Abstract
The production of native α/β tubulin heterodimer in vitro depends on the action of cytosolic chaperonin and several protein cofactors. We previously showed that four such cofactors (termed A, C, D, and E) together with native tubulin act on β-tubulin folding intermediates generated by the chaperonin to produce polymerizable tubulin heterodimers. However, this set of cofactors generates native heterodimers only very inefficiently from α-tubulin folding intermediates produced by the same chaperonin. Here we describe the isolation, characterization, and genetic analysis of a novel tubulin folding cofactor (cofactor B) that greatly enhances the efficiency of α-tubulin folding in vitro. This enabled an integrated study of α- and β-tubulin folding: we find that the pathways leading to the formation of native α- and β-tubulin converge in that the folding of the α subunit requires the participation of cofactor complexes containing the β subunit and vice versa. We also show that sequestration of native α-or β-tubulins by complex formation with cofactors results in the destabilization and decay of the remaining free subunit. These data demonstrate that tubulin folding cofactors function by placing and/or maintaining α-and β-tubulin polypeptides in an activated conformational state required for the formation of native α/β heterodimers, and imply that each subunit provides information necessary for the proper folding of the other.
The cytoskeleton of eukaryotic cells consists of three distinctive networks: microfilaments, intermediate filaments, and microtubules. In common with other proteins, the proper functioning of the subunits from which these networks are assembled depends on their three- dimensional structure. Classical experiments using small enzymes as model systems have established that the formation of correct tertiary structure can occur spontaneously (Anfinsen, 1973) and have led to the idea that all the information required for the proper folding of a protein resides in the amino acid sequence. However, the subunit proteins from which microfilament and microtubule networks are assembled—i.e., actins and tubulins, respectively—do not fold spontaneously; they require the action of cytosolic chaperonin (c-cpn)1 (Chen et al., 1994; Ursic et al., 1994; Vinh and Drubin, 1994), a multisubunit toroidal complex that generates potentially productive folding intermediates via multiple rounds of ATP hydrolysis (Frydman et al., 1992; Gao et al., 1992; Tian et al., 1995a ; Lewis et al., 1996).
Studies on the bacterial homologue of c-cpn, GroEL/ GroES, have led to a wealth of genetic, biochemical, and structural information on the mechanism of facilitated folding (for review see Horwich et al., 1995; Hartl, 1996). In contrast with GroEL/GroES, there is biochemical and genetic evidence that the target range of c-cpn is quite limited (Lewis et al., 1996). Furthermore, neither GroEL/ GroES nor its mitochondrial homologue, Hsp60/Hsp10, can generate productive actin or tubulin folding intermediates (Tian et al., 1995a ). It therefore seems possible that c-cpn may have evolved to overcome specific kinetic traps on the folding pathways of its target proteins.
The in vitro c-cpn–mediated folding of actin and γ-tubulin requires only chaperonin, target protein, and ATP (Gao et al., 1992; Melki et al., 1993). In contrast, the productive folding of α- and β-tubulin requires the additional presence of GTP (both α- and β-tubulin are GTP-binding proteins), a set of protein cofactors, and native tubulin (Gao et al., 1993). Quasinative tubulin folding intermediates (termed IQ) that already contain the GTP-binding pocket are produced via one or more ATP-dependent cycles of interaction with c-cpn (Tian et al., 1995b ); however, in the absence of cofactors, these intermediates cannot proceed to the native state.
In the case of β-tubulin, interaction of c-cpn–generated intermediates with four cofactors (designated A, C, D, and E) and native tubulin leads ultimately to the production of de novo folded β-tubulin subunits that form functional heterodimers (Tian et al., 1996). Yeast homologues of three of these cofactors (A, D, and E) have been identified and shown to give rise to cytoskeletal phenotypes when mutated or deleted. The Saccharomyces cerevisiae homologue of cofactor A, RBL2, can compensate for the lethal overexpression of β-tubulin when it too is overexpressed (Archer et al., 1995). Deletion of CIN1 (Chromosome Instability 1), the gene encoding the yeast homologue of cofactor D, results in supersensitivity to the antimicrotubule drug benomyl, cold sensitivity, and chromosome loss in mitosis (Stearns et al., 1990; Hoyt et al., 1990). The CIN1 protein appears to act in the same pathway as the products of two other yeast genes, CIN2 and CIN4, although neither of these is related to any of the known mammalian tubulin folding cofactors. The PAC2 protein, which was identified as a synthetic lethal in conjunction with the chromosome instability mutant CIN8, is the yeast homologue of cofactor E (Hoyt et al., 1997). No homologue of cofactor C can be discerned via database searches of the yeast genome; however, homologues are identifiable in evolutionarily distant eukaryotes (e.g., Arabidopsis, Caenorhabditis elegans). None of the genes encoding cofactor homologues are essential for viability in yeast, although cofactors are required for the productive folding of an essential protein (tubulin) in vitro.
While cofactors A, C, D, and E participate in the pathway leading to native β-tubulin, these cofactors do not support the efficient in vitro folding of α-tubulin (Tian et al., 1996). Here we describe the isolation, characterization, and functional analysis of cofactor B, a protein cofactor that participates in the efficient generation of native α-tubulin. We demonstrate that the proper folding of α- and β-tubulin is interdependent, and present evidence that this occurs via convergent pathways that result in the formation of a multimolecular α- and β-tubulin and cofactor-containing complex. The reaction cycles that lead to the formation of this complex define a role for cofactor function in generating and maintaining α- and β-tubulins in an activated energy state that is required for the formation of the native α/β tubulin heterodimer.
Materials and Methods
In Vitro Folding Assays
The purification of c-cpn from rabbit reticulocyte lysate and the generation of unfolded 35S-labeled α and β target proteins via their expression in Escherichia coli were done as described previously (Gao et al., 1992, 1993). C-cpn–mediated folding reactions were done at 30°C in folding buffer (Tian et al., 1995a ) and contained 0.2 μM c-cpn, 1 mM ATP, 0.1 mM GTP, and one or more of cofactors A, B, C, D, and E present in varying amounts with respect to c-cpn (see text). Cofactors A–E were either purified from crude extracts of bovine testis as described (cofactor A, Gao et al., 1994; cofactor B, see below; cofactors C–E, Tian et al., 1996), or (in the case of cofactors A–C) from extracts of host Escherichia coli BL21DE3 cells engineered for their expression. Some folding reactions (see text) were supplemented with 2.5 μM native bovine brain tubulin as described previously (Tian et al., 1996).
In some experiments, the reaction products were purified by gel filtration, mixed with native brain tubulin, and assayed for their ability to cocycle through multiple rounds of polymerization and depolymerization as described (Gao et al., 1993). In others, the reaction products were supplemented with 2.5 μM native tubulin and treated by digestion at 30°C for 30 min with 10 μg/ml subtilisin so as to generate carboxy-terminally truncated tubulin (Sackett et al., 1985). The proteolytic reaction was quenched by the addition of PMSF to 5 mM. All in vitro folding reaction products were analyzed on 4.5% nondenaturing polyacrylamide gels containing 0.1 mM GTP as described (Gao et al., 1992).
Purification, Peptide Sequence Analysis, Cloning, and Expression of Cofactor B
Cofactor B was purified from a crude extract of bovine testis tissue (Gao et al., 1994; Tian et al., 1996) by fast performance liquid chromatography using the steps shown in Table I. To improve the efficiency of folding assays containing column fractions, unfolded 35S-labeled target protein was first cycled with c-cpn, ATP, and GTP so as to form quasinative (IQ) intermediates (Tian et al., 1995b ). Peptide sequence analysis of the purified protein was done as described previously (Gao et al., 1994; Tian et al., 1996). A virtual homologue encoding human cofactor B was assembled from the WashU-Merck EST database using the Wisconsin Package, version 8.1 (Genetics Computer Group, Inc., Madison, WI). A full-length cDNA encoding human cofactor B was generated by PCR using a cDNA template prepared from human testis mRNA (Clontech, Palo Alto, CA) following procedures recommended by the supplier. The amplified cDNA product was inserted into the pET23 expression vector, and the recombinant protein was purified after induction of host E. coli BL21DE3 cells. The procedure used for the purification of recombinant human cofactor B was identical to that used for purification of the bovine protein (Table I), except that step 4 was omitted.
Table I.
Purification Scheme for Cofactor B
| Dimension | Matrix | Size*:Buffer‡ | Activity‖ | |||
|---|---|---|---|---|---|---|
| 1 | Q-Sepharose HP | 5/25:I/II | 80–100 mM | |||
| 2 | Hydroxylapatite | 2.6/12:III/IV | nonbinding | |||
| 3 | MonoQ | 1/10:XIV/XV | 0 .22–0.24 M | |||
| 4 | Q-Sepharose HP | 1/10:XIX/XX§ | 0 .25–0.26 M | |||
| 5 | TSKG3000SW | 0.75/60:X |
Size of column bed (width/height) in cm.
Buffer composition as described in Tian et al. (1996)
The composition of buffers used in this dimension was XIX: 20 mM diethanolamine-HCl, 20 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM DTT; XX: same as XIX, except containing 1 M in place of 20 mM KCl.
Counterion concentration containing cofactor B activity.
Isolation of FBα and Glutaraldehyde Cross-linking Experiments
The products of a c-cpn–mediated α-tubulin folding reaction done with unlabeled target protein (Tian et al., 1995b ) and purified bovine cofactor B (equimolar with respect to c-cpn) in the presence of α-[32P]GTP were applied to a 0.75 × 7.5-cm SW gel filtration guard column (TosoHaas, Montgomeryville, PA) equilibrated and run in folding buffer (Tian et al., 1995a ) containing 50 μM GTP and purified rabbit hemoglobin (0.1 mg/ ml) included as a stabilizing agent. The FBα complex emerged from this column at 2.9 ml and was effectively resolved from α-tubulin/c-cpn binary complex. In experiments done to identify the presence of unstable α-tubulin–cofactor complexes, aliquots (15 μl) of this material were incubated for 5 min at 30°C with a fivefold molar excess (with respect to cofactor B) of either cofactor D, E, or both. The products of these reactions were stabilized by the addition of glutaraldehyde to 0.03% and incubation at 30°C for an additional 5 min. The cross-linking reaction was quenched by the addition of ethanolamine to 0.1 M and the reaction products were analyzed on nondenaturing polyacrylamide gels as described (Gao et al., 1993).
In Vitro Translation, Purification of 35S-labeled Tubulin, and Backreactions with Tubulin Folding Cofactors
Full-length mouse α- or β-tubulin cDNAs (Gao et al., 1993) were expressed by coupled transcription/translation in 50 μl of TNT lysate (Promega, Madison, WI) containing [35S]methionine. Native bovine brain tubulin was added to 1 μM, the mixture was incubated for an additional 30 min at 30°C, and the reactions were cleared of particulate material by centrifugation at 200,000 g for 20 min at 4°C in a rotor (TL100; Beckman Instruments, Inc., Fullerton, CA). The concentration of NaCl was adjusted to 0.25 M, and the labeled native tubulin was purified free of cofactors using a microcolumn of DEAE-Sephacel (Pharmacia Fine Chemicals, Piscataway, NJ) (Tian et al., 1996) contained in a glass-wool plugged polypropylene pipette tip. After elution with 0.5 M NaCl, the tubulin was dialyzed to reduce the salt concentration to ∼0.1 M and incubated at 30°C for various times in the presence of nucleotide and one or more purified cofactors. Reaction products were analyzed on nondenaturing polyacrylamide gels as described (Gao et al., 1993).
UV Cross-linking Experiments
A backwards reaction of native tubulin with cofactors (see above) was assembled in folding buffer (Tian et al., 1995a ) containing equimolar amounts of native tubulin heterodimer, cofactors C, D, and E, and 15 μM α–32P-labeled GTP (sp act 200 Ci/mmol). This mixture was incubated for 30 min at 30°C and exposed to UV irradiation at 254 nm for 20 min as described (Eriksson et al., 1982), and the reaction products were analyzed on an 8% SDS polyacrylamide gel.
GTP Hydrolysis Experiments
In experiments to determine the effect of GTPγS in folding reactions, c-cpn–mediated α-tubulin reactions were first done in the presence of 1 mM ATP and 50 μM GTP so as to generate IQ intermediates (Tian et al., 1995b ); this procedure avoided interference of GTPγS in the ATP-dependent cycling of c-cpn. Equimolar amounts of cofactors B, C, D, and E (with respect to c-cpn) and native tubulin (2.5 μM) were then added with or without 1 mM GTPγS and the incubation continued at 30°C for 1 h. In the case of β-tubulin, c-cpn–mediated β-tubulin reactions were done in the presence of 1 mM ATP, 50 μM GTP, and a molar equivalent (with respect to c-cpn) of cofactor D so as to generate FDβ intermediates (Tian et al., 1996). Cofactor C, cofactor E, and native tubulin were then added with or without 1 mM GTPγS and the incubation continued at 30°C for 1 h.
The extent of hydrolysis of α–32P-labeled GTP was measured in folding buffer (Tian et al., 1995a ) by incubation at 30°C for 1 h in reactions containing 15 μM GTP (sp act 200 Ci/mmol), purified native brain tubulin (0.5 μM), and/or one or more cofactors each present at ∼1.5-fold molar excess with respect to tubulin. Reaction products were analyzed by TLC on phospho-ethyleneimine plates as described (Spiegelman et al., 1977), and the yield of labeled GTP and GDP was quantitated using a phosphorimager.
Manipulations in S. cerevisiae
Media for yeast growth and sporulation were as described (Sherman et al., 1983). SGal medium was made as for synthetic dextrose medium except that glucose was replaced with 2% galactose. Benomyl (98.6% pure; maintained in 10 mg/ml in DMSO at −20°C) was a gift from E.I. duPont de Nemours, Inc. (Wilmington, DE) Growth of strains on solid media was assayed by spotting suspensions of cells in water onto plates using a 32-point multipronged inoculating manifold (Dan-Kar Corp., Wilmington, MA). Yeast cells were transformed by the lithium acetate method (Ito et al., 1983); transformants carrying plasmids were selected on synthetic complete medium lacking the appropriate nutrient.
ALF1 was disrupted by the PCR method described by Amberg et al. (1996). Four oligonucleotide primers were synthesized as follows: ALF1.1: CGCAGCTCCACCCATTAATTTGACGC; ALF1.2: GCCTCGAGGGGTCCAACCCTTGGTTT; ALF1.8: ATGGTTAGAGTTGTCATAGAGCAGATTGTACTGAGAG; ALF1.9: TCATCATCGCTCTCCACGTCCTGTGCGGTATTTCACAC. Amplification of ALF1 from S. cerevisiae genomic DNA with ALF1.1 and ALF1.2 was followed by creation of a fusion between sequences flanking ALF1 and either the URA3 or HIS3 gene. The fusion was created by PCR using the plasmids pRS313 and pRS316 (Sikorski and Hieter, 1989) as the source of marker DNAs. These constructs were transformed into a wild-type diploid strain (TPS507; Marschall et al., 1996). The genotypes of two of each of the resulting alf1:: URA3/ALF1 and alf1::HIS3/ALF1 transformants were confirmed by PCR, sporulated, and dissected.
Results
Cofactor B: A Participant in the α-Tubulin Folding Pathway
We originally identified two crude fractions from rabbit reticulocyte lysate that together include the activities required (in addition to c-cpn) to yield correctly folded α- and β-tubulin in in vitro folding reactions (Gao et al., 1993). These crude fractions contain cofactors A, C, D, and E, all of which have been purified and shown to participate in the pathway leading to correctly folded β-tubulin (Tian et al., 1996). However, c-cpn–mediated α-tubulin folding reactions containing these cofactors do not generate appreciable quantities of native α-tubulin, as judged by the very low yield of radiolabel comigrating with authentic tubulin heterodimers on a nondenaturing polyacrylamide gel (Fig. 1 A). We therefore reasoned that one or more additional cofactors must exist that contribute to the efficient production of properly folded α-tubulin.
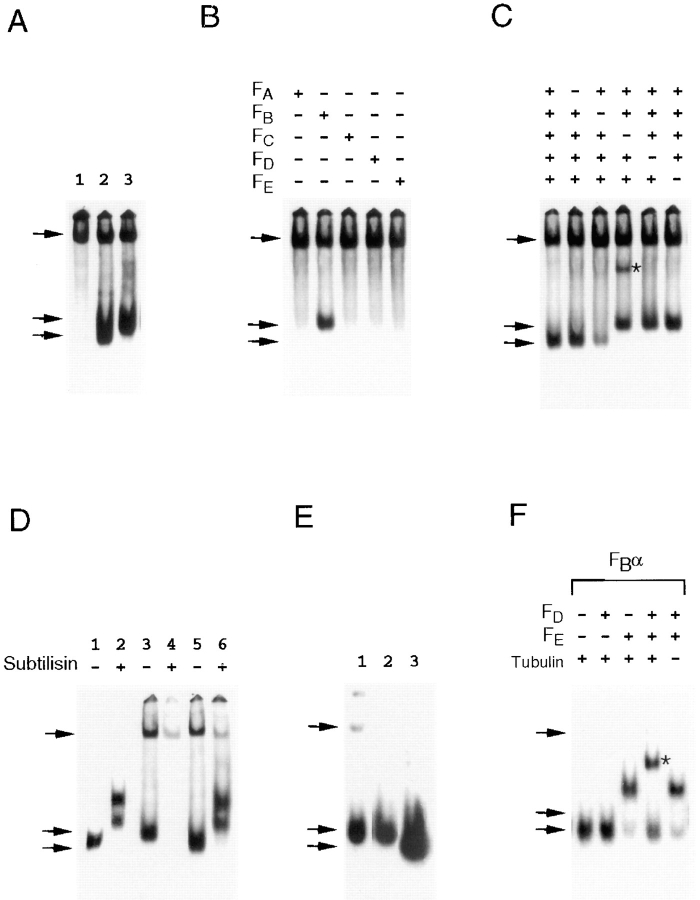
Figure 1.
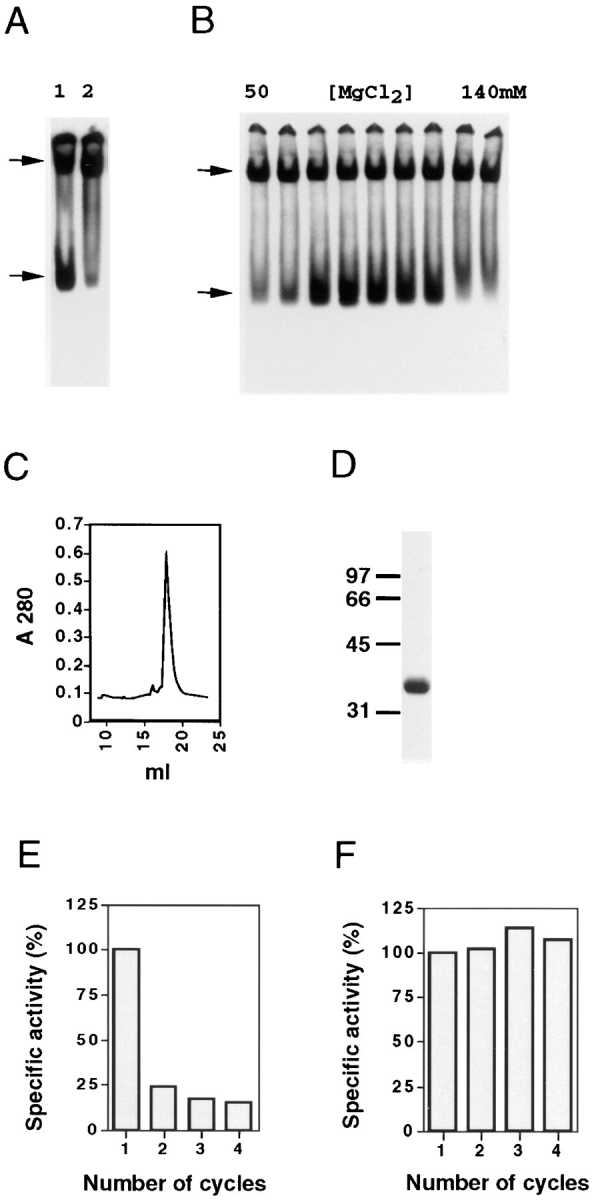
Characterization of cofactor B. (A) The cofactors that support productive β-tubulin folding do not efficiently support α-tubulin folding. Analysis by nondenaturing gel electrophoresis of the products of c-cpn–mediated β-tubulin (lane 1) and α-tubulin (lane 2) folding reactions done with cofactors A, C, D, and E. (B) A novel cofactor (cofactor B) is involved in α-tubulin folding. Analysis of the products of c-cpn–mediated α-tubulin folding reactions done in the presence of aliquots of fractions obtained by anion exchange chromatography of an extract of bovine testis tissue. Only assays of those fractions emerging from the column in the range of 50–140 mM MgCl2 are shown. (C and D) Purification of cofactor B. Absorbance profile of the final step (gel filtration) in the purification of cofactor B (C), together with SDS-PAGE of material contained in the major peak emerging from the column (D). In D, the location of molecular size markers (in kD) is shown. (E and F) The fast-migrating species produced in c-cpn– mediated α-tubulin folding reactions containing purified cofactor B does not cocycle efficiently with native brain tubulin. The products of a c-cpn–mediated α-tubulin folding reaction done in the presence of purified cofactor B alone (E) or cofactors A, B, C, D, and E (F) were subjected to successive cycles of polymerization and depolymerization with added native bovine brain tubulin. The specific activity (relative to the material obtained after the first cycle, taken as 100%) after each cycle is shown. (A and B, upper and lower arrows). Location of α- or β-tubulin/c-cpn binary complexes and either native tubulin (A) or the product generated by bovine cofactor B (B).
To isolate such cofactor(s), we fractionated a crude extract of bovine testis tissue by anion exchange chromatography and assayed the emerging proteins in in vitro c-cpn– mediated α-tubulin folding reactions (see Materials and Methods). Native bovine brain tubulin was added (Gao et al., 1993), and the reaction products were analyzed by nondenaturing polyacrylamide gel electrophoresis. A fast- migrating species was generated in these reactions without supplementation with cofactors A, C, D, and E (Fig. 1 B). We used this assay as a method to purify to homogeneity the protein responsible for the generation of this product. The purified protein (which we termed cofactor B) migrated with an apparent mass of 130 kD upon gel filtration (Fig. 1 C), and consisted of a single polypeptide of 38 kD upon analysis by SDS-PAGE (Fig. 1 D) and 27,561 D by mass spectrometry.
The product of in vitro c-cpn–mediated α-tubulin folding reactions supplemented with purified bovine cofactor B alone comigrated with native tubulin in our gel assay. To see if this material was indeed native, we tested its ability to copurify with unlabeled bovine brain tubulin through several cycles of polymerization and depolymerization and found that it failed to cycle efficiently (Fig. 1 E). However, when a cofactor B–containing α-tubulin folding reaction was supplemented with a mixture of purified β-tubulin– folding cofactors (i.e., cofactors A, C, D, and E), the reaction product (the yield of which was greatly enhanced compared with a parallel reaction lacking cofactor B; see below) cocycled with native tubulin without loss of specific activity, demonstrating that correctly folded α-tubulin was efficiently produced in this experiment (Fig. 1 F).
Cofactors B and E Contain the CLIP-170 Microtubule Binding Motif
We obtained partial amino acid sequence data from purified cofactor B and used this information to search for homologues in the WashU-Merck EST database. This search resulted in the identification of multiple overlapping cDNAs encoding a human protein of 27,394 D, with 80% amino acid identity within the region covered by the bovine peptides sequenced. No other related sequences were identified. To confirm that this homologue indeed encodes cofactor B, we cloned a cDNA encoding the human protein, expressed it in host E. coli cells, purified the recombinant protein, and demonstrated its activity in in vitro c-cpn–mediated α-tubulin folding assays (see below). The complete amino acid sequence encoded by our human cofactor B cDNA (Fig. 2) revealed a region of homology with cofactor E (Tian et al., 1996). This region is also homologous to a motif (the CLIP-repeat) that is tandemly repeated within the microtubule-binding domain of CLIP-170, a microtubule-associated protein that links endocytic vesicles to microtubules (Pierre et al., 1992). The amino acid sequence of cofactor B identifies an open reading frame (YNL148c) encoding an unknown protein in S. cerevisiae with 32% and 52% amino acid sequence identity and similarity, respectively, to human cofactor B. We term this yeast gene ALF1 (Alpha tubulin Folding 1).
Genetic Analysis of the Yeast Homologue of Cofactor B
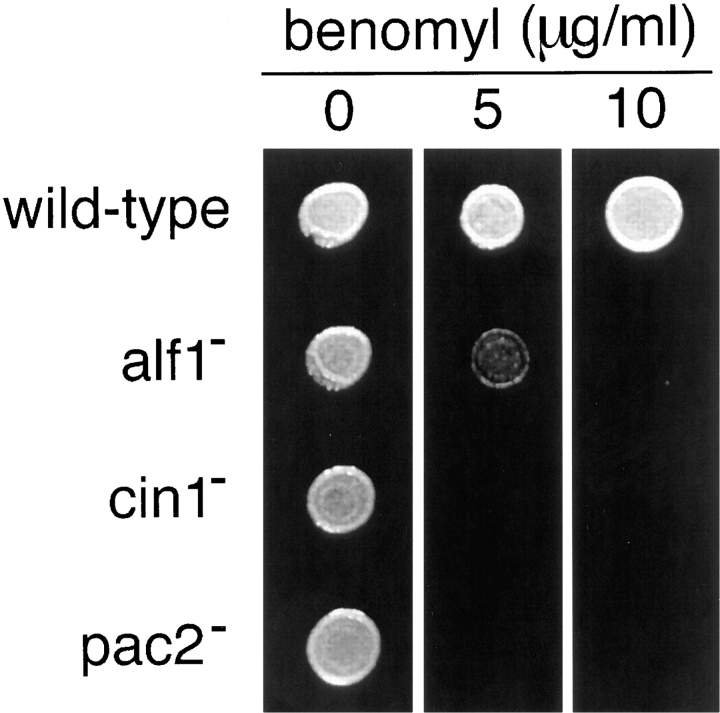
To determine the effect of eliminating its expression in S. cerevisiae, we disrupted the ALF1 gene in a diploid strain and subjected the resulting heterozygote to tetrad analysis. In 16 out of 20 tetrads, all four spores were viable, and the disruption marker segregated 2:2, indicating that ALF1 is not essential for viability. Haploid alf1 null strains were tested for temperature sensitivity and sensitivity to the antimicrotubule drug benomyl; increased sensitivity to benomyl is a common phenotype among mutants affecting the microtubule cytoskeleton (Stearns et al., 1990). In comparison with wild-type, alf1 null strains were supersensitive to benomyl, growing poorly on 5 μg/ml. For comparison, the CIN1, CIN2, and CIN4 genes (the first of these being the yeast homologue of cofactor D) are among the most benomyl-supersensitive mutants known (Hoyt et al., 1990; Stearns et al., 1990), failing to grow on 5 μg/ml benomyl (Fig. 3). alf1 null strains were not cold sensitive.
Figure 3.
Benomyl sensitivity of alf1, cin1, and pac2 mutants. Suspensions of cells were spotted onto yeast extract/peptone/dextrose plates containing either no benomyl (as a control) or benomyl at 5 or 10 μg/ml and grown at 30°C. The strains assayed had null mutations in the genes shown in the figure and were of the same genetic background.
Genetic interactions between ALF1 and other genes involved in microtubule function were tested by making double mutants. alf1::URA3 cin1::HIS3, alf1::URA3 cin2:: LEU2, and alf1::URA3 cin4::LEU2 double mutants were all viable, as was an alf1::URA3 cin1::HIS3 cin2::LEU2 triple mutant. The phenotypes of all these mutants were indistinguishable from the single cin mutants. In contrast, when we attempted to make double mutants between alf1:: URA3 and tub1-1, a mutation in the essential yeast α-tubulin gene, we were unable to recover the double mutant by tetrad analysis. This result indicates that alf1::URA3 and tub1-1 are synthetically lethal.
Cofactor Requirements for Productive α-Tubulin Folding
We compared c-cpn–mediated 35S-labeled α-tubulin folding reactions supplemented with either purified bovine cofactor B or recombinant human cofactor B alone, and analyzed the products on a nondenaturing gel. We found that α-tubulin folding reactions done with either bovine or human cofactor B yielded products that have different electrophoretic mobilities under native conditions; the product of reactions done with bovine cofactor B comigrates with native tubulin, whereas the corresponding product generated in reactions done with human cofactor B runs more slowly (Fig. 4 A). It follows that these species cannot be free α-tubulin molecules; rather, they must be cofactor B/ α-tubulin complexes, which we term FBα.
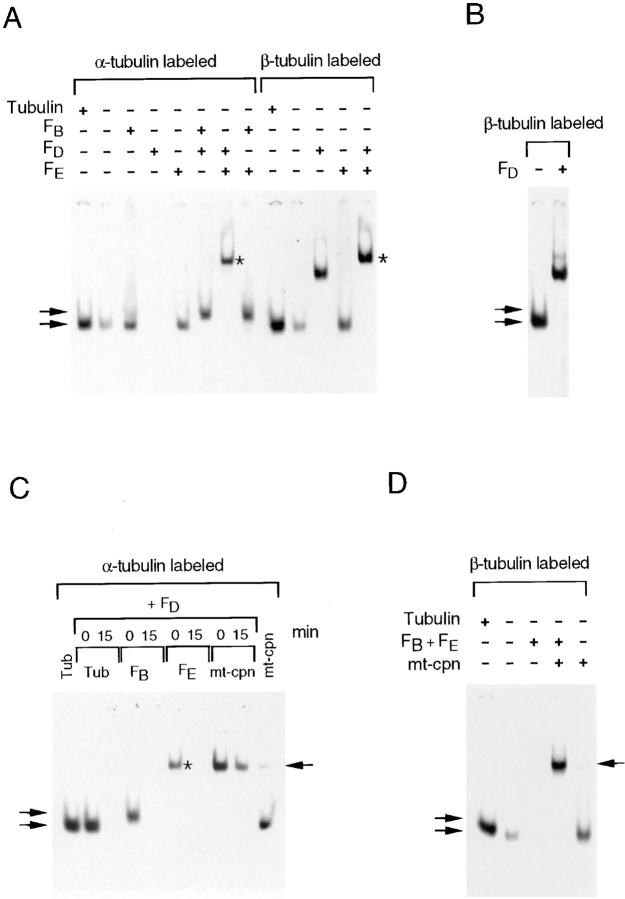
Figure 4.
Cofactors that participate in α-tubulin folding in vitro. (A) Bovine and recombinant human cofactor B form complexes with c-cpn–generated α-tubulin folding intermediates that have different mobilities on a nondenaturing gel. Products of c-cpn– mediated α-tubulin folding reactions done without cofactors (lane 1) or with added purified bovine (lane 2) or recombinant human (lane 3) cofactor B are shown. (B and C) Cofactors required for productive α-tubulin folding in vitro. C-cpn–mediated α-tubulin folding reactions were done in the presence of native tubulin and the cofactors shown in the figure. (D) Native tubulin can be distinguished from the FBα complex by digestion with subtilisin. Analysis of native bovine brain tubulin detected by staining with Coomassie blue (lanes 1 and 2), or the products of in vitro c-cpn–mediated α-tubulin folding reactions containing either cofactor B alone (lanes 3 and 4) or cofactors B, C, D, and E (lanes 5 and 6). Reaction products were either analyzed directly (lanes marked −) or after digestion with subtilisin (lanes marked +). (E) The α-tubulin target protein in FBα contains nonexchangeably bound GTP and can be partitioned to the native state by the action of cofactors C, D, and E, and native tubulin. Analysis of the products of c-cpn–mediated α-tubulin folding reactions done with unlabeled target protein in the presence of α-[32P]GTP and recombinant human cofactor B alone either directly (lane 1), after isolation of the FBα intermediate by gel filtration (lane 2), or after incubation of the latter material in the presence of cofactors C, D, and E (lane 3); all reactions contained added native tubulin. (F) Unstable intermediates formed by reaction of FBα (generated using bovine cofactor B and containing bound 32P-labeled GTP) with cofactors D and E. Nondenaturing gel electrophoresis of reaction products after incubation of isolated FBα with the components shown in the figure. These reaction products were stabilized by brief treatment with glutaraldehyde before application to the gel (see text and Materials and Methods). Note that FBα formed with bovine cofactor B comigrates with native tubulin dimer. (C and F, asterisk). A band of intermediate mobility that appears in reactions containing native tubulin and cofactors D and E (but not C) (see text). (Arrows, top to bottom): the location of α-tubulin/c-cpn binary complexes, the human FBα complex, and either the bovine FBα complex or native tubulin, respectively.
We took advantage of our observation that human FBα and native tubulin migrate differently on nondenaturing gels to determine those cofactors required for c-cpn–mediated α-tubulin folding in vitro. C-cpn–mediated α-tubulin folding reactions containing ATP, GTP, and native brain tubulin were supplemented with subsets of the set of cofactors (A, B, C, D, and E) that we identified as sufficient for proper α-tubulin folding (Fig. 1 F). In reactions done with each of the cofactors alone, we found that c-cpn– mediated α-tubulin folding in the presence of human cofactor B resulted in the generation of the characteristic FBα intermediate, whereas none of the other cofactors on their own yielded a recognizable product (Fig. 4 B). In folding reactions from which each cofactor was omitted in turn, we found that the absence of cofactor A did not influence the production of material that migrated as native tubulin, whereas omission of cofactor B resulted in a greatly reduced yield of this product. In contrast, omission of either cofactors C, D, or E completely eliminated the production of material that migrated as native tubulin, yielding the intermediate characteristic of folding reactions containing (human) cofactor B alone (FBα) and, in reactions containing cofactors D and E but lacking cofactor C, an additional slower moving band (highlighted with an asterisk) (Fig. 4 C). The nature and significance of this species is addressed below.
To obtain further evidence that the product migrating as authentic tubulin in these experiments was indeed native, we took advantage of the observation that native tubulin treated with subtilisin results in truncated molecules that have reduced mobilities on nondenaturing gels, but that nonetheless retain their capacity to polymerize into microtubules (Sackett et al., 1985). A control reaction done with native bovine brain tubulin yielded the expected pair of bands, one formed as a result of subtilisin cleavage of only the β subunit, and one formed as a result of cleavage of both α and β subunits (Bhattacharyya et al., 1985) (Fig. 4 D, lanes 1 and 2). Subtilisin treatment under identical conditions of the products of an α-tubulin folding reaction done in the presence of human cofactor B alone showed that the FBα fast-moving band was completely destroyed by the proteolysis step (Fig. 4 D, lanes 3 and 4). In contrast, the subtilisin-treated products from a c-cpn–mediated α-tubulin folding reaction containing cofactors B, C, D, and E were indistinguishable from those derived from authentic tubulin (Fig. 4 D, lanes 5 and 6). In addition, as in the case of the products of a folding reaction done with cofactors A, B, C, D, and E (Fig. 1 F), the products of a folding reaction containing cofactors B, C, D, and E cocycled efficiently with native brain tubulin through three successive cycles of polymerization and depolymerization (data not shown). We conclude that cofactors B, C, D, and E participate in the pathway leading to correctly folded α-tubulin.
Intermediates in the α-Tubulin Folding Pathway
To see whether FBα contained the nonexchangeable GTP that is associated with native α-tubulin (Spiegelman et al., 1977), we did an α-tubulin c-cpn–mediated folding reaction containing cofactor B using unlabeled target protein in the presence of α-[32P]GTP (Tian et al., 1995b ). Nondenaturing gel analysis of the reaction products showed that the cofactor B–dependent fast-migrating band does indeed contain nonexchangeably bound GTP (Fig. 4 E, lane 1). We conclude that the α-tubulin in this complex is quasi-native in that it contains nonexchangeably bound GTP; its sensitivity to cleavage by subtilisin, on the other hand, demonstrates that it is nonnative.
Does FBα behave as an intermediate in α-tubulin folding? To address this question, we isolated human FBα by gel filtration, and found that it could be efficiently partitioned to the native state by the addition of cofactors C, D, E, and native tubulin, as judged by the mobility of the reaction product on a nondenaturing gel (Fig. 4 E, lanes 2 and 3) and by the ability of this product to cocycle with native brain tubulin through multiple rounds of polymerization and depolymerization without loss of specific radioactivity (data not shown). The greater yield of native product in this experiment compared with the starting material is a reflection of their relative stability during native gel electrophoresis. We conclude that FBα is a bona fide intermediate in the pathway leading to native α-tubulin.
Target protein–cofactor complexes formed between c-cpn–generated folding intermediates and cofactors D or D plus E have been described in the pathway leading to correctly folded β-tubulin (Tian et al., 1996). Since cofactors D and E also participate in the α-tubulin folding pathway (Fig. 4 C), we reasoned that similar complexes might be generated (at least transiently) in α-tubulin folding reactions containing these cofactors. Indeed, α-tubulin folding reactions done in the absence of cofactor C yielded an additional band that might be an intermediate in the α-tubulin folding pathway (Fig. 4 C, asterisk). To further characterize such complexes, we incubated the isolated, labeled FBα intermediate in the presence of cofactors D and E either alone or together, with or without added native tubulin. In initial experiments, we observed very low yields of labeled bands with intermediate mobilities on nondenaturing gels. However, when the products of reactions done with FBα and cofactors D or E (or both) were reacted briefly with glutaraldehyde so as to cross-link and thereby stabilize any complexes that might have formed, analysis of the reaction products on a nondenaturing gel revealed the appearance of additional bands (Fig. 4 F). These data suggest the formation of unstable α-tubulin complexes containing cofactor E (FEα) and cofactors D and E. Moreover, the generation of the complex formed by reaction with cofactors D and E (Fig. 4 F, asterisk) is completely dependent upon the inclusion of native tubulin, suggesting that this complex contains both α- and β-tubulin (see below). The intermediates identified in these experiments might play a part in the post-FBα pathway leading to the generation of native α-tubulin. Indeed, low levels of labeled bands with the same migration properties are evident in kinetic analyses of α-tubulin in vitro translation reactions (Zabala and Cowan, 1992).
Native α- and β-Tubulin Exist in Activated Conformational States
In principle, the pathways going from unfolded to correctly folded tubulins should be reversible, providing an opportunity to examine intermediates generated by the action of cofactors on the native tubulin heterodimer. To study the α- and β-tubulin folding pathways in reverse, we labeled each subunit (separately) with [35S]methionine by translation in vitro. The resulting native tubulin heterodimers were resolved from cofactors on a column of DEAE-Sephacel (Murphy et al., 1977), taking advantage of the fact that native tubulin binds much more tightly than any of the cofactors to the anion exchange resin (Tian et al., 1996). This labeled tubulin was then incubated with various combinations of purified cofactors, and the reaction mixtures were analyzed by native gel electrophoresis (Fig. 5 A).
Figure 5.
Interaction of cofactors with native tubulin subunits. (A) Intermediates produced via reaction of cofactors with native tubulin dimers. Purified native tubulin 35S-labeled in either the α or β subunit was incubated for 1 h at 30°C at a concentration of 0.05 μM with either added unlabeled purified native tubulin (2.5 μM) or with a fivefold molar excess of the cofactors shown in the figure. (B) Purified native tubulin 35S-labeled in the β subunit was incubated for 1 h at 30°C at a concentration of 2.5 μM either without or with a 1.5-fold molar excess of cofactor D. (C) Free α-tubulin subunits generated by sequestration of β-tubulin by cofactor D can be trapped by mitochondrial chaperonin (mt-cpn). Purified native tubulin 35S-labeled in the α subunit was first incubated at 30°C with a 1.5-fold molar excess of cofactor D so as to sequester the β subunits. At t = 0 or t = 15 min thereafter, 2.5 μM purified tubulin, a fivefold molar excess of cofactor B or E, or a 50-fold molar excess of mt-cpn was added, and the incubation continued for an additional 30 min. As a control, native tubulin 35S-labeled in the α subunit was incubated alone with the same molar excess of mt-cpn. (D) Free β-tubulin subunits generated by sequestration of α-tubulin by cofactors B and E are also capturable by an mt-cpn trap. Purified tubulin 35S-labeled in the β subunit was incubated at 30°C for 1 h in the presence of unlabeled tubulin, or a 10-fold molar excess of cofactors B and E without or with a 50-fold molar excess of trap. As a control for the effect of trap on native tubulin itself, the input labeled tubulin was incubated in a parallel reaction with the same molar excess of trap but without addition of cofactors. (A–D) Reaction products were analyzed on 4.5% nondenaturing polyacrylamide gels. Upper and lower arrows on the left denote the location of the human FBα complex and native tubulin heterodimer, respectively; arrows on the right denote the location of mt-cpn. (A and C) Bands produced in reactions done with cofactors D and E are highlighted with an asterisk (see text).
Incubation of native tubulin heterodimer containing 35S-labeled α subunit with a fivefold molar excess of cofactors B or E alone did not result in any shift in the mobility of the radiolabeled species. However, reaction with cofactor D resulted in a complete loss of radiolabeled α-tubulin. Incubation with cofactors D and E caused a quantitative shift of the labeled protein to a species that comigrates with a band that appears in in vitro c-cpn–mediated (i.e., forward) α-tubulin folding reactions (Fig. 4 C, asterisk), as well as with the species seen in a β-tubulin backreaction containing the same cofactors (Fig. 5 A, asterisk). Finally, reaction of tubulin with recombinant human cofactor B and either cofactor D or E resulted in the formation of the FBα intermediate (compare Figs. 4, A–C and 5 A). In the case of reactions done with native tubulin heterodimer containing 35S-labeled β subunit, incubation with cofactor D or with cofactor D plus E resulted in a quantitative shift of label to slower migrating species; these species correspond in mobility to cofactor/β-tubulin complexes formed in in vitro c-cpn–mediated (i.e., forward) β-tubulin folding reactions containing the same cofactors (Tian et al., 1996).
The data shown in Fig. 5 A suggest that cofactor D can interact with native tubulin, disrupting the heterodimer. To rule out the possibility that the tubulin heterodimer might be denaturing during the course of these experiments because of the relatively low tubulin concentration, we repeated the incubation of native heterodimer with a 1.5-fold molar excess of cofactor D using a tubulin concentration of 2.5 μM, i.e., above the reported dissociation constant of 1 μM (Detrich and Williams, 1978). In this case the labeled β-tubulin band was also quantitatively shifted (Fig. 5 B). These data confirm that cofactor D can bind to the native β-tubulin subunit, disrupting the heterodimer and forming a stable (FDβ) complex.
Are free α-tubulin subunits bereft of their β-tubulin partners unstable in solution or merely destabilized during native gel electrophoresis? To address this question, we incubated native tubulin dimer 35S-labeled in its α subunit with a 1.5-fold molar excess of cofactor D; immediately thereafter (which we define as t = 0 min), or after an additional 15 min of incubation at 30°C (defined as t = 15 min) so as to fully form the FDβ complex, the reaction was supplemented with one of several components and the incubation continued for an additional 30 min. When native tubulin was added at t = 0 min, there was no significant loss of labeled tubulin relative to the input counts. However, addition of native tubulin after a 15-min delay resulted in a total loss of counts (Fig. 5 C). We next tested whether free α-tubulin generated in cofactor D–containing reactions could be captured either by other cofactors or by mitochondrial chaperonin (mt-cpn), which we have previously shown to recognize and bind nonnative forms of α-tubulin (Tian et al., 1995a ). Addition of cofactors B or E or mt-cpn at t = 0 min resulted in the binding of α subunits to these proteins. However, when added at t = 15 min, cofactors B or E failed to capture detectable levels of labeled α-tubulin. In contrast, mt-cpn added at t = 15 min was still able to capture about half of the input α-tubulin radioactivity (the remainder may have irreversibly aggregated). Finally, in a control experiment in which native tubulin dimer containing 35S-labeled α-tubulin was incubated with mt-cpn alone, only a small proportion of the α subunit was captured, reflecting a modest amount of denaturation of the input heterodimer. These data demonstrate that, in the absence of the β subunit, the conformation of α-tubulin rapidly decays to a form that is incapable of heterodimerization or interaction with cofactors, and that is recognized as nonnative by mt-cpn. Since cofactor B alone does not interact with native tubulin (Fig. 5 A), the data in Fig. 5 C suggest that it binds to α-tubulin in a conformation intermediate between the native state and the nonnative forms recognized by mt-cpn.
The action of cofactors B and E on native tubulin results in the sequestration of α-tubulin as FBα (Fig. 5 A), suggesting that cofactor E, like cofactor D, interacts with native tubulin. Because cofactor B on its own does not interact with native tubulin, it must acquire its bound α subunit via FEα, which is unstable, but can be stabilized by chemical cross-linking (Fig. 4 F). FBα can also be formed when cofactor D sequesters the β subunit and the remaining α subunit decays to a state recognizable by cofactor B. Since a combination of cofactors B and E results in the formation of a stable FBα complex, this allowed us to investigate the fate of the β subunit in the absence of its partner α-tubulin subunit. We found that incubation of native tubulin dimers with cofactors B and E resulted in the destabilization of β subunits as assayed by their ability to be captured by mt-cpn (Fig. 5 D). We conclude that both α and β subunits in the native tubulin heterodimer exist in metastable conformations; if one subunit is removed by interaction with cofactor(s), the remaining free subunit rapidly decays to a conformational state of lower energy that is incapable of heterodimerization.
GTP Hydrolysis and Tubulin Folding
In the tubulin heterodimer, both α and β subunits bind one molecule of GTP, the latter exchangeably; GTP hydrolysis by β-tubulin is coupled to polymerization (Mitchison and Kirschner, 1986). It has been reported that the slowly hydrolyzable analogue GTPγS inhibits the production of native β-tubulin in in vitro translation cocktails (Fontabla et al., 1993; Paciucci, 1994). We therefore tested the ability of GTPγS to inhibit the folding of α- and β-tubulin using purified components. Both α- and β-tubulin post-c-cpn–mediated reactions were inhibited in the presence of GTPγS, showing that GTP hydrolysis is indeed necessary for cofactor-mediated α- and β-tubulin folding (Fig. 6 A). Under these conditions, both α- and β-tubulin folding intermediates remained complexed with cofactors, appearing as identical doublets upon native gel electrophoresis. We also investigated the conversion to native heterodimer of intermediates generated in the backreaction of native tubulin with cofactors D and E. We first formed tubulin/cofactor complexes in reactions containing cofactors D and E, 10 μM GTP, and native tubulin heterodimer in which either the α or β subunits were 35S-labeled. The addition of cofactor C and unlabeled native tubulin to these intermediates results in the regeneration of native tubulin. This reaction was blocked by the addition of a 100-fold molar excess (relative to GTP) of GTPγS, but not ATPγS (Fig. 6 B). Note that the same labeled doublet appears in the products of both forward (i.e., c-cpn–mediated) and backward α- and β-tubulin folding reactions when these are blocked by GTPγS (compare Fig. 6 A and 6 B). To see which component(s) in this reaction hydrolyzes GTP, we incubated cofactors and tubulin with α–32P-labeled GTP either alone or in various combinations, and assayed for conversion to α-[32P]GDP by TLC (Fig. 6 C). Neither native tubulin heterodimer nor cofactors D, E, or C alone significantly hydrolyzed GTP. Upon coincubation, however, these components hydrolyzed GTP at a rate of at least 0.4 min−1, comparable to the rate of ATP hydrolysis by c-cpn (Melki et al., 1996).
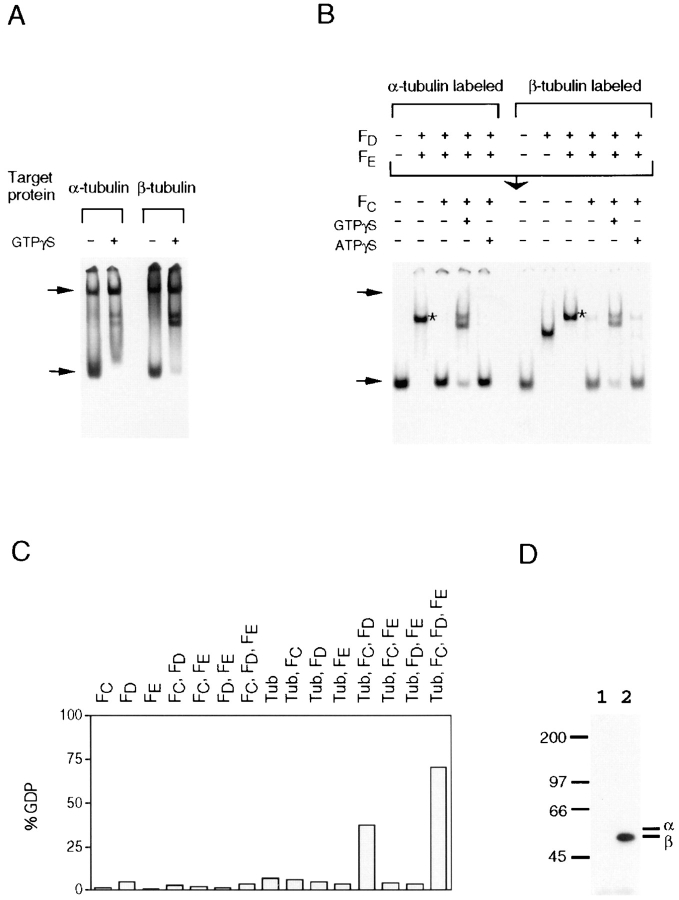
Figure 6.
Role of GTP hydrolysis in tubulin folding. (A) Tubulin folding is accompanied by GTP hydrolysis. In vitro c-cpn–mediated α- or β-tubulin folding reactions were done in the presence of cofactors without or with GTPγS (see Materials and Methods). (B) GTPγS, but not ATPγS, blocks the release of both α- and β-tubulin from complexes containing cofactors D and E. Purified native tubulin heterodimers 35S-labeled in either the α or β subunit were incubated at 30°C for 1 h in the presence of 10 μM GTP with a 1.5-fold molar excess of cofactors D or D plus E to generate tubulin/cofactor complexes (see Fig. 4 A). To complete the reaction, cofactor C and unlabeled tubulin (2.5 μM) were added, and the incubation continued in the presence or absence of GTPγS or ATPγS for an additional hour. (A and B) Reaction products were analyzed on 4.5% nondenaturing polyacrylamide gels. Upper and lower arrows show the location of c-cpn/tubulin binary complex and native tubulin heterodimer, respectively. (C) GTP hydrolysis by cofactors and by tubulin/cofactor complexes. The percentage conversion to GDP is shown. (D) GTP can be cross-linked to β-tubulin in the α/β-supercomplex. Analysis on an 8% SDS–polyacrylamide gel of the UV cross-linked products of a reverse tubulin folding reaction containing cofactors C, D, E, and α–32P-labeled GTP without (lane 1) or with native tubulin heterodimer (lane 2). The location of molecular mass markers (in kD) is shown, together with the location of α- and β-tubulin.
The minimal set of proteins necessary for GTP hydrolysis in these reactions consisted of tubulin and cofactors C and D (Fig. 6 C). When native tubulin was preincubated with a molar excess of cofactor D in this reaction so that no free α-tubulin subunits were present in cofactor-recognizable form (Fig. 5 C), GTP hydrolysis was unimpaired upon addition of cofactor C (data not shown). Thus, the GTP hydrolysis step in tubulin folding requires only β-tubulin and cofactors C and D. To see which of the proteins in the fully constituted active complex binds and hydrolyzes GTP, we incubated native tubulin with a molar excess of cofactors C, D, and E in the presence of α–32P-labeled GTP. Under these conditions, all tubulin subunits are in cofactor complexes, and α-tubulin contains bound, unlabeled GTP, which it does not exchange or hydrolyze throughout the post–c-cpn folding pathway (Fig. 4 E and data not shown). Following UV irradiation, the only species containing labeled, cross-linked GTP was β-tubulin (Fig. 6 D), suggesting that the cofactors stimulate GTP hydrolysis by β-tubulin. It follows that the productive folding of α- and β-tubulin requires the hydrolytic action of β-tubulin in the active complex.
Discussion
Cofactor B Enhances the Folding of α-Tubulin
We recently showed that the c-cpn–mediated folding of β-tubulin in vitro involves the action of four protein cofactors that we named A, C, D, and E (Tian et al., 1996). Here we describe the purification, cloning, and functional characterization of a protein (cofactor B) that acts in the α-tubulin folding pathway (Fig. 1). This cofactor greatly increases the yield of native α-tubulin in c-cpn–mediated folding reactions containing cofactors C, D, and E (Fig. 4 C). Cofactor B acts by capturing quasinative (IQ) intermediates generated by c-cpn in an ATP-dependent reaction (Fig. 4 E and Tian et al., 1995b ); these intermediates (and all subsequent intermediates in the α-tubulin folding pathway) contain nonexchangeably bound GTP, as does native α-tubulin. Reaction with either human recombinant or bovine cofactor B results in species with differing electrophoretic mobilities in our native gel assay (Fig. 4 A); this shows that cofactor B forms a complex with α-tubulin folding intermediates. However, the α-tubulin in these species is not in its native conformation, since it will not exchange into added native tubulin heterodimers and, unlike native tubulin, it is extremely sensitive to proteolysis by subtilisin (Fig. 4 D). The interaction of cofactor B with α-tubulin folding intermediates is consistent with our observation that a null mutation of the yeast cofactor B homologue, ALF1, is lethal in combination with the α-tubulin mutation tub1-1.
Convergence and Symmetry of the α- and β-Tubulin Folding Pathways
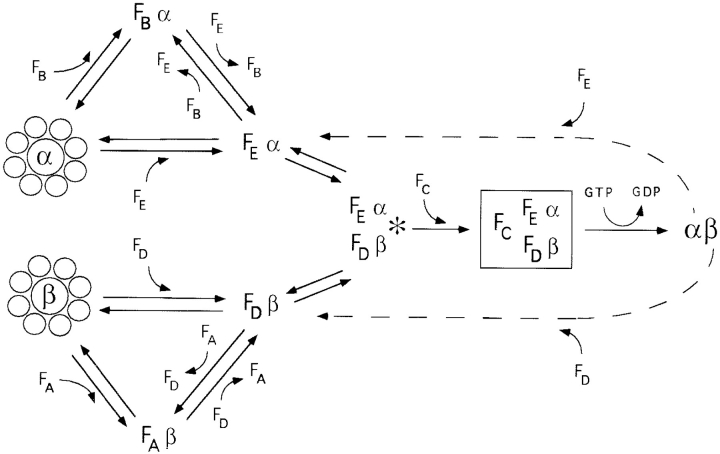
A simple model for tubulin folding that incorporates our data on the interaction of purified cofactors with native tubulin and chaperonin-generated intermediates is presented in Fig. 7. Quasinative α- or β-tubulin folding intermediates generated via ATP-dependent interaction with c-cpn are captured by cofactors B and E (in the case of α-tubulin) or A and D (in the case of β-tubulin; Tian et al., 1996), forming tubulin intermediate/cofactor complexes, i.e., FBα or FEα (in the α-tubulin pathway) or FAβ or FDβ (in the β-tubulin pathway). The FBα and FAβ complexes act as reservoirs, capable of accepting or delivering their target protein to cofactors E and D, respectively. FEα and FDβ interact with each other to form the species (FEα/ FDβ) marked with an asterisk in Figs. 3, 4, and 5. Addition of cofactor C (FC) generates the active entity (the α/β-supercomplex; boxed in Fig. 7), which hydrolyzes GTP and produces native tubulin. The FDβ state is also populated from the backreaction between native β-tubulin subunits and cofactor D, as is FEα from a backreaction between native α-tubulin subunits and cofactor E.
Figure 7.
Convergence and symmetry of the α- and β-tubulin folding pathways (see text). Quasinative α- and β-tubulin folding intermediates produced via ATP-dependent interaction with c-cpn (shown as eight-subunit toroids) interact with a series of protein cofactors (FA, FB, FC, FD, and FE). The pathways converge via the formation of a complex containing α- and β-tubulin and cofactors D and E (asterisk). Entry of cofactor C generates the α/β-supercomplex (boxed); GTP hydrolysis then results in the release of native polypeptides. Broken arrows show the backreaction between native α- or β-tubulin subunits and cofactors E or D. The names of tubulin/cofactor complexes are not intended to reflect their stoichiometry. α, α-tubulin target protein. β, β-tubulin target protein.
Among the tubulin intermediate/cofactor complexes depicted in our model, FDβ, FAβ, and FBα are sufficiently stable to allow their biochemical isolation. The tubulin intermediates in these complexes are convertible to the native state only in the presence of GTP, tubulin itself, and other essential cofactors, i.e., C and E (in the case of FDβ) or C, D, and E (in the case of FAβ and FBα) (Fig. 4 E and Tian et al., 1996). The FEα and FEα/FDβ complexes are unstable during column chromatography and immunoprecipitation; hence the evidence for the existence of these species is necessarily circumstantial, but nonetheless compelling. We infer the existence of FEα from the following considerations: (a) since cofactor B can be omitted from productive α-tubulin folding reactions (albeit with a >90% loss of efficiency) (Fig. 4 C), cofactor E must be capable of capturing some c-cpn–generated intermediates, as it is the only α-tubulin binding cofactor present. (b) FEα can be stabilized and visualized in in vitro α-tubulin folding reactions after chemical cross-linking (Fig. 4 F). FEα interacts with FDβ to form the FEα/FDβ complex (marked with an asterisk in Figs. 4, 5 and 6) whose existence is in turn supported by the following data: (a) because of the high affinity of cofactor D for β-tubulin, in the presence of excess tubulin heterodimer and the absence of cofactor C, cofactor D must be bound to β-tubulin. Thus, the species marked with an asterisk generated in α-tubulin forward and backward reactions containing cofactors D, E, and tubulin must contain β-tubulin in association with cofactor D. Moreover, this species is not formed in α-tubulin folding reactions in which native tubulin is omitted (Fig. 4 F), demonstrating that it is FDβ and not cofactor D alone that interacts with FEα. (b) A species of the same mobility as that seen in α-tubulin folding reactions appears in β-tubulin forward and reverse reactions (Figs. 5 A and 6 B, asterisk) and has been shown to contain cofactors D, E, and β-tubulin (Tian et al., 1996). Since β-tubulin cannot be productively folded without added native tubulin and cofactor E, we conclude that this too is the FEα/FDβ complex.
When cofactor C is added to the FEα/FDβ complex, GTP is hydrolyzed and native tubulin is released. Only this combination of proteins results in the maximal rate of GTP hydrolysis and the production of native tubulin. Thus, all five proteins appear to constitute the GTPase (the α/β-supercomplex) that is active in tubulin folding. Although our model depicts these five proteins as interacting physically in a single complex, we cannot rule out a more elaborate scheme In which subsets of them act in rapid succession.
As presented in our model, the α- and β-tubulin folding pathways are symmetrical. Cofactors A and B act as reservoirs for the sequestration of tubulin folding intermediates; in this capacity, they could serve as buffers that would protect the cell from an unbalanced production of α- or β-tubulin subunits. This notion is consistent with the observation that the otherwise toxic induced overexpression of β-tubulin in yeast can be rescued by a corresponding induced overexpression of the yeast homologue of cofactor A (Archer et al., 1995). In addition, the in vitro folding of α-tubulin is very inefficient in folding reactions that lack cofactor B but that contain native tubulin heterodimer (which is necessary to provide the partner subunit in the α/β-supercomplex) (Fig. 4 C). Similarly, in c-cpn–mediated β-tubulin folding reactions, if native tubulin is added together with cofactors (rather than at the end [Tian et al., 1996]), the yield of native product is extremely low in the absence of cofactor A (Tian, G., and N.J. Cowan, unpublished observations). Thus, cofactors B and A shift the reaction equilibria: cofactors E and D seem to preferentially accept α- or β-tubulin intermediates from FBα and FAβ, respectively, rather than from native heterodimer, favoring de novo folding over the backreaction involving refolding of native subunits.
GTP Hydrolysis and Cofactor Function
The final step in the generation of native tubulin heterodimer involves the hydrolysis of GTP (Figs. 6 and 7). In our previous description of the pathway leading to correctly folded β-tubulin, we found that native β-tubulin folding could be supported by GDP (Tian et al., 1996). However, our c-cpn preparations contained some nucleoside diphosphate kinase activity: hence, in reactions containing ATP and GDP, there was a resulting generation of sufficient levels of GTP to drive the folding reaction. The data shown in Fig. 6 demonstrate conclusively that GTP hydrolysis is necessary for the release of native tubulin from cofactor complexes, whether these are formed in the forward reaction in which urea-denatured tubulin target protein is presented to c-cpn, or in the backreaction, in which native tubulin heterodimer is incubated with cofactors. The combination of tubulin heterodimer and cofactors C, D, and E hydrolyzes GTP at a rate of at least 0.4 min−1 (Fig. 6 C). The continuous cycling of tubulin through cofactor complexes and the concomitant hydrolysis of GTP could serve to stabilize the tubulin subunits; this is consistent with the role of the yeast homologues of cofactors D and E in maintaining microtubule stability in vivo (Stearns et al., 1990; Hoyt et al., 1997). The GTPase formed by these five proteins might serve other cellular functions: e.g., it is possible that cofactors C, D, and E could bind to the GTP cap on polymerizing microtubules, stimulating the hydrolysis of GTP by β-tubulin, and thus influencing microtubule dynamics. It is therefore important to confirm that, in the α/β-supercomplex, one or more of the cofactors is stimulating GTP hydrolysis by β-tubulin, as opposed to the tubulin-dependent stimulation of GTP hydrolysis by a cofactor. Current evidence suggests that this is the case (Fig. 6 D).
Tubulin Cofactor Function In Vitro and In Vivo
When native tubulin dimer is incubated with excess cofactor D, the β-tubulin subunit is efficiently sequestered as FDβ, disrupting the α/β tubulin heterodimer (Fig. 5). Under such conditions, the α-tubulin subunit rapidly loses its ability to reassociate with β-tubulin. In the absence of other cofactors, free α-tubulin simply “disappears,” presumably because of aggregation via exposed hydrophobic surfaces. We confirmed this conclusion by showing that free α-tubulin quickly becomes capturable by mitochondrial chaperonin, which binds nonnative α-tubulin states (Fig. 5 and Tian et al., 1995a ). We conclude that, in the native heterodimer, α-tubulin exists in an activated state. The converse experiment involving sequestration of α subunits by the action of cofactors B and E shows that β-tubulin is also in a metastable conformational state in the heterodimer (Fig. 5 D). The interaction between α and β subunits is therefore critical for the maintenance of functional tubulin heterodimers. This explains why it has never proved possible to biochemically isolate α- or β-tubulins in native form free from their partner subunits.
There are known homologues of cofactors A, B, D, and E (but not C) in S. cerevisiae (Fig. 2 and Stearns et al., 1990; Archer et al., 1995; Hoyt et al., 1990, 1997). Deletion of genes encoding these homologues results in microtubule phenotypes such as supersensitivity to antimicrotubule drugs, temperature sensitivity, and chromosomal instability. These yeast genes are not essential for viability, even though they are single copy. Therefore, since microtubules are indispensable structures, yeast tubulins must be able to reach the native state without the aid of the full complement of cofactors, although there is persuasive evidence that c-cpn is required for tubulin folding in yeast (Chen et al., 1994; Ursic et al., 1994). In that case, the cofactors might be needed only to stabilize native yeast tubulin subunits and to prevent the accumulation of free β subunits that would otherwise be lethal (Archer et al., 1995); this would be consistent with their mutant phenotypes. On the other hand, the participation of cofactors C, D, and E is absolutely required for the proper folding of mammalian α- and β-tubulin in vitro. The differences in cofactor requirements in vitro and in vivo could reflect either the existence of alternative in vivo folding pathways, a critical difference in the folding requirements of yeast and mammalian tubulins, or both.
Figure 2.
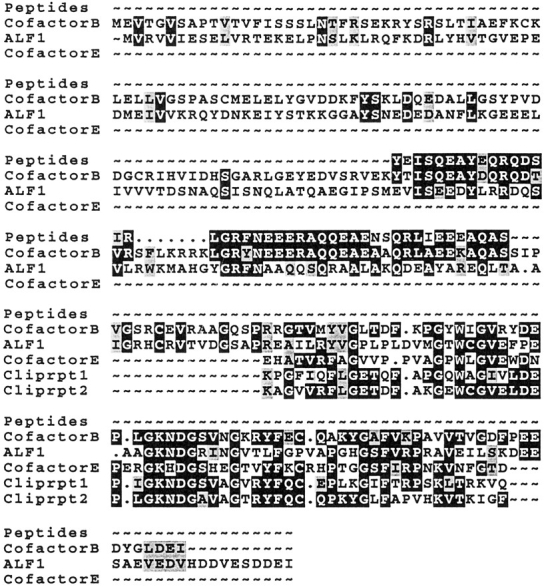
Amino acid sequence of cofactor B: homology with cofactor E and the S. cerevisiae protein Alf1p. Amino acid sequences of peptides derived from purified bovine cofactor B, together with the deduced amino acid sequences of human cofactor B (these sequence data are available from EMBL/GenBank/ DDBJ under accession number AF013488), the S. cerevisiae homologue Alf1p, and the microtubule binding motif in cofactor E. The corresponding repeated motifs in CLIP-170 are shown for comparison.
The complicated and energy-consuming post–c-cpn reactions involving cofactors may have evolved because native tubulin exists as a tight heterodimer. In the case of an unrelated heterodimeric protein, bacterial α- and β-luciferase, if both subunits are allowed to refold from urea separately, they attain stable structures that are unable to dimerize; however, if they are allowed to refold together, they dimerize into an inactive heterodimer, which then isomerizes to the active form of the enzyme (Baldwin et al., 1993; Clark et al., 1993). The luciferase heterodimer therefore appears to act as a kinetic trap on the folding pathways of the individual subunits; the subunits of the dimer are not in their ground state conformation. This contradicts the widely held tenet that all the information required to reach the native state is contained in a protein's amino acid sequence: like the propeptide in some proteases, which can act in cis or in trans to promote correct folding, luciferase is an example of one protein that affects the final conformational state of another. We have shown here that tubulin is another such example: the instability of isolated tubulin subunits implies that the attainment and maintenance of the native conformation of α-tubulin depends on information derived from the β-tubulin subunit and vice versa. We provide strong evidence that the function of the cofactors that act in the post–c-cpn tubulin folding pathways is to bring the α and β subunits together in a supercomplex so that they can achieve this native (i.e., activated) conformation.
Acknowledgments
We thank R. Beavis for mass spectrophotometric analyses, and E. Hammel and N. Kallenbach for stimulating discussions.
This work was supported by grants (to N.J. Cowan and T. Stearns) and a National Research Service Award (to G. Tian) from the National Institutes of Health, and by grants from the Geconcerteerde Onderzoeksactie and the Belgian National Fund for Scientific Research (to C. Ampe). C. Ampe is a Research Associate of the National Fund for Scientific Research.
Abbreviations used in this paper
- c-cpn
cytosolic chaperonin
- mt-cpn
mitochondrial chaperonin
Footnotes
Please address all correspondence to Nicholas J. Cowan, Department of Biochemistry, New York University Medical Center, 550 First Avenue, New York, NY 10016. Tel.: (212) 263-5809. Fax: (212) 263-8166.
References
- Amberg DC, Botstein D, Beasley EM. Precise gene disruption in Saccharomyces cerevisiaeby double fusion polymerase chain reaction. Yeast. 1996;11:1275–1280. doi: 10.1002/yea.320111307. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science (Wash DC) 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Archer JE, Vega LR, Solomon F. Rbl2, a yeast protein that binds to β-tubulin and participates in microtubule function in vivo. Cell. 1995;82:425–434. doi: 10.1016/0092-8674(95)90431-x. [DOI] [PubMed] [Google Scholar]
- Baldwin TO, Ziegler MM, Chaffotte AF, Goldberg ME. Contribution of folding steps involving the individual subunits of bacterial luciferase to the assembly of the active heterodimeric enzyme. J Biol Chem. 1993;268:10766–10772. [PubMed] [Google Scholar]
- Bhattacharyya B, Sackett DL, Wolff J. Tubulin, hybrid dimers, and tubulin S. J Biol Chem. 1985;260:10208–10216. [PubMed] [Google Scholar]
- Chen X, Sullivan DS, Huffaker T. Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc Natl Acad Sci USA. 1994;91:9111–9115. doi: 10.1073/pnas.91.19.9111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AC, Sinclair JF, Baldwin TL. Folding of bacterial luciferase involves a non-native heterodimeric intermediate in equilibrium with native enzyme and the unfolded subunits. J Biol Chem. 1993;268:10773–10779. [PubMed] [Google Scholar]
- Detrich HW, III, Williams RC. Reversible dissociation of the alpha beta dimer of tubulin from bovine brain. Biochemistry. 1978;17:3900–3907. doi: 10.1021/bi00612a002. [DOI] [PubMed] [Google Scholar]
- Eriksson S, Caras IW, Martin DW. Direct photoaffinity labeling of an allosteric site on subunit protein M1 of mouse ribonucleotide reductase by dTTP. Proc Natl Acad Sci USA. 1982;79:81–85. doi: 10.1073/pnas.79.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontabla A, Paciucci R, Avila J, Zabala JC. Incorporation of tubulin subunits into dimers requires GTP hydrolysis. J Cell Sci. 1993;106:627–632. doi: 10.1242/jcs.106.2.627. [DOI] [PubMed] [Google Scholar]
- Frydman J, Nimmesgern E, Erdjument-Bromage H, Wall JS, Tempst P, Hartl F-U. Function in protein folding of TRiC, a cytosolic ring complex containing TCP-1 and structurally related subunits. EMBO (Eur Mol Biol Organ) J. 1992;11:4767–4778. doi: 10.1002/j.1460-2075.1992.tb05582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Thomas JO, Chow RL, Lee G-H, Cowan NJ. A cytoplasmic chaperonin that catalyzes β-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- Gao Y, Vainberg IE, Chow RL, Cowan NJ. Two cofactors and cytoplasmic chaperonin are required for the folding of α- and β-tubulin. Mol Cell Biol. 1993;13:2478–2485. doi: 10.1128/mcb.13.4.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Melki R, Walden P, Lewis SA, Ampe C, Rommelaere H, Vandekerckhove J, Cowan NJ. A novel cochaperonin that modulates the ATPase activity of cytoplasmic chaperonin. J Cell Biol. 1994;125:989–996. doi: 10.1083/jcb.125.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F-U. Molecular chaperones in cellular protein folding. Nature (Lond) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- Horwich AL, Weissman JS, Fenton WA. Kinesis of polypeptide during GroEL-mediated folding. Cold Spring Harbor Symp Quant Biol. 1995;60:435–440. doi: 10.1101/sqb.1995.060.01.048. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiaethat are defective in microtubule-mediated processes. Mol Cell Biol. 1990;10:223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Macke JP, Roberts BT, Geiser JR. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4in a pathway leading to normal microtubule stability. Genetics. 1997;146:849–857. doi: 10.1093/genetics/146.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukada Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis SA, Tian G, Vainberg IE, Cowan NJ. Chaperonin-mediated folding of actin and tubulin. J Cell Biol. 1996;132:1–4. doi: 10.1083/jcb.132.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall L, Jeng R, Mulholland J, Stearns T. Analysis of Tub4p, a yeast γ-tubulin–like protein: implications for microtubule organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Vainberg IE, Chow RL, Cowan NJ. Chaperonin-mediated folding of vertebrate actin-related protein and γ-tubulin. J Cell Biol. 1993;122:1301–1310. doi: 10.1083/jcb.122.6.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Rommelaere H, Leguy R, Vandekerckhove J, Ampe C. Cofactor A is a molecular chaperone required for β-tubulin folding: functional and structural characterization. Biochemistry. 1996;35:10422–10435. doi: 10.1021/bi960788r. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner MW. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Murphy DB, Vallee RB, Borisy GG. Identity and polymerization-stimulatory activity of the non-tubulin proteins associated with microtubules. Biochemistry. 1977;16:2598–2606. doi: 10.1021/bi00631a004. [DOI] [PubMed] [Google Scholar]
- Paciucci R. Role of 300kD complexes as intermediates in tubulin folding and dimerization: characterization of a 25kD cytosolic protein involved in the GTP-dependent release of monomeric tubulin. Biochem J. 1994;301:105–110. doi: 10.1042/bj3010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre P, Scheel J, Rickard JE, Kreis T. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- Sackett D, Bhattacharyya B, Wolff J. Tubulin subunit carboxyl termini determine polymerization efficiency. J Biol Chem. 1985;260:43–45. [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1983. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sikorski RS, Hieter P. A study of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. . Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM, Penningroth SM, Kirschner MW. Turnover of tubulin and the N-site GTP in Chinese hamster ovary cells. Cell. 1977;12:587–600. doi: 10.1016/0092-8674(77)90259-8. [DOI] [PubMed] [Google Scholar]
- Stearns T, Hoyt MA, Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Vainberg IE, Tap WD, Lewis SA, Cowan NJ. Specificity in chaperonin-mediated protein folding. Nature (Lond) 1995a;375:250–253. doi: 10.1038/375250a0. [DOI] [PubMed] [Google Scholar]
- Tian G, Vainberg IE, Tap WD, Lewis SA, Cowan NJ. Quasi-native chaperonin-bound intermediates in facilitated protein folding. J Biol Chem. 1995b;270:23910–23913. doi: 10.1074/jbc.270.41.23910. [DOI] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded β-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Ursic D, Sedbrook JC, Himmel KL, Culbertson MR. The essential yeast TCP1 protein affects actin and microtubules. Mol Biol Cell. 1994;5:1065–1080. doi: 10.1091/mbc.5.10.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh D, Drubin DG. A yeast TCP-1-like protein is required for actin formation in vivo. Proc Natl Acad Sci USA. 1994;91:9116–9120. doi: 10.1073/pnas.91.19.9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabala JC, Cowan NJ. Tubulin dimer formation via the release of α- and β-tubulin monomers from multimolecular complexes. Cell Motil Cytoskeleton. 1992;23:222–230. doi: 10.1002/cm.970230306. [DOI] [PubMed] [Google Scholar]