Abstract
We have identified a new member of the kinesin superfamily in Drosophila, KLP38B (kinesin-like protein at 38B). KLP38B was isolated through its two-hybrid interaction with the catalytic subunit of type 1 serine/threonine phosphoprotein phosphatase (PP1). We demonstrate that recombinant KLP38B and PP1 associate in vitro. This is the first demonstration of direct binding of a kinesin-related protein to a regulatory enzyme.
Though most closely related to the Unc-104 subfamily of kinesin-related proteins, KLP38B is expressed only in proliferating cells. KLP38B mutants show cell proliferation defects in many tissues. KLP38B is required for normal chromatin condensation as embryos from KLP38B mutant mothers have undercondensed chromatin at metaphase and anaphase. This is the first time that a kinesin-related protein has been shown to have such a role. Incomplete lethality of a strong KLP38B allele suggests partial redundancy with one or more additional kinesin-related proteins.
Kinesin-related proteins (KRPs)1, also known as kinesin-like proteins (KLPs) or kinesin family proteins (KIFs), form a superfamily of microtubule-based mechanochemical motors. Kinesin itself, the archetypal member of the family, was discovered in squid axoplasm, suggesting a role in axonal transport (Brady, 1985; Vale et al., 1985). As expected for this role, kinesin heavy chain mutants in Drosophila have defective neuromuscular function (Saxton et al., 1991; Gho et al., 1992). Kinesin-related proteins have since been found in a wide range of organisms, including several fungi, Arabidopsis, Drosophila, Xenopus, and mammals, and are known to be involved in such diverse cellular processes as spindle pole separation, chromosome movement, and neuronal vesicle transport (for review see Goldstein, 1993; Bloom and Endow, 1994, 1995; Moore and Endow, 1996).
The question of how many different kinesin-related proteins there might be in a single organism has been addressed by in situ hybridization studies, using a probe generated by PCR amplification with primers to highly conserved sequences in the motor domain (Endow and Hatsumi, 1991; Stewart et al., 1991). This led to the unexpected conclusion that there are at least 30 members of the kinesin superfamily in Drosophila. It now appears that other higher eukaryotes have a similarly large number of kinesin-related proteins. A number of these have now been sequenced, and their specific functions and regulation are the subject of intense investigation.
Kinesin itself and a number of kinesin-related proteins are involved in the movement of membrane-bound organelles and vesicles. However, a large number of kinesin-related proteins have been found to be involved in various aspects of meiosis and mitosis. This is perhaps not surprising in view of the central role of the microtubule spindle in these processes. Known or postulated roles include all aspects of spindle structure and function, microtubule dynamics, chromatin structure and function, and chromosome movement (Vernos et al., 1995; Walczak et al., 1996; Boleti et al., 1996; for review see Goldstein, 1993; Bloom and Endow, 1995; Moore and Endow, 1996).
Kinesin and KRPs share a conserved motor domain of ∼340 amino acids, which defines the superfamily of kinesin-related proteins. Similarities between the motor domains have been used to construct molecular phylogenies of the KRPs (Moore and Endow, 1996). KRPs with similar motor domains often have similar functions, but this is by no means always the case (see Discussion).
Outside of the motor domains the kinesin-related proteins differ from each other, suggesting that the different cellular roles of these proteins depend on the non-motor sequences. Like kinesin heavy chain, many kinesin-related proteins have a central α-helical region. This region is thought to mediate the homodimerization of the kinesin heavy chain in the native tetrameric protein. The “tail” region is thought to bind the “cargo” and/or accessory proteins. Little is known about the proteins that bind to KRPs, although they are clearly critical to force transduction to the “cargo” and probably also to the regulation of the motor.
Kinesin itself is a tetramer, comprising two heavy chains and two light chains. The heavy chains contain the motor domain at the NH2 terminus and the light chains bind toward the COOH terminus. Another protein, kinectin, is thought to bind to kinesin (Toyoshima et al., 1992; Kumar et al., 1995). Molecular cloning and antibody studies suggest that kinectin is an integral membrane protein anchored in the ER (Futterer et al., 1995; Yu et al., 1995), and thus may function as a molecular link between motor and cargo. Very few associated proteins have been identified for kinesin-related proteins; candidates include Cik1 for Kar3 (Page et al., 1994) and a heterotrimeric sea urchin complex consisting of two different kinesin-related proteins and a non-kinesin protein (Cole et al., 1993).
Little is known of the associated proteins; regulation of motility and cargo binding of kinesin-related proteins is correspondingly poorly understood. Several reports have suggested that phosphorylation may play an important role. Both the heavy and light chains of kinesin are phosphorylated, as is kinectin (Hollenbeck, 1993; Lee and Hollenbeck, 1995). Membrane-associated kinesin heavy chain is more highly phosphorylated than soluble kinesin heavy chain, which suggests that membrane association is regulated by phosphorylation. Several different kinases have been implicated in the phosphorylation of kinesin and KRPs. Pharmacological studies have shown that protein kinase A activation selectively inhibits anterograde axonal transport of vesicles, but not mitochondrial transport or retrograde transport (Okada et al., 1995). This activation induces the phosphorylation of several axonal proteins including kinesin. On the other hand, protein kinase A activation stimulates plus end–directed pigment granule movement in fish chromatophores (Rozdzial and Haimo, 1986; Rodionov et al., 1991; Sammak et al., 1992). The Xenopus kinesin-related protein Eg5 contains a predicted cyclin-dependent kinase (cdk) phosphorylation site in the COOH-terminal tail that is required for spindle association and function (Sawin and Mitchison, 1995; Blangy et al., 1995). Another kinase that may regulate a kinesin-related protein is polo-like kinase (Plk). Plk is a murine protein kinase related to the Drosophila polo, Saccharomyces cerevisiae CDC5, and Schizosaccharomyces pombe plo1 protein kinases, and colocalizes with the kinesin-related protein CHO1/MKLP-1, which it can phosphorylate in vitro (Lee et al., 1995).
Even less is known about the protein phosphatases that must act antagonistically to these kinases. Serine/threonine protein phosphatases are classified into four major classes: types 1, 2A, 2B, and 2C (PP1, PP2A, PP2B, PP2C; for review see Cohen, 1989; Bollen and Stalmans, 1992; Shenolikar, 1994). Okadaic acid, a potent inhibitor of PP2A and PP1, stimulates kinesin motor activity at least twofold, apparently through the hyperphosphorylation of kinesin- associated proteins (McIlvain et al., 1994). Subsequent work has shown that maximal inhibition occurs at 500 nM okadaic acid, implicating PP1 rather than PP2A, which is completely inhibited at much lower concentrations of okadaic acid (McIlvain, J.M., Jr., L. Lindesmith, Y. Argon, and M.P. Sheetz. 1994. Mol. Cell. Biol. 5(Suppl.):31a). This shows that PP1 is present in semipurified motor fractions and regulates motor activity.
In this paper we describe the identification of a Drosophila kinesin-related protein, KLP38B, in a screen for proteins that associate with type 1 serine/threonine phosphoprotein phosphatase (PP1) and show that KLP38B binds PP1 in vitro. KLP38B is expressed in all proliferating cells examined, and is required for normal chromosome condensation in mitosis and for male and female fertility. Incomplete lethality of KLP38B mutants implies a degree of functional redundancy, although KLP38B sequence is not closely related to other known Drosophila KRPs.
Materials and Methods
Two-Hybrid System
Two-hybrid screening for proteins capable of binding to PP1 87B was carried out essentially as in Harper et al. (1993). The start codon of a PP1 87B cDNA (Dombrádi et al., 1989; Axton et al., 1990) was modified to an Nde1 site and subcloned into the Nde1 site of pAS2 to create pAS2-PP1. A Drosophila third instar larval cDNA library, constructed in pACT (Durfee et al., 1993), was a gift from Stephen Elledge (Baylor College of Medicine, Houston, TX), together with the yeast strains and plasmids described in Harper et al. (1993). This library was screened for cDNAs that interact with pAS2-PP1 in the yeast strain Y190 (Harper et al., 1993). 25 cDNAs were isolated that are strong HIS + lacZ + positives in combination with pAS2-PP1 but not with control plasmids (pAS-p53, pAS-cdc2, pAS-lamin). Three of these 25 positives are derived from KLP38B.
Sequence Analysis
Sequence of the 3,605-bp cDNA (see Fig. 1) was determined using cycle sequencing with fluorescent terminators and a semiautomated sequencer (ABI 377; Perkin-Elmer Corp., Norwalk, CT), according to the manufacturer's instructions. Synthetic oligonucleotide sequencing primers were supplied by Hobolth DNA Syntese (Hillerød, Denmark). Other KLP38B cDNAs were sequenced by using a combination of Exo III deletions and primer walking. Sequence analysis and searches of sequence databases were performed using the Genetics Computer Group (GCG) programs (Devereux et al., 1984), PAIRCOIL (Berger et al., 1995), and PEPCOIL (Lupas et al., 1991; Lupas, 1996).
Figure 1.
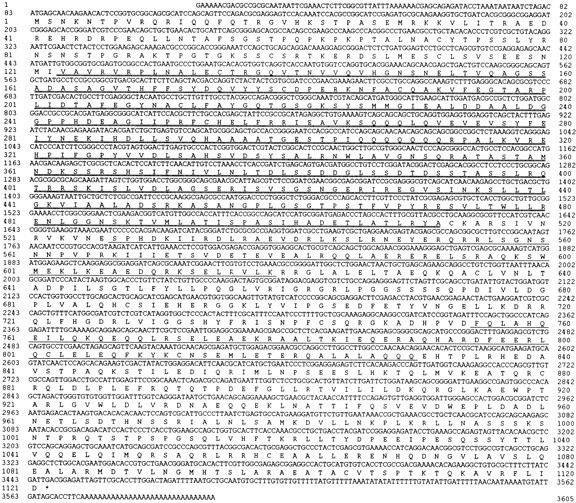
Sequence of the KLP38B cDNA. The sequence of the coding strand of the longest available KLP38B cDNA is depicted with the predicted 1,121– amino acid sequence of its product, based on translation from the first ATG (see Fig. 2 and text). The putative motor domain is shown with a double underline, regions predicted to form coiled-coils by PAIRCOIL with a single underline (see Fig. 2 C). These sequence data are available from EMBL under accession number X99617.
KLP38B cDNA Isolation
cDNAs encoding part of KLP38B were obtained using the two-hybrid system (see above). Longer cDNAs were isolated from the Nick Brown 0–4-h embryonic cDNA library (Brown and Kafatos, 1988) by hybridization, using the longest two-hybrid cDNA as a probe. Two longer cDNAs were isolated, but neither contains the complete KLP38B open reading frame (ORF). Screening a testes cDNA library (kindly provided by T. Hazelrigg, Columbia University, New York) identified another seven cDNAs; again none of them contained the complete KLP38B ORF. Full-length cDNAs were obtained by using a novel PCR-based screening procedure developed for the purpose. XL-2 cells (Stratagene, La Jolla, CA) were transformed with 3 ng of the Nick Brown imaginal disk library. 100 cultures were each inoculated with 500 colony-forming units (CFU) from this transformation. These were grown to mid-log phase. An aliquot from each was stored at −70°C, and the remainder was divided between 10 “row” pools and 10 “column” pools such that each initial culture was represented in a unique pair of pools. DNA was isolated from these pools and used as the template for a PCR reaction between a KLP38B-specific oligo (5′-CCGAGTACGAGTCCACCGAGTGGG-3′) and a vector-specific oligo (5′-GCTCAGAATAAACGCTCAACTTTGGG-3′). The size of the amplified fragments was determined by gel electrophoresis. A 1.1-kb band was observed from a single row pool and a single column pool, unambiguously identifying one of the original 100 cultures as containing a 3.6-kb KLP38B cDNA. This was purified by hybridization with the KLP38B-specific oligo, and the identity of the purified clone was confirmed by PCR. This method is described in detail in Alphey (1997).
KLP38B Cosmid Isolation
A high density gridded filter from the European Drosophila Genome Project representing 19,200 cosmid clones was screened by hybridization with a KLP38B cDNA probe. Construction and screening of this library is described in Siden-Kiamos et al. (1990) and Kafatos et al. (1991). A single positive cosmid, 164E4, was identified and purified.
Identification of KLP38B Mutants
A large-scale screen has identified 2,711 lethal and semilethal P[lacW]- induced mutations on the second chromosome (Török et al., 1993). This modified P element contains a plasmid origin of replication and antibiotic resistance gene, allowing plasmid rescue of DNA flanking the insertion site. Plasmids representing 1,836 lines were independently rescued and pooled in batches of 10 and 100. Pools of 100 plasmids were screened by hybridization with genomic DNA from the KLP38B locus. Hybridizing pools were then narrowed down to single plasmids by screening their constituent subpools and finally individual plasmids, and corresponding mutant lines were obtained. This method is described in detail in Guo et al. (1996). Six pools of 100 plasmids showed cross-hybridization with the KLP38B probe, a 7-kb BamHI/EcoRI fragment from cosmid 164E4 (see Fig. 4). Subdivision of the positive pools identified six insertion lines: l(2)k00802, l(2)k03903, l(2)k04912, l(2)k04914, l(2)k04805, and l(2)k05702. Two additional alleles (KLP38B 24-O and KLP38B 93-E) were gifts from Mike Goldberg (Cornell University, Ithaca, NY). Other stocks were from Bloomington Drosophila Stock Center (Indiana University, Bloomington, IN) and The European Drosophila Stock Center (University of Umea, Umea, Sweden).
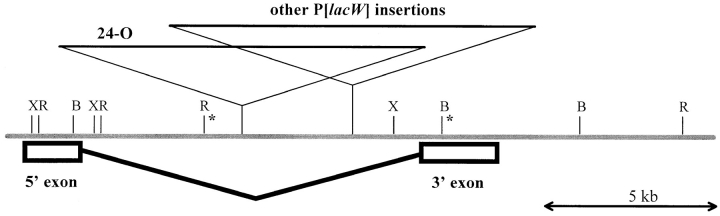
Figure 4.
Molecular map of the KLP38B region. The P[lacW] element insertions in KLP38B mutants are shown in relation to the KLP38B exons and intron. The P[lacW] insertion site of KLP38B 24-O maps ∼3 kb away from the other insertions, which are too close to separate on this scale. Of these other mutants, the insertion site in l(2)k03903 is the most 3′ relative to the KLP38B, while l(2)k04805, l(2)k04912, l(2)k04914, and l(2)k05702 have identical insertion sites 53 bp away and the KLP38B 93-E insertion is another 242 bp further toward the 5′ end of KLP38B. The central gray line represents genomic DNA, the KLP38B exon map is below, and the P element insertion sites are marked above. Asterisks mark the ends of the BamHI–EcoRI fragment used to identify the mutants. B, BamHI; R, EcoRI, X, Xbal.
P[lacW] Reversion
Excision of P[lacW] elements to test for phenotypic reversion was conducted by crossing the P element–bearing lines to the P[Δ2-3]99B transposase source (Robertson et al., 1988; Cooley et al., 1988). Sequences flanking the P element insertions were cloned by plasmid rescue, using the bacterial origin and β-lactamase gene engineered into the P elements (Bier et al., 1989).
Complementation of KLP38B Mutants by a KLP38B cDNA
KLP38B cDNA was excised from the vector pNB40 (Brown and Kafatos, 1988) by digestion with HindIII and NotI. The Drosophila expression vector pW5g26 (Lopez et al., 1994) was linearized with EcoRI. These two molecules were end-filled and ligated together to place the KLP38B cDNA under the control of the hsp26 promoter. This construct was then injected into embryos carrying the P[Δ2-3]99B transposase source (Robertson et al., 1988; Cooley et al., 1988). Transformed progeny of these injected individuals were identified by virtue of the w + marker in pW5g26. A third chromosome insertion was selected. Standard breeding strategies were used to construct flies homozygous or hemizygous for KLP38B mutants and also carrying a single copy of the hsp26-KLP38B transgene. The phenotype of these flies was analyzed in comparison with flies of the same KLP38B genotype but lacking the transgene.
Cytological Preparations
Preparation, fixation, antigen detection, and 4′,6-diamidino-2-phenylindole (DAPI) staining of ovaries, embryos, and larval brains was as described in Gonzalez and Glover (1993). Embryos were incubated in 5 μM taxol for 30 s to stabilize microtubules, as recommended by these authors. The anti-tubulin antibody used was mouse monoclonal TAT-1 (Woods et al., 1989), a gift from Keith Gull (University of Manchester, UK); Texas red– conjugated secondary antibody was purchased from Sigma (Sigma-Aldrich Co., Dorset, UK). RNA in situ hybridization was performed according to Tautz and Pfeifle (1989) and Gonzalez and Glover (1993).
SEM
Adult flies were anesthetized with CO2, mounted on a stub with carbon dag, ventral surface to the stub, and then placed in the low vacuum chamber of an ElectroScan Environmental Scanning Electron Microscope (Cambridge Instruments, Welwyn Garden City, UK). Specimens were observed at 20 kV. Digital images were recorded and manipulated using Adobe PhotoShop (Adobe Systems, Inc., Mountain View, CA).
Microscopy
Fluorescence microscopy was performed using a DM IRBE microscope (Leitz, Wetzlar, Germany). Images were captured using a cooled slow-scan CCD camera (CH250; Photometrics, Ltd., Tucson, AZ), using IP Lab Spectrum software (Signal Analytics, Co., Vienna, VA). This software was also used for pixel counting. Deconvolution was performed using Power Hazebuster software (Vaytek, Inc., Fairfield, IA), and the images were assembled into figures using Adobe Photoshop. Confocal microscopy was performed using a Leitz DM IRBE microscope with Leica TCS 4D confocal system including UV laser.
Protein Expression and In Vitro Binding
A PP1 87B cDNA with the start codon mutagenized to an NdeI site (a gift from David Glover, Dundee University, Dundee, UK) was subcloned into pET-16b (Novagen, Madison, WI). After induction in the expression host cells BLR(DE3) (Novagen), pET-16b-PP1 87B expresses full-length PP1 87B protein with an NH2-terminal fusion encoding His10, allowing affinity purification on immobilized Ni2+ ions. pET-16b with no insert was used as a control; this encodes a short (48-aa) protein including all the vector sequences present in the His10-PP1 87B protein.
35S-labeled KLP38B and procollagen were synthesized by in vitro transcription–translation (Riboprobe and Flexi rabbit reticulocyte lysate kits; Promega, Madison, WI) from full-length KLP38B cDNA and pα1(III)Δ1 (Lees and Bulleid, 1994), respectively. Soluble extracts of BLR(DE3) containing oligo-histidine–tagged PP1 87B or control pET-16b protein were prepared and incubated with 10 μl Ni-NTA Sepharose in buffer A (50 mM Tris-HCl, pH 8, 100 mM KCl, 5 mM MgCl2, 0.1% Triton X-100, 20% glycerol, 10 mM imidazole) for 1 h at 4°C. The Ni-NTA beads were washed four times in this buffer, and then incubated with 1 μl reticulocyte lysate containing 35S-labeled KLP38B or procollagen in 100 μl buffer A for 30 min at room temperature. The beads were washed an additional four times in buffer A supplemented with imidazole to 50 mM, and then eluted with 20 μl of buffer A containing 200 mM imidazole. 10 μl of each eluate was analyzed by SDS-PAGE and phosphoimaging (Fujix Bas 2000; Fuji Photo Film Co., Tokyo, Japan). 0.5 μl of reticulocyte lysate was loaded for comparison (see Fig. 6, lanes 1 and 4).
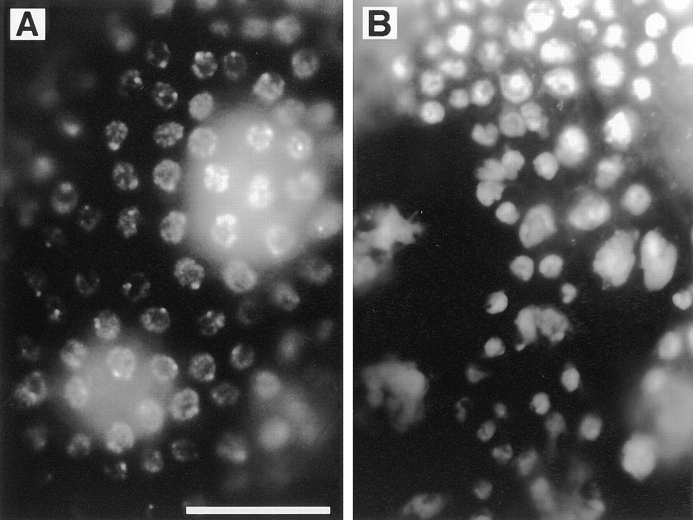
Figure 6.
Follicle cell defects in ovaries from KLP38B mutant females. Wild-type (Oregon R) stage 5–7 egg chambers have an even layer of follicle cells, revealed by DAPI staining (A). Comparable egg chambers from KLP38B 24-O females (B) have less follicle cells, leading to an uneven distribution of the follicle cells and large gaps in the layer. Exposed nurse cells with no overlying follicle cells are clearly visible. The follicle cell nuclei are of uneven size. This may be a consequence of earlier mitotic defects, but it may alternatively indicate uneven polyploidization of these nuclei. Nurse cells are the much larger nuclei beneath the follicle cell layer. Bar, 20 μm.
Results
Isolation of KLP 38B
The PP1 catalytic subunit will dephosphorylate a wide variety of substrates in vitro, but in vivo is found associated with a range of targeting and regulatory subunits that modulate its subcellular localization and substrate specificity (Hubbard and Cohen, 1993). To obtain potential PP1 regulators, we used the yeast two-hybrid interaction trap (for review see Fields and Sternglanz, 1994). This system identifies cDNAs encoding polypeptides that will bind to a “bait” protein in yeast. We used a cDNA library from Drosophila third instar larval RNA (see Materials and Methods). Drosophila has four genes encoding PP1 isoforms (Dombrádi et al., 1990, 1993), but 80% of the total PP1 activity is contributed by the PP1 87B isoform (Axton et al., 1990). Using full-length PP1 87B cDNA fused to GAL4 as the bait, 25 independent cDNAs were identified that interacted with PP1 87B but not with control baits. Sequencing and in situ hybridization to polytene chromosomes revealed that these 25 cDNAs are derived from 16 genes. One of these genes shows strong sequence similarity to NIPP-1, a biochemically characterized mammalian PP1 inhibitor (Van Eynde et al., 1995). Another, identified by three independent cDNAs from the two-hybrid screen, showed sequence similarity to kinesin-like proteins. These cDNAs hybridize to cytological position 38B on chromosome 2L. We refer to this gene as KLP38B (kinesin-like protein at 38B), following established convention (Goldstein, 1993).
Using sequence information from these two-hybrid KLP38B cDNAs, we used a PCR-based screen to isolate a 3.6-kb cDNA from an imaginal disk library (see Materials and Methods). This cDNA contains a single long ORF encoding a kinesin-related protein (Fig. 1). Based on translation from the first ATG, the sequence predicts a protein of 1,121 amino acids with a molecular mass of ∼125 kD and an isoelectric point (pI) of 7.6. Sequence analysis suggests the existence of four distinct domains (Figs. 1 and 2). Amino acid residues 1–122 at the NH2 terminus share no strong similarity with other known sequences. By comparison with other kinesin-related proteins, residues 123 to ∼512 encode the motor domain (Goldstein, 1993), while similarity to most other KRPs ends at residue 528. COOH-terminal to the motor domain is a 210 region of sequence similarity to Unc-104, Kif1A, Kif1B, and OrfW, but not to other known KRPs (Fig. 2). Within this region these five proteins share 39% similarity; 27% of these residues are identical between KLP38B and Unc-104, and 29% between KLP38B and OrfW. The motor domain of KLP38B is also more similar to Unc-104–like KRPs than to others. The remaining 300 residues at the COOH terminus show no significant similarity to other known proteins. On the basis of these comparisons, we place KLP38B in the Unc-104–like subfamily of kinesin-related proteins (Bloom and Endow, 1995; Moore and Endow, 1996; see Discussion). Comparisons of either the motor domain or the entire sequence do not indicate that KLP38B is more closely related to any one Unc-104–like KRP than to another.
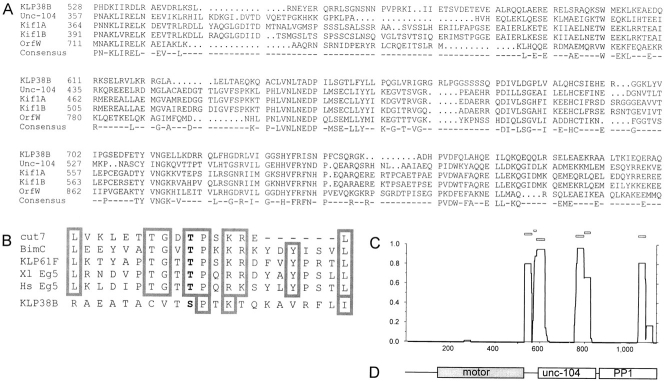
Figure 2.
Sequence alignments. (A) Unc104-related kinesin–related proteins share similarity outside the motor domain. Alignment of KLP38B residues 528–826 with similar kinesin-related proteins, using University of Wisconsin GCG program PILEUP. P528 is the last residue of the conserved region shared between all kinesin–related proteins, containing the motor domain. OrfW (D26361) is the predicted product of cDNA KIAA0042 from Homo sapiens (Nomura et al., 1994), Kif1A (D29951) and Kif1B (D17577) are from Mus musculus (Okada et al., 1995; Nangaku et al., 1994), and Unc-104 (M58582) is from Caenorhabditis elegans (Otsuka et al., 1991). (B) KLP38B potential cdk phosphorylation site is not related to the BimC/Eg5 subfamily. Comparison of the putative cdk phosphorylation site in KLP38B with that of BimC/Eg5-like kinesin-related proteins. Of the residues common to the BimC/Eg5 sequences, only the core cdk consensus (S/T P × K/R) and a leucine are also present in KLP38B. (C) Predicted coiled-coil formation by KLP38B. PEPCOIL output: probabilities of the KLP38B protein forming α-helical coiled-coils as predicted from the algorithms of Lupas et al. (1991), using a 28-residue window. The horizontal axis is position within the protein, and the NH2 terminus is at the origin; the vertical axis is probability of coiled-coil formation. Open boxes above the plot indicate the frame of the coiled-coil repeat. (D) Domain structure of KLP38B. The motor domain, region of extended sequence similarity with Unc-104–related KRPs, and the COOH-terminal PP1-binding region are shown at the same scale as in C. The PP1-binding region is defined by the shortest KLP38B clone from the two-hybrid screen.
Kinesin heavy chains fold into an α-helical coiled-coil in the stalk domain to form a dimer (Yang et al., 1989; de Cuevas et al., 1992). Most kinesin-related proteins are similarly thought to form homodimers, mediated by an extended coiled-coil region, although tetrameric, heterotrimeric, and monomeric kinesin-related proteins have been described (Cole et al., 1993, 1994; Bloom and Endow, 1995). We therefore used the PEPCOIL (Lupas et al., 1991; Lupas, 1996) and PAIRCOIL (Berger et al., 1995) programs to examine the KLP38B sequence for regions likely to form a coiled-coil. The PEPCOIL prediction is shown in Fig. 2 C. The PAIRCOIL output is essentially identical except that there is no predicted coiled-coil region near the COOH terminus (amino acids 1,061–1,077, see Fig. 2 C). This comparatively low level of coiled-coil is comparable to Kif1A (Okada et al., 1995) and Kif1B (Nangaku et al., 1994), which are murine KRPs related to KLP38B and Unc-104 (Fig. 2 A). Recombinant Kif1A and Kif1B exist predominantly as globular monomers, as judged by gel filtration, native PAGE, differential light scattering (Kif1A only), and EM. This suggests that KLP38B may similarly exist predominantly as a monomer.
Sequencing another four cDNAs revealed a number of single base changes relative to the sequence of the 3,605-bp cDNA. These could represent natural polymorphism or mutations induced in the process of cDNA synthesis. The majority of these variants are silent in terms of predicted polypeptide sequence, suggesting that these variants represent natural polymorphism. The single variant affecting the amino acid sequence of the motor domain (S454I) maps to a nonconserved loop. Details of these variants have been deposited in the EMBL database, accession number X99617.
Inspection of the sequence shows a potential cdk phosphorylation site at residue 1,108, close to the COOH terminus. Cdk phosphorylation has been shown to be important for function in Eg5, where phosphorylation is required for spindle association (Sawin and Mitchison, 1995; Blangy et al., 1995; for review see Walczak and Mitchison, 1996). The COOH-terminal cdk sites in BimC/Eg5 subfamily members show sequence similarity near the cdk site (the “BimC box”; Heck et al., 1993). As shown in Fig. 2 B, whereas KLP38B has a potential COOH-terminal cdk phosphorylation site, it does not have the additional residues characteristic of the BimC/Eg5 subfamily. Unc-104, Kif1A, Kif1B, and OrfW also have potential cdk phosphorylation sites; these also share no extended similarity with the BimC/Eg5 subfamily or with each other (not shown).
Transcription Pattern of KLP38B
We have studied the expression pattern of KLP38B by RNA in situ hybridization to various Drosophila tissues and developmental stages (Fig. 3). KLP38B is expressed in proliferating cells at every developmental stage, as well as in the testis and ovary. In the ovarioles, KLP38B is expressed from the germarium onward. Expression is much higher in the nurse cells at later stages, leading to a maternal contribution in the syncytial embryo (not shown). Later in development, KLP38B is expressed in the proliferating cells of the imaginal disks (e.g., eye disk, Fig. 3 B) and central nervous system (Fig. 3 A). In the testis, KLP38B transcripts accumulate during the 90-h growing stage and are degraded shortly after meiosis (Fig. 3 C; see Discussion).
Figure 3.
KLP38B mRNA is found in proliferating cells. (A) Dorsal view of brain from third instar larva. KLP38B transcripts are detected in the inner and outer proliferative centers of the optic lobe and in scattered cells, probably neuroblasts, elsewhere on the optic lobe. (B) Eye-antennal disk from third instar larva. KLP38B transcripts are seen in the proliferative regions: the antennal disk, the most anterior region of the eye disk, and in a broad stripe just behind the morphogenetic furrow. Orientation: posterior is up. (C) Testis from late (dark) pupa. KLP38B transcripts accumulate during the 90-h growing phase when the primary spermatocytes grow before meiosis and are degraded shortly after meiosis.
Isolation of KLP38B Mutants
The major reason for using Drosophila as a model system in which to investigate KRPs and PP1 is the advanced genetics and developmental biology. Gene function can be determined by analyzing the phenotype of mutants defective for the gene. We therefore isolated deficiencies and mutants for KLP38B as follows.
KLP38B was localized to cytological position 38B on chromosome 2L by in situ hybridization of the cDNA to wild-type polytene chromosomes (data not shown). This mapping was further refined by hybridization to deficiency stocks. This showed that KLP38B lies within Df(2L)pr-A20 (38A3-4 to 38B6-C1) and Df(2L)TW9 (37E2-F4 to 38A6-C1). This defines the cytological location as 38A3-38C1, which is consistent with the initial localization to 38B. A PCR-based in situ hybridization experiment aimed at estimating the number and cytogenetic location of genes encoding Drosophila kinesins and kinesin-related proteins had previously suggested that there is such a gene at 38B (Endow and Hatsumi, 1991).
Cosmid 164E4 from the European Drosophila Genome Project (Siden-Kiamos et al., 1990; Kafatos et al., 1991) was identified as containing the KLP38B gene by hybridization to a KLP38B-specific probe. The KLP38B cDNA was mapped onto the cosmid by restriction mapping and further hybridization to determine the gene organization (Fig. 4). A 7-kb BamHI–EcoRI fragment from the middle of the gene was used to identify recessive lethal P element mutants with insertions in the KLP38B gene, using plasmid rescue from a large collection of P element–induced lethal and semilethal mutants (see Materials and Methods). Six such mutants were identified. Two further P element-induced alleles of KLP38B, designated 24-O and 93-E, were generously provided by Mike Goldberg. We refer to these alleles as KLP38B 24-O and KLP38B 93-E. Consistent with the in situ mapping, these P element mutants are all semilethal over Df(2L)pr-A20.
All KLP38B alleles were tested in combination with each other and over Df(2L)pr-A20, a deficiency that removes the KLP38B gene. The observed phenotypes of all the KLP38B alleles, and Df(2L)pr-A20, are entirely recessive. Several of the mutant chromosomes are completely lethal when homozygous, but only semilethal or sterile over other alleles or over Df(2L)pr-A20, indicating that they have one or more additional recessive lethal mutations on the same chromosome. The phenotype of KLP38B 24-O homozygotes is indistinguishable from that of KLP38B 24-O/Df(2L)pr-A20 hemizygotes, so we conclude that KLP38B 24-O has little or no residual KLP38B activity and that KLP38B activity is not absolutely required for viability (see Discussion).
When using mutant chromosomes carrying a P element, there is a possibility that the observed phenotype is not due to the P element insertion, but rather to another mutation on the same chromosome. This can be tested by excising the P element and observing the new phenotype: complete reversion to wild type indicates that the mutant phenotype is entirely due to the P element insertion. Using the strongest mutant, KLP38B 24-O, we found that excision of the P[lacW] element resulted in complete phenotypic reversion, thus demonstrating that the phenotypes observed are due to the insertion of P[lacW] at the KLP38B locus.
Another possibility is that the P element insertions affect one or more genes other than KLP38B. We therefore created transgenic flies carrying the KLP38B cDNA under the control of the hsp26 promoter. Even without heat shock, this construct rescues the sterility of KLP38B 24-O homozygotes and of l(2)k03903/Df(2L)pr-A20 hemizygotes. Similarly, the visible phenotypes are much less severe in these flies. We therefore conclude that the phenotypes we observe in the P element mutants are entirely due to disruption of the KLP38B gene.
The Molecular Basis of the KLP38B Mutations
Plasmid rescue was repeated on each of the P element lines. Restriction maps of the recovered plasmids were compared with the genomic map from cosmid 164E4 to generate a molecular map of the KLP38B locus and mutants. All of the P element insertions map to the KLP38B intron. The recovered plasmids were sequenced using a P element–specific primer to determine the exact insertion sites of the P elements. These data are summarized in Fig. 4.
KLP38B Is Required for Normal Cell Proliferation
The range of phenotypes observed—rough eyes (Fig. 5, A–C), missing bristles (Fig. 5, D–F, and Table I), reduced viability, and male and female sterility—correlates with our RNA in situ data (Fig. 3; unpublished observations) and suggests a role for KLP38B in cell proliferation and gametogenesis (see Discussion). We have also observed an “upheld wing” phenotype in adults, in which the wing blade is held vertically rather than horizontally. Newly eclosed flies (young adults) do not show this phenotype, rather it develops a few days after eclosion. Affected individuals cannot fly. The “upheld wing” phenotype has been observed in mutants affecting development of the indirect flight muscles (DeSimone et al., 1996), but examination of these muscles dissected from KLP38B 24-O homozygotes revealed no obvious abnormalities in muscle structure. Homozygous adults also have poor locomotive ability.
Figure 5.
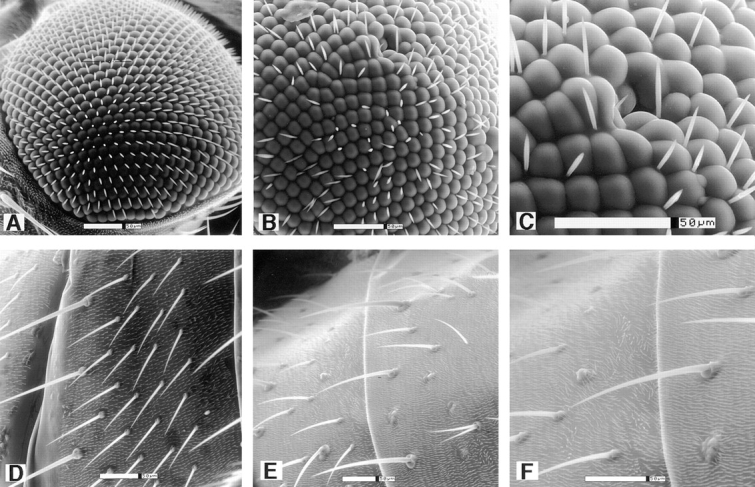
KLP38B mutants show rough eyes and missing bristles. Scanning EM shows clear defects in the eye structure of KLP38B 24-O homozygotes (B and C) compared with the highly ordered wild-type eye (A). Facets are of uneven size and shape and many bristles are missing, consistent with aberrant cell proliferation. Similarly, the wild-type array of abdominal bristles (D) is disrupted in KLP38B 24-O homozygotes (E and F). C and F are details from B and E, respectively. Bars, 50 μm.
Table I.
Scutellar Bristle Defects in KLP38B Mutants
| KLP38B 24–O | l(2)k03903 | l(2)k05702 | Oregon R | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bristle No. | 2 | 1 | 0 | 2 | 1 | 0 | 2 | 1 | 0 | 2 | 1 | 0 | ||||||||||||
| Anterior | 67 | 30 | 3 | 60 | 38 | 2 | 41 | 45 | 14 | 100 | 0 | 0 | ||||||||||||
| Posterior | 76 | 24 | 0 | 82 | 15 | 3 | 45 | 33 | 2 | 100 | 0 | 0 | ||||||||||||
| Duplicated | 0 | 3 | 97 | 0 | 3 | 97 | 0 | 3 | 97 | 0 | 0 | 100 | ||||||||||||
| Extra | 0 | 3 | 97 | 0 | 3 | 97 | 0 | 0 | 100 | 0 | 0 | 100 | ||||||||||||
Wild-type flies have four scutellar bristles, two anterior and two posterior. Disruption of this pattern is frequently observed in cell cycle mutants. Scutellar bristles were counted for 100 homozygous individuals of each of the three KLP38B mutant alleles. Oregon R is a standard wild-type strain. Duplicated bristles are two bristles from the same socket, or two bristles immediately adjacent to each other; extra bristles are bristles not in the characteristic location.
We examined the male sterility of KLP38B 24-O homozygotes in more detail. Squashed preparations of testes from KLP38B 24-O homozygous males were examined by phase-contrast microscopy. This method allows the detection of meiosis and cytokinesis defects by measuring the number of postmeiotic spermatids and the relative size and number of their nuclei and Nebenkerns, which are the mitochondrial derivatives (for examples see Alphey et al., 1992; Williams et al., 1995). No such gross meiotic defect was observed, but no motile sperm were present (not shown).
KLP38B Is Required for Female Fertility and Follicle Cell Proliferation
KLP38B 24-O homozygous females are completely sterile. Mutant ovaries are small, with egg chambers rarely developing past stage 5 of oogenesis (stages according to King, 1970; Mahowald and Kambysellis, 1980; see Spradling, 1993, for review of oogenesis in Drosophila). Further investigation of the mutant ovaries revealed a failure of follicle cell proliferation, so that the larger egg chambers are not completely covered by follicle cells (Fig. 6). In wild-type ovaries, follicle cells divide four or five times during stages 1–5, giving ∼1,000 follicle cells. These then cease cell division but increase in size by polyploidization. At stage 9 the majority of the cells migrate over the expanding oocyte and form a columnar epithelium, while ∼50 remain as a thin layer over the nurse cells and 6–10 migrate between the nurse cells to become the border cells. The majority of KLP38B 24-O egg chambers arrest at stage 5 or earlier. No eggs are laid by these females. DAPI staining clearly shows that these egg chambers have a much lower number of follicle cells than wild type (Fig. 6, A and B). This reduced number of follicle cells fails to maintain a continuous layer as the egg chamber grows, so the larger egg chambers have large gaps in the follicle cell layer. Proliferation of the germ cells in the germarium appears normal, as the mutant egg chambers have the correct number of nurse cells (15 nurse cell nuclei present in 50/50 mutant egg chambers scored).
KLP38B Is Required for Mitotic Chromatin Condensation
As described above, KLP38B 24-O homozygous females are completely sterile and produce no mature oocytes. A weaker allele, l(2)k03903, does lay some eggs but these very rarely hatch. l(2)k03903 also has somewhat less severe rough eyes and bristle defects than KLP38B 24-O and is male fertile. We investigated the basis of the maternal effect lethality in embryos derived from l(2)k03903 females. For clarity, we will refer to these as k03903 embryos.
The majority of k03903 embryos fail to cellularize. Those that do cellularize appear to have uneven cell size and distribution compared with wild type, but gastrulate apparently normally even though they generally fail to hatch. This is not unexpected by comparison with the zygotic phenotype of string and other cell cycle genes, in which failure of mitosis leads to an embryo with far less cells than wild type, which nonetheless gastrulates and develops relatively normally but fails to hatch (Hartenstein and Posakony, 1990; Foe et al., 1993). k03903 embryos fail to form pole cells. They also have an high frequency of “drop-out” nuclei (not shown). These are nuclei that are eliminated from the surface blastoderm layer and fall into the interior, and are indicative of a cell cycle defect (Foe et al., 1993; see Discussion).
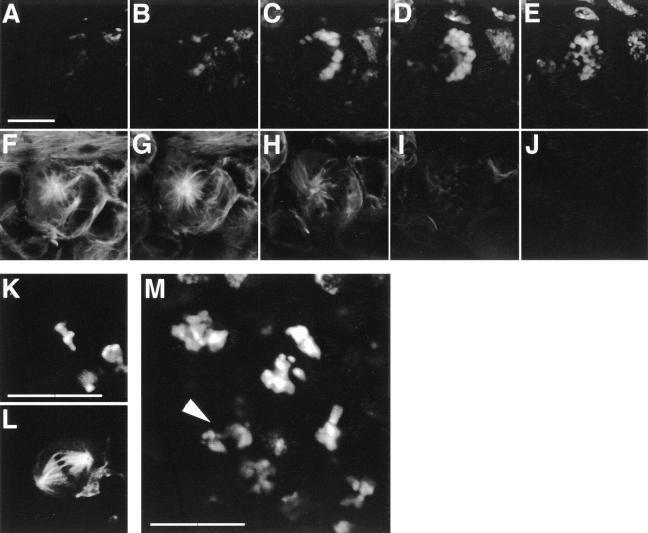
We have examined syncytial and newly cellularized k03903 embryos by DAPI staining and immunofluorescence with anti-tubulin antibodies and find metaphase and anaphase spindles associated with undercondensed chromatin (Fig. 7). This phenotype is clearly distinguishable from wild type and is 100% penetrant: every metaphase or anaphase figure from k03903 embryos has undercondensed chromatin (n = 50 for each of metaphase and anaphase); conversely, every metaphase or anaphase figure from wild-type embryos showed normal chromatin condensation (n = 50 for each of metaphase and anaphase). We have quantified the level of undercondensation at anaphase by counting the number of pixels encompassed by the DAPI staining. Wild-type embryos have mean = 6,935, SD = 365; k03903 embryos have mean = 11,477, SD = 1,904 (n = 10 for each). This is a highly significant difference (t test: t18 = 7.14, P < 0.001).
Figure 7.
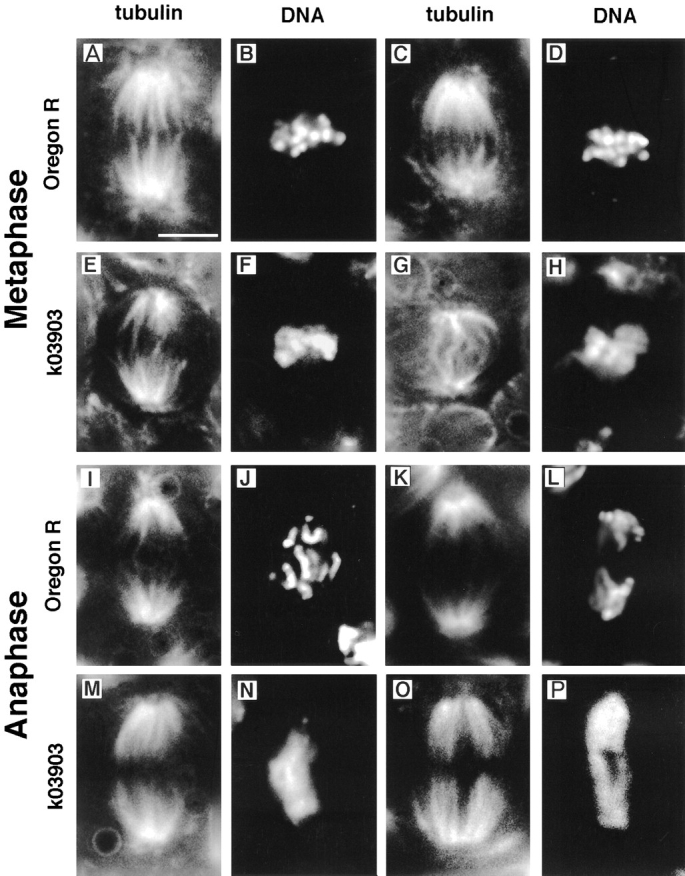
k03903 embryos undergo mitosis with undercondensed chromatin. Metaphase (A–H) and anaphase (I–P) figures from wild-type and k03903 embryos. k03903 embryos have undercondensed chromatin at both metaphase (F and H) and anaphase (N and P) relative to wild type (B and D, and J and L, respectively). k03903 spindles are generally similar to those of wild type, but a range of defects is occasionally seen including kinked spindles (G). Mitotic figures are shown in pairs, with tubulin staining on the left and DAPI on the right. Bar, 5 μm.
In addition to the chromatin condensation defect, we also observed a number of spindle abnormalities in syncytial embryos, including kinked spindles, spindles with some microtubules inappropriately associating with chromatin on an adjacent spindle, and tetrapolar spindles (Fig. 7 G and data not shown). We interpret the relatively low frequency of abnormal spindles as compared with the frequency of abnormal chromatin condensation to indicate that the spindle defects may be a secondary consequence of the chromatin abnormalities (see Discussion).
KLP38B Is Required for Normal Cell Cycle Progression
Cell cycle progression in Drosophila embryos is unusual in a number of respects (for review see Foe et al., 1993). Syncytial embryos have no detectable G1 or G2 phases, and of course have no cytokinesis. They appear to lack many of the checks and controls present in a normal somatic cell cycle. By contrast, neuroblasts in the brain of wandering third instar larvae follow a typical cell cycle of G1-S-G2-M. We therefore examined these cells from KLP38B mutant animals.
The mutant phenotype is characterized by an increased mitotic index (21% of 2,565 cells scored in acetic acid– orcein squashes of 14 KLP38B 24-O homozygous larvae compared with 6.8% for wild-type brains). Approximately two-thirds of these mitotic figures are polyploid. The diploid cells have a metaphase/anaphase ratio not significantly different from wild type (2.6), whereas the polyploid cells have a metaphase/anaphase ratio of 46. Chromatin condensation varies from one cell to another (see Discussion).
We further investigated these abnormal mitotic figures by confocal microscopy of whole brains (Fig. 8). Many of these polyploid cells have chromosomes arranged on monopolar spindles, indicating a centrosome duplication or separation defect. Fig. 8, A–J, is a series of confocal sections through such a cell. Fig. 8, K and L, shows a comparable wild-type metaphase (note the difference in magnification between A–J and K–M). Fig. 8 M shows a field of mitotic cells, illustrating the elevated mitotic index.
Figure 8.
Mitotic defects in KLP38B 24-O neuroblasts. (A–J) Confocal sections through a KLP38B 24-O neuroblast. A–E are DAPI staining; F–J show the tubulin. Sections are at 1-μm intervals. A wild-type metaphase is shown for comparison K and L. M is a confocal section through a field of mitotic cells. Only a single anaphase is present; the remainder are metaphases in various orientations. Bars, 5 μm.
KLP38B Associates with PP1
As described above, KLP38B was originally isolated as a cDNA encoding a protein capable of binding to PP1 87B, the major PP1 catalytic subunit of Drosophila. Three cDNAs were isolated: they differ in length from each other and thus are independent and not derived from clonal amplification of the same original cDNA. All three encode the COOH-terminal region of KLP38B, encoding amino acids 587–1,121, 725–1,121, and 848–1,121, respectively. This demonstrates that the COOH-terminal 273 residues are sufficient to bind PP1.
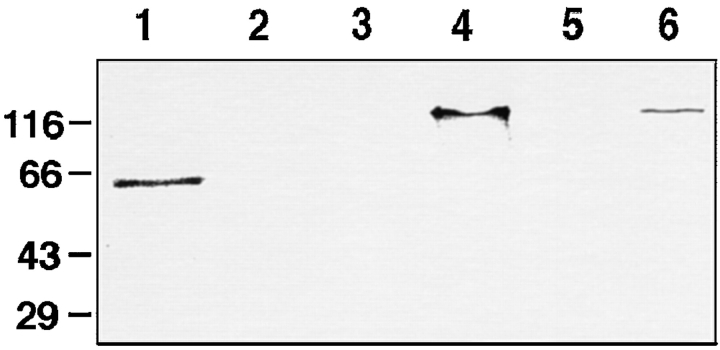
While the two-hybrid system has now been successfully used many times to demonstrate protein–protein binding, it is also known to be capable of producing false positives. We therefore investigated the association of KLP38B and PP1 87B by a completely independent method. We expressed full-length PP1 87B in bacteria, with an oligo-histidine tag to allow affinity chromatography using immobilized Ni2+ ions. We bound PP1 87B to Ni-NTA Sepharose, and then incubated these beads with 35S-labeled KLP38B from an in vitro transcription–translation reaction. After extensive washing we eluted the PP1 87B by increasing the imidazole concentration and analyzed the samples by SDS-PAGE followed by phosphoimaging. We found that 35% of the KLP38B remained bound to the column through the washes and coeluted with PP1 87B (Fig. 9, lane 6). No KLP38B (<0.5%) bound to control beads prepared using extracts from bacteria that cannot synthesize PP1 87B (Fig. 9, lane 5). We conclude that KLP38B can bind to PP1 87B in vitro. A procollagen control does not bind to PP1 under the same conditions. Interestingly, phosphorylation of KLP38B is not required for binding to PP1, as bacterially expressed KLP38B also binds to PP1 (not shown).
Figure 9.
KLP38B binds PP1 in vitro. (Lane 1) Reticulocyte lysate containing 35S-labeled procollagen. This protein was incubated with PP1 beads or control beads. (Lane 2) Eluate from control beads; (lane 3) eluate from PP1 beads. Procollagen does not bind to PP1. (Lane 4) Reticulocyte lysate containing 35S-labeled KLP38B. (Lane 5) Eluate from control beads; (lane 6) eluate from PP1 beads. KLP38B binds to PP1 but not to the control beads. Details of the beads and the binding, washing, and elution conditions are described in Materials and Methods.
Discussion
KLP38B Structure and Function
Structural similarities between kinesin-related proteins in many cases reflect functional similarities. For example, the BimC-related kinesin-related proteins BimC, Eg5, KLP61F, Cin8/Kip1, and Cut7 are all involved in the formation of bipolar spindles (Enos and Morris, 1990; Hagan and Yanagida, 1990; Hoyt et al., 1992; Heck et al., 1993; Kashina et al., 1996a ,b). Loss-of-function mutations of these KRPs are able to duplicate their centrosomes/spindle pole bodies but fail to separate them. Of the Unc-104–like KRPs, Kif1A is a neuron-specific vesicle transporter and is thought to be a functional homologue of Unc-104 (Okada et al., 1995). Kif1B has been found to be associated with mitochondria and is capable of transporting mitochondria along microtubules in vitro (Nangaku et al., 1994). No function is known for OrfW, which was detected in a large-scale sequencing project (Nomura et al., 1994). Loss-of-function unc-104 mutants are paralyzed; they show correct axon formation, but few synapses and synaptic vesicles accumulate in the neuron cell body (Hall and Hedgecock, 1991). This suggests that Unc-104 transports synaptic vesicles along axonal microtubules. The functions ascribed to Kif1B and Unc-104 are clearly very different from the mitotic role we have demonstrated for KLP38B. Drosophila also has another KRP more closely related to Unc-104. For these reasons, we do not believe that KLP38B is a true functional homologue of Unc-104.
The Mitotic Role of KLP38B
The expression pattern of KLP38B in proliferating cells suggests a role for KLP38B in mitosis. This is confirmed by the phenotype of KLP38B mutants. The effect of cell cycle mutants is often seen only in late larval/pupal stages (Gatti and Baker, 1989), as most larval growth is by cell enlargement. Survivors typically show rough eyes and missing bristles, as a consequence of occasional failure of proliferation in the cells from which these structures are derived. Maternal effect lethality is another well-known phenotype of mutation in cell cycle genes: the mother has to provide the embryo with all the components necessary for 13 rapid nuclear divisions, so lack of any gene product required for mitosis will lead to nonviable embryos (Gatti and Baker, 1989; Sullivan et al., 1990; Foe et al., 1993; Gonzalez et al., 1994). KLP38B mutants show all of these phenotypes.
KLP38B 24-O escapers are female sterile: egg chambers do not develop beyond the early stages because of failure of follicle cell proliferation. Embryos derived from l(3)k03903 homozygous females (“k03903 embryos”) have abnormal mitotic figures, with undercondensed chromatin on a metaphase-like spindle. This implies a role for KLP38B in the establishment or maintenance of mitotic chromatin condensation. We cannot rule out the possibility of transient chromatin condensation in these embryos, but we have never observed properly condensed chromatin, so any such condensation cannot last long. Undercondensed chromatin on a metaphase-like spindle could be explained by a metaphase arrest or delay followed by partial decondensation of the chromatin, but we also find that all the anaphase-like figures have undercondensed chromatin, which suggests that these embryos go through a complete mitosis with undercondensed chromatin. These embryos also exhibit a low frequency of spindle abnormalities, which may be a secondary consequence of the abnormal chromatin. We also observe occasional chromatin bridges in cycle 14 (newly cellularized) embryos, as well as some cytokinesis defects, possibly as a consequence of the chromatin bridges.
The syncytial embryo may be incapable of arresting its mitotic cycles in response to problems in chromosome condensation or segregation and instead eliminates the defective nuclei from the blastoderm layer (Sullivan et al., 1990; Foe et al., 1993). Cell divisions later in development have more checkpoints and controls and may be able to delay cell cycle progression until the chromatin is properly condensed. We have not observed undercondensed chromatin in mutant neuroblasts. Many cells, particularly the highly polyploid ones, have overcondensed chromatin. This may be a consequence of spending an extended period in a metaphase-like state (Heck et al., 1993).
Diploid neuroblasts have a wild-type metaphase/anaphase ratio, whereas the polyploid cells have very few anaphases. This suggests that the polyploid cells are almost incapable of completing mitosis, but the high levels of polyploidy imply that these cells are not perfectly arrested but can reenter the cell cycle repeatedly to replicate their chromosomes without chromosome segregation and cytokinesis. Many of the polyploid mitotic figures have a monopolar spindle. This implies a defect in centrosome duplication or segregation. However, not all polyploid figures are monopolar, and we do not see diploid monopolar spindles so this may be a secondary consequence of earlier defects.
The neuroblast phenotype of KLP38B in many ways resembles that of KLP61F (Heck et al., 1993). KLP61F mutant neuroblasts have an elevated mitotic index, no anaphases, and a very high frequency of monopolar spindles. KLP61F is a kinesin-related protein of the bimC subfamily, and thus is not closely related to KLP38B. It forms a two-headed homotetramer and is required for centrosome separation, presumably acting by sliding microtubules past each other (Heck et al., 1993; Kashina et al., 1996a ,b).
We have shown genetically that KLP38B is required for correct chromatin condensation in embryonic divisions. Furthermore, immunofluorescence studies have shown that KLP38B protein is localized to condensed chromatin (Molina, I., and P. Ripoll, personal communication). This does not, however, necessarily imply a direct role for KLP38B in the process of chromatin condensation. For example, KLP38B could be involved in communication between the chromosomes and the spindle, or it could affect the timing of chromatin condensation or spindle function. Differential control of spindle formation and chromatin condensation by cdc25 homologues has been previously observed (White Cooper et al., 1993; Gabrielli et al., 1996). This shows that these two processes are under separate control and suggests the possibility of a cross talk mechanism to allow a degree of coordination. Such a role might also explain the incomplete lethality of KLP38B mutants, as a checkpoint function is required only when things go wrong (see below).
Localization of kinesin-related proteins to chromatin has been described (Afshar et al., 1995; Vernos et al., 1995; Wang and Adler, 1995; Tokai et al., 1996). XKlp1 is chromatin associated throughout the cell cycle (Vernos et al., 1995), and the related chicken KRP chromokinesin is also localized on chromosome arms in mitosis (Wang and Adler, 1995). Vernos et al. (1995) analyzed XKlp1 function by depletion experiments using antisense oligos or anti-XKlp1 antibodies. They concluded that Xklp1 is required for chromosome positioning and bipolar spindle stabilization. The delocalized chromosomes that they observe (Fig. 8 of Vernos et al., 1995) also appear to be unevenly condensed, which might imply a role for XKlp1 in chromatin condensation. Another precedent for KRP involvement in genome organization comes from fission yeast, where microtubule-based movement of chromosomes through attachment of telomeres to the spindle pole body is implicated in homologue recognition (for review see Kohli, 1994).
KLP38B transcripts accumulate in the growing stage of spermatogenesis and are degraded shortly after meiosis. This is very similar to the testis expression pattern of the cdc25 homologue twine (Alphey et al., 1992) and other cell cycle–regulatory genes required for meiosis (Alphey, L., unpublished observations). However, there is little or no postmeiotic transcription in the male germ line (Olivieri and Olivieri, 1965; Gould-Somero and Holland, 1974; for review see Fuller, 1993), so genes involved in postmeiotic differentiation are expected to follow a similar transcription pattern; the gene products are then stored as mRNA or protein until required. Since the male sterility seen in KLP38B mutants does not appear to be associated with severe meiotic defects, we conclude that KLP38B either has no role in male meiosis, or else its role is redundant with another kinesin-related protein.
Does KLP38B Have Nonmitotic Roles?
We have demonstrated a role for KLP38B in mitosis. This does not exclude the possibility that KLP38B may have additional roles. The observation that KLP38B mRNA is present in postembryonic tissues only in cells that are about to divide suggests that KLP38B acts only in mitosis and does not function in nonmitotic microtubule-based transport in these tissues. As noted above, we have observed that males hemizygous or homozygous mutant for KLP38B 24-O are completely sterile. These flies produce no motile sperm but appear to complete meiosis and cytokinesis normally. We therefore deduce a role for KLP38B in postmeiotic spermatid differentiation, in addition to the mitotic role.
Redundancy of KLP38B with Other Kinesin-related Proteins
The phenotypes of KLP38B 24-O/KLP38B 24-O and KLP38B 24-O/ Df(2L)pr-A20 are indistinguishable, indicating that this allele has very little, if any, residual KLP38B activity. Despite this finding, none of our KLP38B alleles are completely lethal. This suggests a degree of functional redundancy with other Drosophila kinesin-related proteins. Such redundancy has been observed before in the case of CIN8/KIP1, two redundant budding yeast kinesin-related proteins (Hoyt et al., 1992). Another possibility is perdurance of the maternal product, which has many precedents among Drosophila cell cycle genes (for review see Gonzalez et al., 1994). We have observed a reduction in the penetrance of the lethality and scutellar bristle phenotypes of KLP38B 24-O in a continuously maintained culture over a 12-mo period, such that the viability is now considerably higher (30–50% relative to heterozygous siblings) and the penetrance of the scutellar bristle defects lower than a year ago (data not shown). This suggests that the stock can readily accumulate modifiers, perhaps subtle allelic variants of genes with overlapping functions. The complete sterility of KLP38B 24-O homozygotes, both males and females, demonstrates an absolute requirement for the KLP38B gene product in gametogenesis. This is reminiscent of KLP3A mutants, which are sterile but viable, even though the KLP3A protein is present in proliferating somatic cells (Williams et al., 1995).
KLP38B and PP1
We have demonstrated binding of KLP38B to PP1 in vitro. Drosophila melanogaster has four genes encoding isoforms of PP1 (Dombrádi et al., 1990, 1993). Strong alleles of PP1 87B show PP1 activity reduced to ∼20% of wild type in extracts of larvae and adults, indicating that this gene is the major isoform at these stages (Baksa et al., 1993). These mutants have a strong cell cycle phenotype: neuroblasts are delayed in mitosis and show aberrant spindle organization and hypercondensed chromatin. Some alleles are also dominant suppressors of position effect variegation, indicating that PP1 has an interphase role in the regulation of chromatin structure as well as a mitotic role. Other roles of PP1 may be masked by redundancy between the isoenzymes and other related protein phosphatases.
If PP1 87B and KLP38B associate in the fly, as suggested by our in vitro experiments, this suggests an explanation for their observed phenotypes: KLP38B is required for normal chromatin condensation and is inhibited by PP1 87B. PP1 has been found in partially purified axonal kinesin, and inhibition of the phosphatase with okadaic acid stimulates kinesin activity (McIlvain et al., 1994 a; McIlvain, J.M., Jr., L. Lindesmith, Y. Argon, and M.P. Sheetz. 1994. Mol. Cell. Biol. 5(Suppl.):31a). However, the true situation may not be so simple. Both PP1 87B and KLP38B are pleiotropic; furthermore, they are both members of gene families with overlapping functions. Axton et al. (1990) suggest that the cell cycle effects of PP1 87B mutants may be due in part to the presence of excess free regulatory subunits interfering with the function of another PP1 isoenzyme or with other proteins essential for mitosis.
Another attractive hypothesis is that KLP38B may be involved in localizing PP1. The PP1 catalytic subunit will dephosphorylate a wide range of substrates in vitro, but in vivo is found associated with targeting and regulatory proteins that modulate its subcellular location and substrate specificity, and it seems likely that there is one or more specific subunits for each cellular function of the enzyme (Hubbard and Cohen, 1993). Association with KLP38B may be involved in the regulation of PP1 regarding chromatin condensation or spindle function.
Regulation of the cell cycle, motor activity, spindle structure, and chromatin condensation by phosphorylation has been found in a variety of systems. We have identified a novel kinesin-related protein (KLP38B) in a screen for PP1-binding proteins and demonstrated for the first time a physical association between a kinesin-related protein and a protein phosphatase. Our phenotypic analysis of mutants defective for KLP38B demonstrates a requirement for this gene for mitotic chromatin condensation and cell proliferation.
Acknowledgments
We are grateful to the following colleagues who sent us reagents: Stephen Elledge for the yeast two-hybrid system, including an unpublished Drosophila cDNA library; Myles Axton and David Glover for PP1 87B cDNA; Istvan Török and Mike Goldberg for P element mutants; Richard Wilson and Neil Bulleid for procollagen cDNA and assistance with in vitro translation. Doug Ruden, Pedro Ripoll, and Sharyn Endow generously shared unpublished data. We thank Robert Saunders for help with cytogenetics, and Trevor Sherwin and Klaus Ersfeld for help with image analysis. Computing facilities at SEQNET and HGMP were made available by the United Kingdom Biotechnology and Biological Sciences Research Council and Medical Research Council, respectively. We also thank Viki Allan, Doug Drummond, and Iain Hagan for helpful discussions and critical reading of the manuscript.
Footnotes
1. Abbreviations used in this paper: cdk, cyclin-dependent kinase; DAPI, 4′,6-diamidino-2-phenylindole; KLP, kinesin-like protein; KRP, kinesin-related protein; ORF, open reading frame.
This work was principally supported by grant SP2290/0101 from the Cancer Research Campaign to L. Alphey, with additional funding from the Royal Society (to L. Alphey) and United Kingdom Biotechnology and Biological Sciences Research Council (to K. Kaiser). Confocal and fluorescence microscopes and semiautomated sequencing facilities were available through the generosity of the Wellcome Trust (grants 045183/Z/95/Z and 044327/Z/95/Z, respectively).
Please address all correspondence to Luke Alphey, School of Biological Sciences, University of Manchester, 2.205 Stopford, Oxford Road, Manchester M13 9PT, United Kingdom. Tel.: (44) 161-275-5111. Fax: (44) 161-275-5082. e-mail: Luke.Alphey@man.ac.uk
References
- Afshar K, Barton NR, Hawley RS, Goldstein LSB. DNA binding and meiotic chromosomal localization of the DrosophilaNod protein. Cell. 1995;81:129–138. doi: 10.1016/0092-8674(95)90377-1. [DOI] [PubMed] [Google Scholar]
- Alphey L. PCR-based method for isolation of full-length clones and splice variants from cDNA libraries. Biotechniques. 1997;22:481–486. doi: 10.2144/97223st02. [DOI] [PubMed] [Google Scholar]
- Alphey L, Jimenez J, White-Cooper H, Dawson I, Nurse P, Glover DM. twine, a cdc25 homologue that functions in the male and female germ-line of Drosophila. . Cell. 1992;69:977–988. doi: 10.1016/0092-8674(92)90616-k. [DOI] [PubMed] [Google Scholar]
- Axton JM, Dombrádi V, Cohen PTW, Glover DM. One of the protein phosphatase 1 isoenzymes in Drosophilais essential for mitosis. Cell. 1990;63:33–46. doi: 10.1016/0092-8674(90)90286-n. [DOI] [PubMed] [Google Scholar]
- Baksa K, Morawietz H, Dombrádi V, Axton JM, Taubert H, Szabó G, Török I, Udvardy A, Gyurkovics H, Szöör B, Glover DM, Reuter G, Gausz J. Mutations in the protein phosphatase gene at 87B can differentially affect suppression of position-effect variegation and mitosis in Drosophila melanogaster. . Genetics. 1993;135:117–125. doi: 10.1093/genetics/135.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Vaessin H, Sheperd S, Lee K, McCall K, Barbel S, Ackerman L, Caretto R, Uemura T, Grell E, Jan LY, Jan YN. Searching for pattern and mutation with a P-lacZ vector. Genes & Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- Blangy A, Lane HA, d'Herin P, Harper M, Kress M, Nigg EA. Phosphorylation by p34cdc2regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell. 1995;83:1159–1169. doi: 10.1016/0092-8674(95)90142-6. [DOI] [PubMed] [Google Scholar]
- Bloom, G.S., and S.A. Endow. 1994. Protein Profile. Vol. 1. Kinesins. Academic Press, NY. 1089–1105. [PubMed]
- Bloom, G.S., and S.A. Endow. 1995. Protein Profile. Vol. 2. Kinesins. Academic Press, NY. 1109–1171.
- Boleti H, Karsenti E, Vernos I. Xklp2, a novel Xenopuscentrosomal kinesin-like protein required for centrosome separation during mitosis. Cell. 1996;84:49–59. doi: 10.1016/s0092-8674(00)80992-7. [DOI] [PubMed] [Google Scholar]
- Bollen M, Stalmans W. The structure, role and regulation of type 1 protein phosphatases. Crit Rev Biochem Mol Biol. 1992;27:227–281. doi: 10.3109/10409239209082564. [DOI] [PubMed] [Google Scholar]
- Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature (Lond) 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- Brown N, Kafatos F. Functional cDNA libraries from Drosophilaembryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Cohen P. The structure and regulation of protein phosphatases. Annu Rev Biochem. 1989;58:453–508. doi: 10.1146/annurev.bi.58.070189.002321. [DOI] [PubMed] [Google Scholar]
- Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature (Lond) 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- Cole DG, Saxton WM, Sheehan KB, Scholey JM. A ‘slow' homotetrameric kinesin-related motor protein purified from Drosophilaembryos. J Biol Chem. 1994;269:22913–22916. [PMC free article] [PubMed] [Google Scholar]
- Cooley L, Kelley R, Spradling A. Insertional mutagenesis of the Drosophilagenome with single P elements. Science (Wash DC) 1988;239:1121–1128. doi: 10.1126/science.2830671. [DOI] [PubMed] [Google Scholar]
- de Cuevas M, Tao T, Goldstein LS. Evidence that the stalk of Drosophilakinesin heavy chain is an α-helical coiled coil. J Cell Biol. 1992;116:957–965. doi: 10.1083/jcb.116.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSimone S, Coelho C, Roy S, VijayRaghavan K, White K. Erect Wing, the Drosophilamember of a family of DNA binding proteins is required in imaginal myoblasts for flight muscle development. Development (Camb) 1996;122:31–39. doi: 10.1242/dev.122.1.31. [DOI] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O , A comprehensive set of sequence analysis programs for VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrádi V, Axton JM, Glover DM, Cohen PTW. Cloning and chromosomal localization of DrosophilacDNA encoding the catalytic subunit of protein phosphatase 1α. Eur J Biochem. 1989;183:603–610. doi: 10.1111/j.1432-1033.1989.tb21089.x. [DOI] [PubMed] [Google Scholar]
- Dombrádi V, Axton JM, Brewis N, Da Cruz e Silva E, Alphey L, Cohen PTW. Drosophilacontains three genes that encode distinct isoforms of protein phosphatase 1. Eur J Biochem. 1990;194:739–745. doi: 10.1111/j.1432-1033.1990.tb19464.x. [DOI] [PubMed] [Google Scholar]
- Dombrádi V, Mann DJ, Saunders RDC, Cohen PTW. Cloning of the fourth functional gene for protein phosphatase 1 in Drosophila melanogasterfrom its chromosomal position. Eur J Biochem. 1993;212:177–183. doi: 10.1111/j.1432-1033.1993.tb17648.x. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A, Lee W-H, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes & Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Endow SA, Hatsumi M. A multimember kinesin gene family in Drosophila. . Proc Natl Acad Sci USA. 1991;88:4424–4427. doi: 10.1073/pnas.88.10.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos AP, Morris NR. Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in A. nidulans. . Cell. 1990;60:1019–1027. doi: 10.1016/0092-8674(90)90350-n. [DOI] [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Foe, V.E., G.M. Odell, and B.A. Edgar. 1993. Mitosis and morphogenesis in the Drosophila embryo: point and counterpoint. In The Development of Drosophila melanogaster. M. Bate and A. Martinez Arias, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 149–300.
- Fuller, M.T. 1993. Spermatogenesis in Drosophila. In The Development of Drosophila melanogaster. M. Bate and A. Martinez Arias, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 71–147.
- Futterer A, Gruppa G, Kramer B, Lemke H, Kronke M. Molecular cloning and characterization of human kinectin. Mol Biol Cell. 1995;6:161–170. doi: 10.1091/mbc.6.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli BG, De Sousa CPC, Tonks ID, Clark JM, Hayward NK, Ellem KAO. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J Cell Sci. 1996;109:1081–1093. doi: 10.1242/jcs.109.5.1081. [DOI] [PubMed] [Google Scholar]
- Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. . Genes & Dev. 1989;3:438–453. doi: 10.1101/gad.3.4.438. [DOI] [PubMed] [Google Scholar]
- Gho M, McDonald K, Ganetzky B, Saxton WM. Effects of kinesin mutations on neuronal functions. Science (Wash DC) 1992;258:313–316. doi: 10.1126/science.1384131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LSB. With apologies to Scheherazade: tails of 1,001 kinesin motors. Annu Rev Genet. 1993;27:319–351. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- Gonzalez, C., and D.M. Glover. 1993. Techniques for studying mitosis in Drosophila. In The Cell Cycle: A Practical Approach. P. Fantes and R. Brooks, editors. IRL Press, Oxford. 163–168.
- Gonzalez C, Alphey L, Glover DM. Cell cycle genes of Drosophila. . Adv Genet. 1994;31:79–138. doi: 10.1016/s0065-2660(08)60396-x. [DOI] [PubMed] [Google Scholar]
- Gould-Somero M, Holland L. The timing of RNA synthesis for spermiogenesis in organ cultures of Drosophila melanogastertestes. Wilhelm Roux's Arch Dev Biol. 1974;174:133–148. doi: 10.1007/BF00573626. [DOI] [PubMed] [Google Scholar]
- Guo Y, Gillan A, Török T, Kiss I, Dow JAT, Kaiser K. Site- selected mutagenesis of the Drosophila second chromosome via plasmid rescue of lethal P-element insertions. Genome Res. 1996;6:972–979. doi: 10.1101/gr.6.10.972. [DOI] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. Novel potential mitotic motor protein encoded by the fission yeast cut7+gene. Nature (Lond) 1990;347:563–565. doi: 10.1038/347563a0. [DOI] [PubMed] [Google Scholar]
- Hall DH, Hedgecock EM. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. . Cell. 1991;65:837–847. doi: 10.1016/0092-8674(91)90391-b. [DOI] [PubMed] [Google Scholar]
- Harper J, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hartenstein V, Rudloff E, Campos-Ortega JA. The pattern of proliferation of the neuroblasts in the wild-type embryo of Drosophila melanogaster. . Roux's Arch Dev Biol. 1987;196:473–485. doi: 10.1007/BF00399871. [DOI] [PubMed] [Google Scholar]
- Heck MMS, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LSB. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. . J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck PJ. Phosphorylation of neuronal kinesin heavy and light chains in vivo. . J Neurochem. 1993;60:2265–2275. doi: 10.1111/j.1471-4159.1993.tb03513.x. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiaekinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard MJ, Cohen P. On target with a new mechanism for the regulation of protein phosphorylation. TIBS. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- Kafatos FC, Louis C, Savakis C, Glover DM, Ashburner M, Link AJ, Siden-Kiamos I, Saunders RDC. Integrated maps of the Drosophilagenome: progress and prospects. Trends Genet. 1991;7:155–161. doi: 10.1016/0168-9525(91)90379-5. [DOI] [PubMed] [Google Scholar]
- Kashina AS, Bashin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature (Lond) 1996a;379:270. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashina AS, Scholey JM, Leszyk JD, Saxton WM. An essential bipolar mitotic motor. Nature (Lond) 1996b;380:550. doi: 10.1038/384225a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, R.C. 1970. Ovarian development in Drosophila melanogaster. Academic Press, New York.
- Kohli J. Telomeres lead chromosome movement. Curr Biol. 1994;4:724–727. doi: 10.1016/s0960-9822(00)00160-3. [DOI] [PubMed] [Google Scholar]
- Kumar J, Yu H, Sheetz MP. Kinectin, an essential anchor for kinesin-driven vesicle motility. Science (Wash DC) 1995;267:1834–1837. doi: 10.1126/science.7892610. [DOI] [PubMed] [Google Scholar]
- Lee KD, Hollenbeck PJ. Phosphorylation of kinesin in vivo correlates with organelle association and neurite outgrowth. J Biol Chem. 1995;270:5600–5605. doi: 10.1074/jbc.270.10.5600. [DOI] [PubMed] [Google Scholar]
- Lee KS, Yuan YL, Kuriyama R, Erikson RL. Plk is an M-phase– specific protein kinase and interacts with a kinesin-like protein, CHO1/ MKLP1. Mol Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees LF, Bulleid NJ. The role of cysteine residues in the folding and association of the COOH-terminal propeptide of types I and III procollagen. J Biol Chem. 1994;269:24354–24360. [PubMed] [Google Scholar]
- Lopez J, Song K, Hirshfeld A, Lin H, Wolfner MF. The fs(1)Ya protein of Drosophila is a component of the lamina of cleavage nuclei, pronuclei and polar bodies in early embryos. Dev Biol. 1994;163:202–211. doi: 10.1006/dbio.1994.1136. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science (Wash DC) 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mahowald, A.P., and M.P. Kambysellis. 1980. Oogenesis. In Genetics and Biology of Drosophila. M. Ashburner and T.R.F. Wright, editors. Academic Press, London. 141–224.
- McIlvain JM, Jr, Burkhardt JK, Hamm-Alvarez S, Argon Y, Sheetz MP. Regulation of kinesin activity by phosphorylation of kinesin-associated proteins. J Biol Chem. 1994;269:19176–19182. [PubMed] [Google Scholar]
- Moore JD, Endow SA. Kinesin proteins: a phylum of motors for microtubule-based motility. Bioessays. 1996;18:207–219. doi: 10.1002/bies.950180308. [DOI] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end- directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Nomura N, Nagase T, Miyajima N, Sazuka T, Tanaka A, Sato S, Seki N, Kawarabayasi Y, Ishikawa K, Tabata S. Prediction of the coding sequences of unidentified human genes. II. The coding sequences of 40 new genes (KIAA0041-KIAA0080) deduced by analysis of cDNA clones from human cell line KG-1. DNA Res. 1994;1:223–229. doi: 10.1093/dnares/1.5.223. [DOI] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 1995;81:769–780. doi: 10.1016/0092-8674(95)90538-3. [DOI] [PubMed] [Google Scholar]
- Olivieri G, Olivieri A. Autoradiographic study of nucleic acid synthesis during spermatogenesis in Drosophila melanogaster. . Mutat Res. 1965;2:366–380. doi: 10.1016/0027-5107(65)90072-2. [DOI] [PubMed] [Google Scholar]
- Otsuka AJ, Jeyaprakash A, Garcia-Anoveros J, Tang LZ, Fisk G, Hartshorne T, Franco R, Born T. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron. 1991;6:113–122. doi: 10.1016/0896-6273(91)90126-k. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz D, Benz WK, Engels WR. A stable source of P element transposase in Drosophila melanogaster. . Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov VI, Gyoeva FK, Gelfand VI. Kinesin is responsible for centrifugal movement of pigment granules in melanophores. Proc Natl Acad Sci USA. 1991;88:4956–4960. doi: 10.1073/pnas.88.11.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozdzial MM, Haimo LT. Bidirectional pigment granule movements of melanophores are regulated by protein phosphorylation and dephosphorylation. Cell. 1986;47:1061–1070. doi: 10.1016/0092-8674(86)90821-4. [DOI] [PubMed] [Google Scholar]
- Sammak PJ, Adams SR, Harootunian AT, Schliwa M, Tsien RY. Intracellular cyclic AMP, not calcium, determines the direction of vesicle movement in melanophores: direct measurement by fluorescence ratio imaging. J Cell Biol. 1992;117:57–72. doi: 10.1083/jcb.117.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawin KE, Mitchinson TJ. Mutations in the kinesin-like protein Eg5 disrupting localization to the mitotic spindle. Proc Natl Acad Sci USA. 1995;92:4289–4293. doi: 10.1073/pnas.92.10.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton WM, Hicks J, Goldstein LSB, Raff EC. Kinesin heavy chain is essential for viability and neuromuscular functions in Drosophila, but mutants show no defects in mitosis. Cell. 1991;64:1093–1102. doi: 10.1016/0092-8674(91)90264-y. [DOI] [PubMed] [Google Scholar]
- Shenolikar S. Protein serine-threonine phosphatases—new avenues for cell regulation. Annu Rev Cell Biol. 1994;10:55–86. doi: 10.1146/annurev.cb.10.110194.000415. [DOI] [PubMed] [Google Scholar]
- Siden-Kiamos I, Saunders RDC, Spanos L, Majerus T, Trenear J, Savakis C, Louis C, Glover DM, Ashburner M, Kafatos FC. Towards a physical map of the Drosophila melanogastergenome: mapping of cosmid clones within defined genomic divisions. Nucleic Acids Res. 1990;18:6261–6270. doi: 10.1093/nar/18.21.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A.C. 1993. Developmental genetics of oogenesis. In The Development of Drosophila melanogaster. M. Bate and A. Martinez Arias, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 71–147.
- Stewart RJ, Pesavento PA, Woerpel DN, Goldstein LSB. Identification and partial characterization of six members of the kinesin superfamily in Drosophila. . Proc Natl Acad Sci USA. 1991;88:8470–8474. doi: 10.1073/pnas.88.19.8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Minden JS, Alberts B. daughterless-abo-like, a Drosophilamaternal-effect mutation that exhibits abnormal centrosome separation during the late blastoderm divisions. Development (Camb) 1990;100:1–12. doi: 10.1242/dev.110.2.311. [DOI] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A nonradioactive in situ hybridisation method for the localisation of specific RNAs in Drosophila embryos reveals translational control of the gene hunchback. . Chromosoma (Berl) 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Tokai N, Fujimoto-Nishiyama A, Toyoshima Y, Yonemura S, Tsukita S, Inoue J, Yamamoto T. Kid, a novel kinesin-like DNA binding protein, is localised to chromosomes and the mitotic spindle. EMBO (Eur Mol Biol Organ) J. 1996;15:457–467. [PMC free article] [PubMed] [Google Scholar]
- Török T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: isolation of lethals with different overgrowth phenotypes. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima I, Yu H, Steuer ER, Sheetz MP. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eynde A, Wera S, Beullens M, Torreskens S, Van Leuven F, Stalmans W, Bollen M. Molecular cloning of NIPP-1, a nuclear inhibitor of protein phosphatase-1, reveals homology with polypeptides involved in RNA processing. J Biol Chem. 1995;270:28068–28074. doi: 10.1074/jbc.270.47.28068. [DOI] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopuskinesin like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ. Kinesin-related proteins at mitotic spindle poles: function and regulation. Cell. 1996;85:943–946. doi: 10.1016/s0092-8674(00)81295-7. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Mitchison TJ, Desai A. XKCM1: a Xenopuskinesin-related protein that regulates microtubule dynamics during mitotic spindle assembly. Cell. 1996;84:37–47. doi: 10.1016/s0092-8674(00)80991-5. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Adler R. Chromokinesin: a DNA-binding, kinesin–like protein. J Cell Biol. 1995;128:861–868. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White-Cooper H, Alphey L, Glover DM. The cdc25 homologue twine is required for only some aspects of the entry into meiosis in Drosophila. . J Cell Sci. 1993;106:1035–1044. doi: 10.1242/jcs.106.4.1035. [DOI] [PubMed] [Google Scholar]
- Williams BC, Reidy MF, Williams EV, Gatti M, Goldberg ML. The Drosophilakinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J Cell Biol. 1995;129:709–723. doi: 10.1083/jcb.129.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, MacRae TH, Baines AJ, Gull K. Definition of individual componenets within the cytoskeleton of Trypanosoma bruceiby a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yang JT, Laymon RA, Goldstein LS. A three-domain structure of kinesin heavy chain revealed by DNA sequence and microtubule binding analyses. Cell. 1989;56:879–889. doi: 10.1016/0092-8674(89)90692-2. [DOI] [PubMed] [Google Scholar]
- Yu H, Nicchitta CV, Kumar J, Becker M, Toyoshima I, Sheetz MP. Characterization of kinectin, a kinesin-binding protein: primary sequence and N-terminal topogenic signal analysis. Mol Biol Cell. 1995;6:171–183. doi: 10.1091/mbc.6.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]