Abstract
Drosophila kelch has four protein domains, two of which are found in kelch-family proteins and in numerous nonkelch proteins. In Drosophila, kelch is required to maintain ring canal organization during oogenesis. We have performed a structure–function analysis to study the function of Drosophila kelch. The amino-terminal region (NTR) regulates the timing of kelch localization to the ring canals. Without the NTR, the protein localizes precociously and destabilizes the ring canals and the germ cell membranes, leading to dominant sterility. The amino half of the protein including the BTB domain mediates dimerization. Oligomerization through the amino half of kelch might allow cross-linking of ring canal actin filaments, organizing the inner rim cytoskeleton. The kelch repeat domain is necessary and sufficient for ring canal localization and likely mediates an additional interaction, possibly with actin.
In Drosophila, kelch is required in the ovary for production of viable eggs. kelch mutants are female sterile as a result of defective cytoplasm transport throughout oogenesis. Kelch is a structural component of the ring canals that provides the intercellular conduits through which cytoplasm is transported from the nurse cells to the oocyte in an egg chamber (for review see Robinson and Cooley, 1996). kelch mutant ring canals have actin filaments that are disorganized and extend into the lumen of the ring canal (Robinson et al., 1994; Tilney et al., 1996). This disorganization apparently causes partial obstruction of the ring canal lumen so that cytoplasm transport is impaired.
Ring canal assembly is initiated with the arrest of mitotic cleavage furrows followed by the addition of several proteins including a protein that immunoreacts with anti-phosphotyrosine antibodies (PY protein), a product of the hu-li tai shao (hts) locus (Yue and Spradling, 1992) called the hts ring canal protein (hts-RC; Robinson et al., 1994), actin filaments, and kelch. Kelch is the last of the known proteins to be localized to the ring canal complex (Robinson et al., 1994), and it does not arrive on all ring canals until after the maximum number of actin filaments has been recruited to the ring canal (Tilney et al., 1996). From the time kelch has reached all ring canals until the end of oogenesis, the ring canals expand from a diameter of 3–4 to 10 μm. In kelch mutants, the ring canals have a normal morphology until the time when kelch would normally reach all of the ring canals. After this time, the ring canals become disorganized. These data suggest that kelch is required to maintain the organization of the actin filaments during the expansion of the ring canal rather than being required for assembly (Robinson et al., 1994; Tilney et al., 1996).
The Drosophila kelch gene produces a single transcript separated into two open reading frames (ORFs) by a UGA stop codon. ORF1 only and full length (ORF1 plus ORF2) kelch proteins are made (Xue and Cooley, 1993; Robinson and Cooley, 1997b ). The ORF1 product is a member of a family of kelch-related proteins that includes several Pox virus ORFs (Koonin et al., 1992; Senkevich et al., 1993), mammalian calicin (von Bülow et al., 1995), and Caenorhabditis elegans spe26 (Varkey et al., 1995). Currently, the protein databases contain four kelch family proteins from C. elegans and at least five mammalian kelch family proteins. Interestingly, the Drosophila kelch ORF1 contains ∼110 amino acids (the NTR; see Fig. 1 A) at the amino terminus not found in other kelch-related proteins. The Drosophila kelch ORF2 domain encodes a protein with no significant homology to known proteins and so far is specific to Drosophila. Although the two kelch protein (ORF1 and full length) motif is conserved in several Drosophila species, the ORF1 protein is sufficient for kelch function (Robinson and Cooley, 1997b ).
Figure 1.
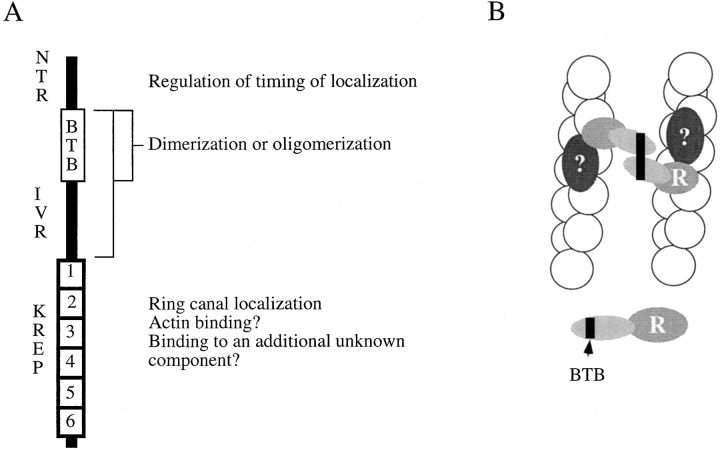
Summary of kelch domain structure and the transgenes studied. (A) Diagram of domain structure of kelch ORF1. NTR includes the amino-terminal of ∼110 amino acids, and the BTB domain includes the next 120 amino acids. The IVR is the 147-amino acid intervening region between the BTB and the six 50-amino acid kelch repeat domain (KREP). (B) Amino-terminally myc epitope-tagged domains expressed in the ovary using the pCOG vector. The number of independent lines generated and the number of lines that showed a high level of expression are listed. The subcellular localization of each protein in various genetic backgrounds is provided.
Kelch ORF1 contains two conserved domains found in other kelch proteins as well as in nonkelch proteins (Fig. 1 A). The first of these, the BTB or POZ domain, is a 120-amino acid motif that is found immediately after the amino-terminal region (NTR)1 in kelch. This domain is also found in several zinc finger-containing transcription factors (Harrison and Travers, 1990; Chardin et al., 1991; DiBello et al., 1991; Zollman et al., 1994) and it has been shown to mediate dimerization in vitro (Bardwell and Treisman, 1994; Chen et al., 1995). A second domain consists of six 50-amino acid repeats known as kelch repeats (see Fig. 1 A; Xue and Cooley, 1993). Kelch repeats are found in several nonkelch proteins including a recently characterized protein in Physarum polycephalum called actin-fragmin kinase (Eichinger et al., 1996). The kelch repeat sequence is predicted to fold into a superbarrel or β-flower structure (Bork and Doolittle, 1994), similar to the repeat sequences in a family of bacterial, fungal, and influenza virus enzymes such as neuraminidase, galactose oxidase, and the sialidases (Varghese et al., 1983; Ito et al., 1994).
Suggestions of the function of the kelch repeat domain come from Limulus proteins called scruin (Tilney, 1975). The Limulus genome contains at least three scruin genes, two of which encode α- and β-scruins (Way et al., 1995a ,b). Each scruin consists of two sets of kelch repeats, one in the amino-terminal half and a second at the carboxy-terminal half of the protein. α-scruin was originally identified as an actin filament cross-linking protein found in the acrosomal actin bundle of the Limulus sperm (Tilney, 1975). Subsequent work has suggested that each kelch repeat domain in α-scruin forms an actin-binding site (Bullitt et al., 1988; Owen and DeRosier, 1993; Schmid et al., 1994; Way et al., 1995b ). C. elegans spe26 is required for a normal actin cytoskeleton during spermatogenesis (Varkey et al., 1995), consistent with the repeat motif providing an interaction with actin. β-scruin, on the other hand, is found in the acrosomal vesicle and may not associate with actin (Way et al., 1995a ). Mammalian calicin localizes to the calyx, a large nonfilamentous cytoskeletal structure that associates with the sperm head nucleus (von Bülow et al., 1995). The calyx does not contain much, if any, filamentous actin. Consequently, it is possible that the kelch repeat domain can mediate a diversity of functions in different kelch family proteins. However, an attractive hypothesis for Drosophila kelch function is that it associates with the ring canal actin, either directly or indirectly, through the kelch repeats, and cross-links the actin filaments by dimerizing through the BTB domain.
To assign functions to the domains of Drosophila kelch, we expressed epitope-tagged kelch ORF1 domains in the ovary and examined their ability to rescue fertility and ring canal morphology of kelch mutants. Their subcellular distribution in wild type and kelch mutant backgrounds was also determined. These experiments uncovered a dominant-negative kelch construct whose behavior suggests that the NTR is required to regulate the timing of kelch localization to the ring canals but is not required otherwise for kelch function. The BTB domain and the 147 amino acids (IVR) intervening between the BTB and kelch repeat domain (collectively called BTBIVR) is required for kelch complex formation. The kelch repeat domain (KREP) is necessary and sufficient for ring canal localization. These data support a model in which kelch ORF1 functions as an oligomeric ring canal actin organizer.
Materials and Methods
Fly Strains
w1118 (Lindsley and Zimm, 1992) flies were used as wild type in these experiments. The hypomorphic kelch mutant, kelneo, was described by Xue and Cooley (1993) and Robinson and Cooley (1997b). The molecular null kelch mutant, kelDE1, was isolated by Schüpbach and Wieschaus (1991) and characterized in Robinson and Cooley (1997b). Transgenic flies expressing untagged kelch ORF1 protein, an alanine-substituted (Alanine), serine-substituted (Serine), and UGA-deleted (ΔUGA) full length kelch proteins were described by Robinson and Cooley (1997b). Fly stocks were maintained under standard conditions.
Construct Generation
To express epitope-tagged domains in the fly ovary, we prepared a myc epitope tagging cassette (KNmyc; Robinson and Cooley, 1997b ) that begins with an HpaI site that is found 81 bp upstream of the first ATG in the kelch 5′UTR and includes the first 19 amino acids of kelch that include two potential start methionines. The myc 9E10 epitope (AEEQKLISEEDLN; Evan et al., 1985) was placed after kelch residue 19. The following polylinker in the following reading frame was placed after the myc epitope:
| EcoRI | NdeI | XhoI | BglII | Stop | NotI |
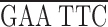
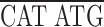
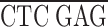 GC
GC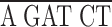 G TAA
G TAA 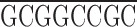
The cassette was subcloned into pCOG, a germline expression vector (Robinson and Cooley, 1997b ). All truncations of kelch were made by preparing the appropriate cDNA fragments with back-to-back EcoRI and NdeI sites at the 5′ end of the fragments and a BglII or BamHI sites at the 3′ end of the cDNA fragment. We found that this combination of sites in this reading frame allowed us to move the cDNA fragments easily between pCOG and expression vectors, which allows us to express the proteins in Drosophila Schneider cells, bacteria, or Pichia.
The cDNA fragments were prepared by PCR amplification and subcloned. The subcloned fragments were analyzed by dideoxynucleotide sequencing to make sure no unwanted mutations had been introduced. The following kelch domains were prepared. The amino acid numbers correspond to the sequence in Xue and Cooley (1993): ORF1: S17 - M688; ORF1-R: S17 - P401; BTB IVR: Q120 - P401; BTB: Q120 - L252; IVR: D253 - P401; IVR KREP: D253 - M688; KREP: M400 - M688; ΔNORF1: Q120 - M688.
Generation of Transgenic Animals
Transformation plasmids were microinjected along with the Δ2-3 transposase helper plasmid into w1118 syncytial blastoderm embryos according to standard techniques. Transgenic flies were identified by the complementation of the white eye phenotype by the white mini-gene in pCOG. P element insertions were maintained in a wild-type background as well as in kelneo and kelDE1 backgrounds.
Western Analysis
Western analysis of Drosophila ovarian extracts was performed as described in Robinson and Cooley (1997b). For anti-kelch monoclonal antibodies, we used anti-kelch 1B (Xue and Cooley, 1993). For anti–hts-RC, we used anti-hts 655 4B (Robinson et al., 1994). For anti-myc staining, we used anti-myc 9E10 (Evan et al., 1985). For anti-KREP, we used mouse polyclonal sera #5-6 (Xue and Cooley, 1992).
Immunolocalization and Confocal Imaging
Immunocytochemical analysis of whole ovaries was performed as described in Robinson and Cooley (1997b). For actin visualization, ovaries were stained with rhodamine-conjugated phalloidin (Molecular Probes, Inc., Eugene, OR). For antibody staining, ovaries were immunostained with hts-RC antibody hybridoma supernatant, anti–hts-RC 655 4B (1:1 dilution; Robinson et al., 1994), kelch hybridoma supernatant, anti-kelch 1B (1:1 dilution; Xue and Cooley, 1993), or anti-myc 9E10 hybridoma supernatant (1:1 dilution; Evan et al., 1985).
Nuclear Staining
Nuclear staining was performed as described in Robinson et al. (1997). We dissected and fixed ovaries from 10–20 females per genotype as described for immunocytochemistry. During the third from final wash in PBT (1 × PBS, 0.3% Triton-X 100, 0.5% BSA), DAPI (Molecular Probes, Inc.) was added to the PBT at 1 μg/ml. The specimens were washed two more times and were examined and photographed using a microscope (Axiophot; Zeiss, Inc., Thornwood, NY) with a 40× objective (1.3 NA).
Expression, Purification, and Analysis of Recombinant BTB and BTBIVR Proteins
To prepare His-tagged BTB (his:BTB) and BTBIVR (his:BTBIVR) proteins, the respective cDNA fragments were cloned into pET 14b (Novagen, Madison, WI). Proteins were expressed using the HMS174: DE3::pLys S bacterial strain (Novagen). The bacteria were lysed using a French press in a medium salt buffer. The proteins were extensively purified by affinity chromatography using Ni2+/NTA agarose resin (Qiagen, Chatsworth, CA).
The molecular weight of his:BTB was determined by SDS–polyacrylamide gel electrophoresis and by dynamic light scattering on a Dynapro-801 instrument. His:BTB and his:BTBIVR proteins were also analyzed by gel filtration on a superose 12 column using an FPLC (Pharmacia Fine Chemicals, Piscataway, NJ) system. For gel filtration, retention times were compared to those of bovine serum albumin (65 kD), ovalbumin (43 kD), carbonic anhydrase (29 kD), and lactalbumin (19.3 kD).
Results
Expression of myc Epitope-Tagged Domains
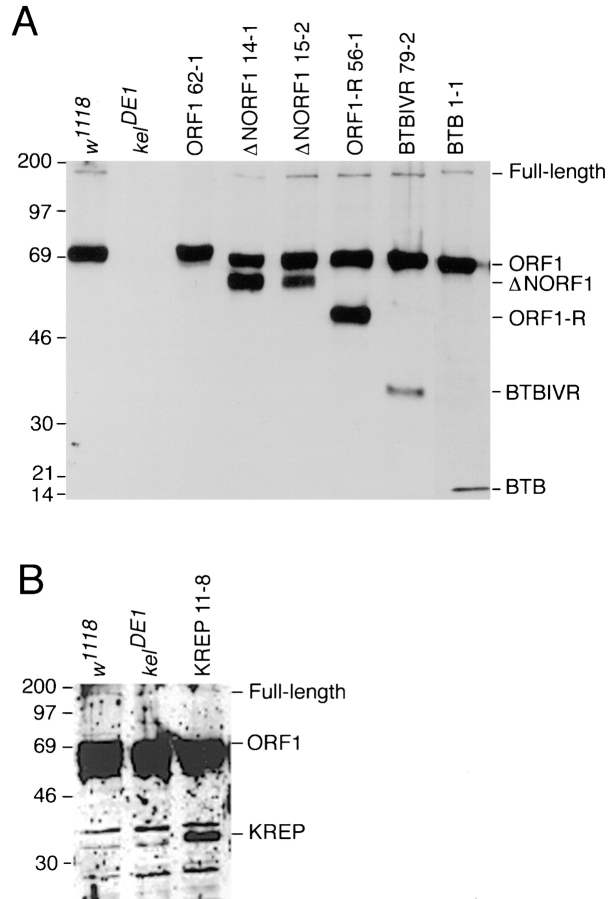
To investigate the mechanism by which kelch organizes the ring canal actin cytoskeleton, myc epitope-tagged domains were expressed in the ovary using the pCOG vector. A panel of myc epitope-tagged, truncated kelch domain constructs was engineered, and transgenic lines were established (Fig. 1 B). Western analysis of the lines using anti-kelch 1B antibodies (Xue and Cooley, 1993) and anti-kelch repeat polyclonal sera showed the expression of proteins of the expected size (Fig. 2). Myc:ORF1, myc: ΔNORF1, myc:ORF1-R, myc:BTBIVR, and myc:BTB produced proteins that could be recognized by anti-kelch 1B, which recognizes the BTB domain. Myc:KREP is not recognized by anti-kelch 1B but is recognized by polyclonal sera generated against the repeat region of kelch (Xue, 1992). For some of the transgenes, we had no difficulty isolating lines expressing high levels of protein. However, for some of the transgenes encoding truncated proteins, we generated over 20 inserts to obtain a single useful line (Fig. 1 B). We attribute the difficulty of isolating high expressing lines of some of the proteins to instability of the truncated proteins. We tested the subcellular localization of each transgenically produced protein in the presence or absence of the endogenous kelch proteins using either wild-type, kelneo (expresses a small amount of endogenous kelch), or the kelDE1 (expresses no detectable kelch) mutant backgrounds (Robinson and Cooley, 1997b ). We also tested whether the truncated proteins could rescue the kelch mutant defects or whether the proteins had any dominant-negative activity (summarized in Fig. 1 B; presented below).
Figure 2.
Expression of myc epitope-tagged domains verified by Western analysis. (A) The endogenous 76-kD ORF1 and 160-kD full length kelch proteins are detected in wild-type (w1118) egg chambers; both proteins are absent in kelDE1 egg chambers. An ∼80-kD myc:ORF1 product is detected in kelDE1 egg chambers. A 65-kD myc:ΔNORF1 (two representative lines, 14-1 and 15-2, are shown), a 47-kD myc:ORF1-R, a 35-kD myc:BTBIVR, and a 17-kD myc:BTB protein are detected in wild-type egg chambers by anti-kelch 1B antibodies. (B) A 33-kD myc:KREP product is detected in one transgenic line with unpurified anti-KREP polyclonal antibodies. Endogenous kelch proteins are also detected, although the ORF1 product is significantly obscured by a prominent group of nonkelch proteins with which the polyclonal antibodies cross-react. These cross-reacting proteins are also detected in kelDE1extracts. Only the expected 33-kD KREP protein is unique to the myc:KREP line.
Deletion of the NTR Region
Removal of the NTR region (myc:ΔNORF1) produced a protein that had severe dominant-negative effects on egg chamber morphology and ring canal stability. As a positive control, myc:ORF1 rescued all kelch mutant defects, and the protein could be seen on ring canals in a wild-type or kelch mutant background (Fig. 3, A and B; Robinson and Cooley, 1997b ). In addition, no deleterious effects were observed when the myc:ORF1 protein was expressed in a wild-type background. In contrast, we examined carefully several independent lines expressing myc:ΔNORF1 in a wild-type background, all of which showed a dramatic loss of nurse cell plasma membrane integrity (Fig. 3, C and D). This dominant-negative effect began around stage 6 of oogenesis (not shown). The defect was proportional to the level of expression of the myc:ΔNORF1 transgene product; lines expressing lower levels of protein had less dramatic effects, and very low expressing lines had no dominant-negative effects (not shown). Ring canals became difficult to detect after stage 6, and those that did persist were often thin with a larger diameter than normal or had a much reduced diameter. Of those that persisted, myc: ΔNORF1 and actin were seen in the ring canal (Fig. 3, E and F). The loss in membrane stability caused multinucleate nurse cells, and nurse cell nuclei frequently moved into the oocyte. The number of nuclei transported into the oocyte ranged from 0 to 15 with an average of ∼4–7 nuclei per oocyte depending on the line (Fig. 3, G–I).
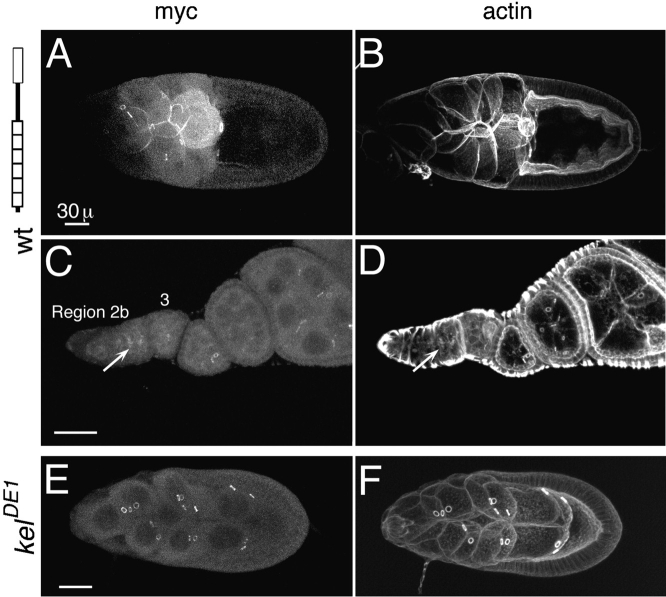
Figure 3.
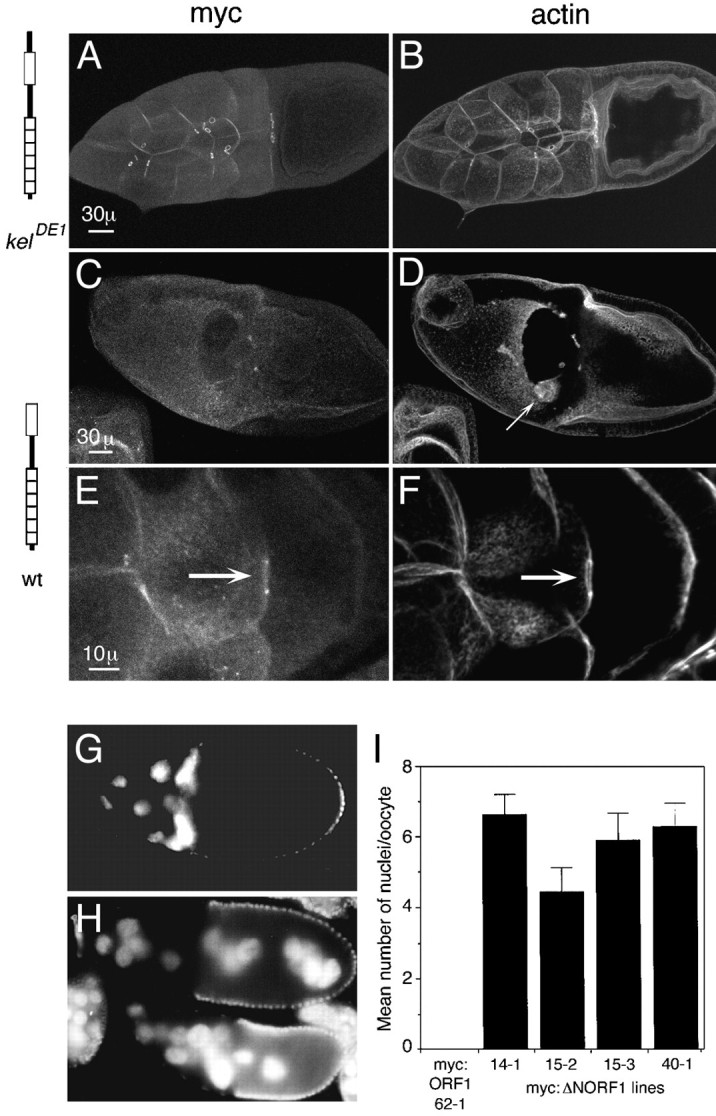
Dominant-negative effect caused by myc:ΔNORF1. (A) Myc:ORF1 localizes normally to ring canals. Some protein is also observed localizing to the cortical cytoskeleton. (B) The ring canals and nurse cell plasma membranes in kelDE1 egg chambers that express myc:ORF1 are intact and normal, as seen by visualizing filamentous actin. (C–F) Wild-type egg chambers expressing myc:ΔNORF1. (C) In wild-type stage 10 egg chambers that express myc:ΔNORF1, the nurse cell plasma membranes are severely disrupted and in this case missing. No ring canals are observed by staining for myc:ΔNORF1. (D) Actin staining of the same egg chamber as in C verified that the nurse cell membranes are completely disrupted. The border cells (arrow) have migrated toward the oocyte but are mislocalized, probably because there is no place for them to dock. (E) An occasional ring canal persists in this stage 9 egg chamber, and myc:ΔNORF1 (arrow) can be seen localizing to it. (F) Actin is also localized to the same thin ring canal. Cortical actin is also seen on some of the nurse cell membranes. (A, C, and E) Anti-myc staining to detect the transgene product. (B, D, and F) Rhodamine-conjugated phalloidin staining to detect filamentous actin. (G) A wild-type egg chamber expressing myc:ORF1 has normal positioning of the nuclei in the nurse cells. (H) Wild-type egg chambers expressing myc: ΔNORF1 have nuclei transported into the oocyte. (G and H) Nuclei are detected by DAPI staining. (I) Comparison of the number of nuclei transported into the oocytes in four independent wild-type lines that express myc:ΔNORF1. No nuclei were observed transported into the oocytes in wild-type egg chambers that express myc:ORF1 (line 62-1). 17–30 stage 10 or older egg chambers from 5–10 females of each line were counted for the averages. Histograms represent the means, and the error bars represent the standard error of the mean. The phenotype was observed in several additional trials although they were not quantitated. Bars: (A and C) 30 μm; (E) 10 μm.
The NTR domain controls timing of kelch localization to ring canals. In wild-type egg chambers and in kelDE1 egg chambers expressing myc:ORF1, kelch protein began to localize to ring canals in stage 1 egg chambers, but kelch was not readily visible on all ring canals until around stage 4 (Fig. 4, A and B). However, myc:ΔNORF1 began to localize to ring canals much earlier beginning in region 1 in the germarium (Fig. 4 C). This is very early in ring canal development at a time before hts-RC and actin begin to form the ring canal inner rim (Fig. 4 D; Robinson et al., 1994). Localization of myc:ΔNORF1 to ring canals in region 1 occurred in wild-type and kelch mutant ovaries. The amount of protein detected on ring canals at this stage was proportional to the level of expression of the transgene (not shown).
Figure 4.
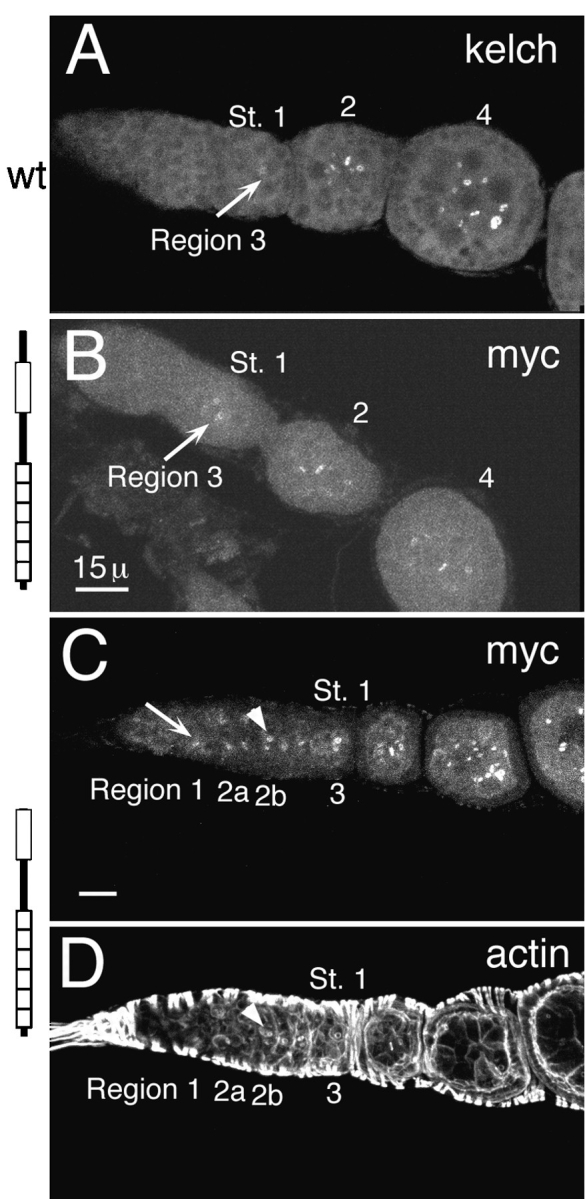
Myc:ΔNORF1 localizes to ring canals earlier than normal. (A) Endogenous kelch localized to ring canals (arrow) beginning in region 3/stage 1. It takes until stage 4 for it to be detected on all of the ring canals in the egg chamber. This ovariole was stained with anti-kelch 1B antibodies. (B) Myc:ORF1 began to localize to ring canals (arrow) also in region 3/stage 1 egg chambers. It also took a few stages to reach all of the ring canals in the egg chamber. (C) Myc:ΔNORF1 began to localize to region 1 ring canals (arrow). The ring canals identified by the arrowhead correspond to those identified by the arrowhead in D. B and C were stained with anti-myc 9E10 antibodies to recognize the transgene products. (D) The same germarium as in C stained with rhodamine-conjugated phalloidin reveals the accumulation of the robust inner rim of actin filaments. The inner rim of actin filaments begin to accumulate by region 2a but are not really obvious until region 2b (arrowhead). The germaria in B–D are kelDE1 mutants expressing the respective transgene. Bars, 15 μm.
One myc:ΔNORF1 line, called myc:ΔNORF1 15-2, had a less dramatic dominant-negative effect and was capable of rescuing the ring canal morphology and fertility of kelDE1 mutants. This was surprising since the other lines expressing myc:ΔNORF1 at high levels had very similar dominant-negative effects on ring canal and plasma membrane stability in kelneo and kelDE1 backgrounds. In egg chambers carrying myc:ΔNORF1 15-2 in a wild-type background, many of the plasma membranes remained intact, and egg chamber morphology was frequently quite normal (Fig. 5, A and B). The egg chambers did have some plasma membrane disruption as indicated by the number (around 4) of nurse cell nuclei observed in the oocyte (Fig. 3 I). Myc:ΔNORF1 protein localization to ring canals (Fig. 5 A) began earlier than normal at germarial region 2b (Fig. 5, C and D) but did not accumulate as much or as rapidly as in the higher expressing lines. This line rescued the fertility of kelDE1 mutants and rescued ring canal morphology (Fig. 5, E and F). We did not observe inappropriate transport of nurse cell nuclei into the oocytes in egg chambers from kelDE1 flies carrying myc:ΔNORF1 15-2 (not shown).
Figure 5.
Myc:ΔNORF1 expressed at a low level can rescue kelDE1 mutants. (A) Myc:ΔNORF1 15-2 expresses myc: ΔNORF1 at a lower level and localizes to ring canals of a wild-type stage 10 egg chamber. (B) The same egg chamber as in A stained for actin shows intact nurse cell membranes and many intact ring canals. (C) The same line myc:ΔNORF1 15-2 in a wild-type background expressed myc: ΔNORF1 that localized to region 2b ring canals (arrow), which is earlier than normal, but the protein was not as abundant. (D) Actin staining of the same germarium as in C also showed actin localization to ring canals (arrow) at the same time as myc:ΔNORF1. (E) Myc:ΔNORF1 15-2 in a stage 9 kelDE1 egg chamber rescued the ring canals and egg chamber morphology. Myc:ΔNORF1 protein localized to ring canals normally. (F) Actin staining of the same egg chamber in E revealed intact nurse cell membranes and ring canals with a robust, well organized actin inner rim. A, C, and E were stained with anti-myc 9E10 antibodies. Bars, 30 μm.
KREP Is Necessary and Sufficient for Ring Canal Localization
To examine the requirements for ring canal localization, we expressed ORF1 minus the kelch repeat domain (myc: ORF1-R) and ORF1-R minus the NTR (myc:BTBIVR) in the ovary. Each of these proteins localized to ring canals in wild-type flies but failed to localize in kelneo or kelDE1 flies (Fig. 6, B–E). ORF1-R also localized to ring canals in kelDE1 flies when nonepitope-tagged ORF1-only protein or full length kelch proteins, generated by substituting an alanine (alanine full length) or serine (serine full length) residue for the UGA stop codon or by deleting the stop codon (ΔUGA), were provided transgenically (Table I). This indicates that ORF1-R and BTBIVR proteins require a KREP-containing kelch protein for ring canal localization. To test whether KREP is sufficient for localization, we expressed the kelch repeat domain (myc:KREP). We did not detect localization of myc:KREP to ring canals in wild-type egg chambers, but in kelneo and kelDE1 egg chambers, it localized to ring canals (Fig. 6, F and G). This indicates that KREP is sufficient for ring canal localization. A myc:IVRKREP protein also localized to ring canals only in kelch mutants (not shown). Finally, we asked whether ORF1-R could interact with the KREP domain or if it interacted with the amino half of kelch ORF1. We generated kelDE1 flies that expressed myc:ORF1-R and myc:KREP. Anti-myc antibodies gave ring canal staining as expected for myc:KREP. However, antibodies to the BTB domain (anti-kelch 1B) failed to stain ring canals, showing that myc:ORF1-R did not localize to ring canals (Table I). This indicates that ORF1-R interacts with the amino half of an intact kelch ORF1 protein to localize to ring canals.
Figure 6.
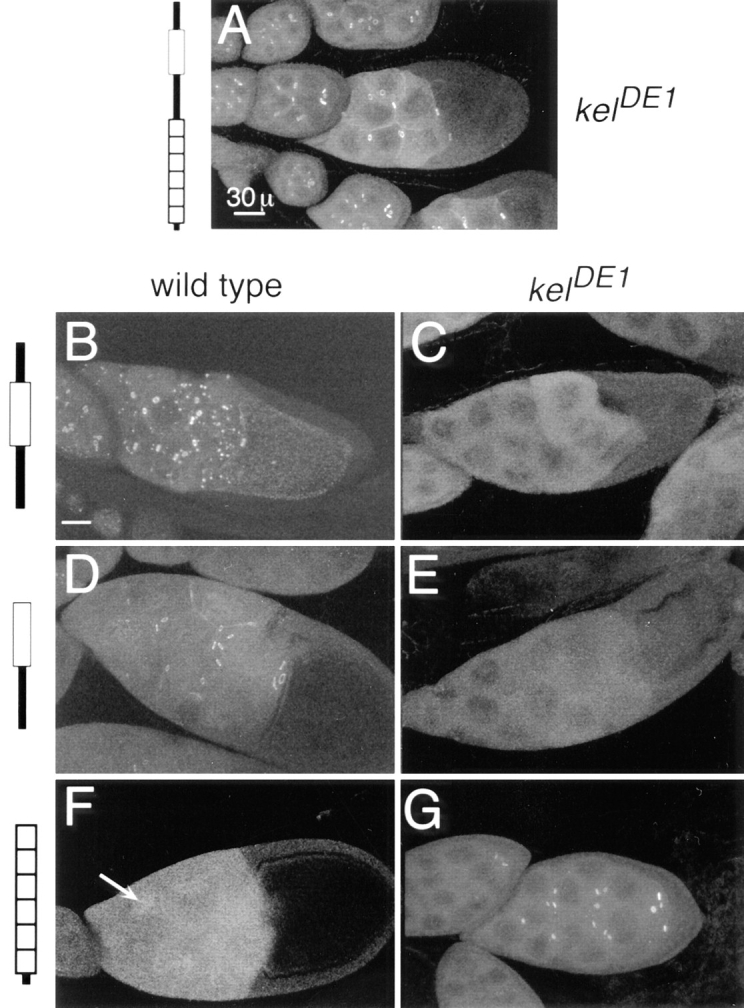
The BTBIVR region depends on an intact ORF1 for ring canal localization. The KREP domain is necessary and sufficient for ring canal localization. Myc:ORF1 localizes to and rescues kelDE1 ring canals (A). Myc:ORF1-R localized to ring canals in wild-type egg chambers (B) but not to ring canals in kelDE1 egg chambers (C). In wild-type egg chambers (B), myc:ORF1-R also formed punctate structures in the cytoplasm of the nurse cells. These structures contain no detectable PY protein, hts-RC, or actin (not shown) and appeared to be dependent on the dosage of myc:ORF1-R. Similar structures were seldomly observed in kelDE1 egg chambers (C). Myc:BTBIVR localized to ring canals in wild-type egg chambers (D) but not to ring canals in kelDE1 egg chambers (E). Myc:KREP was not detected on ring canals in wild-type egg chambers (F) but did localize to ring canals in kelDE1 egg chambers (G). Some nuclear localization (F, arrow) of KREP was observed in wild-type egg chambers. All egg chambers were stained with anti-myc 9E10 antibodies. Bars: (A) 30 μm; (B–G) 30 μm.
Table I.
Myc:ORF1-R Localization Summary
| Genetic background | myc:ORF1-R | |
|---|---|---|
| Wild type | Ring canals | |
| kelneo | Cytoplasm | |
| kelDE1 | Cytoplasm | |
| ORF1 alone | Ring canals | |
| Alanine full length | Ring canals | |
| ΔUGA full length | Ring canals | |
| Serine full length* | Ring canals | |
| KREP | Cytoplasm |
Serine full length protein was tested in a kelneo/kelDE1background; all other transgene products were tested in a kelDE1/kelDE1 background.
In wild-type egg chambers expressing myc:ORF1-R, the myc:ORF1-R protein localized to slightly aberrant ring canals characterized by separations in the inner rim. This was most easily observed by anti-myc staining to detect the myc:ORF1-R protein. The actin and anti–hts-RC staining did not show dramatic defects in the inner rim structure, only occasional separations in the inner rim (not shown; Fig. 1 B).
BTBIVR Is Required for Kelch Function
We examined the morphology of the ring canals from kelDE1 mutant egg chambers expressing each of the truncated kelch proteins that localize to ring canals to see what domains are required to organize the actin cytoskeleton. Wild-type ring canals have actin-rich inner rims that define an open lumen through which cytoplasm is transported during oogenesis (Fig. 7 A). The kelch mutant ring canals have disorganized actin inner rims that partially occlude the lumen (Fig. 7 B). Myc:ORF1 localized to and rescued the morphology of kelch mutant ring canals (Fig. 7, C and D). In kelch mutants expressing a rescuing dose of myc:ΔNORF1 (from myc:ΔNORF1 15-2), myc:ΔNORF1 localized to and rescued the mutant ring canal morphology (Fig. 7, E and F). This suggests that the NTR region of kelch is not required for kelch's ring canal organizational activity. The BTBIVR region was required since myc: KREP localized to ring canals but was not capable of rescuing the kelch mutant ring canal morphology (Fig. 7, G and H).
Figure 7.
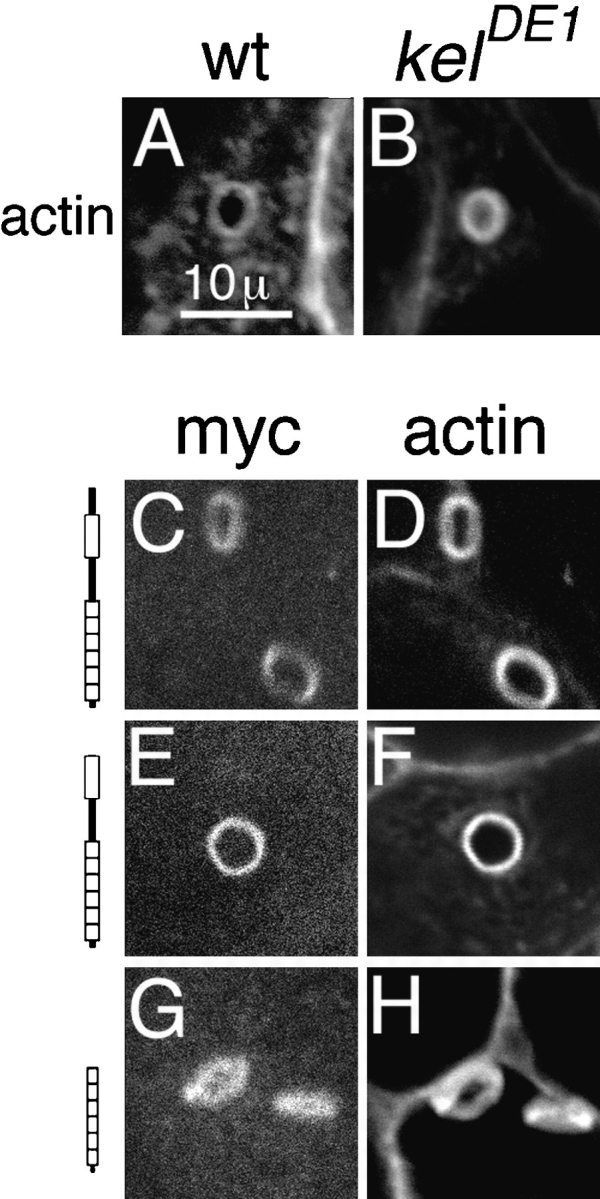
The BTBIVR region is required for kelch's ring canal organizer activity. Actin filaments in wild-type ring canals (A) are neatly organized in the ring canal inner rim, while actin filaments in kelDE1 ring canals (B) are disorganized and extend into the lumen of the ring canal. Myc:ORF1 localized to kelDE1 mutant ring canals (C) and organized the inner rim actin (D). Myc:ΔNORF1 produced from the myc:ΔNORF1 15-2 line also localized to kelDE1 mutant ring canals (E) and organized the actin filaments in the inner rim (F). Myc:KREP also localized to kelDE1 mutant ring canals (G); however, it was unable to rescue ring canal morphology (H).
Both BTB and IVR Regions Are Required for BTBIVR Localization
Since myc:BTBIVR localized to ring canals in wild-type egg chambers, it was possible that either the BTB or the IVR domains were required for this activity. We examined 47 independent lines that carry a myc:BTB transgene (Fig. 1 B). Several of the lines expressed myc:BTB as determined by Western analysis. However, none of the 47 lines showed ring canal localization of myc:BTB (not shown). We generated 25 independent lines carrying a myc:IVR transgene and did not detect any localization of myc:IVR to ring canals in any of the 25 lines (Fig. 1 B). It was more difficult to demonstrate expression of myc:IVR by Western immunoblotting because we do not have an antibody directly against the domain, and we find that relatively high expression is required to detect the myc epitope by Western analysis using the anti-myc 9E10 antibodies (Evan et al., 1985). Some lines did have low level nuclear staining that was above background, suggesting that the transgene was expressed (not shown).
BTB Domain Is Sufficient for Dimerization In Vitro
The localization of myc:BTBIVR to ring canals in an ORF1-dependent manner is consistent with BTBIVR dimerizing with ORF1 through the BTB domain. This interpretation is consistent with the findings that the BTB domain (also called POZ domain) from several zinc finger-containing transcription factors dimerizes in vitro (Bardwell and Treisman, 1994; Chen et al., 1995). To test whether the kelch BTB domain can mediate dimerization, we expressed histidine-tagged BTB (his:BTB) and BTBIVR (his:BTBIVR) domains in Escherichia coli and purified the recombinant proteins by nickel (Ni2+) chromatography. Both proteins were expressed and could be extensively purified in one step using this recombinant expression system (Fig. 8).
Figure 8.
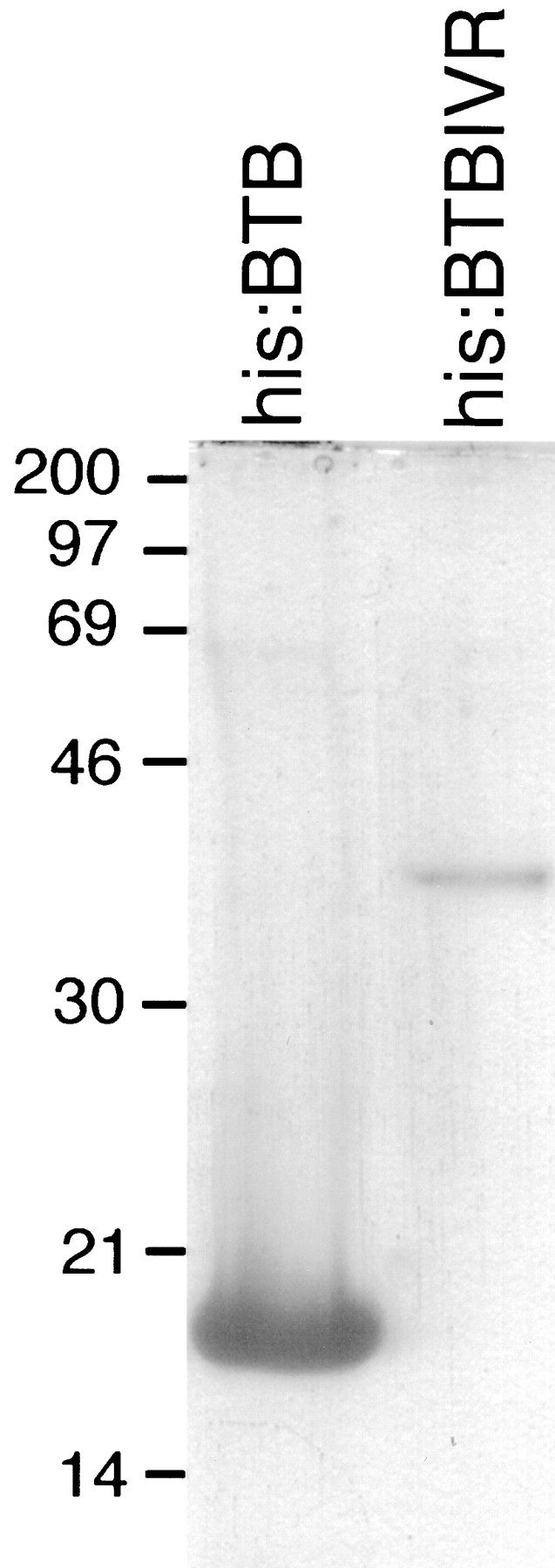
Purified his:BTB and his:BTBIVR proteins. The extensively purified proteins were analyzed by SDS– polyacrylamide gel electrophoresis followed by Coomassie brilliant blue staining. His:BTB had an apparent molecular mass of ∼17 kD, and the his:BTBIVR protein had an apparent molecular mass of ∼35 kD.
We tested his:BTB for dimerization using dynamic light scattering. The predicted and the apparent (determined by SDS-PAGE) molecular masses of the his:BTB domain are each ∼17 kD. Dynamic light scattering indicated that the protein sample was monodisperse with a mean translational diffusion coefficient of 792.5. This gave a Stokes radius of 2.9 nm and an estimated molecular weight of 41 kD. This is consistent with the protein forming dimers in solution. We examined the protein by gel filtration chromatography and compared its retention time to protein standards. Most of the protein had a relative molecular weight of 43 kD, which is again consistent with the protein existing as dimers. A small population of protein (<15%) had an apparent molecular weight comparable to tetramers.
His:BTBIVR was more difficult to analyze since it had a tendency to form aggregates and could not be concentrated >0.4 mg/ml. By gel filtration chromatography, his: BTBIVR separated into three populations. One population eluted from the column with a retention time consistent with monomers, a second population eluted as dimers, and a third population eluted in the void volume of the column consistent with either extremely large molecular weight forms (>500 kD) or aggregates.
Discussion
A Simple Model for Kelch Function
kelch mutant ring canals are highly disorganized and have additional actin filaments that extend into the canal partially obstructing cytoplasm transport (Robinson et al., 1994; Tilney et al., 1996). Although Drosophila kelch ORF1 is a member of a large family of kelch proteins, the biochemical functions for this family have not yet been discerned. However, two domains in kelch are also found in diverse nonkelch proteins. By comparison to the nonkelch proteins, a simple model for kelch function is that there are three interaction domains (Fig. 9). First, kelch might bind to ring canal actin filaments through KREP. Second, it dimerizes through the BTB domain, thereby crosslinking the actin filaments in the ring canal into the well organized inner rim. Finally, since kelch localizes to the ring canals, we would hypothesize that there is a third interaction of kelch that allows it to bind to the ring canal actin specifically. Here, we have performed a structure– function analysis to test various aspects of this model.
Figure 9.
Kelch is an oligomeric ring canal actin organizer. (A) Summary of domain functions. (B) A proposed model for how kelch organizes the ring canal actin filaments. Through the KREP domain (R), kelch localizes to the ring canal possibly by binding to an unknown ring canal component (?). It likely binds to another component (possibly the actin filaments) also through the KREP domain. The protein forms cross-links through the BTBIVR region of the protein.
NTR Function
Because the NTR region is not present in other kelch family members, we tested whether the NTR is required for kelch function. The myc:ΔNORF1 15-2 line that expresses moderate amounts of protein was able to rescue kelch mutant sterility, restoring ring canal morphology. This indicates that the NTR domain is not strictly required for kelch's ring canal organizational activity. However, in highly expressing lines, myc:ΔNORF1 had a strong dominant-negative effect on ring canal stability, indicating that it has a very important function. There was a dramatic loss of plasma membrane stability in stage 6 and older egg chambers. Although it is possible that ΔNORF1 disrupts plasma membrane directly, we believe membrane instability is due to a specific defect in ring canal assembly. First, myc:ΔNORF1 is specifically localized to ring canals in early stages of oogenesis. It is not obviously localized to the cortical cytoskeleton until the time when the ring canals begin to break down. We have also detected some cortical localization of myc:ORF1 that does not result in a breakdown in plasma membrane integrity. Second, myc: ΔNORF1 shows earlier than normal ring canal localization correlating with myc:ΔNORF1's dominant-negative effect being due to a defect in ring canal assembly. Since myc: ΔNORF1 localized to region 1 ring canals, we speculate that the protein disrupts initial assembly of the ring canals so that they are destabilized later. Supporting this idea, myc:ΔNORF1's localization to region 1 ring canals is even earlier than hts-RC and the robust inner rim of actin that normally begins to accumulate by region (Robinson et al., 1994). These results suggest that kelch function is fairly tightly controlled and is not required, and in fact is not desirable, until after the initial assembly of the ring canal.
The NTR is apparently required to regulate the localization of kelch to ring canals. Since the ovarian tumor gene promoter in the pCOG vector provides germline expression beginning in the stem cell, one possible explanation for myc:ΔNORF1's early localization is that it was transcribed earlier than normal. However, myc:ORF1 expressed using the same pCOG vector had a wild-type time course for localization to ring canals. A second possible explanation is that there might be negative regulators of translation in the NTR region allowing myc:ΔNORF1 to be translated earlier than normal. However, in high expressing lines of other kelch proteins that contain the NTR domain, we could detect the expression of unlocalized proteins in the cytosol in region 2 egg chambers (Robinson, D.N., and L. Cooley, unpublished observations). Therefore, the best explanation for myc:ΔNORF1's early localization is that the NTR regulates the timing of protein localization. The NTR might accomplish this by interacting with an unknown protein that sequesters kelch in the cytosol until the time to localize. Alternatively, intramolecular interactions between the NTR and another domain in kelch could cause the protein to fold in such a way that it cannot bind to ring canals until the protein is activated. There are no known proteins with domains that have a high sequence homology to the NTR. The NTR is rich in asparagines, glutamines, and histidines having two stretches of polyglutamines including one six residues long and a second eight residues long. These stretches might mediate some of the interactions of the NTR. For example in the human protein, huntingtin, which has a long polyglutamine stretch, the polyglutamine tract is involved in protein–protein interactions (Li et al., 1995; Burke et al., 1996).
In addition to myc:ΔNORF1, a cheerio hypomorphic mutant (cher2) allows early assembly of the ring canals followed by ring canal degeneration (Robinson et al., 1997). This mutant allele also displays loss of nurse cell membrane integrity resulting in aberrant transport of nurse cell nuclei into the oocyte. However, the mechanisms of the defects in cher2 compared to myc:ΔNORF1 are different since in cher2 egg chambers, kelch does not localize earlier than normal. Finally, myc:ΔNORF1 has a much more dramatic and penetrant effect on ring canal and plasma membrane integrity than the cher2 mutant has.
BTBIVR Mediates Oligomerization
The BTBIVR region was the minimal unit that localized to ring canals in an ORF1-dependent manner. Since ORF1-R failed to localize to ring canals that contained only KREP, the BTBIVR region must interact with the amino half of kelch. One model suggests that the BTBIVR region mediates oligomerization. Consistent with this, there are subtle defects in the ring canal inner rims of wild-type egg chambers expressing myc:ORF1-R, perhaps because some nonproductive complexes form. However, the in vivo experiments cannot distinguish between direct and indirect interactions. In vitro, purified recombinant kelch BTB and BTBIVR domains are capable of mediating dimerization. The fact that the BTB domain was not sufficient to bind ring canals in vivo might indicate that kelch BTB-mediated dimerization is a relatively low affinity interaction, and additional residues in the IVR region are required to form a high affinity interaction domain that can promote dimerization in vivo.
BTB (also called POZ) domain oligomerization has been analyzed in vitro for some of the transcription factors that have this domain. In gel shift assays, the proteins produced a shift expected for dimerization (Bardwell and Treisman, 1994; Chen et al., 1995). The amino-terminal 50 amino acids in the bric à brac (bàb) BTB domain were sufficient to mediate dimerization (Chen et al., 1995). However, in another zinc finger protein called ZID, the first 69 amino acids of its BTB domain were insufficient to dimerize (Bardwell and Treisman, 1994), indicating that there are differences in the way different BTB domains interact. Furthermore, some BTB domains have different specificities. For example, the tramtrack (ttk; Harrison and Travers, 1990) BTB domain formed homodimers or heterodimers with the GAGA transcription factor BTB domain (Soeller et al., 1993), while the ZID BTB domain did not interact with the ttk BTB domain (Bardwell and Treisman, 1994). The BTB domain is predicted to be largely α-helical, and α-helical wheel modeling reveals a hydrophobic face that could mediate the dimerization interaction (Chen et al., 1995). Two highly conserved, charged residues (corresponding to Drosophila kelch residues: D163 and R171; Xue and Cooley, 1993) map to the hydrophobic face. Mutations that changed the charge of these residues in the bàb BTB domain disrupted in vitro dimerization, while mixed populations with compensatory mutations in trans restored the interaction, suggesting that these residues are involved in electrostatic interactions (Chen et al., 1995). Clearly there are differences in the way this interaction domain functions in different proteins. Comparative structural studies are needed to elucidate the basis of BTB domain interaction.
In vivo, some BTB fusion proteins behave in a dominant-negative manner. Three alleles of Drosophila pipsqueak are caused by inappropriate splicing of the transcript that when translated produces a truncated BTB fusion protein. The truncated protein appears to be worse than having no protein at all (Horowitz and Berg, 1995) and can act dominant negatively if overexpressed from a transgene (Horowitz and Berg, 1996). In a rare translocation variant in humans that leads to acute promyelocytic leukemia, a BTB domain-containing zinc finger protein PLZF is fused to the retinoic acid receptor α (RARα). The PLZF-RARα acts dominant negatively by disrupting normal RARα function in retinoic acid-sensitive myeloid cells (Chen et al., 1993, 1994). The BTB domain fusions might sequester proteins in nonproductive complexes or mediate other aberrant interactions contributing to their pathogenesis.
KREP Domain Function
The KREP domain is both necessary and sufficient for localization to ring canals. From sequence comparisons with α-scruin, which has two KREP domains (Way et al., 1995b ) that each bind actin (Tilney, 1975; Bullitt et al., 1988; Owen and DeRosier, 1993; Schmid et al., 1994), we speculate that the kelch KREP domain might bind to ring canal actin filaments. Preliminary data indicates that the KREP domain can bind to actin filaments in vitro (Robinson, D.N., and L. Cooley, unpublished results). Since the KREP domain specifically binds to ring canals, we propose that there is at least one additional interaction with an unknown factor that promotes specificity for ring canals. Without the specificity interaction, it is unlikely that KREP would preferentially bind ring canal actin filaments over other actin in the nurse cells since ring canals contain a very small proportion of the actin in the cell. In addition, because myc:ΔNORF1 can localize to ring canals as early as region 1, the unknown factor is present on ring canals before the addition of hts-RC and the actin filaments that form the inner rim. Since cheerio is required for kelch to localize to ring canals (Robinson et al., 1997), it could encode the factor that allows KREP to bind to ring canals.
Another example of a protein in which a kelch repeat domain might mediate two interactions is the Physarum polycephalum actin-fragmin kinase (AFK). In this protein, the 35-kD amino half of the protein is a novel protein kinase domain, and the 35-kD carboxy half consists primarily of six kelch repeats. This protein specifically phosphorylates actin in the actin–fragmin complex (Eichinger et al., 1996). Perhaps the AFK KREP domain recognizes sites on both actin and fragmin, giving it its specificity so that the kinase domain can phosphorylate its actin substrate.
Kelch's Ring Canal Organization Activity
Our data suggest that kelch has at least three activities: ring canal localization, dimerization, and ring canal organization. We have separated the ring canal organizational activity from the ring canal localization and dimerization functions using nonrescuing, full length kelch proteins. Previously, we expressed full length kelch proteins by creating alanine-substituted (alanine full length), serine-substituted (serine full length), and UGA-deleted (ΔUGA full length), full length kelch transgenes (Robinson and Cooley, 1997b ). The alanine full length kelch was partially functional and was able to rescue the hypomorphic kelneo allele but not the molecular null kelDE1 allele. Serine and ΔUGA full length kelch proteins could not rescue either kelch allele. All three of the mutant full length proteins were able to localize to ring canals, indicating that the ring canal specificity interaction is intact. Here, we observed each of the full length kelch proteins interacted with myc: ORF1-R allowing myc:ORF1-R to localize to ring canals. This indicates that the full length kelch proteins are capable of dimerizing normally. Consequently, the full length proteins are defective in a third interaction required for ring canal organization. Perhaps the 84-kD ORF2 domain in the full length protein sterically disrupts the proposed ability of the KREP domain to interact with actin, and the degree of steric hindrance depends on the residue substituted at the stop codon.
Ring Canal Growth
It has been speculated that ring canals increase their diameter from 3–4 μm (stage 4) to 10 μm (stage 11) by sliding the actin filaments with respect to one another (Robinson et al., 1994; Tilney et al., 1996). The density of actin filaments and the filament number per cross-sectional area are relatively constant (∼720) during these stages of ring canal development (Tilney et al., 1996). This implies that existing actin filaments must increase their length or additional actin filaments are gradually added during expansion to maintain the filament density. During actin filament sliding, cross-linking proteins could break and reform their associations with the actin filaments to facilitate expansion while maintaining ring canal organization. Since the concentration of actin in the ring canal inner rim is so high (millimolar range; Robinson and Cooley, 1997a ), low affinity interactions with Kd's in the μM range can easily promote the reversibility of these interactions. Kelch is a good candidate for doing just this, since in kelch mutants the ring canals become disorganized during these stages (Robinson et al., 1994; Tilney et al., 1996). The interactions that allow KREP to localize and BTBIVR to dimerize might be relatively high affinity, while the putative actin interaction site might form the hypothesized low affinity interaction site that maintains ring canal organization during growth.
Conclusion
These in vivo studies of kelch function support and extend our model for kelch activity. First, the dominant-negative protein that resulted from the deletion of the NTR was unexpected and points out how important temporal control and order of addition during ring canal assembly are. Identification of proteins that interact with the NTR should provide great insight into the events that regulate the ring canal cytoskeleton assembly. Second, our in vivo and in vitro data support a model in which the BTB domain mediates dimerization. However, in vivo, the BTB domain is not sufficient and requires sequences in the IVR region. Certainly, more complex interactions can be envisioned to explain the data, such as having an additional factor that promotes binding between the BTBIVR domains. In addition, since there are differences in the requirements for BTB domain dimerization from different proteins, further mutagenesis and structural studies are needed to elucidate the molecular basis of this interaction. Third, there are two remaining functions that can be assigned to kelch: ring canal localization and ring canal organization. Our results verify that the KREP domain provides ring canal localization and suggest that it might mediate the ring canal organizational function. Discovery of the binding partner that promotes ring canal localization may identify new ring canal components and will facilitate many new studies on how proteins are added to the ring canals. Ring canal organizational activity likely involves a direct or indirect interaction with the ring canal actin filaments. Experiments are in progress to test direct interactions with actin. Comparative functional studies on the KREP domain would be particularly informative since this motif is wide spread and there is considerable sequence divergence in many kelch-related proteins.
Acknowledgments
We thank Jesper Christensen and the Tattersall lab for access to the FPLC equipment. We appreciate the help of Elias Lolis with dynamic light scattering and comments on the manuscript. We also thank the Artavanis-Tsakonas lab and the Howard Hughes Medical Institute for generously sharing their confocal microscope. We thank the members of the Cooley lab for a fun and enthusiastic environment in which to work.
This work was supported by grants from the National Institutes of Health, the Pew Charitable Trusts, and the American Cancer Society.
Abbreviations used in this paper
- KREP
kelch repeat domain
- NTR
amino-terminal region
- ORF
open reading frame
Footnotes
Please address all correspondence to Lynn Cooley, Department of Genetics, Yale University School of Medicine, 333 Cedar St., New Haven, CT 06510. Tel.: (203) 785-5067; Fax: (203) 785-6333; E-mail: lynn.cooley@yale.edu
Douglas Robinson's current address is Department of Biochemistry, Beckman Center, Stanford University, Stanford, CA 94305.
References
- Bardwell VJ, Treisman R. The POZ domain: a conserved protein-protein interaction motif. Genes Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Bork P, Doolittle RF. Drosophilakelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Bullitt ESA, DeRosier DJ, Coluccio LM, Tilney LG. Three- dimensional reconstruction of an actin bundle. J Cell Biol. 1988;107:597–611. doi: 10.1083/jcb.107.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JR, Enghild JJ, Martin ME, Jou Y-S, Myers R R, Roses AD, Vance JM, Strittmatter WJ. Huntingtin and DRPLA proteins selectively interact with the enzyme GAPDH. Nat Med. 1996;2:347–350. doi: 10.1038/nm0396-347. [DOI] [PubMed] [Google Scholar]
- Chardin P, Courtois G, Mattei M-G, Gisselbrecht S. The KUPgene, located on human chromosome 14, encodes a protein with two distant zinc fingers. Nucleic Acids Res. 1991;19:1431–1436. doi: 10.1093/nar/19.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Brand NJ, Chen A, Chen S-J, Tong J-H, Wang Z-Y, Waxman S, Zelent A. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-α locus due to a variant t(11;17) translocation associated with acute promyelocytic leukemia. EMBO (Eur Mol Biol Organ) J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Guidez F, Rousselot P, Agadir A, Chen S-J, Wang Z-Y, Degos L, Zelent A, Waxman S, Chomienne C. PLZF-RARα fusion proteins generated from the variant t(11;17)(q23:q21) translocation in acute promyelocytic leukemia inhibit ligand-dependent transactivation of wild-type retinoic acid receptors. Proc Natl Acad Sci USA. 1994;91:1178–1182. doi: 10.1073/pnas.91.3.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zollman S, Couderc J-L, Laski FA. The BTB domain of bric àbrac mediates dimerization in vitro. Mol Cell Biol. 1995;15:3424–3429. doi: 10.1128/mcb.15.6.3424. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- DiBello PR, Withers DA, Bayer CA, Fristrom JW, Guild GM. The Drosophila Broad-Complexencodes a family of related proteins containing zinc fingers. Genetics. 1991;129:385–397. doi: 10.1093/genetics/129.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Bomblies L, Vandekerckhove J, Schleicher M, Gettemans J. A novel type of protein kinase phosphorylates actin in the actin-fragmin complex. EMBO (Eur Mol Biol Organ) J. 1996;15:5547–5556. [PMC free article] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SD, Travers AA. The tramtrack gene encodes a Drosophila finger protein that interacts with the ftztranscriptional regulatory region and shows a novel embryonic expression pattern. EMBO (Eur Mol Biol Organ) J. 1990;9:207–216. doi: 10.1002/j.1460-2075.1990.tb08097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H, Berg CA. Aberrant splicing and transcription termination caused by P element insertion into the intron of a Drosophilagene. Genetics. 1995;139:327–335. doi: 10.1093/genetics/139.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz H, Berg CA. The Drosophila pipsqueakgene encodes a nuclear BTB-domain-containing protein required early in oogenesis. Development (Camb) 1996;122:1859–1871. doi: 10.1242/dev.122.6.1859. [DOI] [PubMed] [Google Scholar]
- Ito N, Phillips SEV, Yadav KDS, Knowles PF. Crystal structure of a free radical enzyme, galactose oxidase. J Mol Biol. 1994;238:794–814. doi: 10.1006/jmbi.1994.1335. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Senkevich TG, Chernos VI. Protein sequence motifs: a family of DNA virus genes that consists of fused portions of unrelated cellular genes. Trends Biochem Sci. 1992;17:213–214. doi: 10.1016/0968-0004(92)90379-n. [DOI] [PubMed] [Google Scholar]
- Li X-J, Li S-H, Sharp A, Nucifora FC, Jr, Schilling G, Lanahan A, Worley P, Snyder SH, Ross CA. A huntingtin-associated protein enriched in brain with implications for pathology. Nature (Lond) 1995;378:398–402. doi: 10.1038/378398a0. [DOI] [PubMed] [Google Scholar]
- Lindsley, D.L., and G.G. Zimm. 1992. The Genome of Drosophila melanogaster. Academic Press, San Diego. 1133 pp.
- Owen C, DeRosier D. A 13-Å map of the actin-scruin filament from the Limulusacrosomal process. J Cell Biol. 1993;123:337–344. doi: 10.1083/jcb.123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Stable intercellular bridges in development: the cytoskeleton lining the tunnel. Trends Cell Biol. 1996;6:474–479. doi: 10.1016/0962-8924(96)84945-2. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Genetic analysis of the actin cytoskeleton in the Drosophilaovary. Annu Rev Cell Dev Biol. 1997a;13:147–170. doi: 10.1146/annurev.cellbio.13.1.147. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Cooley L. Examination of the function of two kelch proteins generated by stop codon suppression. Development (Camb) 1997b;124:1405–1417. doi: 10.1242/dev.124.7.1405. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Cant K, Cooley L. Morphogenesis of Drosophilaovarian ring canals. Development (Camb) 1994;120:2015–2025. doi: 10.1242/dev.120.7.2015. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Smith-Leiker TA, Sokol NS, Hudson AM, Cooley L. Formation of the Drosophila ovarian ring canal inner rim depends on cheerio. . Genetics. 1997;145:1063–1072. doi: 10.1093/genetics/145.4.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Agris JM, Jakana J, Matsudaira P, Chiu W. Three-dimensional structure of a single filament in the Limulusacrosomal bundle: scruin binds to homologous helix-loop-β motifs in actin. J Cell Biol. 1994;124:341–350. doi: 10.1083/jcb.124.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T, Wieschaus E. Female sterile mutations on the second chromosome of Drosophila melanogaster.II. Mutations blocking oogenesis or altering egg morphology. Genetics. 1991;129:1119–1136. doi: 10.1093/genetics/129.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Muravnik GL, Pozdnyakov SG, Chizhikov VE, Ryazankina OI, Shchelkunov SN, Koonin EV, Chernos VI. Nucleotide sequence of XhoI O fragment of ectromelia virus DNA reveals significant differences from vaccinia virus. Virus Res. 1993;30:73–88. doi: 10.1016/0168-1702(93)90017-h. [DOI] [PubMed] [Google Scholar]
- Soeller WC, Oh CE, Kornberg TB. Isolation of cDNAs encoding the DrosophilaGAGA transcription factor. Mol Cell Biol. 1993;13:7961–7970. doi: 10.1128/mcb.13.12.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG. Actin filaments in the acrosomal reaction of Limulussperm. J Cell Biol. 1975;64:289–310. doi: 10.1083/jcb.64.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Tilney MS, Guild GM. Formation of actin filament bundles in the ring canals of developing Drosophilafollicles. J Cell Biol. 1996;133:61–74. doi: 10.1083/jcb.133.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, Laver WG, Colman PM. Structure of the influenza virus glycoprotein antigen neuraminidase at 2.9 Å resolution. Nature (Lond) 1983;303:35–40. doi: 10.1038/303035a0. [DOI] [PubMed] [Google Scholar]
- Varkey JP, Muhlrad PJ, Minniti AN, Do B, Ward S. The Caenorhabditis elegans spe-26gene is necessary to form spermatids and encodes a protein similar to the actin-associated proteins kelch and scruin. Genes Dev. 1995;9:1074–1086. doi: 10.1101/gad.9.9.1074. [DOI] [PubMed] [Google Scholar]
- von Bülow M, Heid H, Hess H, Franke WW. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219:407–413. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- Way M, Sanders M, Chafel M, Tu Y-H, Knight A, Matsudaira P. β-scruin, a homologue of the actin crosslinking protein scruin, is localized to the acrosomal vesicle of Limulussperm. J Cell Sci. 1995a;108:3155–3162. doi: 10.1242/jcs.108.10.3155. [DOI] [PubMed] [Google Scholar]
- Way M, Saunders M, Garcia C, Sakai J, Matsudaira P. Sequence and domain organization of scruin, an actin-cross-linking protein in the acrosomal process of Limulussperm. J Cell Biol. 1995b;128:51–60. doi: 10.1083/jcb.128.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, F., 1992. Molecular analysis of kelch, a gene necessary for nurse cell cytoplasm transport in Drosophila oogenesis. Ph.D. thesis. Yale University, New Haven, CT.
- Xue F, Cooley L. kelch encodes a component of intercellular bridges in Drosophilaegg chambers. Cell. 1993;72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- Yue L, Spradling AC. hu-li tai shao, a gene required for ring canal formation during Drosophilaoogenesis, encodes a homolog of adducin. Genes Dev. 1992;6:2443–2454. doi: 10.1101/gad.6.12b.2443. [DOI] [PubMed] [Google Scholar]
- Zollman S, Gödt D, Privé GG, Couderc J-L, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. . Proc Natl Acad Sci USA. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]