Abstract
In homozygous rolling stone embryos, the fusion of myoblasts to syncytial myotubes is diminished. Nevertheless, the visceral mesoderm, the heart mesoderm, and few somatic muscles are properly formed. Thus, we postulate a central role of rolling stone for the fusion process within the somatic mesoderm. We have cloned the rolling stone gene, and the deduced protein sequence is in accordance with a transmembrane protein, which agrees with the enrichment of Rost in the membrane fraction of Drosophila embryos. No homologous genes have been described so far. rolling stone is expressed in the embryonic nervous system and cells of the somatic mesoderm, most notable in muscle founder cells. To elucidate the function of rolling stone for myoblast fusion, we applied a knock-out strategy. The expression of an antisense rolling stone transcript specifically within the mesoderm of wild-type embryos results in fusion defects of myoblasts, proving that the rolling stone expression in the mesoderm is responsible for the rolling stone phenotype. We suggest that rolling stone is a member of a group of genes that are necessary for the fusion process during myogenesis.
The formation of the larval muscles in Drosophila melanogaster during embryogenesis was described by Bate (1990). Two remarkable events are the specification of muscle precursor cells for each of the larval muscles, and the fusion of myoblasts with these precursor cells to establish mature myotubes. It was suggested that myogenesis begins with the segregation of single founder cells within the somatic mesoderm. These founder cells may in some way direct the fusion process to precursors, and later on into mature myotubes (Bate, 1990, 1993; Dohrmann et al., 1990).
Since a couple of molecular markers for specific muscle precursors are available, e.g., apterous (Bourgouin et al., 1992), nautilus (Michelson et al., 1990; Paterson et al., 1990), and S59 (Dohrmann et al., 1990), some aspects of precursor formation within the mesoderm can be studied in detail. Most of the identified genes code for transcription factors, and their expression appears in overlapping sets of muscle precursors (for review see Abmayr et al., 1995). It might well be that every founder or precursor cell has its characteristic set of factors that specify and maintain its identity, as it is well studied for genes specifying the neural fate in the developing nervous system (for review see Jimenez and Modolell, 1993). If a single factor or a combination of factors is missing, the founder or precursor cell might be unspecified and probably fails to recruit fusion-competent cells from the surrounding mesoderm, or fusion occurs but the developing muscle is effected in its location, routing, or attachment. This view is supported by the observation that mutations in genes like apterous and single-minded, which are likely involved in the specification of muscle precursors, result in developmental defects characterized by the loss of specific muscles, misrouting, or abnormal size of muscles (Bourgouin et al., 1992; Lewis and Crews, 1994).
Beside those genes that are specific for a subset of precursors, genes like mef2, which show a ubiquitous expression within the somatic mesoderm, were described recently (Lilly et al., 1994, 1995; Nguyen et al., 1994; Taylor et al., 1995). Mutations in mef2 reveal strong developmental muscle defects. In homozygous mef2 mutants, the initial myogenic steps, including the formation of nautilus- and even-skipped–expressing precursors, appear normal, although later during development multinucleated nau- expressing syncytial muscles are not formed. Interestingly, in the mef2 mutants, the myoblasts detectable with an antibody against β3 tubulin remain unfused (Bour et al., 1995; Lilly et al., 1995, Ranganayakulu et al., 1995). The role of Mef2 might be to maintain the myogenic pathway and to establish the expression of genes, like myosin heavy chain (MHC)1, integrin, tropomyosin, and in part β3 tubulin, rather than to specify muscle precursors (Bour et al., 1995; Lilly et al., 1995; Ranganayakulu et al., 1995; Lin et al., 1996; Renkawitz-Pohl, R., personal communication).
A further group of genes should be necessary for the formation of muscles during embryogenesis. These genes code for proteins needed directly for the fusion process of myoblasts with the muscle precursors to form the myotubes. This process includes cell–cell recognition between the precursor and the fusion-competent cell, cell adhesion between the fusing cells, and the fusion event itself. So far, only a small number of genes that seem to be involved directly in myoblast fusion in Drosophila have been characterized: Drac1 (Luo et al., 1994), myoblast city (mbc; Rushton et al., 1995), rolling stone (rost; Paululat et al., 1995), blown fuse (blow; Doberstein et al., 1997), and at least one more on the X-chromosome (Drysdale et al., 1993). Mutations in mbc, blow, or rost or overexpression of a mutated Drac1 gene results in specific defects in the fusion process. Drac1 encodes a G-protein that is involved in signal transduction, myoblast city is not characterized on the molecular level so far, and blown fuse encodes a novel cytoplasmic protein. The molecular analysis presented here shows that rost very likely encodes a transmembrane protein.
At least two different fundamental processes may lead to fusion defects of myoblasts: First, single muscle founders or precursors are not formed or not specified. As a consequence, fusion-competent cells cannot fuse because the master cell that directs this process is missing. Otherwise, these cells could be occupied by other precursors in the neighborhood. The nem-mutant (Burchard et al., 1995) may serve as an example. The phenotype of this mutation shows the loss of some specific muscles (e.g., muscles 21– 24) and the duplication of others (dorsal oblique or dorsal acute muscles, SBM [muscle 8]; Knirr S., S. Breuer, A. Paululat, and R. Renkawitz-Pohl, manuscript submitted for publication). Secondly, muscle precursors are specified, but the myoblasts are not competent for fusion. mbc and rost seem to be mutants of this group, in which a high number of unfused myoblasts are observable (see also Drysdale et al., 1993). Stainings with the molecular markers nautilus (Paululat et al., 1995), even-skipped (Paululat, A., unpublished data), vestigial, and S59 (Rushton, E., personal communication) show clearly that the early muscle precursor cells are properly formed in the rost mutant. Therefore, in mbc as well as in rost homozygous mutants, the muscle precursors seem to be specified. The cytological analysis of late embryos confirm these results. Besides the rounded myoblasts, another group of elongated and β3 tubulin– or MHC-positive cells appear in both mutants, which exhibit all properties of muscle precursors, like attachment to the apodemes or contact with motoneurons (Rushton et al., 1995; Paululat, A., unpublished results). In mbc, as well as in rost, precursors are differentiated, but fusion with myoblasts does not occur.
To analyze the function of rost during myoblast fusion, we cloned and characterized the rost gene on the molecular level. In this paper we present data showing that the mesodermal expression of rost gene is essential for myoblast fusion and that the rost gene codes for a transmembrane protein.
Materials and Methods
Drosophila Stocks
For a detailed description of the P-element–induced allele, the EMS alleles, and the characterization of the rost phenotype, see Paululat et al. (1995).
Generation of UAS-antisense-rost Strains
To create the UAS-antisense-rost construct, a XhoI-XbaI fragment from a rost-Bluescript SKII (Stratagene, La Jolla, CA) cDNA subclone, containing nearly the complete coding region of the rost gene, was cloned in antisense orientation into the vector pUAST (Brand and Perrimon, 1993). Transgenic flies containing the UAS-antisense-rost construct were generated by injection of w-embryos, according to published procedures (Rubin and Spradling, 1982; Spradling and Rubin, 1982). Four independent transformants were obtained and expanded into homozygous stocks. The twi-Gal4 driver line was generously provided by Alan Michelson. Stocks homozygous for twi-Gal4 were crossed at 24°C to the described UAS-antisense-rost stocks. All progeny contain one copy of the driver and the UAS construct. The embryos were analyzed using a β3 tubulin (Leiss et al., 1988) or MHC (Kiehart and Feghali, 1986) antibody.
Promoter Construct and lacZ Expression
For testing promoter fragments, we constructed a rost promoter–lacZ P-element transformation vector. The clone that we used for the fusion construct contains ∼400 bp 5′ of the putative rost ATG. This clone was generated by PCR because of the lack of appropriate restriction site, and as a control, different independent PCR products were sequenced and compared with the original genomic sequences. One correct PCR product was blunt end cloned into Bluescript and afterwards transferred into a P-element transformation vector (pCaSpeR, w+) that contains a lacZ reporter gene behind the multiple cloning site. Flies were transformed using standard techniques. Nine independent insertions were isolated and tested for reporter gene expression using rabbit or mouse anti–β-Gal antibodies (Vector Labs, Burlingame, CA). The variation was high in the level of expression but not in the tissue specificity. In particular, older embryos reveal a consistent reporter gene activity in muscles. The strongest expression was observed in a third chromosome insertion (M6), which we used for all experiments shown here.
Immunohistological Staining of Embryos
Eggs laid by flies of the appropriate genetic constitution were collected on agar–apple juice plates. Eggs were collected over an appropriate course of time to obtain an age distribution enabling the visualization of different stages of muscle development. Eggs were dechorionated, permeabilized, and fixed essentially as described by Leiss et al. (1988). After washing and blocking in BBT (0.15% crystalline BSA, 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 40 mM MgCl2, 20 mM glucose, 50 mM sucrose, 0.1% Tween 20)— alternatively we used PBT buffer—the eggs were incubated overnight with a dilution of the appropriate antibody. For staining of mesodermal derivatives, the anti–β3 tubulin antibody was used (Leiss et al., 1988). The bound antibody was detected with a biotinylated secondary antibody and stained with the Vectastain ABC Elite-kit (Vector Labs) using diaminobenzidine as detection agent. Double stainings were performed as described by Lawrence et al. (1987). The stained embryos were embedded in Epon, and photos (Ektar 25, Ektachrome 64T, or Ektachrome 160T film; Kodak, Rochester, NY) were taken under Nomarski optics with a Zeiss Axiophot microscope (Jena, Germany).
Plasmid Rescue and Chromosomal Walking to Isolate the rost Genomic Region
Plasmid rescue was carried out using standard molecular biology methods. DNA from 50 flies with the genotype rostP20/CyO was cut with appropriate restriction enzymes, ligated overnight, and transformed to commercially available high efficiency ultra competent cells (DH5 α, BRL). DNA from single colonies were tested with different restriction enzymes to identify overlapping clones. A Canton-S genomic lambda gt11 library was screened using an EcoRV fragment from the plasmid rescue procedure. Several overlapping phages were isolated and used for further screenings. Overall, we pulled out phage clones encompassing ∼31 kb. Different DNA probes spanning this region were used to screen a 0–16-h random primed embryonic cDNA library, kindly provided by Bernd Hovemann (Bochum, Germany). Several cDNAs, coding for three different transcription units, were identified and characterized in detail.
Subcloning and Sequencing
The cDNA inserts were subcloned using the EcoRI cloning site into the Bluescript plasmid vector (Stratagene) or amplified by PCR, using primers deduced from the lacZ gene sequence, flanking the insertion site of lambda gt11, as the EcoRI sites used to establish the library were not always intact. The cDNA sequence as well as genomic sequences were determined by sequencing both strands of overlapping clones using the dideoxy chain-termination method (T7 sequencing kit; Pharmacia LKB Biotechnology, Piscataway, NJ). Exon–intron boundaries were analyzed by sequence comparison of cDNA and genomic clones.
Rapid Amplification of cDNA Ends (RACE)
The RACE amplification was performed mainly following the procedure of Frohmann (1990), with some modifications. 1 mg poly (A)+ RNA was randomly primed and reverse transcribed using 10 U of SuperscriptII Reverse Transcriptase (BRL). After polyadenylation with Terminal Transferase, 5′ ends of the cDNA were preamplified in a PCR reaction for 15 cycles using a polyT VN adapter primer (GACTCGAGTCGACATCGA TTTTTTTTTTTTTTTVN) and a gene-specific primer. In a second PCR reaction, an aliquot of the preamplified cDNA was further amplified for 30 cycles using the adapter primer (GACTCGAGTCGACATCGA) and a second nested gene-specific primer. The gene-specific primers were derived from the putative 5′ region of the identified rost sequence.
RNA Preparation and Northern Blots
Developmental Northern blots were prepared with ∼10 μg of staged poly(A)+ RNA per lane. RNA was fractionated on formaldehyde-agarose gels and transferred to Hybond N (Amersham Corp., Arlington Heights, IL) by capillary blotting. Following standard protocols, the hybridization was carried out at 42°C in 5× SSC and 50% formaldehyde and 5× Denhardt's solution. After washing in 2× and 1× SSC for at least 1 h, the filter was exposed for ∼1–2 wk.
DNA Preparation and Southern Hybridization, Polytene Chromosome Hybridization
These general methods for molecular biology were performed according to Sambrook et al. (1989).
Whole Mount In Situ Hybridizations
Whole mount in situ hybridization of embryos were carried out as described by Tautz and Pfeifle (1989) with some minor modification. In situ hybridization of dissected ovaries were done following the procedure of Montell et al. (1992).
Antibody Production, Protein Preparations, Isolation of Membrane Fractions and Western Blot Analysis
To produce an antibody against Rost, we took advantage from the hydrophilic structure of the NH2-terminal part of the protein to synthesize a peptide. The peptide with the sequence K S F N K E L Q R A N F G F C corresponds to the NH2 terminus of the deduced Rost protein (amino acids 9–22; see Fig. 2) and was synthesized including an additional Cys at the COOH-terminal end, coupled to KLH and injected into rabbits using the commercial service of Eurogentec (Belgium). The serum was purified using an affinity chromatographic column (Affigel).
Figure 2.

Nucleotide and predicted amino acid sequence of the rolling stone transcription unit. Three different overlapping cDNAs encompassing 1,212 bp were selected for sequencing. The longest cDNA that we have isolated so far did not contain the putative start codon. Translated into an amino acid, this cDNA is missing the first two amino acids shown in the figure. Therefore, we completed the open reading frame by sequencing genomic clones using synthetic oligonucleotides. The genomic sequence (from nucleotide 1 to 399) shown in this figure begins with a SacI restriction site. This site is identical to the SacI site left from the P-element insertion site shown in Fig. 1 A. The sequence between nucleotide 400 and 1612 is based on cDNAs. The first methionine of the deduced Rost amino acid sequence is numbered 1. The positions of the three introns were marked with symbols (T). The P-element insertion is labeled with an asterisk. These sequence data are available from GenBank/EMBL/DDBJ under accession number AF006955.
For characterization of the Rost protein, 5 g of staged embryos (8–12 h) were collected, homogenized in 10 mM Tris-HCl, pH 8, 1 mM EDTA, 20 μg/ml PMSF, and 1 μg protease inhibitor mix (Antipain), and centrifuged at 300 g twice for 10 min each to remove debris. The supernatant was homogenized again and centrifuged twice for 10 min at 4,000 g to remove further cellular debris. The remaining supernatant was applied to a sucrose step gradient (2 M sucrose, 1.4 M sucrose, and 0.25 M sucrose in the buffer used for homogenization). The gradient was centrifuged for 1 h at 100,000 g, and the pellet was resuspended in 100 μl PBS including 20 μg/ ml PMSF. OD280 was adjusted to 15–20 OD280/ml. 10 μl per slot were applied to Laemmli gels. A further purification was applied by centrifugation for 1 h at 100,000 g through a 0.25 M sucrose cushion in 0.1 M Na2CO3, pH 11. The pellet was again resuspended and adjusted at a concentration described above. However, staining the gel after blotting with Coomassie blue (see lanes 1* and 2* in Fig. 1 D) shows that lane 1 contained about 1/10 of protein loaded in lane 2. The preparation of membrane proteins described above follows the method described by Hortsch (1994) with minor modifications.
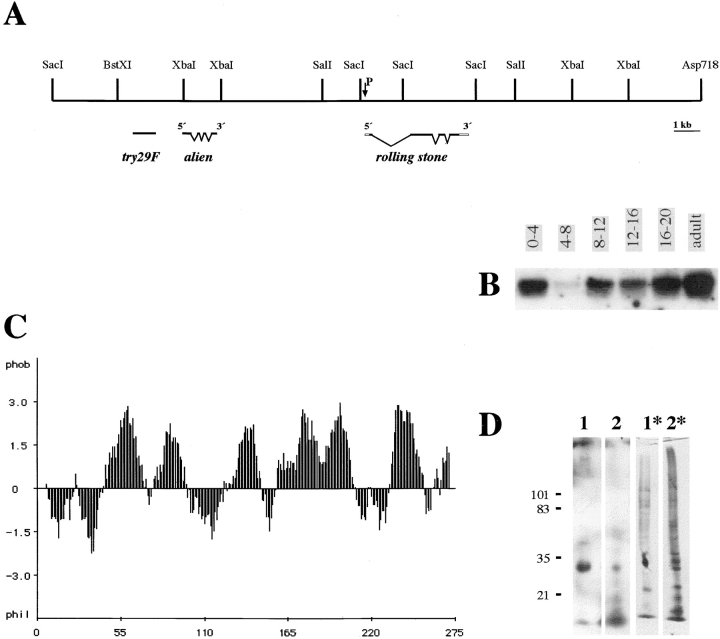
Figure 1.
(A) Molecular map of the rolling stone locus. The horizontal line represents ∼31 kb genomic DNA, which we isolated by a combination of plasmid rescue and chromosome walking. The insertion site of the P-element in rostP20 is marked by an arrow. Overlapping phage clones were used as probes for screening of cDNA libraries, and isolated cDNA clones were mapped on the genomic DNA by hybridization, restriction analysis, and sequencing. Below the genomic map, the locations of the isolated transcription units are diagrammed with closed boxes representing coding regions. The P-element is located 248 bp upstream of the putative ATG of rost gene. alien lies at about 6–7 kb upstream of the P-element insertion site in the orientation as it is assigned (Goubeaud et al., 1996). try29F (Paululat, 1996) lies upstream of alien. Positions of the introns of rost were determined by sequencing of the corresponding genomic DNA. (B) Northern blot analysis of the rolling stone transcript. A rost cDNA clone hybridized to poly(A)+-RNA reveals one single transcript of ∼1.4 kb. Note the strong signal with RNA from 0–4-h-old embryos. RNA extracts from 4–8 h-old embryos reveal a very weak signal. Beyond 8–12 h in embryonic development, the expression of rost persists on a higher level. The equal amount of RNA in each lane was controlled with Rp49 (O'Connell and Rosbash, 1984; data not shown). (C) Hydrophobicity analysis of the putative Rost protein. Six or seven possible transmembrane domains are notable. The first part of the deduced protein reveals a hydrophilic stretch of amino acids. From this part of the Rost protein, we selected 15 amino acids to generate a synthetic peptide that was used to produce antibodies. (D) Accumulation of Rost protein in membrane-enriched embryonic protein extracts. Western analysis of embryonic protein extracts isolated from 4–26-h-old embryos following a protocol for isolation of membrane proteins (lane 1) and a crude, unpurified protein extract (lane 2). Lane 1 was loaded with ∼1/10 of the protein amount compared to lane 2, which we verified by staining the gel with Coomassie blue after blotting (lanes 1* and 2*).
The SDS-PAGE was performed according to the method of Laemmli (1970). After blotting, the membrane was washed three times with PBT and blocked with PBT containing 5% BSA for 1 h at room temperature and then washed again three times with PBT.
Incubation with the peptide antibody (diluted 1:100) was done overnight at 4°C with agitation. The secondary antibody was an anti-rabbit IgG peroxidase conjugate (Sigma Chemical Co., St. Louis, MO) diluted 1:5,000. The staining was done with ECL detection reagents (Amersham Corp.).
Results
The rolling stone Mutant Phenotype
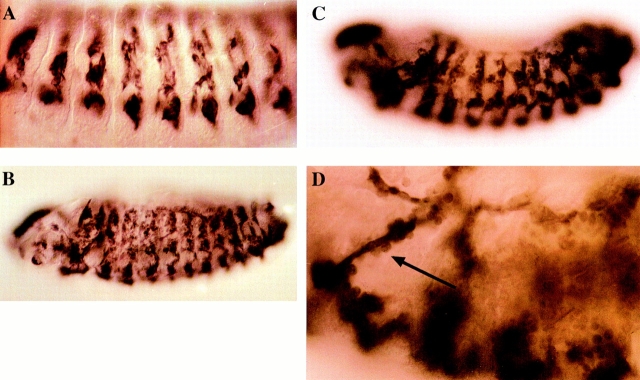
We aim to characterize components that are essential for the development of the somatic musculature of Drosophila melanogaster. Previously, we described the mutant rost in a genetic analysis (Paululat et al., 1995). Homozygous rost mutants show a severe reduction of the body wall musculature because of the inability of many myoblasts to fuse to myotubes. The defects during myogenesis cover the majority of the somatic muscles in head, thoracic, and abdominal muscles. The pharyngeal muscles, consisting of a very regular row of myotubes, each of which normally originates from the fusion of three or four cells, also shows the fusion defect (not shown). Besides structural proteins like Tubulin and MHC, muscle-specific transcription factors like Mef2 are also expressed within the unfused myoblasts (data not shown). Heart and visceral mesoderm develop normally. Interestingly, some somatic muscles, e.g., muscles of the ventral longitudinal group, are often not affected in either any of the analyzed EMS alleles or the P-element–induced allele (Paululat et al., 1995). This indicates that some precursors must use other genes for the fusion process (see Discussion). We want to mention that the severity of the fusion phenotype in the strongest rost alleles that we isolated are weaker than the fusion defects in mbc mutants. For instance, the number of properly formed myofibers is higher in rost then in mbc; we will discuss this point later.
The genetic and cytological characterization of rost mutants showed that this gene is essential for most myoblasts to enter fusion. Analysis of the epidermis, the central and peripheral nervous system, and the outgrowth of motoneurons reveals no detectable developmental abnormalities of these tissues as far as it is detectable with the used markers (Paululat et al., 1995). Moreover, the early muscle precursors are also properly formed. This was examined previously using nautilus as a marker for some of the well-characterized precursors (Paululat et al., 1995) and S59, vestigial (we thank Emma Rushton, Cambridge, UK, for testing S59 and vestigial expression), and even-skipped, which stains the muscle 1 precursor (Paululat, A., unpublished data). Later in embryogenesis, elongated unfused myoblasts are visible. These cells are the remaining founder cells, as was shown previously for the mbc mutant (Rushton et al., 1995), which exhibits a phenotype similar to that found in rost. Interestingly, the motoneurons in the rost mutant seem to have contact to elongated unfused myoblasts, as well as to some rounded myoblasts from the postulated fusion-competent cell pool, but we have not proven that this contact has a functional relevance.
A P-element Insertion at 29F/30A Causes the rolling stone Phenotype in the rost P20 Allele
In situ hybridizations to polytene chromosomes revealed that a single P-element maps to 29F/30A on the left arm of the second chromosome (data not shown). To verify that the P-element localized to this chromosomal region is indeed responsible for the rost phenotype, we mobilized the P-element from this strain. Excision of the P-element reverted the lethal phenotype to vitality; staining with the β3 tubulin antibody furthermore revealed that the muscles develop properly in these revertants (data not shown). We performed Southern blot experiments to prove that a single P-element at 29F/30A indeed causes the rost phenotype and not another smaller P-element that we did not detect with the in situ hybridization to polytene chromosomes. For the excision of the P-element, both inverted repeats have to be present. Therefore, genomic Southern blots with a small probe (500 bp), including the inverted repeat, give only a single signal in the fly strain carrying the rost mutation, showing that only one intact P-element with inverted repeats is integrated in the genome.
Isolation of the rolling stone Gene by Plasmid Rescue and Genomic Walking
As a first step to analyze the function of the rost gene, we cloned and sequenced the gene. The original rost mutant was generated by a P-element mutagenesis, with the P-element vector pUCsneo, which can be excised from the P-strain and transformed into bacterial hosts (Steller and Pirrotta, 1985; Cooley et al., 1988). Digestion of genomic DNA from rost P20/CyO flies with SalI and subsequent ligation and transformation resulted in clones comprising genomic DNA 3′ of the P-element insertion, whereas digestion with XhoI leads to the isolation of 5′ sequences. EcoRV does not cut in pUCsneo and was used to isolate 1.9 kb of genomic sequences flanking the insertion on both sides. The genomic sequences from the EcoRV rescue clone were taken to recover a larger genomic region of 31 kb from a genomic DNA library. We mapped the pUCsneo insertion site by Southern hybridization and sequence comparison of the rescued DNA clones with genomic clones (Fig. 1 A). To prove that the derived genomic clones are in fact from the P-insertion site of the rost P20/CyO flies, we performed in situ hybridizations to polytene chromosomes with genomic clones as well as with the pUCsneo containing rescue clones. Both probes hybridize to the same chromosomal position 29F/30A.
Transcription Units Localized in the Cloned Genomic Region
Genomic clones, spanning ∼15 kb to the left and 15 kb to the right of the P-element insertion, were used as probes to isolate cDNAs from a 0–16-h embryonic library. Three different transcripts were located within this region (Fig. 1 A). The 5′ end of one transcription unit is placed in close proximity to the P-element (see below) and thus was favored as corresponding to the rost gene. Several overlapping cDNAs of the presumptive rost transcription unit were isolated from a randomly primed 0–16-h embryonic cDNA library, kindly provided by B. Hovemann. By combining the overlapping cDNAs, 1,212 base pairs of the transcript are available. Additional 5′ sequences were isolated by using the 5′ RACE technique. Using cDNAs as probes, we found a single transcript of 1.4 kb (Fig. 1 B) in Northern hybridization experiments. As we will show below, the P-element in rost P20 is located in the leader sequence of the presumptive rost gene.
The second identified gene maps 12 kb to the left of the P-element insertion. Its expression is restricted to the posterior midgut during late embryogenesis. This gene, which we named try29F, encodes a protein with homology to trypsin-like proteins. This feature placed try29F very likely into the trypsin gene family (Paululat, 1996). In addition, with the expression pattern it can be excluded that try29F is involved in muscle formation.
The third identified gene, called alien, maps at about 6–7 kb to the left of the P-element insertion site and is expressed in muscle attachment sites during embryogenesis. The embryonic Alien protein distribution, detected using an antibody against Alien, is unaffected in homozygous rost P20 embryos and thus not responsible for the rost phenotype (Goubeaud et al., 1996).
rolling stone Encodes a Putative Transmembrane Protein
We sequenced the putative rost gene and determined the open reading frame that reveals the capacity to code for a 275–amino acid protein (Fig. 2). Database search with the deduced amino acid sequence revealed no significant homologies to known proteins. The extremely hydrophobic structure, including several putative transmembrane domains and a hydrophilic NH2 terminus (Fig. 1 C), indicates that Rost could be a membrane-spanning protein. This structure may explain the failure to express rost in Escherichia coli to generate an antibody against a bacterial Rost fusion protein. Based on the deduced Rost amino acid sequence and the hydrophobicity analysis (Fig. 1 C), we used a synthetic peptide from the NH2-terminal part to generate an antibody (see Materials and Methods). Western blots with embryonic protein extracts revealed that the Rost antiserum recognizes a single protein. Therefore, we used this serum to prove our suggestion that rost encodes an integral membrane protein. Western blots using whole embryonic protein extracts and protein samples enriched for membrane fractions (see Materials and Methods) show that Rost protein is highly concentrated in the membrane protein extract (compare lanes 1 and 2 with lanes 1* and 2* in Fig. 1 D). This result demonstrates clearly that Rost is a membrane protein or at least a membrane-bound or -associated protein.
Mutations in rolling stone Cause the Myoblast Fusion Defect
To prove that the described gene is indeed responsible for the observed fusion defects in rost mutants, we localized the P-element insertion site in rost P20 by sequencing of the genomic rescue clone that we used for isolation of rost. The exact position of the P-element insertion is marked in Fig. 2. Using 5′ race techniques to amplify 5′ sequences of rost, we were able to isolate a rost leader sequence that includes the P-element insertion position. Therefore, rost is indeed needed for myoblast fusion, as the P-element is located in the 5′ untranslated leader region of rost and responsible for the fusion phenotype. Together with our antisense strategy (see below) and the observed results in that assay, we argue that the presented gene is indeed responsible for the rost phenotype.
The rolling stone Gene Is Expressed in Ectodermal and Mesodermal Cells
Developmental Northern blot experiments showed that the transcript is present in 0–4-h-old embryos (Fig. 1 B). A higher level of expression is furthermore visible between 8–12 h and persists during late embryonic development. Only a weak signal is visible in RNA isolated from 4–8-h-old embryos, which may be a hint that the early rost mRNA (0–4 h) is a maternal component. Furthermore, rost is expressed in adult tissues (see Fig. 1 B). In comparison to the expression level of Rp49 (O'Connell and Rosbash 1984), which codes for a ribosomal subunit and serves as a control for comparable amounts of loaded RNA, the level of rost mRNA is extremely low (data not shown). To determine the distribution and localization of rost transcripts at the cellular level, we performed whole mount in situ hybridization experiments using rost cDNAs as a probe. Fig. 3 shows the dynamic expression pattern during oogenesis (Fig. 3 A) and embryogenesis (Fig. 3, B–I). Maternal RNA is detectable in nurse cells at a high expression level. This result corresponds very well with the hybridization signal found in the Northern blot using RNA from 0–4-h embryos and from adults. Likely the signal is caused by the ovarian expression. Zygotic rost transcripts are first detected at the beginning of germband retraction (Fig. 3, B–D) in segmental repeated clusters of cells close to the midline (Fig. 3, B and C, arrows) and in cell groups and rows in lateral positions (Fig. 3 E). Further staining is visible in the head region. During late germband retraction (stage 13), expression is detected in segmental arranged groups of cells in dorsal, lateral, and ventral positions (Fig. 3, E and I, show the same embryo at higher magnification). These cells are very likely mesodermal cells (see Discussion). In a ventral view of a stage 13 embryo at higher magnification (Fig. 3 G), a staining in the central nervous system (CNS) is visible as well. These cells are located at the midline, one row of cells in a medial position and one row of cells lateral. Shortly thereafter the expression in the mesodermal tissue fades out rapidly but remains visible in the CNS (Fig. 3 H). Later on, the expression of rost becomes restricted to some cells of the CNS, the brain, and the gonads (Fig. 3 I, arrow).
Figure 3.
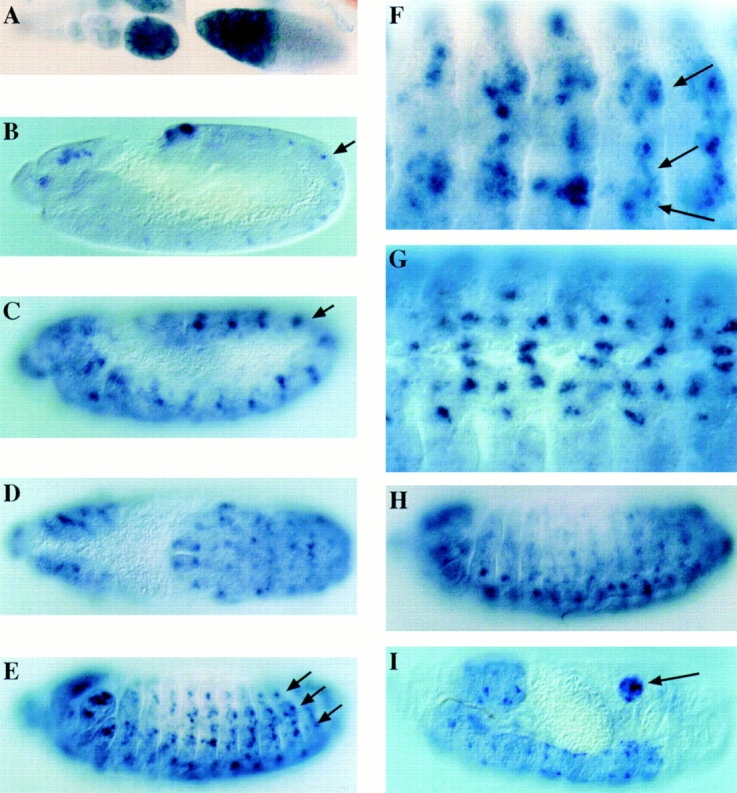
Localization of rost transcripts in wild-type embryos. The spatial distribution of rost transcripts were determined by whole mount in situ hybridization with a digoxygenin-labeled cDNA probe as described by Tautz and Pfeifle (1989). Embryonic stages were determined according to Campos-Ortega and Hartenstein (1985). Embryos are oriented so that dorsal is to the top, ventral to the bottom, anterior to the left, and posterior to the right. (A) Expression of rost in the nurse cells of dissected ovaries. (B) A lateral view of an early stage 12 embryo at the beginning germband retraction. Rows of cells expressing rost are visible in a segmental repeated pattern (arrow). Cells in the lateral and dorsal head region are labeled, too. (C) A stage 12 embryo reveals a specific rost staining in small groups of cells in every segment (arrow). In comparison to B, the number of rost-expressing cells increases. (D) A dorsal view of a stage 12 embryo demonstrates that rost-expressing cell clusters appear close to the midline and in more lateral regions. (E) At stage 13, when the germband retraction is nearly completed, many segmental arranged cells in dorsal, lateral, and ventral positions (arrows) express rost. At higher magnification (F), the rost-expressing cells appear clustered (arrows). At this stage, the fusion of myoblasts is nearly complete, and muscle fibers form tightly packed cell masses. The rost-expressing cell clusters prefigure the positions of the developing muscle fibers perfectly. (G) A comparable embryo in a ventral view. An additional expression of rost is seen in a high number of cells within the CNS. (H) After stage 14, the mesodermal rost expression fades out rapidly. (I) At stage 16/17, the expression of rost becomes restricted to some cells of the CNS, the brain, and the gonads (arrow).
The Developmental Expression of a rost–lacZ Fusion Gene under the Control of a Short rolling stone Promoter Fragment
Because the in situ hybridization method did not allow us to follow rost expression as a marker for the terminal cell phenotype, we used 5′ genomic sequences upstream of the putative ATG codon and fused them to lacZ gene in a P-element vector with the idea that the cytoplasmic antigen expressed under the control of rost regulatory elements could be followed into later stages and with higher resolution. With antibodies against β-galactosidase, the reporter protein is first seen at stage 10–11 in a pattern similar to the results observed with the rost cDNA as a probe for in situ hybridization (Fig. 4, A and B). The pattern reflects the segmental arrangement of muscle founders in the somatic mesoderm. At higher magnification, these cells reveal the very typical shape of muscle founders (Fig. 4 D), as was described by Bate (1990). The first and strongest expression of the reporter is seen in the dorsal somatic mesoderm, where the most dorsal muscles will develop (Fig. 4 B). In a focus that shows the overlaying tissue, a number of smaller cells show a weaker reporter gene expression (Fig. 4 C). By position and the time, when this expression is visible, these cells are probably myoblasts from the fusion-competent cell pool. At later stages, cells in dorsal, lateral, and ventral positions express the reporter gene (Fig. 4 E). At higher magnification, it becomes visible that the reporter gene is expressed in growing muscle fibers at the time the fusion occurs. The remaining founder area, which contains the highest reporter protein concentration, is still obvious in the syncytial myofibers (Fig. 4 F). At stage 15–16, single myotubes can be identified, e.g., the segmental border muscle (muscle 8) and two of the three ventral oblique muscles group (muscle 15–17), as well as some dorsal and dorsal-lateral muscles (Fig. 4, G and H). The muscles mentioned above are affected in strong rost alleles (see Paululat et al., 1995). In comparison to the observed results with the rost cDNA used in the in situ hybridization experiment, the reporter gene expression allows us to identify rost expression in the early mesoderm and to follow the expression into mature muscle fibers. It should be noted that the short rost promoter drives the reporter gene only in some of the 30 muscle founders per hemisegment. The in situ hybridization data obtained with a cDNA probe indicates an expression of rost in a higher number of muscle founder cells. Obviously, additional regulatory elements are needed to give rise to the complete mesodermal rost expression pattern.
Figure 4.
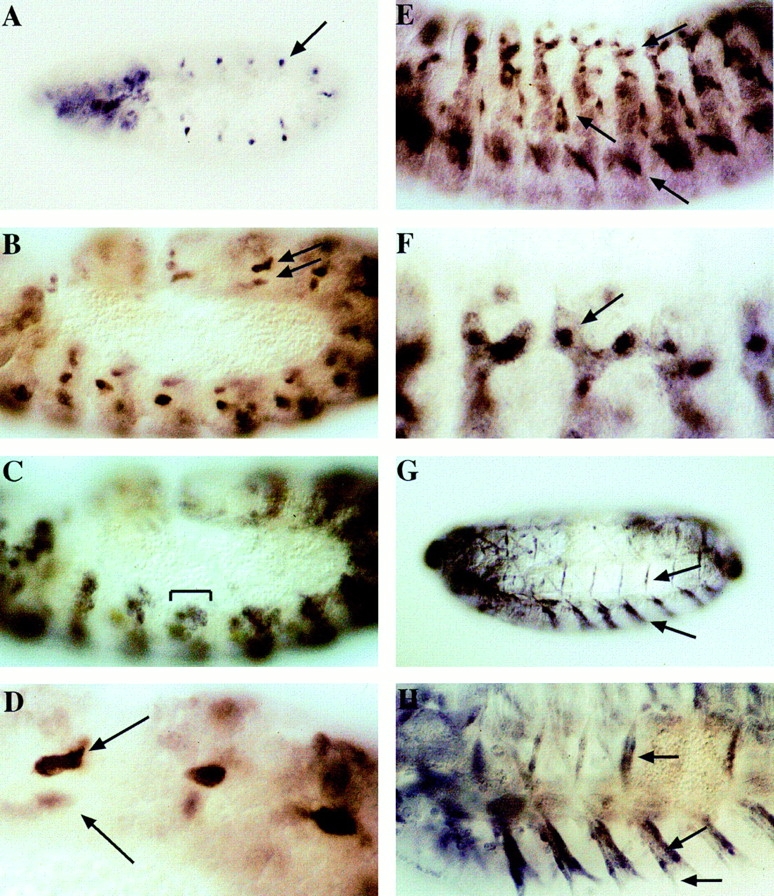
Developmental expression of a rost–lacZ fusion gene under the control of a rost promoter. All stainings were done using an antibody against β-galactosidase. (A) The reporter gene product is first seen at the extended germband stage in single cells in the thoracic and abdominal segments (arrow). (B) Shortly thereafter, the number of rost–lacZ–expressing cells increases. The pattern reflects exactly the segmental arrangement of dorsal muscle founders in the mesoderm (arrows). (C) The same embryo as in B, but focused on the immediately neighboring tissue. A group of smaller cells (bar) that accompanied the underlying larger rost positive cell show reporter gene expression. By the position and time where this expression is visible, these cells are probably myoblasts from the fusion-competent cell pool. (D) rost-expressing founder cells at higher magnification (arrows). These cells reveal the very typical shape of muscle founders as it was described by Bate (1990). (E) At stage 13, reporter gene expression is detectable in growing muscle fibers in dorsal, lateral, and ventral positions (arrows). This is much better seen at a higher magnification (F) of the dorsal region of the same embryo. The fusion of myoblasts to build a muscle fiber has occurred, and the remaining founder area, which contains the highest reporter protein concentration, is still obvious in the syncytial myofibers (arrow). (G) When the mature muscle system is developed, reporter gene activity becomes restricted to some single myotubes, which can be identified. The stage 16 embryos shown in G and H reveal the strongest reporter expression in some ventral muscles (arrow), muscle 8 (arrow), and a weaker staining in a few lateral and dorsal muscles. (H) Stage 16 embryo at higher magnification. Muscle 8 and two ventral muscles are labeled with arrows.
rost expression in the CNS and the gonads, seen with the cDNA probe, is not visible with the lacZ reporter gene. This may be due to the short rost regulatory sequence that we used for the reporter construct. Other regulatory elements in the upstream region or enhancer elements may be necessary for the CNS expression.
Expression of Antisense rost RNA within the Mesoderm Results in Defects in the Fusion of Myoblasts to Myotubes
We used two different approaches to prove finally whether the cloned rost gene is essential for myoblast fusion. First, we tried to rescue the mutant phenotype by introducing 11 kb of genomic DNA, including the wild-type rost gene, into the homozygous mutant background using germline transformation. Approximately 3.9 kb upstream and 4 kb downstream sequences flanking the rost gene were cloned into the pCaSpeR4 transformation vector. None of the four established lines exhibit a complete rescue into adult viability. To look for a partial embryonic rescue, we tested the four independent rost rescue lines using the β3 tubulin antibody to visualize muscle development. Few embryos are detectable showing a weaker fusion defect than the rost 23 allele that was used for the injection to establish the transgenic stocks. Thus, we conclude that 3.9 kb upstream sequences are not sufficient to achieve a high level of rost expression necessary for a significant rescue.
Our second approach to prove that rost is essential for myoblast fusion was to reduce the level of embryonic Rost protein using an antisense strategy with the Gal4-UAS system, developed by Brand and Perrimon (1993). A 1.0-kb cDNA fragment that contains nearly the complete coding region was cloned in antisense orientation into the pUAST transformation vector. Four independent transgenic fly lines were established containing the UAS-antisense-rost construct. To drive the transcription of the UAS-antisense-rost construct specifically in mesoderm, we used the twi-Gal4 driver line in which Gal4 is expressed under the control of the twist promoter (kindly provided by Dr. A. Michelson). Crossings between the independent UAS-antisense-rost flies and the driver lines were analyzed using the β3 tubulin antibody to monitor embryonic muscle development. Fig. 5, C and D, shows representative embryos from crossings of two different UAS-antisense-rost lines with the twi-Gal4 line. The specific phenotype of these embryos corresponds very well to the phenotype of the rost mutant (Fig. 5 B). Unfused myoblasts are arranged in groups at sites where, in wild-type embryos, the muscles are formed (Fig. 5 C). Some myoblasts have contact to elongated myotubes, but they remain unfused (Fig. 5 D). The twi-Gal4 line alone exhibits no muscle abnormalities (data not shown). This result demonstrates very clearly that the identified gene is essential in mesodermal differentiation. Nevertheless, we observed some differences between the twi-Gal4/anti-rost embryos and the homozygous rost embryos (Paululat et al., 1995). The muscles which are affected in these kinds of experiments are mainly some of the lateral longitudinals, the lateral oblique muscles, and some of the more ventral muscles, whereas in rost alleles (e.g., rost 23; Paululat et al., 1995), the majority of muscles are affected. Overall, the twi-Gal4/ antisense-rost embryos reveal a weaker phenotype than the rost alleles. An explanation for the differences could be that the twi-Gal4 line does not express Gal4 uniformly in the mesoderm during that time at which rost is expressed, or that the Gal4 expression level is not high enough in every muscle. Nevertheless, the rost gene, which encodes a presumptive transmembrane protein, is essential for myoblast fusion during embryogenesis.
Figure 5.
Expression of antisense rolling stone RNA within the mesoderm results in a fusion defect of myoblasts. (A) A wild-type embryo at stage 13/14. The fusion of myoblasts into myotubes is nearly completed. (B) A rost 23 embryo at stage 14 that reveals a high number of unfused myoblasts. (C) A typical embryo at stage 14/15 from a crossing between the driver line twi-Gal4 and one of our UAS-antisense-rost (line 14.3.4.4) lines. In wild-type, the muscles are well formed at this stage. Note the irregularities in muscle arrangement and the many unfused myoblasts that are located in groups. The embryo shown here exhibits virtually the same phenotype as was shown for the rost EMS- and P-element–induced alleles (Paululat et al., 1995). (D) At higher magnification, an antisense embryo demonstrates the unfused myoblasts together with some myotubes. Interestingly, the single myoblasts are attached to the myotube (arrow). The embryo shown here is the result of a crossing between twi-Gal4 and UAS antisense-rost (line 10.8.7.1). All embryos were stained by an antibody against β3 tubulin, which we used as a marker for muscle development (Leiss et al., 1988; Buttgereit et al., 1996).
Discussion
The Drosophila body wall muscles are morphologically well characterized. In the abdominal segments A2–A7, 30 muscles per hemisegment form a characteristic pattern, and the other segments show some modifications of this basic pattern (Crossley, 1978; Campos-Ortega and Hartenstein, 1985; Bate, 1990). Bate (1990) proposed that individual muscle founder cells exist that recruit other myoblasts to fuse to the individual founders, leading first to muscle precursors and second, after addition of further myoblasts, to myotubes. The analysis of genes that act directly in myoblast fusion is important to understand the cellular process that results in mature myofibers. The electron microscopic study of Doberstein et al. (1997) is the first attempt to elucidate the fusion process on the cellular level. By taking advantage of the existing fusion mutants in comparison to the wild-type situation, the authors were able to separate the fusion process into different essential steps, which include recognition and adhesion of myoblasts, the formation of a prefusion complex, plaque formation along the membranes of attached cells, cell alignment and membrane apposition, and plasma membrane breakdown. In this view, it is absolutely necessary to isolate genes that are highly specifically involved in the described cellular events for understanding myoblast fusion during development. So far, only a limited number of such genes or mutants acting in this developmental step were described: mbc (Rushton et al., 1995), rost (Paululat et al., 1995; this study), blow (Doberstein et al., 1997), and at least one more on the X-chromosome (Drysdale et al., 1993). Besides blow, which encodes a novel cytoplasmic myoblast component, rost is the only one that was cloned so far, and the present data may be of particular interest with respect to membrane turnover during the fusion process.
rolling stone Is Necessary within the Mesoderm to Promote Fusion
The rost phenotype raises the question at which cellular level, e.g., in all myoblasts or only in muscle founders, Rost is essential and at which subcellular level the gene product acts. By cloning and sequence analysis, the deduced open reading frame of the rost gene was determined. The computer-supported sequence analysis revealed no homology to known gene products. The structural prediction, however, implicates a role of Rost in cell–cell recognition, cell communication, or membrane turnover, as the Rost protein is very hydrophobic (Fig. 1 C) and most likely a membrane-bound or membrane-passing protein. This view is supported by our finding that Rost can be isolated by extracting membrane-enriched embryonic protein fractions (Fig. 1 D). Thus, the Rost protein could fulfill its biological action on the side of the muscle founder cells, by mediating a signal, or its biological function might be on the side of fusion-competent myoblasts, as Rost may receive a signal that enables the recognition of the founder cell and starts cell fusion. In the case of being a receptor, we would expect that many myoblasts express Rost to be able to receive the signal. If rost is acting on the site of the signal, we would expect that only a limited number of cells are synthesizing this protein. Another possible role of Rost is implicated by the electron microscopic study of Doberstein et al. (1997). One important step in myoblast fusion is the formation of a prefusion complex that is accompanied by a dramatic concentration of vesicles near the cytoplasmic face of each of the juxtaposed plasma membranes. The authors suggest that the vesicles fuse with the plasma membrane, forming an electron microscopic dense plaque. Shortly thereafter, a plasma membrane breakdown results finally into fused syncytial myotubes. In rost, the formation of vesicles as well as the plaques still occurs between the myoblast, but the following membrane breakdown is affected. As Rost is an extremely hydrophobic membrane component, the described cellular phenotype indicates that Rost may play a direct role in membrane turnover during the fusion process. If so, we would be not surprised if rost is expressed in muscle founders as well as in cells from the fusion competent cell pole, so in both cell types rost is needed for the fusion.
The whole mount in situ hybridization shows that the rost gene is expressed in mesodermal cells, which appear in a regular pattern in every segment. The expression level of rost is very low, and transcripts disappear during the fusion process. To identify the rost-expressing cells, we performed a rost-lacZ reporter assay. We found reporter activity early in the mesoderm in single muscle founder and precursor cells. Interestingly, a weaker reporter activity, which comes a bit later, was observed in a tissue that neighbors the precursor. We assume that these cells are mesodermal cells that belong to the “fusion-competent cell” pool, but the exact identification remains to be clarified. Later in embryogenesis, the reporter activity becomes restricted to single muscle fibers, most likely because of the stability of the β-galactosidase, which we detected with an antibody. Only a subset of the 30 muscles per hemisegment shows reporter gene activity, which may be explained by the short promoter element that we used and which may be not long enough to give a complete mesodermal pattern. rost transcripts are not detectable in mature myotubes. Nevertheless, the late staining of the reporter gene allowed us to prove that Rost is indeed in muscles. As β-galactosidase is very stable, this must not be true for the Rost protein. The early expression of rost in single founder cells and shortly thereafter in neighboring cells is more important in the view of the myoblast fusion process. It means that Rost is indeed needed directly in founders just before they undergo the fusion process. In rost mutants, founders are well developed but fail to fuse. A possible explanation that rost activity becomes obvious with a short delay in the surrounding mesodermal cells might be that these cells have to receive a signal from the founder cell to become recruited. The genetic program, which is activated then, may include the expression of rost.
The functional role of rost in the mesoderm is confirmed by our results using the Gal4/UAS system to inhibit the translation of rost with an antisense-rost construct that was driven within the mesoderm. This leads to defects in the fusion process (Fig. 5) and finally in a phenotype that is very similar to the original rost mutant. Most importantly, this experiment shows the function of rost directly in the mesoderm. The postulated membrane localization of Rost and the expression of rost in the mesoderm, showed by the reporter gene assay, and the phenotype analyzed in the EM study of Doberstein et al. (1997) show that Rost is the first protein directly involved in the fusion of myoblasts.
From the data presented in this paper, we propose that Rost is essential in muscle founder cells to fuse with surrounding myoblasts into a syncytium but not for determination of the founders, as we know from the expression of founder cell markers, like even-skipped, etc., in the rost mutant (data not shown). Thus, we propose the idea that rost is expressed and needed in specified muscle founder cells. It is very likely that Rost is localized within the membrane and is involved directly in the fusion with myoblasts from the fusion-competent cell pool.
The fusion phenotypes of the myoblast city mutant (Rushton et al., 1995) and blown fuse (Doberstein et al., 1997) implicate that these genes may act in the same cascade. The fourth known gene involved in myoblast fusion is Drac1 (Luo et al., 1994). The EM analysis of Doberstein et al. (1997) indicates that the rost gene plays a direct role within the fusion process, at a mechanistic position between earlier acting genes like mbc and blow and later acting players like Drac1. The overexpression of a mutated form of this G-protein–encoding gene results in a similar fusion defect and may also be involved in the same cascade. To understand the role of Rost in mesoderm differentiation, it will be important to analyze the cascade of interactions during the fusion of myoblasts with muscle founders. The molecular analysis of the genes mentioned above and their possible interactions will be necessary to elucidate the fusion process. Further characterization of rost may be helpful to understand myogenesis. Because of the evolutionary conservation of this fundamental process, we would not be surprised if homologues of rost were detected in vertebrates and other phyles in the future.
Acknowledgments
We thank T. Orr-Weaver and A. Spradling for sending us P-element– induced lethals and B. Hovemann for the embryonic cDNA library. We thank R. Hyland for excellent technical assistance. We thank A. Brand and N. Perrimon for providing us the pUAST vector and A. Michelson for the twi-Gal4 line. We thank E. Rushton for checking Vestigial and S59 protein expression in the rost 15 allele and G. Korge for his help with polytene chromosome in situ hybridizations.
This work was supported by the Deutsche Forschungsgemeinschaft by grants to R. Renkawitz-Pohl (Re628/7-2 and Re628/7-3) and the Fond der Chemischen Industrie.
Abbreviations used in this paper
- blow
blown fuse
- CNS
central nervous system
- mbc
myoblast city
- MHC
myosin heavy chain
- RACE
rapid amplification of cDNA ends
- rost
rolling stone.
Footnotes
Address all correspondence to Dr. Renate Renkawitz-Pohl, Zoologie-Entwicklungsbiologie, FB Biologie, Philipps-Universität, Karl-von-Frisch Str., 35032 Marburg, FRG. Fax: 49-6421-281538. E-mail: paululat@mailer.uni-marburg.de
References
- Abmayr SM, Erickson MRS, Bour BA. Embryonic development of the larval body wall musculature of Drosophila melanogaster. . Trends Genet. 1995;11:153–159. doi: 10.1016/s0168-9525(00)89030-7. [DOI] [PubMed] [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. . Development (Camb) 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- Bate, M. 1993. The Development of Drosophila melanogaster. Vol. II. M. Bate and A.M. Arias, editors. Cold Spring Habor Laboratory Press, Cold Spring Harbor, NY. 1013–1090.
- Bour BA, O'Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. DrosophilaMEF2, a transcription factor that is essential for myogenesis. Genes Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Bourgouin C, Lundgren SE, Thomas JB. apterous is a DrosophilaLIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9:549–651. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of alternating cell fates and generating dominant phenotypes. Development (Camb) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Burchard S, Paululat A, Hinz U, Renkawitz-Pohl R. The mutant not enough muscles (nem) reveals reduction of the Drosophilaembryonic muscle pattern. J Cell Sci. 1995;108:1443–1454. doi: 10.1242/jcs.108.4.1443. [DOI] [PubMed] [Google Scholar]
- Buttgereit D, Paululat A, Renkawitz-Pohl R. Muscle development and attachment to the epidermis is accompanied by β3 and β1 tubulin isotypes, respectively. Int J Dev Biol. 1996;40:189–196. [PubMed] [Google Scholar]
- Campos-Ortega, J.A., and V. Hartenstein. 1985. The Embryonic Development of Drosophila melanogaster. Springer-Verlag, Berlin. 227 pp.
- Cooley L, Kelly R, Spradling A. Insertional mutagenesis of the Drosophilagenome with single P-elements. Science (Wash DC) 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- Crossley, A.C. 1978. The morphology and development of the Drosophila muscular system. In The Genetics and Development of Drosophila. Vol. 2b. M. Ashburner, and T. Wright, editors. Academic Press, New York. 499–560.
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuseis required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann C, Azpiazu N, Frasch M. A new Drosophilahomeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes Dev. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- Drysdale R, Rushton E, Bate M. Genes required for embryonic muscle development in Drosophila melanogaster. . Roux's Arch Dev Biol. 1993;202:276–295. doi: 10.1007/BF00363217. [DOI] [PubMed] [Google Scholar]
- Frohmann, M.A. 1990. RACE: Rapid amplification of cDNA ends. In PCR Protocols. A Guide to Methods and Applications. M.A. Innis, D.H. Gelfand, J.J. Sninsky, and T.J. White. Academic Press, San Diego, CA. 28–38.
- Goubeaud A, Knirr S, Renkawitz-Pohl R, Paululat A. The Drosophila gene alien is expressed in the muscle attachment sites during embryogenesis and encodes a protein highly conserved between plants, Drosophilaand vertebrates. Mech Dev. 1996;57:59–68. doi: 10.1016/0925-4773(96)00532-1. [DOI] [PubMed] [Google Scholar]
- Hortsch M. Preparation and analysis of membranes and membrane proteins from Drosophila. . Methods Cell Biol. 1994;44:289–301. doi: 10.1016/s0091-679x(08)60920-6. [DOI] [PubMed] [Google Scholar]
- Jimenez F, Modolell J. Neural fate specification in Drosophila. . Curr Opin Genet Dev. 1993;3:626–632. doi: 10.1016/0959-437x(93)90099-b. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R. Cytoplasmic myosin from Drosophila melanogaster. . J Cell Biol. 1986;103:1517–1525. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lawrence PA, Johnston P, MacDonald P, Struhl G. Borders of parasegments in Drosophila embryos are delimited by the fushi tarazu and even-skippedgenes. Nature (Lond) 1987;328:440–442. doi: 10.1038/328440a0. [DOI] [PubMed] [Google Scholar]
- Leiss D, Hinz U, Gasch A, Mertz R, Renkawitz-Pohl R. β3 tubulin expression characterises the differentiating mesodermal germ layer during Drosophilaembryogenesis. Development (Camb) 1988;104:525–531. doi: 10.1242/dev.104.4.525. [DOI] [PubMed] [Google Scholar]
- Lewis JO, Crews ST. Genetic analysis of the Drosophila single-mindedgene reveals a central nervous system influence on muscle development. Mech Dev. 1994;48:81–91. doi: 10.1016/0925-4773(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle lineages during Drosophilaembryogenesis. Proc Natl Acad Sci USA. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. . Science (Wash DC) 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin MH, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosingene muscle expression. Proc Natl Acad Sci USA. 1996;93:4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Liao YJ, Jan LY, Jan YN. Distinct morphogenetic functions of similar small GTPases: DrosophilaDrac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 1994;8:1787–1802. doi: 10.1101/gad.8.15.1787. [DOI] [PubMed] [Google Scholar]
- Michelson AM, Abmayr SM, Bate M, Martinez A, Arias, Maniatis T. Expression of a MyoD family member prefigures muscle pattern in Drosophilaembryos. Genes Dev. 1990;4:2086–2097. doi: 10.1101/gad.4.12a.2086. [DOI] [PubMed] [Google Scholar]
- Montell DJ, Roth P, Spradling AC. slow border cells, a locus required for a developmentally regulated cell migration during oogenesis, encodes DrosophilaC/EBP. Cell. 1992;71:51–62. doi: 10.1016/0092-8674(92)90265-e. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA. D-mef2: A Drosophilamesoderm-specific MADS box-containing gene with a biphasic expression profile during embryogenesis. Proc Natl Acad Sci USA. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell P, Rosbash M. Sequence, structure, and codon preference of the Drosophilaribosomal protein 49 gene. Nucleic Acids Res. 1984;12:5495–5513. doi: 10.1093/nar/12.13.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson BM, Walldorf U, Eldridge J, Dübendorfer A, Frasch M, Gehring W. The Drosophilahomologue of vertebrate myogenic-determination genes encodes a transiently expressed nuclear protein marking primary myogenic cells. Proc Natl Acad Sci USA. 1990;88:3782–3786. doi: 10.1073/pnas.88.9.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paululat A. try29F (Drosophila trypsin-related protein), a new member of the Drosophilatrypsin-like protease gene family, is specifically expressed in the posterior midgut. Gene. 1996;172:245–247. doi: 10.1016/0378-1119(96)00163-1. [DOI] [PubMed] [Google Scholar]
- Paululat A, Burchard S, Renkawitz-Pohl R. Fusion from myoblasts to myotubes is dependent on the rolling stone gene (rost) of Drosophila. . Development (Camb) 1995;121:2611–2620. doi: 10.1242/dev.121.8.2611. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in the larval and adult myogenesis in Drosophila. . Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophilawith transposable element vectors. Science (Wash DC) 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophilamuscle development. Development (Camb) 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophilagerm line chromosomes. Science (Wash DC) 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Steller H, Pirrotta V. A transposable P vector that confers selectable G418 resistance to Drosophilalarvae. EMBO (Eur Mol Biol Organ) J. 1985;4:167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveal translational control of the segmentation gene hunchback. . Chromosoma (Berl) 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Taylor MV, Beatty KE, Hunter HK, Baylies MK. Drosophila MEF2 is regulated by twistand is expressed in both the primordia and differentiated cells of the embryonic somatic, visceral and heart musculature. Mech Dev. 1995;50:29–41. doi: 10.1016/0925-4773(94)00323-f. [DOI] [PubMed] [Google Scholar]