunits are held together via several different sorts of hydrophobic interactions, hydrogen bonds, and salt bridges (Sixma et al., 1993). Presumably, analogous noncovalent interactions hold the larger VacA oligomers together.
The original description of VacA activation by acidification reported that changes occurred in its circular dichroism and fluorescence spectra as well as in its proteolytic fragmentation pattern but reported that no change occurred in its EM structure (de Bernard et al., 1995). We now conclude that in fact a major change in the EM structure of VacA does occur upon acidification and that the observed dissociation of the VacA oligomer could account, at least in part, for the reported changes in its circular dichroism and fluorescence spectra. We further speculate that acid-induced loosening of the oligomeric VacA structure could result in increased surface exposure of previously hidden hydrophobic VacA domains. As such, acidified VacA could more readily bind to and/or penetrate membranes. This may help to explain the observation that acidification of certain recombinant VacA fragments enhanced their capacity to penetrate lipid vesicle membranes and form ion-conductive channels (Moll et al., 1995). Similarly, when exposed to acidic conditions, several other bacterial protein toxins and tumor necrosis factor α are known to undergo conformational changes that permit them to insert into membranes and form ion-conductive channels (Boquet and Duflot, 1982; Hoch et al., 1985; Farahbakhsh et al., 1987; Dumont and Richards, 1988; Papini et al., 1988; Milne and Collier, 1993; Milne et al., 1994; Montecucco et al., 1994; Narhi et al., 1996).
Current evidence suggests that within acidic intracellular compartments, the B (binding) subunits of A-B type toxins insert into membranes, which facilitates entry of enzymatically active A subunits into the cytosol (Montecucco et al., 1994). Presumably, similar acid activation and membrane insertion of VacA also may occur within endosomes. Morphologic studies have demonstrated that VacA is indeed internalized by cells (de Bernard et al., 1995; Garner and Cover, 1996), and VacA-induced vacuolation of cells apparently depends upon the proper functioning of endosomal Rabs (Papini et al., 1997). However, H. pylori is unique in that it resides in the mucus layer of the human stomach, where it is exposed to a range of pHs, ranging from pH 7 at the depths of the mucus layer at the surface of the epithelial cells to pH 2 at the lumenal surface of the gastric mucus layer (Quigley and Turnberg, 1987). Thus, VacA may well be exposed to acidic pH and disassemble before contacting or entering its target cells. The capacity of VacA to disassemble at acid pHs is likely to be an important feature of its activation and mechanism of action.
Acknowledgments
We thank Beverly Hosse (Vanderbilt University) for technical assistance with the VacA preparation, Robyn Roth (Washington University) for producing all the deep-etch replicas and for much of the EM, and J. Hiroshi Morisaki (Washington University) for generating the final computer images of individual molecules in Figs. 10 and 11. Special thanks also to Dr. Pietro Lupetti (Department of Evolutionary Biology, University of Siena) for many stimulating discussions and suggestions, and also for providing the replicas that were photographed to generate Fig. 3.
Footnotes
Address all correspondence to Timothy L. Cover, Division of Infectious Diseases, A3310 Medical Center North, Vanderbilt University School of Medicine, Nashville, TN 37232. Tel.: (615) 322-2035. Fax: (615) 343-6160.
This project was supported by U.S. Public Health Service grants GM-29647 (J. Heuser), AI-39657, and R29 DK-45293 (T. Cover) by Human Frontiers Science Program grant RG355/94 (J. Heuser), by the Department of Veterans Affairs (T. Cover), and by a postdoctoral fellowship from the Helen Hay Whitney Foundation (P. Hanson).
References
- Atherton JC, Cao P, Peek RM, Jr, Tummuru MKR, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- Austin JW, Doig P, Stewart M, Trust TJ. Macromolecular structure and aggregation states of Helicobacter pyloriurease. J Bacteriol. 1991;173:5663–5667. doi: 10.1128/jb.173.18.5663-5667.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin JW, Doig P, Stewart M, Trust TJ. Structural comparison of urease and a GroEL analog from Helicobacter pylori. . J Bacteriol. 1992;174:7470–7473. doi: 10.1128/jb.174.22.7470-7473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ. The bacteria behind ulcers. Sci Am. 1996;274:92–97. doi: 10.1038/scientificamerican0296-104. [DOI] [PubMed] [Google Scholar]
- Boquet P, Duflot E. Tetanus toxin fragment forms channels in lipid vesicles at low pH. Proc Natl Acad Sci USA. 1982;79:7614–7618. doi: 10.1073/pnas.79.24.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL. The vacuolating cytotoxin of Helicobacter pylori. . Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. . J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- Cover TL, Blaser MJ. Helicobacter pyloriinfection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med. 1996;41:85–117. [PubMed] [Google Scholar]
- Cover TL, Puryear W, Perez-Perez GI, Blaser MJ. Effect of urease on HeLa cell vacuolation induced by Helicobacter pyloricytotoxin. Infect Immun. 1991;59:1264–1270. doi: 10.1128/iai.59.4.1264-1270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cover TL, Tummuru MKR, Cao P, Thompson SA, Blaser MJ. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pyloristrains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- de Bernard M, Papini E, de Filippis V, Gottardi E, Telford J, Manetti R, Fontana A, Rappuoli R, Montecucco C. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. J Biol Chem. 1995;270:23937–23940. doi: 10.1074/jbc.270.41.23937. [DOI] [PubMed] [Google Scholar]
- Domenighini M, Pizza M, Jobling MG, Holmes RK, Rappuoli R. Identification of errors among database sequence entries and comparison of correct amino acid sequences for the heat-labile enterotoxins of Escherichia coli and Vibrio cholerae. . Mol Microbiol. 1995;15:1165–1167. doi: 10.1111/j.1365-2958.1995.tb02289.x. [DOI] [PubMed] [Google Scholar]
- Dumont ME, Richards FM. The pH-dependent conformational change of diphtheria toxin. J Biol Chem. 1988;263:2087–2097. [PubMed] [Google Scholar]
- Farahbakhsh ZT, Baldwin RL, Wisnieski BJ. Effect of low pH on the conformation of Pseudomonas exotoxinA. J Biol Chem. 1987;262:2256–2261. [PubMed] [Google Scholar]
- Figura N, Guglielmetti P, Rossolini A, Barberi A, Cusi G, Musmanno RA, Russi M, Quaranta S. Cytotoxin production by Campylobacter pyloristrains isolated from patients with peptic ulcers and from patients with chronic gastritis only. J Clin Microbiol. 1989;27:225–226. doi: 10.1128/jcm.27.1.225-226.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner JA, Cover TL. Binding and internalization of Helicobacter pyloricytotoxin by epithelial cells. Infect Immun. 1996;64:4197–4203. doi: 10.1128/iai.64.10.4197-4203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy SJS, Holmgren J, Johansson S, Sanchez J, Hirst TR. Coordinated assembly of multisubunit proteins: oligomerization of bacterial enterotoxins in vivo and in vitro. Proc Natl Acad Sci USA. 1988;85:7109–7113. doi: 10.1073/pnas.85.19.7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylik SJ, Wasilko DJ, Haskell SL, Gootz TD, Lee SE. Bisulfite or sulfite inhibits growth of Helicobacter pylori. . J Clin Microbiol. 1994;32:790–792. doi: 10.1128/jcm.32.3.790-792.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE. Procedure for freeze-drying molecules adsorbed to mica flakes. J Mol Biol. 1983;169:155–195. doi: 10.1016/s0022-2836(83)80179-x. [DOI] [PubMed] [Google Scholar]
- Heuser JE. Development of the quick-freeze, deep-etch, rotary-replication technique of sample preparation for 3-D electron microscopy. Prog Clin Biol Res. 1989;295:71–83. [PubMed] [Google Scholar]
- Heuser JE, Zhu Q, Clarke M. Proton pumps populate the contractile vacuoles of Dictyostelium amoebae. . J Cell Biol. 1993;121:1311–1327. doi: 10.1083/jcb.121.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch DH, Romero-Mira M, Ehrlich BE, Finkelstein A, DasBupta BR, Simpson LL. Channels formed by botulinum, tetanus, and diphtheria toxins in planar lipid bilayers: relevance to translocation of proteins across membranes. Proc Natl Acad Sci USA. 1985;82:1692–1696. doi: 10.1073/pnas.82.6.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LT, Mobley HLT. Purification and NH2-terminal analysis of urease from Helicobacter pylori. . Infect Immun. 1990;58:992–998. doi: 10.1128/iai.58.4.992-998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, Dallai R, Rappuoli R, Telford JL. Oligomeric and subunit structure of the Helicobacter pylorivacuolating cytotoxin. J Cell Biol. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti R, Massari P, Burroni D, de Bernard M, Marchini A, Olivieri R, Papini E, Montecucco C, Rappuoli R, Telford JL. The Helicobacter pyloricytotoxin: importance of native conformation for induction of neutralizing antibodies. Infect Immun. 1995;63:4476–4480. doi: 10.1128/iai.63.11.4476-4480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti M, Arico B, Burroni D, Figura N, Rappuoli R, Ghiara P. Development of a mouse model of Helicobacter pyloriinfection that mimics human disease. Science (Wash DC) 1995;267:1655–1658. doi: 10.1126/science.7886456. [DOI] [PubMed] [Google Scholar]
- Martin RG, Ames BN. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961;236:1372–1379. [PubMed] [Google Scholar]
- Milne JC, Collier RJ. pH-dependent permeabilization of the plasma membrane of mammalian cells by anthrax protective antigen. Mol Microbiol. 1993;10:647–653. doi: 10.1111/j.1365-2958.1993.tb00936.x. [DOI] [PubMed] [Google Scholar]
- Milne JC, Furlong D, Hanna PC, Wall JS, Collier RJ. Anthrax protective antigen forms oligomers during intoxication of mammalian cells. J Biol Chem. 1994;269:20607–20612. [PubMed] [Google Scholar]
- Moll G, Papini E, Colonna R, Burroni D, Telford J, Rappuoli R, Montecucco C. Lipid interaction of the 37-kDa and 58-kDa fragments of the Helicobacter pyloricytotoxin. Eur J Biochem. 1995;234:947–952. doi: 10.1111/j.1432-1033.1995.947_a.x. [DOI] [PubMed] [Google Scholar]
- Montecucco C, Papini E, Schiavo G. Bacterial protein toxins penetrate cells via a four-step mechanism. FEBS Lett. 1994;346:92–98. doi: 10.1016/0014-5793(94)00449-8. [DOI] [PubMed] [Google Scholar]
- Narhi LO JS, Philo, Li T, Zhang M, Samal B, Arakawa T. Induction of α-helix in the β-sheet protein tumor necrosis factor-α: acid-induced denaturation. Biochemistry. 1996;35:11454–11460. doi: 10.1021/bi952767n. [DOI] [PubMed] [Google Scholar]
- Papini E, Sandona D, Rappuoli R, Montecucco C. On the membrane translocation of diphtheria toxin: at low pH the toxin induces ion channels on cells. EMBO (Eur Mol Biol Organ) J. 1988;7:3353–3359. doi: 10.1002/j.1460-2075.1988.tb03207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papini E, de Bernard M, Milia E, Bugnoli M, Zerial M, Rappuoli R, Montecucco C. Cellular vacuoles induced by Helicobacter pylorioriginate from late endosomal compartments. Proc Natl Acad Sci USA. 1994;91:9720–9724. doi: 10.1073/pnas.91.21.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papini E, Satin B, Bucci C, de Bernard M, Telford JL, Manetti R, Rappuoli R, Zerial M, Montecucco C. The small GTP binding protein rab7 is essential for cellular vacuolation induced by Helicobacter pyloricytotoxin. EMBO (Eur Mol Biol Organ) J. 1997;16:15–24. doi: 10.1093/emboj/16.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley EMM, Turnberg LA. pH of the microclimate lining human gastric and duodenal mucosa in vivo: studies in control subjects and in duodenal ulcer patients. Gastroenterology. 1987;92:1876–1884. doi: 10.1016/0016-5085(87)90619-6. [DOI] [PubMed] [Google Scholar]
- Ruddock LW, Ruston SP, Kelly SM, Price NC, Freedman RB, Hirst TR. Kinetics of acid-mediated disassembly of the B subunit pentamer of Escherichia coliheat-labile enterotoxin: molecular basis of pH stability. J Biol Chem. 1995;270:29953–29958. doi: 10.1074/jbc.270.50.29953. [DOI] [PubMed] [Google Scholar]
- Sixma TK, Kalk KH, van Zanten BAM, Dauter Z, Kingma J, Witholt B, Hol WGL. Refined structure of Escherichia coliheat-labile enterotoxin, a close relative of cholera toxin. J Mol Biol. 1993;230:890–918. doi: 10.1006/jmbi.1993.1209. [DOI] [PubMed] [Google Scholar]
- Telford JL, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce MF, Censini S, Covacci A, Xiang Z, et al. Gene structure of the Helicobacter pyloricytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994a;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Covacci A, Ghiara P, Montecucco C, Rappuoli R. Unravelling the pathogenic role of Helicobacter pyloriin peptic ulcer: potential new therapies and vaccines. Trends Biotechnol. 1994b;12:420–426. doi: 10.1016/0167-7799(94)90031-0. [DOI] [PubMed] [Google Scholar]
- Williams RC, Wyckoff RWG. Applications of metallic shadow-casting to microscopy. J Appl Physics. 1946;17:23–33. [Google Scholar]