Abstract
SR proteins are required for constitutive pre-mRNA splicing and also regulate alternative splice site selection in a concentration-dependent manner. They have a modular structure that consists of one or two RNA-recognition motifs (RRMs) and a COOH-terminal arginine/serine-rich domain (RS domain). We have analyzed the role of the individual domains of these closely related proteins in cellular distribution, subnuclear localization, and regulation of alternative splicing in vivo. We observed striking differences in the localization signals present in several human SR proteins. In contrast to earlier studies of RS domains in the Drosophila suppressor-of-white-apricot (SWAP) and Transformer (Tra) alternative splicing factors, we found that the RS domain of SF2/ASF is neither necessary nor sufficient for targeting to the nuclear speckles. Although this RS domain is a nuclear localization signal, subnuclear targeting to the speckles requires at least two of the three constituent domains of SF2/ASF, which contain additive and redundant signals. In contrast, in two SR proteins that have a single RRM (SC35 and SRp20), the RS domain is both necessary and sufficient as a targeting signal to the speckles. We also show that RRM2 of SF2/ASF plays an important role in alternative splicing specificity: deletion of this domain results in a protein that, although active in alternative splicing, has altered specificity in 5′ splice site selection. These results demonstrate the modularity of SR proteins and the importance of individual domains for their cellular localization and alternative splicing function in vivo.
Numerous protein and ribonucleoprotein components are required to catalyze pre-mRNA splicing, which occurs within a macromolecular complex, the spliceosome. The major components of the spliceosome, which assembles on individual pre-mRNAs, are the small nuclear ribonucleoprotein particles (snRNPs)1 U1, U2, and U4/6·U5; the polypeptides that associate with hnRNA to form hnRNP particles (hnRNP proteins); and a large set of non-snRNP protein splicing factors, which includes the SR family of proteins (for reviews see Dreyfuss et al., 1993; Fu, 1995; Krämer, 1996; Manley and Tacke, 1996).
The SR proteins are essential splicing factors and also regulate alternative splicing of many pre-mRNAs by affecting the selection of 5′ splice sites (Ge and Manley, 1990; Krainer et al., 1990a ,b; Zahler et al., 1993; for review see Horowitz and Krainer, 1994). Their activity in alternative splicing is antagonized by members of the hnRNP A/ B family of proteins (Mayeda and Krainer, 1992; Mayeda et al., 1994). Thus, the counteracting activities of these two families of antagonistic factors can regulate alternative splicing, both in vitro and in vivo (Mayeda and Krainer, 1992; Cáceres et al., 1994; Yang et al., 1994). SR proteins are characterized by the presence of one or two copies of the RNA-recognition motif (RRM) and a COOH-terminal domain rich in arginine and serine residues (RS domain) (Zahler et al., 1992; for review see Birney et al., 1993). These proteins are thus very closely related in their domain structure, primary sequence, and functional properties. Individual SR proteins interact with pre-mRNA during the early stages of spliceosome assembly (Krainer et al., 1990b ; Fu, 1993; Staknis and Reed, 1994; Zahler and Roth, 1995) and stimulate binding of U1 snRNP particles to the 5′ splice site (Eperon et al., 1993; Kohtz et al., 1994) and binding of U2AF to the 3′ splice site (Zuo and Maniatis, 1996).
Splicing factors are distributed nonrandomly within the nucleus. Immunofluorescence localization studies showed that snRNP particles and SR proteins are organized in the interphase nucleus in a characteristic speckled pattern. Two morphologically distinct nuclear structures have been identified as constituents of the nuclear speckles by electron microscopy: interchromatin granule clusters and perichromatin fibrils (for review see Spector, 1993). The speckled pattern comprises 20–50 regions that are highly concentrated in splicing factors (Spector, 1990; Huang and Spector, 1992). Immunofluorescence staining and immunoelectron microscopy studies showed that the splicing factor SC35, a member of the SR family of proteins, colocalizes with snRNPs within the nuclear speckle domains (Fu and Maniatis, 1990; Spector et al., 1991). In some cell types, snRNPs can be additionally found in another discrete nuclear structure, the coiled body (for review see Lamond and Carmo-Fonseca, 1993).
The nuclear organization of splicing factors is dynamic, as shown by the fact that inhibition of transcriptional and/ or splicing activity causes reorganization of the nuclear speckles (Spector et al., 1983; O'Keefe et al., 1994). The most intense speckled regions are adjacent to, but not coincident with, transcriptionally active sites, as defined by fluorescent labeling of nascent RNAs (Wansink et al., 1993) and by in situ hybridization (Huang and Spector, 1991; Xing et al., 1993, 1995). Likewise, the interchromatin granule clusters are not sites of tritiated uridine incorporation (for review see Fakan and Puvion, 1980). The fact that splicing does not appear to take place at the majority of sites where splicing factors are most concentrated (Mattaj, 1994; Zhang et al., 1994; Huang and Spector, 1996) led to the suggestion that splicing factors shuttle between interchromatin granule clusters (sites of storage and/or reassembly) and perichromatin fibrils (sites of active transcription and splicing) (Jiménez-García and Spector, 1993). Many introns, though not all, are spliced as nascent transcripts in vivo (Beyer and Osheim, 1988; Bauren and Wieslander, 1994; Kiseleva et al., 1994). An intimate connection between transcription and splicing is further suggested by the observation that a subpopulation of hyperphosphorylated RNA polymerase II localizes to the nuclear speckle domains (Bregman et al., 1995; Mortillaro et al., 1996). The hyperphosphorylated COOH-terminal domain of the polymerase large subunit appears to mediate interactions with components related to splicing (Mortillaro et al., 1996; Yuryev et al., 1996; Du and Warren, 1997; McCracken et al., 1997).
Several protein kinases capable of phosphorylating SR proteins on serine residues in vitro have been described: SRPK1 (Gui et al., 1994a ,b), Clk/Sty (Colwill et al., 1996), a lamin-B receptor-associated kinase (Nikolakaki et al., 1996), and unexpectedly, DNA topoisomerase I (Rossi et al., 1996). In addition, it has been shown that addition of SRPK1 to permeabilized cells (Gui et al., 1994a ) or overexpression of Clk/Sty (Colwill et al., 1996) results in a diffuse distribution of splicing factors, probably because of hyperphosphorylation of their RS domains. It has been proposed that the level of phosphorylation may control the subnuclear distribution of SR proteins in interphase cells and the reorganization of the speckle domains during mitosis (Colwill et al., 1996; Gui et al., 1994a ; Misteli and Spector, 1996).
Previous studies showed that the RS domains of two Drosophila splicing regulators, suppressor-of-white-apricot (SWAP) and Transformer (Tra), direct these splicing factors to the nuclear speckles (Li and Bingham, 1991; Hedley et al., 1995). We sought to investigate the role of the structural domains of SR proteins in cellular and subnuclear localization and to study the requirement for individual domains of these proteins for modulation of alternative splicing in vivo. To this end, we transiently overexpressed in HeLa cells several epitope-tagged SR protein cDNAs encoding either the wild-type proteins or several mutant derivatives thereof. We then determined the cellular distribution of the tagged proteins, as well as their activity in regulating alternative splicing of transcripts expressed from cotransfected reporter genes. We found that SR proteins that have either one or two RRMs differ in their requirements for the RS domain as a nuclear speckle targeting signal. We also demonstrate that the presence of RRM2 can affect the alternative splicing specificity of SF2/ASF in vivo, suggesting that the modular structure of SR proteins is important for regulated splicing.
Materials and Methods
Epitope-tagged Expression Plasmids
Oligonucleotide primers were purchased from GIBCO BRL (Gaithersburg, MD). PCR conditions using Vent polymerase (New England Biolabs, Beverly, MA) were as previously described (Krainer et al., 1991). The epitope-tagged, full-length SF2/ASF expression plasmid was constructed by amplifying an SF2/ASF cDNA (Krainer et al., 1991) with specific primers and subcloning of the resulting PCR product as an XbaI-BamHI fragment into the pCGTHCFFLT7 expression vector (Wilson et al., 1995). The resulting vector, pCGT7-SF2, like the previously described pCG-SF2 vector, is under the control of the cytomegalovirus enhancer/ promoter (Tanaka and Herr, 1990; Cáceres et al., 1994) but also includes an NH2-terminal epitope tag, MASMTGGQQMG. This epitope tag corresponds to the first 11 residues of the bacteriophage T7 gene 10 capsid protein and is recognized by the T7 tag monoclonal antibody (Novagen, Inc., Madison, WI). The SF2/ASF mutants and domain deletions were previously described (Cáceres and Krainer, 1993) and were subcloned into the same epitope-tagged expression vector as wild-type SF2/ASF. For the SRp40, SC35, SRp20, and hnRNP A1 constructs, PCR products were amplified with specific primers and subcloned into the same expression vector. In the case of the SRp20 constructs, because of the presence of an internal XbaI site in the SRp20 cDNA, the amplified fragments were designed with SpeI and BamHI sites and were subcloned into the XbaI-BamHI sites of pCGT7-SF2. In the case of the SRp40 constructs, because of the presence of an internal BamHI site in the SRp40 cDNA, the amplified fragments were designed with XbaI and BclI sites and were subcloned into the XbaI-BamHI sites of pCGT7-SF2. The SRp40-ΔRS protein comprises residues 2–183; SC35-ΔRS comprises residues 2–94; and SRp20-ΔRS comprises residues 2–85. Construction of the A1-RS protein was previously described (Mayeda et al., 1994); this chimeric protein comprises amino acids 1–185 from hnRNP A1 and 195–248 from SF2/ASF.
For the nucleoplasmin fusions, the vector pCGT7-NPc was constructed by amplifying a Xenopus nucleoplasmin cDNA (Dingwall et al., 1987) with specific primers and cloning the resulting PCR product into the XbaI and BamHI sites of the pCGTHCFFLT7 expression vector. This procedure results in the insertion of amino acids 2–149 of nucleoplasmin followed by an XbaI site and stop codon, COOH-terminal to the T7 epitope. This nucleoplasmin fragment is also known as the nucleoplasmin core domain, NPc. PCR fragments comprising residues 198–248 from the RS domain of SF2/ASF or 88–164 from the RS domain of SRp20 were subcloned downstream of NPc to generate NPc-RSSF2 and NPc-RSSRp20, respectively.
Cell Culture and Transfections
HeLa cells were grown in DME supplemented with 10% FCS and transfected with 1 μg of plasmid DNA per 60-mm dish of 60–75% confluent cells, in the presence of 20 μg lipofectin (GIBCO BRL) (Cáceres et al., 1994).
Indirect Immunofluorescence
Cells were fixed for immunofluorescence assays 24 h after transfection. Cells were washed with PBS and incubated with 3% paraformaldehyde, 0.3% Triton (in PBS) for 5 min at room temperature, followed by incubation with 3% paraformaldehyde for 30 min. The fixed cells were incubated for 1 h at room temperature with 1:500 anti-T7 monoclonal antibody (Novagen, Inc.). The cells were washed three times with PBS and incubated for 1 h at room temperature with 1:500 fluorescein-conjugated goat anti–mouse IgG (Cappel Laboratories, Malvern, PA). Double immunofluorescence labeling was performed by incubation for 1 h at room temperature with 1:500 anti-T7 monoclonal antibody (Novagen, Inc.) and 1: 2,000 human anti-Sm serum. After washing, the cells were incubated with 1:50 Texas red–conjugated antimouse IgG and 1:50 fluorescein-conjugated antihuman IgG (Cappel Laboratories). Samples were observed on a microscope (model Axiovert 405M; Carl Zeiss, Inc., Thornwood, NY), and images were acquired with a cooled CCD camera (model NU200; Photometrics, Inc., Woburn, MA) using Oncor Image software (Gaithersburg, MD). For confocal microscopy, a confocal laser scanning microscope (model LSM410; Carl Zeiss, Inc.) was used. For localization of the endogenous SF2/ASF protein, a monoclonal antibody against SF2/ASF was used as culture supernatant at a 1:5 dilution.
Quantitation of Fluorescence Images
Images were acquired under identical conditions, ensuring that the maximal signal was not saturating, and were subjected to contrast stretching, excluding the top and bottom 1% of pixels. Acquisition of images and measurements were performed using Oncor Image software 2.0.04. To measure the intensity of speckles, a modified version of a point-hit method was used (Weibel et al., 1969). Random test lines were drawn over the cell nucleus, several pairs of random test points on the test line were chosen arbitrarily, and the absolute intensity value of the test points was measured (X 1, X 2). A value S for the intensity of speckles was determined as the ratio between the sum of differences and the average intensity value of all points on the test line (X av):
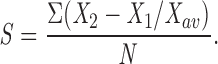
N represents the number of pairs of test points used for one sample. Typically, four to six pairs of test points were chosen arbitrarily on a test line, and a minimum of three test lines were used for every cell nucleus examined. For each sample, at least four nuclei were examined. All results represent values obtained from two separate experiments, and the results are average values ± standard deviation from the pooled data.
In Vivo Analysis of Alternative Splicing
Transfections of HeLa cells and purification of total RNA were as previously described (Cáceres et al., 1994). Briefly, 1 μg of expression plasmid was cotransfected into HeLa cells with 6 μg of the adenovirus E1A reporter plasmid pMTE1A (Zerler et al., 1986) in the presence of 20 μg of Lipofectin (GIBCO BRL). The cells were grown to 60–75% confluence in 60-mm dishes and harvested 24 or 48 h after transfection, and 200 ng of total RNA was analyzed by reverse transcriptase PCR (RT-PCR), as described (Screaton et al., 1995). Briefly, first-strand oligo(dT)-primed cDNA synthesized with superscript II (GIBCO BRL) from 200 ng of total RNA was amplified with Taq DNA polymerase (Perkin-Elmer Corp., Norwalk, CT) for 25 cycles, using 5′-end–labeled forward primer. Amplified products separated by urea-PAGE were detected by autoradiography and quantitated by PhosphorImage analysis (Fujix, BAS2000; Fuji Photo Film Co., Tokyo, Japan). The PCR primers were previously described (Cáceres et al., 1994).
Results
Role of the Structural Domains of SR Proteins in Cellular Distribution and Subnuclear Localization
First, we analyzed the localization of the endogenous SF2/ ASF protein using an anti-SF2/ASF monoclonal antibody (Krainer, A.R., unpublished). This antibody does not cross- react with other SR proteins and detects all phosphorylation variants of SF2/ASF; its detailed characterization will be published elsewhere. We observed a typical nuclear speckled pattern and also a diffuse nuclear signal, but no nucleolar staining (Fig. 1). This immunofluorescence pattern resembles that of endogenous U2B′′ and Sm snRNP proteins (for review see Spector, 1993). No staining of coiled bodies was observed, in agreement with previous data on SC35 localization (Spector et al., 1991).
Figure 1.
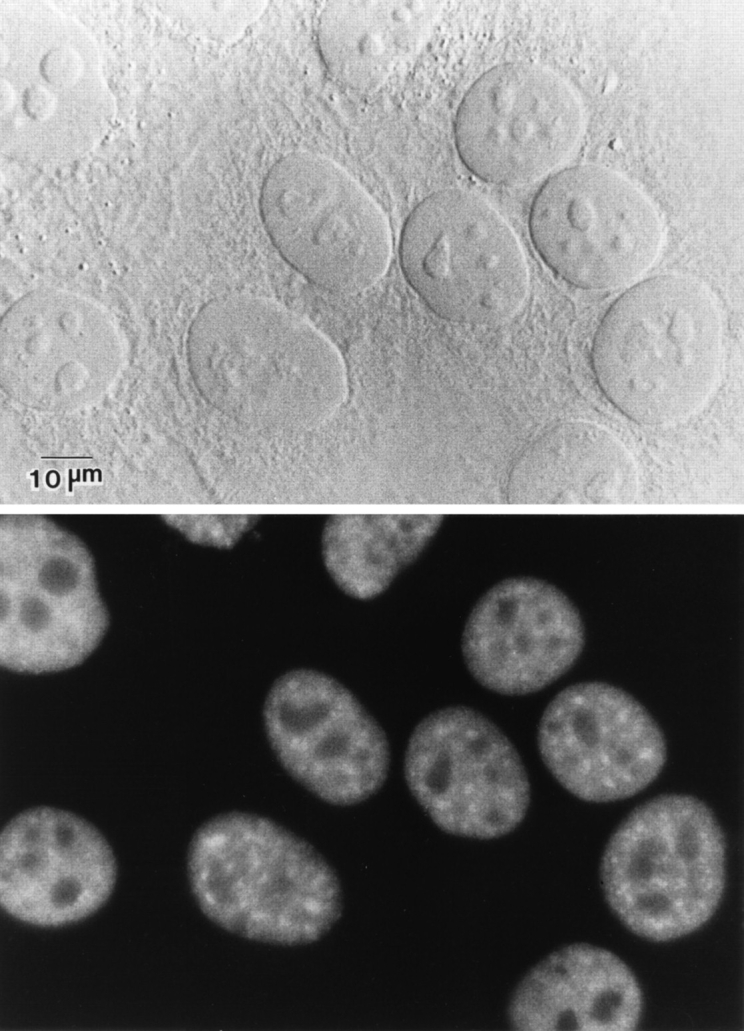
Localization of endogenous SF2/ASF. HeLa cells were fixed, and the cellular localization of SF2/ASF was determined by indirect immunofluorescence with an anti-SF2/ASF monoclonal antibody (Krainer, A.R., unpublished). The protein localizes in a speckled nuclear pattern and is also diffusely distributed throughout the nucleoplasm. (Top) DIC image; (bottom) fluorescence image of the same field. Bar, 10 μm.
To determine the role of individual domains of SF2/ASF and other SR proteins in nuclear and subnuclear localization, we transiently overexpressed in HeLa cells several epitope-tagged SR protein cDNAs encoding either the wild-type proteins or several mutant derivatives and determined the cellular distribution and subnuclear localization of the tagged proteins by indirect immunofluorescence microscopy. We verified the expression of all the transfected cDNAs by Western blot analysis of whole-cell lysates. All constructs encoded proteins with a bacteriophage T7 gene 10 (T7) epitope tag at their NH2 termini (see Materials and Methods), allowing detection of the exogenous proteins with a monoclonal antibody that recognizes this epitope. All expressed proteins accumulated to similar levels in transfected HeLa cells and appeared to be full length; in most cases, doublets were detected, which may represent different states of phosphorylation (data not shown). In the case of SF2/ASF wild-type and mutant proteins, the use of an anti-SF2/ASF monoclonal antibody allowed us to compare the levels of exogenous and endogenous SF2/ASF in the cell population. After normalizing for transfection efficiency, we estimate that the transiently expressed proteins accumulate in the transfected cells at 5–10-fold higher levels than endogenous SF2/ASF, which is in good agreement with our previous results (Cáceres et al., 1994; and data not shown).
Indirect immunofluorescence using the T7 tag antibody showed that 24 h after transfection, the transiently expressed wild-type SF2/ASF protein localized exclusively in the nucleus, with a typical speckled pattern (Fig. 2 a). 48 h after transfection, SF2/ASF overexpression resulted in the formation of nuclear aggregates: the speckles were fewer in number and appeared enlarged (data not shown), in agreement with previous reports (Hedley et al., 1995; Romac and Keene, 1995). The remaining localization data were obtained at the shorter time point, when the nuclear staining pattern of the transiently expressed wild-type protein coincides with that of the endogenous protein in untreated cells. This concordance suggests that transient expression is a valid approach to study the localization signals of SR proteins. Although the transfected proteins are expressed at higher levels, we note that the natural abundance of individual SR proteins fluctuates considerably in different cell types and that modulation of the levels of these proteins appears to be the key to their role in alternative splicing (Ge and Manley, 1990; Krainer et al., 1990a ; Zahler et al., 1993; Cáceres et al., 1994).
Figure 2.
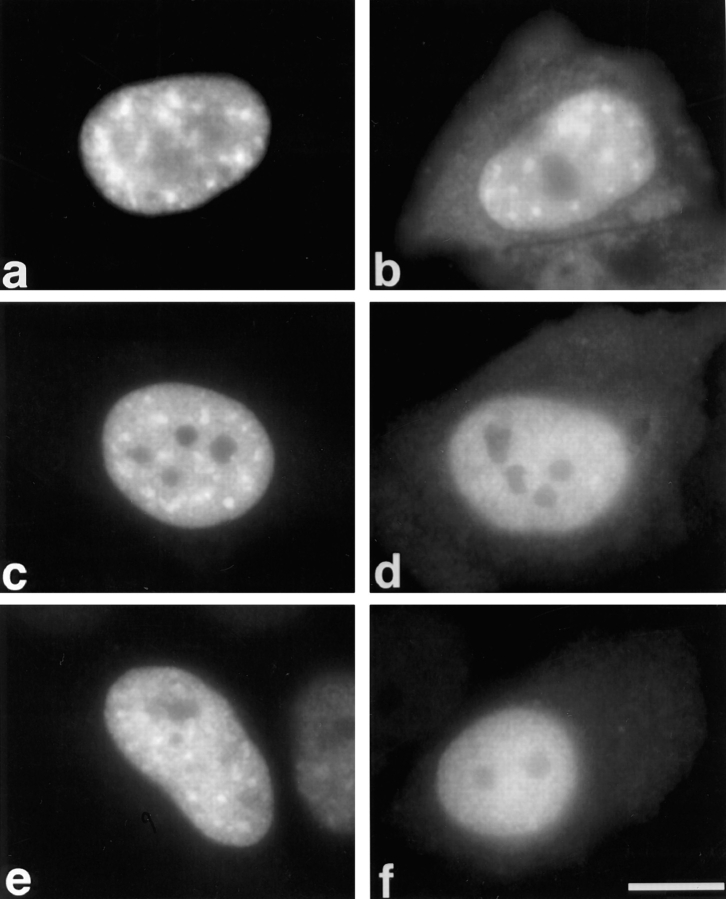
Role of SF2/ASF structural domains in cellular localization and distribution in the nuclear speckles. The structure of the SF2/ASF domain-deletion mutants was previously described (Cáceres and Krainer, 1993). (a) SF2-WT, (b) SF2-ΔRS, (c) RRM1/RS, (d) RRM2/RS, (e) RRM1, (f) RRM2. HeLa cells were transfected with plasmids encoding the respective T7-tagged proteins and fixed 24 h after transfection. The localization of the expressed proteins was determined by indirect immunofluorescence with anti-T7 monoclonal antibody and FITC-conjugated secondary antibody. Bar, 5 μm.
The localization of several transiently expressed SF2/ ASF domain-deletion mutants was then compared to that of the wild-type protein (SF2-WT). The structures of the mutant proteins, designated ΔRS, RRM1/RS, RRM2/RS, RRM1, and RRM2, have been previously described (Cáceres and Krainer, 1993). (RRM2 has sometimes been referred to as pseudo-RRM, ΨRRM, or RRM-homology, because of its lack of canonical RNP-1 and RNP-2 submotifs; see Birney et al., 1993.) In contrast to wild-type SF2/ ASF, the ΔRS mutant protein localized mostly in the nucleus (excluding the nucleoli) but also in the cytoplasm; unexpectedly, nuclear speckles were still clearly detectable (Fig. 2 b). This observation demonstrates that the RS domain of SF2/ASF, although required for exclusive nuclear localization, is not essential for subnuclear localization to the speckles. The RRM1/RS and RRM2/RS mutant proteins displayed a similar cellular distribution to that observed with the ΔRS mutant protein, and they also localized to speckles (Fig. 2, c and d). In contrast, when individual domains were expressed (RRM1 or RRM2), the mutant proteins localized throughout the cell, and no nuclear speckles were detected (Fig. 2, e and f). Expression of just the epitope-tagged RS domain from SF2/ASF was very inefficient and could not be detected by immunofluorescence (data not shown).
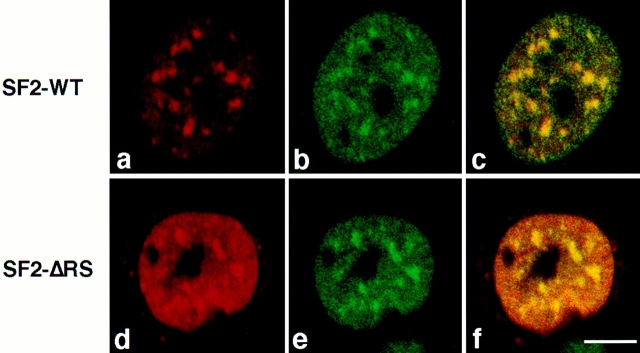
The transiently expressed SF2-WT and SF2-ΔRS proteins colocalized with snRNP particles in the endogenous speckles, as shown by confocal laser scanning microscopy using the anti–T7 tag mouse monoclonal antibody (red) (Fig. 3, a and d) and a human autoimmune serum specific for the Sm core proteins of snRNP particles (green) (Fig. 3, b and e). Colocalization of SF2/ASF proteins and snRNPs results in a yellow color (Fig. 3, c and f). We conclude that transiently expressed SF2-WT and a fraction of SF2-ΔRS proteins localize properly to nuclear speckles containing endogenous snRNPs.
Figure 3.
Colocalization of transiently expressed SF2/ASF, with or without its RS domain, with endogenous snRNPs. HeLa cells were transiently transfected with plasmids expressing T7-tagged, wild-type SF2/ASF (SF2-WT; top row) or a derivative lacking the RS domain (SF2-ΔRS; bottom row). The cells were fixed 24 h after transfection and analyzed by double-label immunofluorescence using laser scanning confocal microscopy. SF2/ASF or SF2-ΔRS were detected using an anti-T7 mouse monoclonal antibody followed by Texas red–conjugated secondary antibody (a and d). Endogenous snRNPs were detected in the same cell using an anti-Sm human serum followed by FITC-conjugated secondary antibody (b and e). Both SF2-WT and SF2-ΔRS colocalized with endogenous snRNPs in nuclear speckles, as shown by the yellow color when the two signals were superimposed (c and f). Bar, 5 μm.
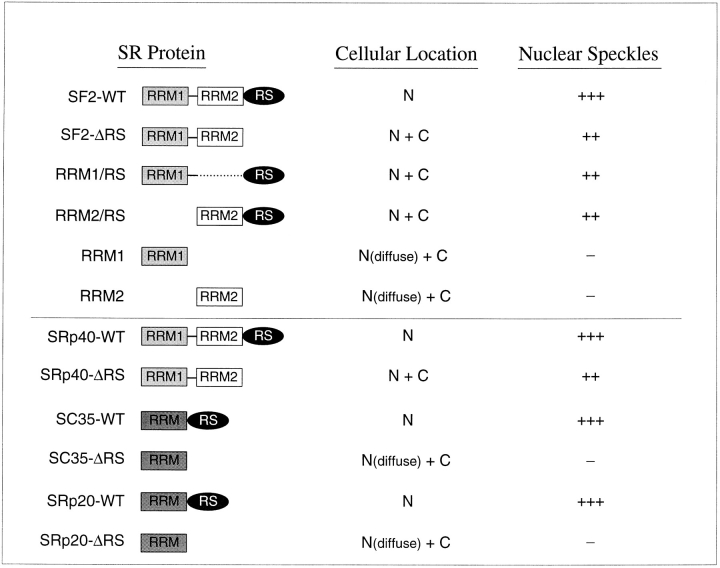
These experiments demonstrate that all three structural domains of SF2/ASF are required for exclusive nuclear localization since deletion of the RS domain, or of either RRM, resulted in proteins with both nuclear and cytoplasmic distribution. However, there appears to be functional redundancy in the speckle localization signals within SF2/ ASF. At least two domains are necessary for SF2/ASF localization to nuclear speckles: either two copies of the RRM or one of the two RRMs together with the RS domain.
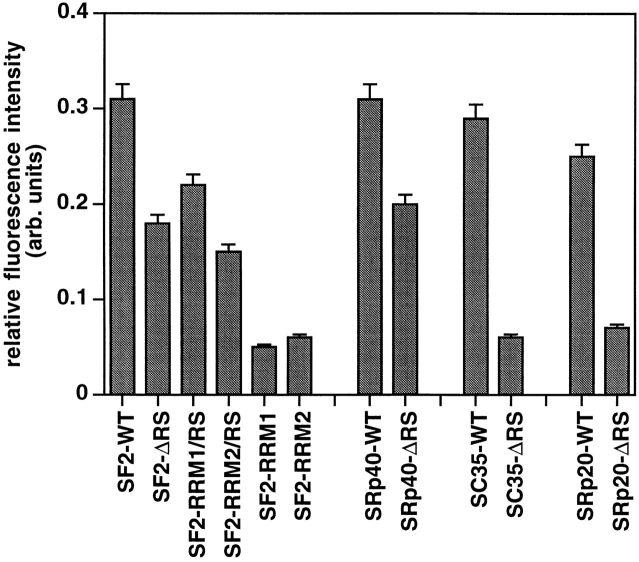
Next, we analyzed the effect of deleting the RS domain from another SR protein that has two RRMs (SRp40), and from two SR proteins that have a single RRM (SC35 and SRp20). To this end, these epitope-tagged proteins (either wild-type proteins or mutant proteins lacking the RS domain) were transiently expressed in HeLa cells, and their cellular localization was analyzed by indirect immunofluorescence. Wild-type SRp40 localized exclusively to the nucleus and showed a typical speckled pattern (Fig. 4 a). Deletion of its RS domain resulted in localization both in the nucleus (excluding the nucleoli) and in the cytoplasm; however, nuclear speckles were clearly detected (Fig. 4 b), as was the case with SF2/ASF lacking its RS domain. Both SC35 and SRp20 wild-type proteins also localized exclusively to the nucleus and displayed a typical nuclear speckled pattern (Fig. 4, c and e, respectively). In contrast, SC35ΔRS and SRp20ΔRS localized throughout the cytoplasm and nucleoplasm, and no nuclear speckles were detected (Fig. 4, d and f); this cellular distribution is very similar to that observed with the RRM1 and RRM2 single-domain mutants of SF2/ASF (Fig. 2). These observations indicate a significant difference in the localization signals present in two SR proteins that have two copies of the RRM (SF2/ ASF and SRp40), and two SR proteins that have only one RRM (SC35 and SRp20), since only in the latter case is the RS domain absolutely required for localization to the speckle domains. Quantitation of the fluorescence signals confirmed these qualitative observations. The relative intensity of nuclear speckles was reduced by 30% in constructs lacking one of the three domains and further reduced by 80% in those constructs lacking two domains. Loss of one domain from two-domain SR proteins had the same effect as loss of two domains from a three-domain SR protein. The relative intensity values were similar for wild-type SRp40, SC35, and SRp20 as compared to SF2/ASF, and were similarly decreased for SC35-ΔRS and for SRp20-ΔRS, as compared to SF2/ASF RRM1 or RRM2 mutants (Fig. 5).
Figure 4.
Cellular localization of other transiently expressed SR proteins with or without their RS domains. HeLa cells were transfected with plasmids encoding the following epitope-tagged proteins: (a) wild-type SRp40, (b) SRp40-ΔRS, (c) wild-type SC35, (d) SC35-ΔRS, (e) wild-type SRp20, and (f) SRp20-ΔRS. The cells were fixed 24 h after transfection and the localization of the expressed proteins was determined as in Fig. 1. Bar, 5 μm.
Figure 5.
Quantitative analysis of localization of transfected proteins in nuclear speckles. The relative fluorescence intensity was calculated as described in Materials and Methods. Note that the relative intensity of speckles decreased by ∼30% for SF2/ASF lacking one of its three domains, and by ∼80% when two of the three domains were missing. Results represent average values ± standard deviation from the pooled data of at least two experiments.
The finding that the RS domain of SF2/ASF is not required for speckle localization was unexpected since previous work showed that the RS domains of Drosophila Tra and SWAP (which have no RRMs) are necessary and sufficient for targeting to speckles (Li and Bingham, 1991; Hedley et al., 1995).
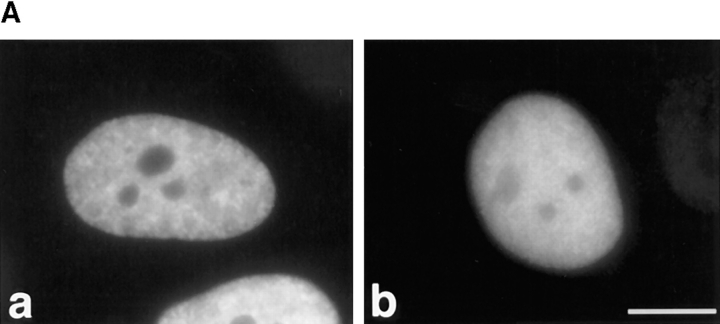
Although the RS domain of SF2/ASF is not required to target SF2/ASF to the speckles, we tested whether it can target a different protein to this subnuclear domain. We assayed the subnuclear localization of a chimeric protein, A1-RS, which consists of both RRMs from hnRNP A1 and the RS domain from SF2/ASF replacing the natural COOH-terminal glycine-rich domain (G-domain) of hnRNP A1 (Mayeda et al., 1994). This domain of hnRNP A1 contains a short signal for bidirectional transport between the nucleus and the cytoplasm (Michael et al., 1995; Siomi and Dreyfuss, 1995; Weighardt et al., 1995). The hnRNP A1 wild-type protein gave a diffuse nucleoplasmic staining rather than a speckled pattern (Fig. 6 A, a), in agreement with published data (for review see Piñol-Roma and Dreyfuss, 1993). Replacing the G-domain of hnRNP A1 by the RS domain of SF2/ASF did not target the chimeric protein to the nuclear speckles (Fig. 6 A, b), demonstrating that, at least in this context, the RS domain of SF2/ASF is not sufficient to direct a heterologous protein to the nuclear speckles. On the other hand, the A1-RS protein localized exclusively in the nucleus, whereas hnRNP A1 lacking a COOH-terminal domain is known to localize throughout the cell (Siomi and Dreyfuss, 1995). Therefore, the RS domain of SF2/ASF is a nuclear localization signal, but not a subnuclear speckle localization signal.
Figure 6.
Cellular localization of transiently expressed chimeric proteins. HeLa cells were transfected with plasmids encoding the following epitope-tagged proteins: (A) (a) hnRNP A1; (b) A1-RS, a chimeric protein with both RRMs from hnRNP A1 fused to the COOH-terminal RS domain of SF2/ASF. (B) (a) NPc, the nucleoplasmin core domain; (b) NPc-RSSF2, a chimera consisting of NPc fused to the RS domain from SF2/ASF; (c) NPc-RSSRp20, NPc fused to the RS domain of SRp20. The cells were fixed 24 h after transfection and the localization of the expressed proteins was determined as in Fig. 1. (C) Colocalization of transiently expressed NPc-RSSRp20 with endogenous snRNPs. HeLa cells were transiently transfected with plasmid expressing T7-tagged NPc-RSSRp20. The cells were fixed 24 h after transfection and analyzed by double-label immunofluorescence using confocal laser scanning microscopy. NPc-RSSRp20 was detected using an anti-T7 mouse monoclonal antibody followed by Texas red–conjugated secondary antibody (a). Endogenous snRNPs were detected in the same cell using an anti-Sm human serum followed by FITC-conjugated secondary antibody (b). NPc-RSSRp20 colocalized with endogenous snRNPs in nuclear speckles, as shown by the yellow color when the two signals were superimposed (c). Bar, 5 μm.
Next, we compared the nuclear localization signals as well as the subnuclear targeting properties of both types of RS domain, i.e., those present in SR proteins with either one or two copies of the RRM. To this end, we constructed chimeric proteins in which the RS domain of either SF2/ASF or SRp20 was fused to a protein reporter, the nucleoplasmin core domain. This protein fragment by itself displays a cytoplasmic distribution (Fig. 6 B, a) since it lacks the nuclear localization signal present in wild-type nucleoplasmin (Dingwall et al., 1987). Both types of RS domain act as nuclear localization signals, as demonstrated by the fact that both NPc-RSSF2 and NPc-RSSRp20 localized to the nucleus (Fig. 6 B, b and c). However, a clear difference was evident in the ability of these RS domains to target a protein reporter to the nuclear speckles. Whereas a diffuse nuclear staining was observed with the SF2/ASF RS domain (Fig. 6 B, b), the RS domain of SRp20 was sufficient to direct the NPc reporter to the nuclear speckles (Fig. 6 B, c). The nuclear speckles observed with the NPc reporter fused to the RS domain of SRp20 colocalized with endogenous human Sm (Fig. 6 C), as shown by confocal laser microscopy using the mouse anti–T7 tag monoclonal antibody (red) (Fig. 6 C, a) and human autoimmune serum specific for the Sm core proteins of snRNP particles (green) (Fig. 6 C, b). Colocalization of NPc-RSSRp20 protein and snRNPs results in a yellow color (Fig. 6 C, c). We conclude that the transiently expressed NPc-RSSRp20 protein localizes properly in nuclear speckles containing endogenous snRNPs, demonstrating that this RS domain is sufficient to determine subnuclear localization in the speckle domain. These experiments demonstrate the existence of two distinct pathways for localization of different SR proteins to the nuclear speckles and also show the unique properties of individual RS domains.
Role of SF2/ASF Domains in Alternative Splicing Regulation In Vivo
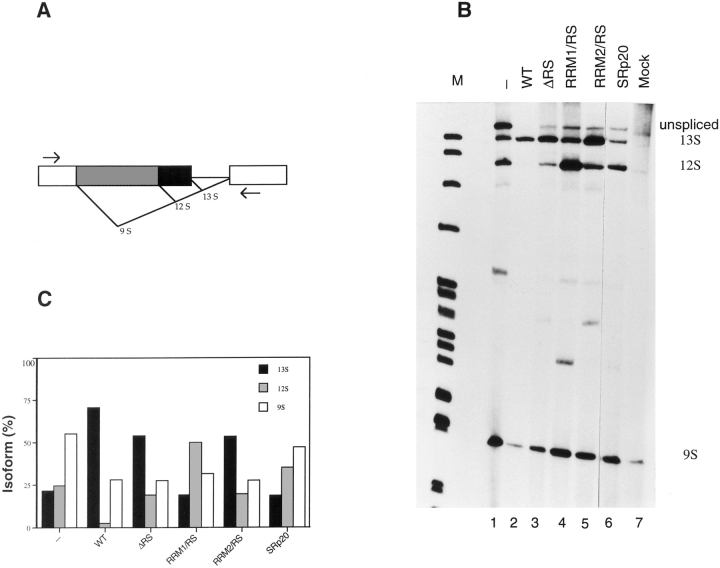
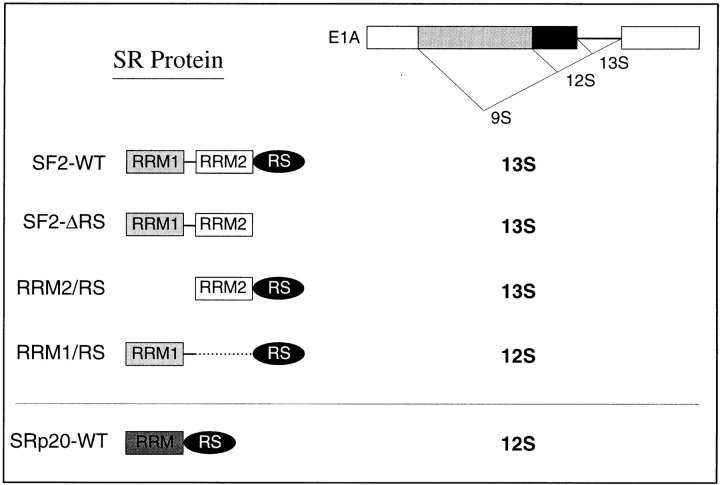
In addition to their function as general splicing factors, SR proteins regulate alternative splicing in a concentration-dependent manner (Ge and Manley, 1990; Krainer et al., 1990a ; Zahler et al., 1993). We sought to investigate the role of individual domains of SF2/ASF in the regulation of alternative splicing in vivo. To this end, we overexpressed several of the constructs described above and assayed changes in the patterns of alternative splicing of an adenovirus E1A splicing reporter. It was previously shown that SF2/ASF strongly activates the use of the proximal 13S E1A 5′ splice site in vivo (Cáceres et al., 1994; Wang and Manley, 1995) and that overexpression of hnRNP A1 results in activation of the distal 9S 5′ splice site (Cáceres et al., 1994; Yang et al., 1994). Compared to wild-type SF2/ ASF, which gave the expected activation of the 13S 5′ splice site (Fig. 7 B, lane 2), the ΔRS protein led to only slightly lower relative levels of 13S mRNA (Fig. 7 B, lane 3), demonstrating that the RS domain of SF2/ASF is not required for alternative splicing modulation in vivo. This result is consistent with previous findings (Wang and Manley, 1995), except that in the earlier study the 9S mRNA levels remained constant for both wild-type and ΔRS proteins, whereas in our hands they decreased, as expected from in vitro studies. The SF2/ASF mutants with a single RRM and the RS domain had strikingly different functional properties: both proteins influenced alternative splicing, but whereas RRM2/RS strongly activated the 13S 5′ splice site (Fig. 7 B, lane 5), RRM1/RS strongly and reproducibly stimulated the 12S 5′ splice site (Fig. 7 B, lane 4). The unexpected altered specificity observed upon deletion of RRM2 demonstrates that the nature of the RRM can influence the selection of a particular 5′ splice site. Activation of the E1A 12S 5′ splice site by the RRM1/RS mutant is similar to the effect of SRp20, which naturally lacks RRM2 (Fig. 7 B, lane 6; Screaton et al., 1995). Quantitation of the relative use of the E1A 5′ splice sites upon overexpression of the different proteins is shown in Fig. 7 C.
Figure 7.
Role of SF2/ASF structural domains in regulating alternative splicing of adenovirus E1A pre-mRNA. (A) Diagram of the E1A reporter gene. The alternative 5′ splice sites and splicing events that generate 13S, 12S, and 9S mRNAs are shown schematically. The location of the exon primers used for RT-PCR analysis is shown. (B) Alternative splicing activity of the SF2/ASF domain-deletion mutants. Each of the indicated SF2/ ASF mutant and control proteins was overexpressed from plasmids cotransfected with the E1A reporter gene. RNA was harvested 24 h after transfection and analyzed by RT-PCR with a labeled forward primer, denaturing PAGE, and autoradiography, as described in Materials and Methods. The positions of the unspliced pre-mRNA, and of 13S, 12S, and 9S spliced mRNAs are indicated on the right. The background products obtained from mock-transfected cells are shown in lane 7. End-labeled DNA size markers are shown in lane M. (C) Quantitation of E1A mRNA isoforms in transfected cells. The relative amounts of 13S, 12S, and 9S E1A mRNAs were calculated from the data in B, using a PhosphorImager, and the percentage of each isoform is shown.
Both constructs with a single RRM and the RS domain (RRM1/RS and RRM2/RS) were also active in regulating alternative splicing of a β-thalassemia splicing reporter, leading in both cases to activation of the most proximal cryptic 5′ splice site (data not shown); this is a similar pattern as that obtained with wild-type SF2/ASF (Cáceres et al., 1994). We and others previously reported that deletion of RRM1 or RRM2 in SF2/ASF abolished the alternative splicing activity of this protein in vitro (Cáceres and Krainer, 1993; Zuo and Manley, 1993). This apparent discrepancy with our present results may reflect incorrect folding of the mutant proteins in vitro, but not in vivo, and/or the different pre-mRNA substrates used in the two types of assays. Wang and Manley (1995) reported that small deletions within RRM2 inactivated the protein in vivo; since we find that deletion of the entire domain imparts a novel specificity on the protein in a similar assay, it is likely that the small deletions disrupted the folding of the RRM, which has a highly conserved tertiary structure (for review see Birney et al., 1993).
Taken together, the cotransfection results suggest that RRM2 of SF2/ASF plays a critical role in the specificity of alternative splicing with certain substrates, such as the E1A pre-mRNA. Thus, all the functional SR protein constructs that include RRM2 (SF2/ASF WT, ΔRS, and RRM2/RS) favored the 13S 5′ splice site, whereas those that lack a second RRM (RRM1/RS and SRp20) selected the 12S 5′ splice site (Fig. 8). We conclude that there is a different functional requirement for individual domains of SR proteins in the regulation of alternative splicing in living cells. Whereas the RS domain is dispensable for this activity, as was previously demonstrated in vitro (Cáceres and Krainer, 1993; Zuo and Manley, 1993), the presence of a particular RRM can promote selection of a specific splice site and also influence substrate specificity.
Figure 8.
Summary of the role of SR protein domains on E1A alternative splicing specificity in vivo. The domain structures of SF2/ASF and domain-deletion mutants thereof, and that of SRp20, are shown schematically. The predominant E1A isoform generated upon overexpression of each protein is shown on the right. The presence of RRM2 of SF2/ASF correlates with use of the proximal 13S 5′ splice site, whereas proteins that lack this atypical RRM cause preferential use of the 12S 5′ splice site.
Discussion
We have studied the role of individual domains of SR proteins in cellular distribution and subnuclear localization, as well as in alternative splicing activity in vivo. Unexpected differences were uncovered in the localization pathways for different SR proteins, despite the close conservation of structure and biochemical properties among members of this protein family. SR proteins with a single RRM require an RS domain for proper localization in the nuclear speckles, whereas SR proteins with two RRMs do not; in the latter, weak speckle targeting signals are present in each of the three constituent domains and function additively. The fact that single-domain deletion variants of SF2/ASF localized properly made it possible to determine the role of each domain in alternative splicing in vivo. Each of the three domains could be deleted individually without abrogating the ability of SF2/ASF to modulate alternative splice site selection. Remarkably, however, deletion of RRM2 imparted a distinct activity on the protein, such that it promoted the selection of a different alternative 5′ splice site in the adenovirus E1A pre-mRNA.
Nuclear Localization of SR Proteins
The SR protein SF2/ASF has a modular structure, consisting of two RRMs and a COOH-terminal RS domain. Here we showed that each of these three domains contributes additively to the nuclear localization of the protein since deletion of individual domains results in proteins that, unlike the wild-type protein, no longer have an exclusively nuclear distribution (Fig. 9). In particular, the RS domain of SF2/ASF contributes to its nuclear localization since deletion of this domain causes nuclear and cytoplasmic distribution of the resulting mutant protein. In agreement with this finding, when the RS domain of SF2/ASF was fused to the NPc protein reporter (which on its own localizes in the cytoplasm), the fusion protein (NPc-RSSF2) localized in the nucleus (Table I). Thus, the SF2/ASF RS domain is a nuclear targeting signal.
Figure 9.
Summary of the localization of SR protein variants. The domain structures of the SF2/ASF wild-type and mutant proteins are shown schematically above the line, and those of SRp40, SC35, and SRp20, with or without an RS domain, are shown below the line. The observed cellular location of the expressed proteins is indicated as N (nuclear) or C (cytoplasmic). In all cases, the expressed proteins were excluded from nucleoli. Subnuclear accumulation in speckles is indicated by + or − signs, with the number of + signs reflecting the quantitations shown in Fig. 5. Proteins that did not accumulate in speckles gave a diffuse nuclear pattern.
Table I.
Summary of Localization Of Chimeric Proteins
| Protein | Cellular location | Nuclear speckles | ||
|---|---|---|---|---|
| A1-RS | N | − | ||
| NPc | C | − | ||
| NPc-RSSF2 | N | − | ||
| NPc-RSSRp20 | N | + |
The A1-RS protein consists of the two RRMs from hnRNP A1 fused to the COOH-terminal RS domain of SF2/ASF. NPc is the nucleoplasmin core domain (Dingwall et al., 1987). This domain was fused to the SF2/ASF RS domain (NPc-RSSF2) or to the SRp20 RS domain (NPc-RSSRp20). The observed cellular distribution was either nuclear (N) or cytoplasmic (C). Subnuclear accumulation in speckles is indicated by + or − signs.
We do not know at present whether the nuclear and cytoplasmic distribution observed with several mutants represents incomplete nuclear import and/or incomplete retention of these proteins in the nucleus. Two alternative explanations, which are not mutually exclusive, can be proposed on the basis of our findings. In the first model, there are multiple nuclear localization signals distributed throughout SF2/ASF. These signals have additive effects, such that deletion of either RRM, or of the RS domain, reduces the overall steady-state accumulation in the nucleus. Thus, when all three structural domains are present, the protein has an exclusively nuclear distribution. When only two of the three domains are present, a fraction of the protein accumulates in the cytoplasm, and when only one domain is present, the proportion of cytoplasmic protein greatly increases. In the second model, partition between the nucleus and the cytoplasm can be attributed to incomplete nuclear retention of SF2/ASF mutants lacking either RRM repeat or the RS domain. In support of this model, we have shown that fusing the RS domain to a cytoplasmic reporter, NPc, directs the fusion protein exclusively to the nucleus. NPc is thought to have a propensity to be retained in the nucleus, once it gets there, because of oligomerization (Michael et al., 1995). In contrast, the SF2/ASF mutant proteins that lack one of the two RRMs (RRM1/RS, RRM2/RS) display mostly nuclear but also cytoplasmic localization, and they have been shown to have reduced RNA binding, compared to the wild-type protein. SF2/ASF RRM domains expressed individually distribute evenly throughout the whole cell (Fig. 2) and bind RNA very poorly (Cáceres and Krainer, 1993; Zuo and Manley, 1993).
In the case of SR proteins that in their natural form have a single RRM (e.g., SC35 and SRp20), deletion of the RS domain resulted in mutant proteins with nuclear and cytoplasmic distribution (Fig. 9). Therefore, the nuclear localization mechanism appears to be different for the two SR protein subfamilies: in the case of SC35 and SRp20, a single RRM together with the RS domain resulted in exclusive nuclear localization. In contrast, in the case of SF2/ASF, either RRM together with the RS domain resulted in nuclear and cytoplasmic distribution (RRM1/RS and RRM2/RS; Fig. 2), and exclusive nuclear localization was only achieved when both RRMs were present together with the RS domain.
Localization of SR Proteins to Nuclear Speckles
The process of nucleo-cytoplasmic transport has been extensively studied, and several components of this pathway have been identified (for reviews see Silver, 1991; Dingwall and Laskey, 1991; Görlich and Mattaj, 1996). In contrast, the mechanisms involved in localization of proteins within specific subnuclear regions are poorly understood. They may involve active transport mediated by subnuclear targeting signals, or alternatively, passive diffusion and binding to the nuclear matrix or its components. It has been proposed that the speckle domains are anchored directly or indirectly to the nuclear matrix (Spector et al., 1983). In agreement with this hypothesis, a role for the nuclear matrix in splicing has been postulated (Zeitlin et al., 1987), and antibodies raised against components of the nuclear matrix cross-react with SR proteins (Blencowe et al., 1994, 1995).
The RS domain is the most prominent feature shared by splicing factors that localize in a speckled pattern. As such, it is the best candidate domain to mediate this subnuclear localization, as first proposed and tested with Drosophila proteins by Bingham and coworkers (Li and Bingham, 1991). In the case of human SF2/ASF, we have shown that the RS domain is neither necessary nor sufficient for localization to the speckles. In marked contrast, deleting the RS domain of two human SR proteins that have a single RRM caused the mutant proteins to distribute throughout the cell, and accumulation in the speckled region was no longer observed. A similar result was obtained when expressing individual domains of SF2/ASF (RRM1 and RRM2 proteins). In addition, by fusing the RS domain of SRp20 to a cytoplasmic protein reporter, we demonstrated that this RS domain is necessary and sufficient to target a protein to the nuclear speckle domains (Table I). These results demonstrate the existence of two different mechanisms for localization to the speckle domains, based on the different behavior of two types of RS domains.
The lack of speckle-targeting signals in the RS domain of human SF2/ASF was unexpected, in light of previous findings with the RS domains of the Drosophila SWAP and Tra splicing regulators, which are necessary and sufficient to target reporter proteins to the speckled region (Li and Bingham, 1991). A recent study further defined the specific elements within the Tra RS domain required for localization in the speckled region (Hedley et al., 1995). In addition to a classical bipartite nuclear localization sequence, a novel motif was defined, which is necessary and sufficient for subnuclear localization. The subnuclear targeting signal of Tra and homologous motifs present in Drosophila Tra2 and SWAP, and in human U1-70K and SC35, consist of three or four basic amino acids (generally arginine and histidine) followed by a run of RS dipeptides (Hedley et al., 1995). Interestingly, this motif is present in human SR proteins that have a single RRM (SC35, SRp20 and 9G8) (Ayane et al., 1991; Fu and Maniatis, 1992; Vellard et al., 1992; Cavaloc et al., 1994), but it is absent from some, though not all, of the SR proteins with two RRMs (SF2/ASF, SRp40 and SRp30c) (Ge et al., 1991; Krainer et al., 1991; Screaton et al., 1995).
The Drosophila Tra and SWAP alternative splicing factors have RS domains, but they lack RRMs. In contrast, the modular structure of SR proteins, with a COOH-terminal RS domain of variable length and one or two RRMs, is likely to allow multiple protein–protein and protein–RNA interactions. The mechanism of nuclear speckle localization may be a complex process, by analogy to the pathways described for localization in coiled bodies or in nucleoli (Schmidt-Zachmann and Nigg, 1993; Bohmann et al., 1995; Scheer and Weisenberger, 1995), which involve complex signals rather than simple sequence motifs.
Studies with the Drosophila splicing regulators suggested a mechanism for subnuclear localization based on protein–protein interactions mediated by the RS domains. Recent studies pointed to a role for the RS domains of Drosophila and/or human Tra, Tra2, U2AF35, U1-70K, and those of several SR proteins in mediating specific protein–protein interactions, which may be modulated by phosphorylation of many of the serine residues (Wu and Maniatis, 1993; Amrein et al., 1994; Kohtz et al., 1994; Xiao and Manley, 1997). In particular, the RS domains of SC35 and SF2/ASF are thought to mediate specific interactions with the RS domains of both the U1-70K polypeptide and with the 35-kD subunit of the splicing factor U2AF (Wu and Maniatis, 1993; Kohtz et al., 1994). These interactions are thought to be involved in defining and bridging the splice sites during spliceosome assembly, but in principle they could also play a role in specifying subnuclear localization. Although both snRNP polypeptides and SR proteins colocalize in a speckled distribution, their localization appears to involve different molecular interactions since the speckled distribution of snRNP proteins is sensitive to RNase A treatment, while the SC35 distribution is not affected (Spector et al., 1991).
Proteins that lack RS domains, such as the splicing factor PSF, are thought to localize to the speckled region by interacting with snRNPs or with RS domain-containing proteins (Patton, J., personal communication; Hedley et al., 1995). Deletion mutants of Tra lacking the RS domain can still localize in the speckles, provided that they are able to interact with Tra2 (Hedley et al., 1995). Thus, two mechanisms may operate to target splicing components to the nuclear speckles: a direct one, in which the presence of a targeting signal determines the proper subnuclear localization, and an indirect one, in which proteins lacking a targeting signal localize to the speckle domains by binding to splicing components that have a targeting signal. If SF2/ ASF is targeted to the speckles indirectly, by protein–protein or RNA–protein interactions, the interaction regions must be redundant since we showed that each of the three constituent domains can be deleted individually without a complete loss of localization in the speckle domains. Currently, the RS domain of SF2/ASF is the only region of the protein known to be involved in protein–protein interactions, but this domain is neither necessary nor sufficient for speckle localization of SF2/ASF. Targeting by interaction with RNA is a distinct possibility; to explain the present localization data, this model would require that a single RRM derived from an SR protein be unable to interact stably with RNA, and that either a second RRM or an RS domain be required to stabilize interactions with RNA. In vitro RNA-binding experiments showed that these are indeed the properties of SF2/ASF (Cáceres and Krainer, 1993; Zuo and Manley, 1993; Jamison et al., 1995). The direct targeting model is more likely for single-domain SR proteins, such as SC35, whose localization is RNase resistant (Spector et al., 1991), and which appear to have short sequence motifs that are sufficient for correct subnuclear localization (Hedley et al., 1995).
Alternative Splicing Activity of SR Proteins
We analyzed the role of the structural domains of SF2/ ASF in regulation of alternative splicing in living cells (Fig. 8). We found that the RS domain of SF2/ASF is not required for regulation of alternative splicing in vivo, since a mutant protein lacking this domain regulates alternative splice site selection of an E1A splicing reporter in a manner very similar to the wild type. This finding is consistent with previous observations made in vitro and in vivo (Cáceres and Krainer, 1993; Zuo and Manley, 1993; Wang and Manley, 1995). Whereas the in vitro studies also showed that the RS domain of SF2/ASF is required for its full activity in constitutive splicing, the observation that it is not required for concentration-dependent effects on alternative splicing may suggest that for this particular function, the lack of the RS domain can be compensated by interactions with other SR proteins. As noted above, however, the only protein–protein interaction region of the protein known so far is, in fact, the RS domain (Wu and Maniatis, 1993; Amrein et al., 1994; Kohtz et al., 1994).
We showed that the presence of a particular domain, RRM2 of SF2/ASF, confers selectivity for a specific alternative 5′ splice site. Thus, all the constructs we analyzed that included RRM2 favored the most proximal 5′ splice site in E1A pre-mRNA (13S), whereas natural (SRp20) or mutant (SF2-RRM1/RS) proteins lacking RRM2 selected the middle 5′ splice site (12S). This remarkable difference in splice site selection demonstrates that the RRMs of SF2/ASF function as modules that contribute to specificity in alternative splicing, and that the functions of the two RRM modules can be separated. Moreover, this effect is substrate specific since both the mutant SF2/ASF lacking RRM2 and SRp20 had altered specificity (compared to the other proteins) with the E1A pre-mRNA but showed normal specificity with a β-thalassemia pre-mRNA (Screaton et al., 1995; data not shown). We conclude that the selection of alternative splice sites depends on both the properties of the pre-mRNA as well as on the presence of particular domains in the SR proteins. However, not all SR proteins that possess or lack RRM2 will necessarily have the same alternative splicing specificities as SF2/ASF and SRp20, respectively. For example, overexpression of SC35, which has a single RRM, fails to activate 12S splicing (Wang and Manley, 1995; data not shown). These substrate differences among SR proteins are consistent with the notion that they may function as global regulators of alternative splicing for distinct classes of pre-mRNAs in vivo (Screaton et al., 1995).
The mechanistic relation between general or specific RNA binding by SR proteins and their activity in alternative splicing regulation has not been established. Although both RRMs in SF2/ASF are required for efficient binding to RNA (Cáceres and Krainer, 1993; Zuo and Manley, 1993) and to high affinity sites (Tacke and Manley, 1995), our present data show that mutants of SF2/ASF lacking either RRM1 or RRM2 are nevertheless active in alternative splicing in vivo. The fact that alternative splicing activity can be observed in vivo with SF2/ASF mutants lacking any one of its three constituent domains suggests that this activity of the protein does not require highly sequence-specific binding to RNA. On the other hand, the different splice site activation specificity of the SF2/ASF mutants, depending on which of the two RRMs was deleted, suggests that sequence-specific interactions may play a role in selecting particular splice sites for activation. The alternative splicing activity of SF2/ASF has been shown to correlate with its ability to promote multiple occupancy of alternative 5′ splice sites by U1 snRNP (Eperon et al., 1993). Interestingly, SF2/ASF mutants lacking either RRM are still able to form a ternary complex with U1 snRNP and pre-mRNA containing a functional 5′ splice site (Jamison et al., 1995). The activity of similar mutants in alternative splicing in vivo is consistent with this finding.
In summary, we have shown that the modular structure of SR proteins has profound consequences for their cellular localization, alternative splicing activity, and also their specificity in alternative 5′ splice site selection.
Acknowledgments
We thank Angus Wilson for the T7-expression vector, C. Dingwall for the nucleoplasmin cDNA, and Mario Gimona and Sui Huang for helpful discussions.
Abbreviations used in this paper
- G-domain
glycine-rich domain
- RRM
RNA-recognition motif
- RS domain
arginine/serine-rich domain
- RT-PCR
reverse transcriptase PCR
- snRNP
small nuclear ribonucleoprotein particle
- SWAP
suppressor-of-white-apricot
- Tra
Transformer
Footnotes
T. Misteli was supported by the Swiss National Science Foundation and the Human Frontiers Science Program. G.R. Screaton was supported by the Wellcome Trust and the Arthritis and Rheumatism Council. D.L. Spector is supported by a grant from NIH/NIGMS (GM 42694). J.F. Cáceres and A.R. Krainer were supported by grants GM42699 from NIH/ NIGMS and CA13107 from NCI and by the Pew Charitable Trusts.
Please address all correspondence to Adrian R. Krainer, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, NY 11724-2208. Tel.: (516) 367-8417. Fax: (516) 367-8453. E-mail: krainer@cshl.org
J.F. Cáceres' present address is MRC Human Genetics Unit, Western General Hospital, Crewe Road, Edinburgh EH4 2XU, United Kingdom.
References
- Amrein H, Hedley ML, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- Ayane M, Preuss U, Köhler G, Nielsen PJ. A differentially expressed murine RNA encoding a protein with similarities to two types of nucleic acid binding motifs. Nucleic Acids Res. 1991;19:1273–1278. doi: 10.1093/nar/19.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Nickerson JA, Issner R, Penman S, Sharp PA. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Kim J, McCaw P, Sharp PA. New proteins related to the Ser-Arg family of splicing factors. RNA. 1995;1:852–865. [PMC free article] [PubMed] [Google Scholar]
- Bohmann K, Ferreira J, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol. 1995;131:817–831. doi: 10.1083/jcb.131.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres JF, Krainer AR. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO (Eur Mol Biol Organ) J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres, J.F., S. Stamm, D.M. Helfman, and A.R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science (Wash. DC). 265:1706–1709. [DOI] [PubMed]
- Cavaloc Y, Popielarz M, Fuchs J-P, Gattoni R, Stévenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO (Eur Mol Biol Organ) J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI. The Clk/Sty protein kinase phosphorylates SR protein factors and regulates their intracellular distribution. EMBO (Eur Mol Biol Organ) J. 1996;15:265–275. [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Dilworth SM, Black SJ, Kearsey SE, Cox LS, Laskey RA. Nucleoplasmin cDNA reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO (Eur Mol Biol Organ) J. 1987;6:69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Du L, Warren SL. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eperon IC, Ireland DC, Smith RA, Mayeda A, Krainer AR. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO (Eur Mol Biol Organ) J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Puvion E. The ultrastructural visualization of nucleolar and extranucleolar RNA synthesis and distribution. Int Rev Cytol. 1980;65:255–299. doi: 10.1016/s0074-7696(08)61962-2. [DOI] [PubMed] [Google Scholar]
- Fu X-D. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature (Lond) 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature (Lond) 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science (Wash DC) 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- Ge H, Manley JL. A protein factor, ASF, controls alternative splicing of SV40 early pre-mRNA in vitro. . Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science (Wash DC) 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Gui J-F, Lane WS, Fu X-D. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature (Lond) 1994a;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- Gui J-F, Tronchere H, Chandler SD, Fu X-D. Purification and characterization of a kinase specific for the serine and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci USA. 1994b;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley ML, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine rich splicing factor. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS, Krainer AR. Mechanisms for selecting 5′splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10:100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. U1 and U2 small nuclear RNAs are present in nuclear speckles. Proc Natl Acad Sci USA. 1992;89:305–308. doi: 10.1073/pnas.89.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Spector DL. Intron-dependent recruitment of pre-mRNA splicing factors to sites of transcription. J Cell Biol. 1996;133:719–732. doi: 10.1083/jcb.133.4.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Pasman Z, Wang J, Will C, Lührmann R, Manley JL, Garcia-Blanco M. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-García LF, Spector DL. In vivo evidence that transcription and splicing are coordinated by a recruiting mechanism. Cell. 1993;73:47–59. doi: 10.1016/0092-8674(93)90159-n. [DOI] [PubMed] [Google Scholar]
- Kiseleva E, Wurtz T, Visa N, Daneholt B. Assembly and disassembly of spliceosomes along a specific pre-messenger RNP fiber. EMBO (Eur Mol Biol Organ) J. 1994;13:6052–6061. doi: 10.1002/j.1460-2075.1994.tb06952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz JD, Jamison SF, Will CL, Zuo P, Lührmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature (Lond) 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990a;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. Purification and characterization of SF2, a human pre-mRNA splicing factor. Genes Dev. 1990b;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Krainer AR, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Li H, Bingham PM. Arginine/Serine rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Mattaj IW. Splicing in space. Nature (Lond) 1994;372:727–728. doi: 10.1038/372727a0. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–372. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Cáceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO (Eur Mol Biol Organ) J. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature (Lond) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Protein phosphorylation and nuclear organization of pre-mRNA splicing. Trends Cell Biol. 1996;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E, Simos G, Georgatos SD, Giannakouros AT. A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J Biol Chem. 1996;271:8365–8372. doi: 10.1074/jbc.271.14.8365. [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñol-Roma S, Dreyfuss G. hnRNP proteins: localization and transport between the nucleus and the cytoplasm. Trends Cell Biol. 1993;3:151–155. doi: 10.1016/0962-8924(93)90135-n. [DOI] [PubMed] [Google Scholar]
- Romac JM-J, Keene JD. Overexpression of the arginine-rich carboxy-terminal region of U1 snRNP 70K inhibits both splicing and nucleocytoplasmic transport of mRNA. Genes Dev. 1995;9:1400–1410. doi: 10.1101/gad.9.11.1400. [DOI] [PubMed] [Google Scholar]
- Rossi F, Labourier E, Forné T, Divita G, Derancourt J, Riou JF, Antoine E, Cathala G, Brunel C, Tazi J. Specific phosphorylation of SR proteins by mammalian DNA topoisomerase I. Nature (Lond) 1996;381:80–82. doi: 10.1038/381080a0. [DOI] [PubMed] [Google Scholar]
- Scheer U, Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994;6:354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Schmidt-Zachmann MS, Nigg EA. Protein localization to the nucleolus: a search for targeting domains in nucleolin. J Cell Sci. 1993;105:799–806. doi: 10.1242/jcs.105.3.799. [DOI] [PubMed] [Google Scholar]
- Screaton GR, Cáceres JF, Mayeda A, Bell MV, Plebanski M, Jackson DG, Bell JI, Krainer AR. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO (Eur Mol Biol Organ) J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver PA. How proteins enter the nucleus. Cell. 1991;64:489–497. doi: 10.1016/0092-8674(91)90233-o. [DOI] [PubMed] [Google Scholar]
- Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Higher order nuclear organization: three-dimensional distribution of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1990;87:147–151. doi: 10.1073/pnas.87.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO (Eur Mol Biol Organ) J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO (Eur Mol Biol Organ) J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct–2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- Vellard M, Sureau A, Soret J, Martinerie C, Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci USA. 1992;89:2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Manley JL. Overexpression of the SR proteins ASF/SF2 and SC35 influences alternative splicing in vivo in diverse ways. RNA. 1995;1:335–346. [PMC free article] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel ER, Stäubli W, Gnägi HR, Hess FA. Correlated morphometric and biochemical studies on the liver cell. I: Morphometric model, stereological methods, and normal morphometric data for rat liver. J Cell Biol. 1969;42:69–91. doi: 10.1083/jcb.42.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weighardt F, Biamonti G, Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Peterson MG, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Dobner PR, Lawrence JB. Higher level organization of individual gene transcription and RNA splicing. Science (Wash DC) 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- Xing Y, Johnson CV, Moen PT, McNeil J, Lawrence JB. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC35-domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bani MR, Lu S-J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. . Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Roth MB. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neugebauer KM, Lane WS, Roth MB. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science (Wash DC) 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- Zeitlin SA, Parent A, Silverstein S, Efstratiadis A. Pre-mRNA splicing and the nuclear matrix. Mol Cell Biol. 1987;7:111–120. doi: 10.1128/mcb.7.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerler B, Moran B, Maruyama K, Moomaw J, Grodzicker T, Ruley EH. Adenovirus E1A coding sequences that enable ras and pmt oncogenes to transform cultured primary cells. Mol Cell Biol. 1986;6:887–899. doi: 10.1128/mcb.6.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature (Lond) 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]
- Zuo P, Manley JL. Functional domains of the human splicing factor ASF/SF2. EMBO (Eur Mol Biol Organ) J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]