Abstract
Cofilin is an actin depolymerizing protein found widely distributed in animals and plants. We have used electron cryomicroscopy and helical reconstruction to identify its binding site on actin filaments. Cofilin binds filamentous (F)-actin cooperatively by bridging two longitudinally associated actin subunits. The binding site is centered axially at subdomain 2 of the lower actin subunit and radially at the cleft between subdomains 1 and 3 of the upper actin subunit. Our work has revealed a totally unexpected (and unique) property of cofilin, namely, its ability to change filament twist. As a consequence of this change in twist, filaments decorated with cofilin have much shorter ‘actin crossovers' (∼75% of those normally observed in F-actin structures). Although their binding sites are distinct, cofilin and phalloidin do not bind simultaneously to F-actin. This is the first demonstration of a protein that excludes another actin-binding molecule by changing filament twist. Alteration of F-actin structure by cofilin/ADF appears to be a novel mechanism through which the actin cytoskeleton may be regulated or remodeled.
The actin-based cytoskeleton plays an important role in cell locomotion. As cells move in response to external signals, the cytoskeleton is continuously remodeled: actin filament networks are formed at the frontal lamella, which attaches to the underlying substratum through focal adhesions, thereby providing adhesive contacts for the traction necessary for retraction of the cell body. For this process to continue, actin subunits must be continuously recycled to the leading edge (for review see Small, 1995; Welch et al., 1997). Although the assembly rates of actin in vitro are extremely rapid (Drenckhahn and Pollard, 1986), the pointed end disassembly rate constant measured in vitro (∼0.3 s−1) is far too slow to recycle the monomers necessary for this process to continue. Moreover, rates nearly 100 times slower than this have been measured under physiological conditions (Coluccio and Tilney, 1983). This compares with required rates in excess of 7 s−1 for the turnover of actin filaments in the lamellipods of macrophages and 10-fold higher rates that might be necessary to account for the locomotion of leukocytes at 30 μm/min (Zigmond, 1993). The most obvious way to increase the disassembly rate is to sever filaments to expose new barbed ends, where the dissociation rate constant for ADP-actin subunits is much higher (>7 s−1; Pollard, 1986). The cofilin/ADF family of proteins are the most obvious candidates to do this, particularly because of their apparent ability to sever filamentous (F)1-actin without capping (Hawkins et al., 1993; Hayden et al., 1993).
ADF (also called destrin) was first isolated as an actin depolymerizing factor in chick embryo brain and is widely distributed in animal and plant tissues (Bamburg et al., 1980; Bamburg and Bray, 1987; Lopez et al., 1996). A highly homologous protein, cofilin (70% identical to ADF in sequence), has been shown to modulate actin filament disassembly in a pH-dependent manner (Yonezawa et al., 1985). Related proteins have since been identified in a number of other organisms (Cooper et al., 1986; Moon et al., 1993; McKim et al., 1994; Gunsalus et al., 1995). Actin binding is inhibited by phosphorylation of Ser3 (Agnew et al., 1995) and by binding to phosphatidyl inositol 4,5-bis phosphate (Yonezawa et al., 1990). Taken together these results suggest that the ADF/cofilin family of proteins is composed of stimulus-responsive modulators of actin dynamics (for review see Moon and Drubin, 1995; Theriot, 1997).
The mechanism by which these proteins accelerate actin filament dynamics is still controversial. Actophorin and ADF were classified as actin filament-severing proteins based on visualization by light microscopy (Maciver et al., 1991; Hawkins et al., 1993). However, high concentrations of protein gave minimal severing, and attempts to quantitate the severing activity by biochemical methods suggested that it is <0.1% that of gelsolin. Like cofilin, ADF binds preferentially to filaments <pH 7.3, and promotes disassembly at higher values (Hawkins et al., 1993; Hayden et al., 1993). Because these proteins consist of a single domain (Hatanaka et al., 1996; Federov et al., 1997; Leonard et al., 1997), the switch from G- to F-actin binding must involve subtle differences in the cofilin/actin interface at the two pH values. Like actophorin (Maciver and Weeds, 1994), ADF binds preferentially to ADP-actin monomers, which may in part explain its preferential binding to filament subunits (Maciver, S.K., B. Pope, and A. Weeds, unpublished observations). Recent work has cast doubt on the idea that disassembly is due solely to filament severing: rather ADF appears to modulate the rate constants of assembly and disassembly at the ends of filaments (Carlier et al., 1997; Maciver, S.K., B. Pope, and A. Weeds, unpublished observations).
Here we report our studies on the interaction of cofilin with F-actin by electron cryomicroscopy and image reconstruction. Our analysis shows that cofilin binds cooperatively between two actin subunits along the filament. The most startling finding of this work is that cofilin induces a substantial change in the twist of F-actin. One ramification of this structural change is to alter the phalloidin-binding site, thereby preventing its binding and making it ineffective as a probe for F-actin. This is the first demonstration of an actin-binding protein that “competes” for binding by altering F-actin's twist. The implications of this structure on cofilin's function in the cell and the structure of the actin filament will be discussed.
Materials and Methods
Protein Preparations and Binding Studies
Recombinant human cofilin was expressed in Escherichia coli as previously described for ADF (Hawkins et al., 1993). It was purified from the bacterial lysate on DEAE-cellulose at pH 8.0, where it was not retarded (Hawkins et al., 1993). After adjusting the pH to 6.6, the protein was further purified on CM-cellulose (10 × 2.5 cm) in 10 mM sodium succinate, pH 6.6, 0.5 mM dithiothreitol, 1 mM NaN3 and eluted with a gradient to 0.2 M NaCl. The purified protein was dialyzed into 10 mM Tris-HCl, pH 8.0, 0.2 mM EGTA, 2 mM dithiothreitol, 1 mM NaN3, concentrated by Centricon ultrafiltration and stored at −80°C after drop freezing into liquid nitrogen.
Rabbit muscle actin was prepared as described previously (Harris and Weeds, 1983) or obtained from Cytoskeleton (Denver, CO) and stored at 10 mg/ml in 2.0 mM Tris-HCl, pH 8.0, 0.2 mM CaCl2, 0.2 mM Na2ATP, 0.005% NaN3 at −80°C. Human platelet actin was also obtained from Cytoskeleton. Binding to F-actin was measured by cosedimentation in an ultracentrifuge (TL; Beckman Instr., Fullerton, CA) at 220,000 g as described previously (Hawkins et al., 1993).
Sample Preparation for Electron Microscopy
For structural studies, fully decorated actin filaments were produced by incubating 2.38 μM platelet F-actin with 11.9 μM cofilin in F-buffer (50 mM NaCl, 2 mM MgCl2, 0.2 mM CaCl2, 1 mM DTT, 0.2 mM ATP), containing 15 mM Pipes, pH 6.6 (“low pH buffer”), for 30–90 min on ice. Samples were plunged in a room maintained at 18°C and <30% humidity. Approximately 7 μL of filaments was placed on 400 mesh copper grids prepared with holey carbon films, blotted with filter paper, rapidly frozen in ethane slush cooled with liquid nitrogen, and maintained in liquid nitrogen until use. Plunged samples were pelleted for 15 min at 100,000 g in an airfuge (Beckman Instr.) and SDS-PAGE gels run to confirm cofilin binding to actin. Partial decoration of muscle F-actin was achieved using 2 μM F-actin with 6 μM cofilin in “low pH buffer” for 30–90 min at room temperature. The effect of cofilin binding at higher pH was examined using 2 μM F-actin and 6 μM cofilin in F-buffer containing 10 mM Tris, pH 8.2, for 30–90 min at room temperature. Grids of F-actin were prepared as controls.
Electron Microscopy
Transmission electron microscopy of frozen-hydrated samples was performed at 100 kV using an electron microscope (1200EX; Jeol, Ltd., Tokyo, Japan) with a Gatan anti-contaminator cooled to −179°C with liquid nitrogen. Grids were maintained at −167°C using a Gatan cryotransfer system cooled with liquid nitrogen. Images were recorded on Kodak SO-163 film at a nominal magnification of 30,000 and an electron dose of ∼8 e−/Å2. Electron microscopy of filaments negatively stained with uranyl acetate was performed using a transmission electron microscope (301; Philips, Eindhoven, The Netherlands, CT) at 80 kV.
Image Processing and Analysis
Electron cryomicrographs with a good distribution of filaments and exhibiting minimal drift or charging were scanned on a densitometer (Perkin-Elmer Corp., Norwalk, CT) at 5.3 Å per pixel. The defocus of the micrographs used for the structural analysis was determined by incoherent averaging of calculated diffraction patterns obtained from either regions of adjacent carbon or protein embedded in ice (Zhou et al., 1996). Images of cofilin-decorated platelet actin were recorded at 1.4–1.6 μm underfocus (first node of the contrast transfer function, 1/22.5–1/24.5 Å−1) and those of undecorated platelet actin at 1.5–2.1 μm underfocus (first node, 1/24–1/29 Å−1). Scanned images were selected for their straightness, distribution, and length (minimum of 8 crossovers). Image preparation for reconstruction was performed using Phoelix run on a Silicon Graphics (Mountain View, CA) workstation (Whittaker et al., 1995b ; Schroeter and Bretaudiere, 1996). Regions of cofilin-decorated filaments analyzed ranged in length from 0.22 to 1.41 μm (mean = 0.58 μm). Those from F-actin ranged from 0.29 to 2.01 μm (mean = 0.82 μm).
Particle Indexing
Computed power spectra of decorated filaments revealed a movement from the usual position of the first layerline for F-actin at 1/(365 ± 15) Å−1 to 1/(270 ± 6) Å−1. Some of the best preserved particles had visible reflections in the computed diffraction patterns past 1/27.5 Å−1. Cofilin-decorated filaments were indexed manually after computationally straightening the particles and computing their Fourier transforms. Layerline heights (l) and radial positions of the first peaks (R) were measured from the calculated diffraction patterns. In addition, layerlines were identified and their positions measured from synthetic “phase display” patterns, which incorporate information about the phase differences across the meridians of the particles weighted by the amplitudes (display-phase program courtesy of M. Schmid, Baylor College of Medicine, Houston, TX). Values of l and R for layerlines for which there was clear phase behavior were used to index the patterns (DeRosier and Moore, 1970; Stewart, 1988). The simplest selection rule most consistent with the particles was found to be 20 units in 9 turns (twist of 2.222 subunits per turn), assuming that the hand of the actin filament was unchanged. Small variations in the exact symmetries of the filaments studied were observed as is usual for actin structures. The mean twist for the population of filaments included in the reconstruction was 2.218, which corresponds to a selection rule of 264 units in 119 turns. However, all layerlines collected were brought to the simpler selection rule before merging and performing the three-dimensional reconstruction.
Layerline Collection, Merging, and Calculation of Three-dimensional Reconstructions
Layerline collection, merging, and helical reconstructions were done by standard methods (DeRosier and Moore, 1970; Amos, 1975; Whittaker et al., 1995b ). After correcting for the phase origin and particle tilt, layerlines were separated into near- and far-side datasets and subjected to three rounds of alignment against successive averages. For the final round of alignment the following layerlines (n [Bessel order], l) were used from F-actin: (2,4), (4,8), (−5,17), (−3,21), (−1,25), (1,29), (3,33), and (5,37). The equivalent layerlines were used for the cofilin-decorated F-actin dataset. All data points along the layerlines (excluding those at the meridian) up to a resolution of 1/26 Å−1 for the decorated filaments and 1/30 Å−1 for the actin filaments were used in the alignments.
Datasets showing disagreement in polarity assigned to the two sides, low (<10°) up/down difference in phase residuals, high (>50°) phase residuals for both sides, or uncharacteristically large tilts and/or shifts during correction for phase origin were excluded. The mean phase residuals calculated from the F-actin and cofilin/F-actin datasets during the last round of alignment were 29.4° (up/down difference = 25.6°) and 38.6° (up/down difference = 20.5°), respectively. The cofilin/F-actin reconstruction included 13 layerlines (highest l = 42; largest n = 7) averaged from 38 datasets representing the contributions of ∼4,380 actin/cofilin subunits. The F-actin reconstruction was calculated from an average of 20 datasets (16 layerlines; highest l = 20; largest n = 7) containing a total of 3,294 actin subunits. The two reconstructions were brought to a common phase origin by aligning preliminary averages of each against a reconstruction of rabbit muscle F-actin. The polarity of the filament was determined by alignment against a reconstruction of myosin S1-decorated F-actin (Whittaker et al., 1995a ).
Visualization of Maps
Three-dimensional density maps were calculated by Fourier-Bessel inversion of averaged layerline data. The F-actin reconstruction was calculated at both 54/25 and 20/9 symmetries for comparison to the cofilin/F-actin reconstruction. The significance of features within the reconstructions was assessed by generating t-maps between near- and far-side averages of each reconstruction (Milligan and Flicker, 1987). These maps revealed no significant differences between the near- and far-side averages for either reconstruction, indicating that the features in the maps are reliable. Atomic models and electron density maps were displayed and manipulated using IRIS Explorer (Numerical Algorithms Group, Ltd., Oxford, UK) and O version 5.9 (Jones et al., 1991). Ribbon diagrams were generated using Ribbons 2.65 (Carson and Bugg, 1986), saved as Inventor format files, and displayed in IRIS Explorer.
Results
Appearance of Actin Filaments Decorated with Cofilin
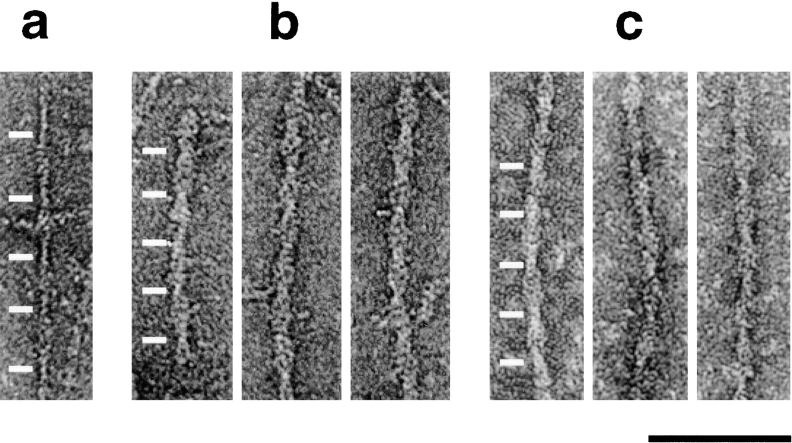
Fig. 1 a shows a muscle actin filament of approximately five crossovers. The crossovers are ∼365 Å in length, which is typical for F-actin. Actin filaments decorated with human cofilin and ADF are shown in Fig. 1, b and c, respectively. At pH 6.5, both cofilin and ADF bind filaments without destabilizing them. The decorated filaments are thicker, owing to the presence of cofilin. Surprisingly, however, they have substantially shorter crossover lengths (∼270 Å on average). Therefore, a major effect of binding by either cofilin or ADF to actin is to change the helical twist.
Figure 1.
Cofilin and ADF change the twist of rabbit muscle F-actin. Electron micrographs of negatively stained F-actin (a) and filaments decorated with cofilin (b) and ADF (c) at pH 6.5. Both cofilin and ADF induce a 25% reduction in crossover length. White bars indicate crossover positions. Bar, 0.1 μm.
Fig. 2 shows cryomicrographs of platelet actin at pH 6.6, in the presence and absence of cofilin. A single computationally straightened filament without cofilin, ∼100 Å diam, is shown in Fig. 2 a. The fine features are difficult to discern owing to the imaging conditions used to produce these cryomicrographs, but the calculated diffraction pattern (Fig. 2 c) shows that the filament is highly ordered. The strongest layerlines have been labeled with their helical index parameters: n (Bessel order) and l (layerline number). (These values are analogous to the indices used to describe discrete reflections in the diffraction patterns of crystalline objects.) The spacings of layerlines n = 2, l = 4 and n = −1, l = 27 occur at 1/366 Å−1 and 1/59 Å−1, consistent with their positions in diffraction patterns of muscle F-actin, in which they correspond to the periodicities of the long pitch double helix of actin and the left-handed, short-pitch helix, respectively. Thus, muscle and nonmuscle actin filaments are indistinguishable at this resolution.
Figure 2.
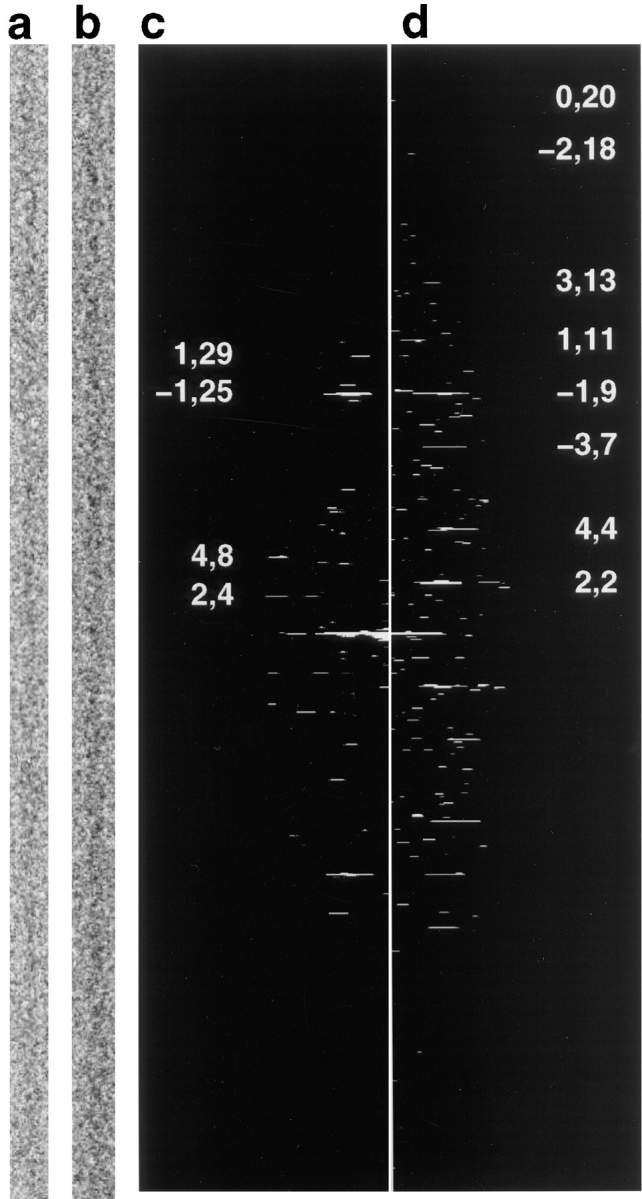
Cofilin changes the twist of platelet F-actin. (a) Portion of a computationally straightened F-actin filament; (b) portion of a computationally straightened actin filament decorated with cofilin. The defocus of both micrographs shown was 1.5 μm. Decorated filaments are ∼120–130 Å diam with accentuated ‘crossovers' relative to control actin. (c) Computed diffraction pattern calculated from the actin filament shown in (a). The length of the filament used to calculate this pattern was 1.44 μm. (d) Computed diffraction pattern calculated from the cofilin-decorated actin filament shown in (b). The length of the filament used to calculate this pattern was 1.41 μm. Prominent layerlines in each diffraction pattern are labeled with values of n and l. The axial position of layerline n = 2, l = 4 in the F-actin diffraction pattern is 1/366 Å−1 and that of n = 2, l = 2 in the cofilin/F-actin diffraction pattern is 1/272 Å−1.
The filament in Fig. 2 b was obtained after incubating platelet F-actin with a 4-fold excess of human cofilin at pH 6.6. Under these conditions all of the filaments were fully decorated with cofilin. They differ from F-actin in two major respects: (a) they are ∼20 to 30% larger in diameter, presumably owing to the additional mass of cofilin; (b) they differ in the length of the “crossover,” which is a characteristic of F-actin. This is more apparent in the computed diffraction patterns (Fig. 2 d). The height of layerline n = 2, l = 2 indicates that the crossover length of this filament is 272 Å. This movement relative to the equivalent layerline in the undecorated filament (n = 2, l = 4) dramatically illustrates that cofilin changes the twist of nonmuscle actin also. The position of the first meridional shows that the axial rise per subunit is virtually unchanged by cofilin binding. Thus, the effect of cofilin is to reduce the rotation per subunit (running along the short-pitched, left-handed helix) by about 4 to 5° while causing little or no change in the rise per subunit (for a review of actin's helical symmetry see Aebi et al., 1986; Trachtenberg et al., 1986). Therefore, although the crossover length is reduced, the change in twist does not affect the overall length of the filaments. The range of crossover lengths observed for cofilin-decorated filaments was between 264 and 276 Å (n = 30), well below the normal range for muscle actin and the range of mean crossover lengths we observed for platelet actin (347 to 370 Å; based on 70 measurements).
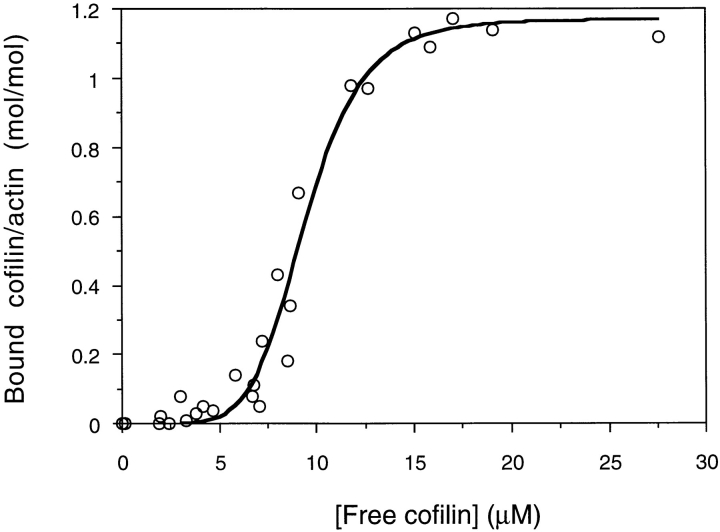
It has been reported that ADF binds muscle F-actin in a highly cooperative manner in vitro (Hawkins et al., 1993; Hayden et al., 1993). Cosedimentation experiments with F-actin demonstrate that cofilin binds to F-actin in a similar manner (Fig. 3). The Hill constant calculated by nonlinear least squares fitting of 6.4 confirms the high degree of cooperativity.
Figure 3.
Cooperativity of cofilin binding to rabbit muscle F-actin at pH 6.5. 6 μM F-actin was cosedimented with cofilin (0–35 μM). Supernatant and pellets were analyzed by SDS-PAGE and concentrations of cofilin and actin estimated by densitometry. Bound cofilin is expressed as a molar ratio to actin subunits. Nonlinear least-squares fitting shows a maximum binding at 1.16 cofilin/actin and a Hill constant of 6.4.
Fig. 4 presents cryomicrographs of muscle actin incubated with a 2-fold excess of cofilin at pH 6.6. The lower cofilin to actin ratio was used to prepare a mixture of both decorated and undecorated filaments to be imaged in the same micrograph (Fig. 4 a). The diffraction patterns in Fig. 4, d and e were calculated from the boxed filaments in Fig. 4 a. The heights of the n = 2 layerlines (at 1/352 and 1/275 Å−1, respectively) confirm that this field contains both types of filament, lending support to the idea that cofilin binds cooperatively.
Figure 4.
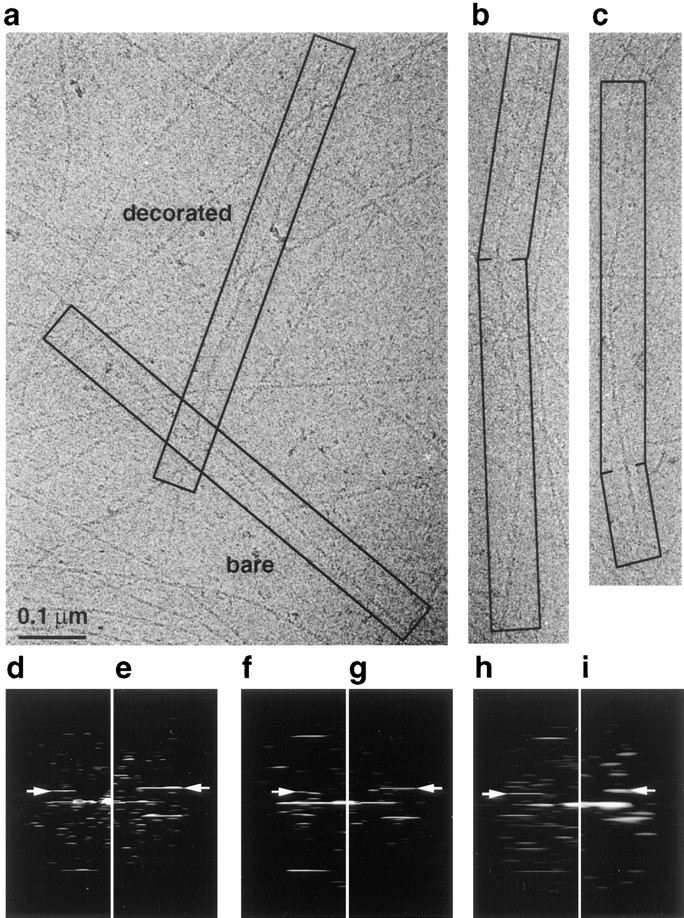
Cooperativity of cofilin binding to rabbit muscle F-actin shown by electron cryomicroscopy. (a) Micrograph of frozen- hydrated filaments decorated with cofilin at subsaturating conditions. (b and c) Micrographs of partially decorated filaments: the top region of each filament is thinner, bare F-actin, whereas the lower region is decorated with cofilin. (d and e) Computed diffraction patterns calculated from the bare F-actin and decorated F-actin images in a. (f and g) Computed diffraction patterns from the undecorated and decorated halves of filament b. (h and i) Computed diffraction patterns from the undecorated and decorated halves of filament c. In this case the decorated region of the filament is only about four crossovers long. This results in a broadening of the layerline data i. Arrowheads indicate the positions of equivalent (J2) layerlines.
We also obtained images of partially decorated filaments. This is a very rare event at a 3:1 cofilin to actin ratio (the conditions used to produce the micrographs in Fig. 4) probably because of the highly cooperative nature of cofilin binding to filaments. Fig. 4 b shows a partially decorated filament that was computationally divided into its undecorated (upper) and decorated (lower) halves. Calculated diffraction patterns revealed that the undecorated half has a mean crossover length of 371 Å, whereas the decorated half has a crossover length of 278 Å (Fig. 4, f and g). Another example of a partially decorated filament is shown in Fig. 4 c. In this case the undecorated half has a mean crossover length of 356 Å, and the decorated half has a mean crossover length of 278 Å. These results indicate that the change in twist occurs only in those regions of the filament where cofilin is bound and is not propagated along the rest of the filament.
F-actin from rabbit muscle was also incubated with a 2-fold excess of cofilin at pH 8, to see whether filaments were also decorated under conditions in which depolymerization is strongly favored. We found a dramatic difference in the appearance and distribution of the filaments in the cryomicrographs (results not shown). From 93 micrographs taken at 30,000×, 77 had few or no filaments in the fields. In some cases (16 micrographs) we found short actin filaments that were laterally aggregated into disorganized “bundles,” which ranged in size from 60 to 120 nm diam and 0.75 to 2.1 μm in length. In contrast, at pH 6.6, only one of these lateral aggregates was observed (out of 68 micrographs taken). Some single filaments were also observed at the higher pH. These appeared to be decorated based on their diameters and short crossover lengths. Thus, the change in twist is induced by cofilin and is independent of pH.
Features in the Reconstructions
Our structural analysis of cofilin/actin was pursued further using the images obtained from platelet actin. Decorated filaments diffracted strongly compared to their undecorated counterparts (Fig. 2, c and d). In addition to the change in twist that affected the heights of various layerlines, other differences were clearly visible relative to undecorated F-actin. The radial positions of the peaks in the diffraction pattern for the first n = 2 and 4 were moved inward owing to the increased filament diameter, and their intensities were stronger. This was expected based on the enhanced appearance of the crossovers which correspond to the two-start actin helix. In addition, the intensity of the n = −1 was dampened while other layerlines (n = −3, 3, −2, and 0), which are difficult to see in F-actin diffraction patterns, were strengthened in the decorated filaments. Ultimately, we were able to reconstruct the cofilin-decorated filaments to higher resolution (27.5 Å axially) than that attained from undecorated filaments (35 Å axially). Nevertheless, the major differences that exist between the two reconstructions were still present when the resolution of the cofilin/actin reconstruction was truncated to match the F-actin map (results not shown).
The final mean phase residuals calculated from the controls and decorated actin datasets were 29.4 and 38.6°, respectively. These phase residuals compare very favorably to those reported for other actin structures calculated in a similar way (that is, calculated using the entire layerlines rather than just the peaks; McGough et al., 1994; Orlova et al., 1994; Owen and DeRosier, 1993; Bremer et al., 1994; Lehman et al., 1994). The somewhat higher phase residual obtained for the decorated filaments may be a function of the smaller defocus used to obtain these images and the inclusion of higher resolution data during the alignment.
Three-dimensional density maps were computed by Fourier-Bessel inversion of the averaged layerline data after truncation of the layerlines at 1/30 Å−1 (for F-actin) and 1/25 Å−1 (for cofilin/actin) to take into account the position of the first node in the contrast transfer function. Fig. 5 presents surface renderings of the platelet F-actin and cofilin/F-actin reconstructions showing several crossovers of each. Each filament shown contains 40 actin subunits to emphasize that although the crossover length has been reduced by cofilin binding, the overall length of the filament is unchanged.
Figure 5.
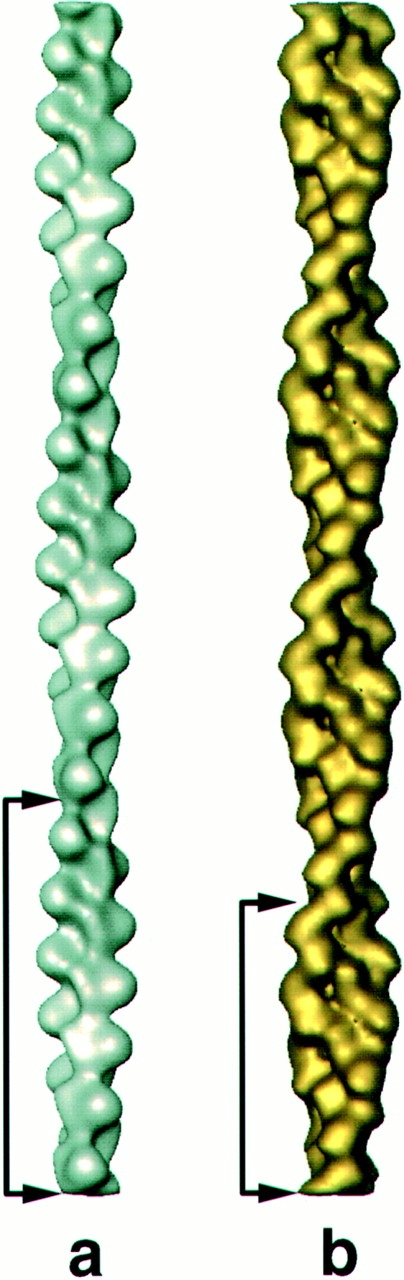
Reconstructions of F-actin and cofilin/actin filaments demonstrating the change in crossover length. (a) Platelet F-actin exhibiting 54/25 symmetry, equivalent to 2.160 subunits per turn. (b) Cofilin/F-actin exhibiting 20/9 symmetry (2.222 subunits per turn). Arrows mark the crossover length. There are 40 actin subunits in each filament.
Identification of the Cofilin-binding Site
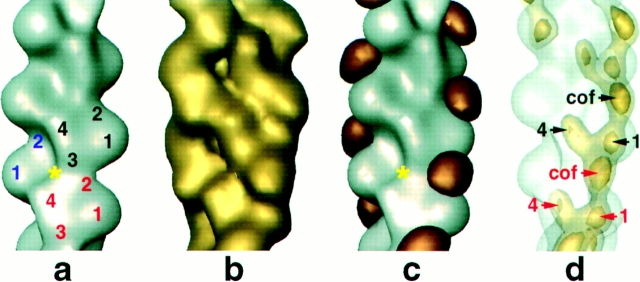
When we brought the F-actin layerline data to the twist of the cofilin/F-actin, we saw little change in the actin subunit. Fig. 6 a presents the reconstruction with the approximate locations of the four subdomains of actin indicated on the filament. The surface view of platelet F-actin is equivalent to reconstructions of muscle actin. Comparison of the reconstruction of the decorated filament (Fig. 6 b) with that for F-actin shows several structural features in common. Both filaments display two twisting strands of the long-pitch helix that are half staggered relative to each other and connected by a bridge of density running along the one-start, left-handed helix, connecting subdomain 4 of one subunit with subdomain 1 of the next. The cofilin-decorated filament differs from F-actin because of the extra density at the junction between two longitudinally associated actin subunits.
Figure 6.
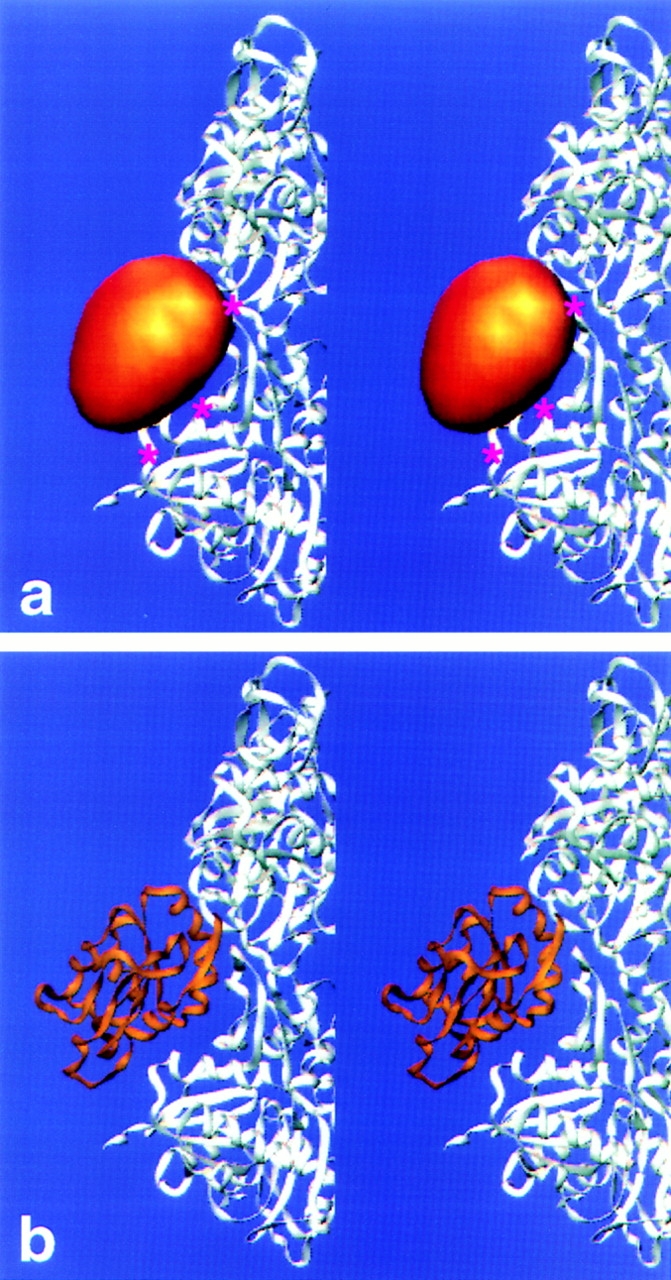
Identification of the cofilin-binding site by difference mapping. (a) Platelet actin reconstruction brought to the twist of the cofilin/actin filament (2.222 subunits per turn). The subdomains of actin and the phalloidin-binding site (yellow asterisk) are indicated. (b) Cofilin/actin reconstruction at the same orientation as in (a). (c) Positive difference density (copper) calculated by subtracting a from b. The contour level chosen encloses 100% (nominal) molecular volume. (d) Effects of contouring the cofilin/actin reconstruction using high thresholds. The actin reconstruction is shown as a transparent surface (silver) and the cofilin-decorated filament (gold) at two different molecular volumes (40 and 10%) to emphasize the strongest densities in the map.
A difference map was computed in real space after aligning the reconstructions to a common phase origin. Contouring the positive differences to enclose the nominal molecular volume of cofilin reveals a single rugby ball-shaped mass of dimensions 32 × 37 × 43 Å (copper-colored mass in Fig. 6 c). These values agree well with the overall dimensions of destrin, which is closely related to cofilin (Hatanaka et al., 1996). The major axis of cofilin makes a 30° angle with the plane normal to the helical axis of the filament. The difference map shows that cofilin is centered (axially) at about the position of subdomain 2 of the lower actin subunit and radially at the cleft between subdomains 1 and 3 of the upper actin subunit.
Analysis of Filament Structures Rendered at Different Contour Levels
Because of the limited resolution of most EM reconstructions, there is some ambiguity in assigning the precise boundary separating the particle from the embedding medium. Nonetheless, reconstructions are generally displayed as volume renderings having definite molecular boundaries. There are a number of ways to choose the boundary (for review see Frank, 1996). We have found it helpful in our analysis to display the reconstructions at a variety of contour levels as has been suggested and done by others (DeRosier and Moore, 1970; Milligan and Flicker, 1987; Bremer et al., 1994). Fig. 6 a presents the F-actin reconstruction contoured at a level that encloses the Lorenz model of the actin filament (nominal molecular volume = 150%). The cofilin/F-actin reconstruction in Fig. 6 b is contoured at a molecular volume (nominally 145%) at which the actin portion of the decorated filament best matches that of the undecorated filament. In the composite map (Fig. 6 d) the F-actin reconstruction is shown as a transparent surface (silver) and the cofilin-decorated filament (gold) at two different molecular volumes (40 and 10%) to emphasize the strongest densities in the map. This reveals that the strongest feature corresponds to cofilin, while the next strongest corresponds to subdomains 1 and 4 of actin, respectively. The strongest intrafilament contacts are actin–cofilin–actin contacts along the two-start helix, followed by the actin–actin contacts running along the one-start, left-handed helix. The longitudinal actin– actin contacts are the weakest. The same type of exercise on F-actin (results not shown) reveals that the strongest contact is along the one-start helix and the strongest density in subdomain 1. Subdomain 4 is not resolvable as a separate mass in the F-actin reconstruction. It is important to bear in mind, however, that the nominal resolution of the F-actin and cofilin/F-actin reconstructions differ. In addition, the F-actin reconstruction contained images that were further from focus than the cofilin/F-actin images. Therefore, it is not clear if there is any biological significance to the relative strengths of these contacts (Bremer et al., 1994).
Discussion
Cofilin Changes the Twist of Actin Filaments
The most startling finding of this work is that cofilin induces a substantial change in the twist of F-actin. This is the first protein found to do so. Cofilin brings filaments to a new mean twist of 2.218 units/turn resulting in a shorter crossover length of 269 Å, ∼72% that of F-actin. Our micrographs showing both decorated and undecorated filaments in the same field (Fig. 4) clearly demonstrate that the difference in crossover length is not attributable to large variations in the imaging conditions. This structural change probably accounts for cofilin's cooperative binding to F-actin (Fig. 3).
It has long been noted that F-actin exhibits a natural variation in twist (Hanson et al., 1967). The range in mean twists normally observed is from 2.154 to 2.167 subunits per turn, resulting in mean crossover lengths ranging from 385 to 358 Å. The two prevailing models describing actin's natural variation in twist are the “angular disorder” model (Egelman et al., 1982; Egelman and DeRosier, 1992) and the “lateral slipping” model (Aebi et al., 1986; Bremer et al., 1991). These models differ in a number of key respects, including whether changes in twist are coupled with changes in diameter and whether they are cumulative or compensatory. Analysis of the cofilin/F-actin filaments may shed light on this issue, since these filaments represent an unprecedented shift in actin twist. We have compared the positions of subdomains 1 and 4 (using high contour levels) for the cofilin/F-actin reconstruction with those from reconstructions of αA1-2/F-actin (McGough et al., 1994) and S1-decorated actin (Whittaker et al., 1995a ). We found them to be at identical radii from the filament axis (results not shown). This suggests that changes in F-actin twist are not coupled with changes in diameter. In addition, the observation that the crossover lengths in bare regions of actin adjacent to stretches of decorated filament are not unusually long (Fig. 4), would tend to argue against a compensatory mechanism underlying F-actin twist. It is interesting to note that the change in rotation per subunit induced by cofilin binding to actin (∼4 to 5°) falls within the estimates of the magnitude of the angular component in the random angular disorder model (Egelman and DeRosier, 1992).
Whatever the source of the disorder, it has been suggested that there is a biological advantage in having an actin filament with variable twist, particularly with regards to actin's ability to incorporate into macromolecular assemblies such as bundles (Trachtenberg et al., 1986). Some actin-binding proteins, including myosin and scruin, have been shown to reduce this disorder (Trachtenberg et al., 1986; Stokes and DeRosier, 1987) as also does the fungal toxin phalloidin (Bremer et al., 1991). However, in none of these cases is the crossover length outside the normal range for actin.
It has been reported that binding of ADF to F-actin is inhibited by phalloidin in vitro (Hayden et al., 1993; Carlier et al., 1997), and rhodamine–phalloidin will not bind to cofilin/actin rods in vivo (Moriyama et al., 1987; Moon et al., 1993). One interpretation of this is that ADF/cofilin and phalloidin compete for the same site on actin. The Lorenz model of F-actin proposed that phalloidin is positioned to contact 3 actin subunits (Lorenz et al., 1993; Fig. 6 a, asterisk). Our work shows that the phalloidin-binding site is 15 to 20 Å away from cofilin on the filament; thus the mutual exclusion of binding cannot be attributed to overlapping sites. Rather, the results suggest that the change in twist induced by cofilin alters the phalloidin-binding site and thereby prevents its binding. Conversely, once phalloidin has bound and stabilized the filament structure, cofilin is no longer able to interact. This is analogous to the effect of allosteric regulators, which act mainly by altering the affinity for the binding of substrates. The fact that cofilin–actin rods (induced by heat shock and other stresses to the cell) fail to bind phalloidin (Moriyama et al., 1987; Moon et al., 1993) suggests that this change in twist is also present inside cells. This supports the idea that the twist change induced by cofilin is biologically relevant.
The binding of cofilin to F-actin is highly co-operative, which we believe reflects its nonrandom association with F-actin. Recently, Carlier et al. (1997) have argued that the apparent co-operativity of binding of ADF from Arabidopsis thaliana to rabbit muscle actin is a consequence of the relative affinities of this protein for G-ADP actin over F-ADP. The balance of these affinities was such that only partial depolymerization was achieved at saturating ADF concentrations, varying little with pH. Our results with human ADF and cofilin have shown complete depolymerization at saturating ADF concentrations at pH 8.0, indicating a much higher affinity for G-ADP actin than F-ADP actin as compared to the Arabidopsis ADF (Hawkins et al., 1993). By contrast, there is very little depolymerization by ADF or cofilin at pH 6.5, suggesting a reversal of the relative affinities at the lower pH value. Thus we would attribute the high degree of co-operativity to the structural transition rather than to the relative affinities for monomeric and polymeric actins.
The ability of cofilin and ADF to bind ADP-actin subunits and alter the twist of F-actin may explain its acceleration of the dissociation rate of subunits from the pointed ends of filaments (Maciver, S.K., B. Pope, and A. Weeds, unpublished observations; Carlier et al., 1997). If the strongest intrafilament contacts are between actin–cofilin– actin contacts along the two start helix, then labeled, terminal subunits or adjacent, unlabeled subunits may be less tightly bound. This may also explain the apparent weak severing activity (Hawkins et al., 1993; Hayden et al., 1993; Maciver, S.K., B. Pope, and A. Weeds, unpublished observations). Light microscopic observations showed that curved segments of actin had a higher probability of breaking in the presence of actophorin (Maciver et al., 1991). It seems reasonable that the change in twist increases the distortion in the filaments, which might result in fragmentation. It has recently been shown that actin filaments are more easily broken when they are twisted: when 10-μm filaments were turned through 90°, the breaking force was reduced from 600 to 320 pN (Tsuda et al., 1996). Thus, although much of the depolymerizing effect of these proteins might arise from an increase in the dissociation rate constant at the pointed end (Maciver, S.K., B. Pope, and A. Weeds, unpublished observations; Carlier et al., 1997), the low levels of fragmentation observed by microscopy could be an adventitious consequence of the structural changes induced in the filaments.
Relationship to Other Actin-binding Proteins
Electron microscopy has been used to study the binding sites of a variety of actin-binding proteins including myosin, tropomyosin, α-actinin, gelsolin, and scruin. The cofilin-binding site appears to overlap the sites for myosin (Rayment et al., 1993; Schroder et al., 1993), tropomyosin (Vibert et al., 1993; Lehman et al., 1994, 1995), caldesmon-containing thin filaments (Hodgkinson et al., 1997), and α-actinin (McGough et al., 1994). This is in agreement with published biochemical data indicating competition between cofilin and these proteins (Bernstein and Bamburg, 1982; Nishida et al., 1984; Yonezawa et al., 1988). Gelsolin also binds F-actin in a similar location as cofilin (McGough, A., W. Chiu, and M. Way, unpublished observations). With the possible exception of myosin, which is still a matter of some debate (Rayment et al., 1993; Schroder et al., 1993), all F-actin–binding proteins have been found to interact with at least two subunits in the filament. We propose that it is this feature that confers filament specificity on F-actin–binding proteins. Thus, any change in the filament geometry could modulate the binding of other proteins and thereby control the hierarchy of interactions that must exist in cells where many actin-binding proteins occur together. As such, one previously overlooked function of cofilin may be its action as an “F-actin sequestering protein,” providing the cell with a pool of polymerized actin that is distinct from the actin cytoskeleton in that this pool might not bind cross-linking or motor proteins.
Towards Building a Model for Cofilin/F-Actin Interaction
We first examined whether the cofilin-dependent change in rotation introduced bad contacts in the atomic model of F-actin. A model of F-actin at the new twist was built using the Lorenz model of actin (Lorenz et al., 1993) and applying a rotation between adjacent monomers of −162° about the z-axis. Examination of the model at the new twist revealed only two possibly bad contacts (atoms <2 Å apart), both of which occur at the interface between subdomain 2 of the lower and 3 of the upper subunit (Val45 with Lys291; Arg39 with Glu167). Residues 39 and 45 are located in the DNase I–binding loop, which is postulated to be a variable domain of actin (Orlova and Egelman, 1992; Lorenz et al., 1993; Bremer et al., 1994; Tirion et al., 1995). Because no changes in the actin subunits were required to accommodate the new symmetry, we have interpreted our results using a simple rigid body rotation to the Lorenz model. However, it is possible (although not necessary) that the actin subunits undergo conformational changes in addition to the large change in twist. Unfortunately, differences in the imaging conditions used to obtain the micrographs of F-actin and cofilin/F-actin are likely to produce changes in the molecular envelopes of the two maps; therefore, it would not be appropriate to use these structures for a detailed investigation of domain movements. We are currently working to improve the resolution of the F-actin reconstruction to match that of the cofilin/F-actin structure.
The cofilin/F-actin reconstruction provides the first direct structural framework into which cofilin's interactions with actin may be modeled. As a first step in this process, we have interactively fit the actin and destrin models to the molecular envelope of F-actin/cofilin. An interesting feature of all three of the atomic structures from members of the cofilin family (Hatanaka et al., 1996; Federov et al., 1997; Leonard et al., 1997) is that the helix that has been proposed to bind to G-actin (Hatanaka et al., 1996) is distorted. One consequence of this distortion in the helix is that the destrin molecule is now able to fit more readily into the pocket in the atomic model of F-actin that is formed between the two longitudinally associated actin subunits. (This is in stark contrast to the clashes that would exist if, for example, the gelsolin domain 1 structure is placed in the same location.) Our reconstruction and difference mapping show that this pocket is the region on F-actin with which cofilin interacts (Fig. 6 c). We rotated and translated the destrin coordinates to both fit this pocket on F-actin and match the molecular envelope from our reconstruction. Additional biochemical data concerning residues involved in the interaction were also considered, in particular the involvement of two lysine residues at the NH2-terminal end of the helix (Yonezawa et al., 1991; Moriyama et al., 1992). After completing the fit, two loops in the destrin structure (Ser24–Ile29 and Gly61– Ile64) were not accounted for by the electron density map of cofilin/F-actin. In addition, a region of the so-called hydrophobic plug (Ser265–Glu270) is not enclosed by the cofilin/F-actin structure.
The results of this manual fitting are shown in Fig. 7. Cofilin is positioned to make interactions with two actin subunits at the cleft between subdomains 1 and 3 of the upper and at subdomains 2 and 1 of the lower subunits. Residues in the upper actin monomer that fall in or near the cofilin-binding site include: Tyr143–Thr149, Ile345– Leu346, Leu349–Thr351, and Gln354. Residues in the lower actin monomer include: Phe21, Arg28, Gly36–Pro38, His40–Val43, Gln49–Val54, Glu57, Lys61, His87–His88, Phe90–Val96, and His101. Unfortunately, unlike actin or myosin subfragment 1 which are both readily fitted into EM maps (Schroder et al., 1993; Rayment et al., 1993; Bremer et al., 1994; McGough et al., 1994), cofilin (ADF/ destrin) is a relatively symmetrical object at low resolution. Therefore, although the long axis of the molecule can be oriented with a fair amount of confidence, the surface of destrin that actually contacts actin is far from certain, based on the electron cryomicroscopy data alone. Our model is intended to serve as a starting point from which experiments may be designed to determine precise information about the actin/cofilin interface. Further electron cryomicroscopy studies in which cofilin has been labeled at specific residues should also aid in this process. Residues in destrin that could be involved in the interaction with actin based on the placement shown in Fig. 7 b include: Ser1–Val5, Cys12–Lys22, and Leu111–Lys132.
Figure 7.
| 0.8515 | 0.5072 | 0.1331 | |||
| −0.3293 | 0.7147 | −0.6170 | |||
| −0.4081 | 0.4815 | 0.7756 | |||
| −1.7062 | −12.3144 | −12.5306 |
One property of cofilin that might be explained from its location on F-actin is the pH sensitivity of filament binding. After positioning the destrin coordinates on the actin filament, we investigated the resulting placement of histidine residues at the putative interface, reasoning that these might be important in the pH-mediated switch. Histidines are not conserved in the cofilin family, and comparison of the locations of the two histidines in destrin with the three in actophorin, when mapped onto the structure shown in Fig. 7, shows that none is at the actin interface. On the other hand, there are four actin histidines near the cofilin-binding site (40, 87, 88, 101) that might be important in cofilin/actin interactions. Both cofilin and ADF/destrin bind preferentially to filaments <pH 7.3, and depolymerize actin at higher pH values (Yonezawa et al., 1985; Hawkins et al., 1993; Hayden et al., 1993). By contrast, actophorin cosedimentation with F-actin is not pH-dependent (Maciver, S.K., B. Pope, and A. Weeds, unpublished observations). This dichotomy argues against a simple explanation of the pH switch being mediated solely by histidines.
Another property of cofilin that might be explained by the cofilin/F-actin structure is cofilin's sensitivity to the nucleotide state of actin (Maciver and Weeds, 1994). There is growing evidence that the nucleotide state of actin affects the position of subdomain 2 (Orlova and Egelman, 1992, 1993; Strzelecka-Golaszewska et al., 1993; Tirion et al., 1995). Our work shows that the cofilin-binding site includes subdomain 2. In addition, analysis of the atomic model of F-actin shows that the twist change introduces a strain at the longitudinal actin–actin contact (between subdomains 2 and 3 of two actin subunits). This strain might be more easily accommodated in ADP–F-actin because of increased flexibility in this form of actin (Orlova and Egelman, 1993). One or both of these factors might provide the structural basis for cofilin's sensitivity to the nucleotide state of actin. However, α-actinin also binds near subdomain 2 (McGough et al., 1994), and there is no evidence for its binding being regulated in this way.
Conclusions
Our reconstruction provides the first direct information about how the cofilin family of proteins binds actin. Cofilin binds F-actin between adjacent subunits along the two-start helix. Binding by both cofilin and ADF (destrin) results in a substantial reduction in the crossover length of the filament. This structural change probably accounts for the co-operative nature of binding and represents a novel mechanism to prevent filament binding by other actin binding proteins. In addition, the cofilin-induced twisting of actin filaments may introduce strains in the actin filament that contributes to the increase in filament dynamics and fragmentation observed by others.
Acknowledgments
We thank Sue Whytock (MRC-LMB, Cambridge, UK) for the negatively stained images in Fig. 1; S. Almo (Albert Einstein College of Medicine, New York, NY), H. Hatanaka (Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan), P. McLaughlin (University of Edinburgh, Edinburgh, Scotland), and K. Holmes (Max-Planck Institut für Medizinsche Forshung, Heidelberg, Germany) for providing atomic coordinates; and R. Milligan (Scripps Research Institute, La Jolla, CA) for the actoS1 layerline data. We also thank D. DeRosier (Brandeis University, Waltham, MA), R. Diaz (Florida State University, Tallahassee, FL), M. Sherman (Baylor College of Medicine, Houston, Texas), P. Thuman-Commike (Rice University, Houston, Texas), M. Stewart, and T. Bullock (MRC-LMB, Cambridge, UK) for helpful discussions.
This work was supported by a Grant-in-Aid from the American Heart Association (to A. McGough), grants from National Institutes of Health (RR02250) and the National Science Foundation (BIR9413229 and BIR9412521) to W. Chiu, and the W.M. Keck Center for Computational Biology (Houston, TX).
Abbreviations used in this paper
- F-actin
filamentous actin
- G-actin
globular actin
- l
layerline height
- n
Bessel order
- R
radial position of the first peak
Footnotes
Please address all correspondence to Dr. Amy McGough, Verna and Marrs McLean Department of Biochemistry, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030. Tel.: (713) 798-6989; Fax: (713) 796-9438.
References
- Aebi U, Millonig R, Salvo H, Engel A. The three-dimensional structure of the actin filament revisited. Ann NY Acad Sci. 1986;483:100–119. doi: 10.1111/j.1749-6632.1986.tb34502.x. [DOI] [PubMed] [Google Scholar]
- Agnew BJ, Minamide LS, Bamburg JR. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J Biol Chem. 1995;270:17582–17587. doi: 10.1074/jbc.270.29.17582. [DOI] [PubMed] [Google Scholar]
- Amos LA. Combination of data from helical particles: correlation and selection. J Mol Biol. 1975;99:65–73. doi: 10.1016/s0022-2836(75)80159-8. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Bray D. Distribution and cellular localization of actin depolymerizing factor. J Cell Biol. 1987;105:2817–2825. doi: 10.1083/jcb.105.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, Harris HE, Weeds AG. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS (Fed Eur Biochem Soc) Lett. 1980;121:178–182. doi: 10.1016/0014-5793(80)81292-0. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin depolymerizing factor (ADF) Cell Motil. 1982;2:1–8. doi: 10.1002/cm.970020102. [DOI] [PubMed] [Google Scholar]
- Bremer A, Millonig RC, Sutterlin R, Engel A, Pollard T. The structural basis for the intrinsic disorder of the actin filament: the “lateral slipping” model. J Cell Biol. 1991;115:689–703. doi: 10.1083/jcb.115.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer A, Henn C, Goldie K, Engel A, Smith PR, Aebi U. Towards atomic interpretation of F-actin filament three-dimensional reconstructions. J Mol Biol. 1994;741:683–700. doi: 10.1006/jmbi.1994.1617. [DOI] [PubMed] [Google Scholar]
- Carlier M, Santolini J, Laurent V, Didry D, Yan H, Chua N, Pantaloni D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J Cell Biol. 1997;136:1307–1323. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson M, Bugg CE. Algorithm for ribbon models of proteins. J Mol Graphics. 1986;4:121–122. [Google Scholar]
- Coluccio L, Tilney L. Under physiological conditions actin disassembles slowly from the nonpreferred end of an actin filament. J Cell Biol. 1983;97:1629–1634. doi: 10.1083/jcb.97.5.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Blum JD, Williams RCJ, Pollard TD. Purification and characterization of actophorin, a new 15,000-Dalton actin-binding protein from Acanthamoeba castellanii. . J Biol Chem. 1986;261:477–485. [PubMed] [Google Scholar]
- DeRosier DJ, Moore PB. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J Mol Biol. 1970;52:355–369. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- Drenckhahn D, Pollard T. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J Biol Chem. 1986;261:12754–12758. [PubMed] [Google Scholar]
- Egelman EH, DeRosier DJ. Image analysis shows that variations in actin crossover spacings are random, not compensatory. Biophys J. 1992;63:1299–1305. doi: 10.1016/S0006-3495(92)81716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH, Francis N, DeRosier DJ. F-actin is a helix with a random variable twist. Nature (Lond) 1982;298:131–135. doi: 10.1038/298131a0. [DOI] [PubMed] [Google Scholar]
- Federov A, Lappalainen P, Federov E, Drubin D, Almo S. Structure determination of yeast cofilin. Nat Struct Biol. 1997;4:366–369. doi: 10.1038/nsb0597-366. [DOI] [PubMed] [Google Scholar]
- Frank, J. 1996. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. Academic Press, San Diego. 342 pp.
- Gunsalus KC, Bonaccorsi S, Williams E, Verni F, Gatti M, Goldberg ML. Mutations in twinstar, a Drosophilagene encoding a cofilin/ADF homologue, result in defects in centrosome migration and cytokinesis. J Cell Biol. 1995;131:1243–1259. doi: 10.1083/jcb.131.5.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson J. Axial period of actin filaments. Nature (Lond) 1967;213:353–356. [Google Scholar]
- Harris HE, Weeds AG. Plasma actin depolymerizing factor has both calcium-dependent and calcium-independent effects on actin. Biochemistry. 1983;22:2728–2741. doi: 10.1021/bi00280a022. [DOI] [PubMed] [Google Scholar]
- Hatanaka H, Ogura K, Moriyama M, Ichikawa S, Yahara I, Inagaki F. Tertiary structure of destrin and structural similarity between two actin-regulating protein families. Cell. 1996;85:1047–1055. doi: 10.1016/s0092-8674(00)81305-7. [DOI] [PubMed] [Google Scholar]
- Hawkins M, Pope B, Maciver S, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- Hayden SM, Miller PS, Brauweiler A, Bamburg JR. Analysis of the interactions of actin depolymerizing factor (ADF) with G- and F-actin. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- Hodgkinson JL, El-Mezgueldi M, Marston S, Craig R, Vibert P, Lehman W. 3D reconstruction of smooth muscle thin filaments: contribution of caldesmon and calponin to filament structure. Biophys J. 1997;72:2398–2404. doi: 10.1016/S0006-3495(97)78885-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature (Lond) 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan C, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Lehman W, Craig R, Vibert P. Ca2+-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature (Lond) 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- Lehman W, Vibert P, Uman P, Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J Mol Biol. 1995;251:191–196. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]
- Leonard S, Gittis A, Petrulla E, Pollard T, Lattman E. Crystal structure of the actin-binding protein actophorin from Acanthamoeba. . Nat Struct Biol. 1997;4:369–373. doi: 10.1038/nsb0597-369. [DOI] [PubMed] [Google Scholar]
- Lopez I, Anthony R, Maciver S, Jiang C-J, Khan S, Weeds AG, Hussey P. Pollen specific expression of maize genes encoding actin depolymerizing factor-like proteins. Proc Natl Acad Sci USA. 1996;93:7415–7420. doi: 10.1073/pnas.93.14.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Popp D, Holmes KC. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Weeds AG. Actophorin preferentially binds monomeric ADP-actin over ATP-bound actin: consequences for cell locomotion. FEBS (Fed Eur Biochem Soc) Lett. 1994;347:251–256. doi: 10.1016/0014-5793(94)00552-4. [DOI] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. . J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Way M, DeRosier D. Determination of the α-actinin binding site on actin filaments by cryoelectron microscopy and image analysis. J Cell Biol. 1994;126:433–443. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K, Matheson C, Marra M, Wakarchuk M, Baillie D. The Caneorhabditis elegansunc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol Gen Genet. 1994;242:346–357. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- Milligan RA, Flicker PF. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon A, Drubin D. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol Biol Cell. 1995;6:1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AL, Janmey PA, Louie KA, Drubin D. Cofilin is an essential component of the yeast cortical cytoskeleton. J Cell Biol. 1993;120:421–435. doi: 10.1083/jcb.120.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Nishida E, Yonezawa N, Sakai H, Matsumoto S, Nishida E, Iida K, Yonezawa N, Koyasu S, Yahara I, et al. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc Natl Acad Sci USA. 1987;84:5262–5266. doi: 10.1073/pnas.84.15.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama K, Yonezawa N, Sakai H, Yahara I, Nishida E. Mutational analysis of chimeric proteins between cofilin and destrin. J Biol Chem. 1992;267:7240–7244. [PubMed] [Google Scholar]
- Nishida E, Maekawa S, Sakai H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry. 1984;23:5307–5313. doi: 10.1021/bi00317a032. [DOI] [PubMed] [Google Scholar]
- Orlova A, Egelman EH. Structural basis for the destabilization of F-actin by phosphate release following ATP hydrolysis. J Mol Biol. 1992;227:1043–1053. doi: 10.1016/0022-2836(92)90520-t. [DOI] [PubMed] [Google Scholar]
- Orlova A, Egelman EH. A conformational change in the actin subunit can change the flexibility of the actin filament. J Mol Biol. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- Orlova A, Yu X, Egelman EH. Three-dimensional reconstruction of a complex of F-actin with antibody Fab fragments to actin's amino terminus. Biophys J. 1994;66:276–285. doi: 10.1016/s0006-3495(94)80791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen C, DeRosier DJ. A 13 Å map of the actin–scruin filament from the Limulus acrosomal process. J Cell Biol. 1993;123:337–344. doi: 10.1083/jcb.123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I, Holden HM, Whittaker M, Yohn M, Lorenz M, Holmes KC, Milligan RA. Structure of the actin–myosin complex and its implications for muscle contraction. Science (Wash DC) 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J, Agnew BJ, Abe H, Bamburg JR, Mitchison TJ. Xenopusactin depolymerizing factor/cofilin (XAC) is responsible for the turnover of actin filaments in Listeria monocytogenes tails. J Cell Biol. 1997;136:1323–1332. doi: 10.1083/jcb.136.6.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder RR, Manstein DJ, Jahn W, Holden H, Rayment I, Holmes KC, Spudich JA. Three-dimensional atomic model of F-actin decorated with Dictyosteliummyosin S1. Nature (Lond) 1993;364:171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- Schroeter JP, Bretaudiere J-P. SUPRIM: easily modified image processing software. J Struct Biol. 1996;116:131–137. doi: 10.1006/jsbi.1996.0021. [DOI] [PubMed] [Google Scholar]
- Small JV. Getting the actin filaments straight: nucleation-release or treadmilling. Trends Cell Biol. 1995;5:52–55. doi: 10.1016/s0962-8924(00)88939-4. [DOI] [PubMed] [Google Scholar]
- Stewart M. Computer image processing of electron micrographs of biological structures with helical symmetry. J Electron Micros Tech. 1988;9:325–358. doi: 10.1002/jemt.1060090404. [DOI] [PubMed] [Google Scholar]
- Stokes DL, DeRosier DJ. The variable twist of actin and its modulation by actin-binding proteins. J Cell Biol. 1987;109:1005–1017. doi: 10.1083/jcb.104.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot JA. Accelerating on a treadmill: ADF/cofilin promotes rapid actin filament turnover in the dynamic cytoskeleton. J Cell Biol. 1997;136:1165–1168. doi: 10.1083/jcb.136.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirion, M.M., D. ben-Avraham, M. Lorenz, and K.C. Holmes. 1995. Normal modes as refinement parameters for the F-actin model. Biophys. J. 68:5–12. [DOI] [PMC free article] [PubMed]
- Trachtenberg S, Stokes D, Bullitt E, DeRosier D. Actin and flagellar filaments: two helical structures with variable twist. Ann NY Acad Sci. 1986;483:89–99. doi: 10.1111/j.1749-6632.1986.tb34500.x. [DOI] [PubMed] [Google Scholar]
- Tsuda Y, Yasutake H, Ishijima A, Yanagida T. Torsional rigidity of single actin filaments and actin–actin bond breaking force under torsion measured directly by in vitro micromanipulation. Proc Natl Acad Sci USA. 1996;93:12937–12942. doi: 10.1073/pnas.93.23.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vibert P, Craig R, Lehman W. Three-dimensional reconstruction of caldesmon-containing smooth muscle filaments. J Cell Biol. 1993;123:313–321. doi: 10.1083/jcb.123.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Mallavarapu A, Rosenblatt J, Mitchison T. Actin dynamics in vivo. Curr Opin Cell Biol. 1997;9:54–61. doi: 10.1016/s0955-0674(97)80152-4. [DOI] [PubMed] [Google Scholar]
- Whittaker M, Wilson-Kubalek EM, Smith JE, Faust L, Milligan RA, Sweeney HL. A 35-Å movement of smooth muscle myosin on ADP release. Nature (Lond) 1995a;378:748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- Whittaker M, Carragher BO, Milligan RA. PHOELIX: a package for automated helical reconstruction. Ultramicroscopy. 1995b;58:245–250. doi: 10.1016/0304-3991(95)00057-8. [DOI] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Sakai H. pH control of actin polymerization by cofilin. J Biol Chem. 1985;260:14410–14412. [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Iida K, Yahara I, Sakai H. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease-i with actin by phosphoinositides. J Biol Chem. 1990;265:8382–8386. [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Maekawa S, Sakai H. Studies on the interaction between actin and cofilin purified by a new method. Biochem J. 1988;251:121–127. doi: 10.1042/bj2510121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Iida K, Kumagai H, Yahara I, Sakai H. Inhibition of actin polymerization by a synthetic dodecapeptide patterned on the sequence around the actin-binding site of cofilin. J Biol Chem. 1991;266:10485–10489. [PubMed] [Google Scholar]
- Zhou ZH, Hardt S, Wang B, Sherman MB, Jakana J, Chiu W. CTF determination of images of ice-embedded single particles using a graphics interface. J Struct Biol. 1996;116:216–223. doi: 10.1006/jsbi.1996.0033. [DOI] [PubMed] [Google Scholar]
- Zigmond SH. Recent quantitative studies of actin filament turnover during cell locomotion. Cell Motil Cytoskeleton. 1993;25:309–316. doi: 10.1002/cm.970250402. [DOI] [PubMed] [Google Scholar]