Abstract
Nuclear dots containing PML and Sp100 proteins (NDs) play a role in the development of acute promyelocytic leukemia, are modified after infection with various viruses, and are autoimmunogenic in patients with primary biliary cirrhosis (PBC). PML and Sp100 gene expression is strongly enhanced by interferons (IFN). Based on immunostaining with a monoclonal antibody (mAb C8A2), a third protein, nuclear dot protein 52 (NDP52), was recently localized in NDs. Here we analyzed the cellular localization, expression, and structure of NDP52 in more detail. Our NDP52-specific sera revealed mainly cytoplasmic staining but no ND pattern, neither in untreated nor in IFN-treated cells. Cells transfected with NDP52 expression vectors showed exclusively cytoplasmic staining. In subcellular fractionation experiments, NDP52 was found in cytoplasmic and nuclear fractions. Unlike as described for Sp100 and PML, NDP52 mRNA and protein levels were only marginally enhanced by IFN γ and not enhanced at all by IFN β. NDP52 homodimerization but no heterodimerization with Sp100 or PML could be demonstrated. None of the 93 PBC sera tested contained autoantibodies against NDP52. Finally, mAb C8A2 reacted not only with NDP52 but also with a conformation-dependent epitope on the Sp100 protein. These data imply that NDP52 forms homodimers but no heterodimers with Sp100 and PML, lacks autoantigenicity in PBC, localizes mainly in the cytoplasm, and is associated with the nucleus, but not with NDs. Finally, unlike Sp100 and PML, NDP52 expression is neither markedly enhanced nor localization detectably altered by type I and II IFNs.
The nucleus of eukaryotic cells is a highly complex structure that consists of different domains as defined by structural and/or functional characteristics (Strouboulis and Wolffe, 1996). Nuclear dots (NDs)1 are structures of punctate shape within the cell nucleus and belong to the heterogeneous group of nuclear bodies (Brasch and Ochs, 1992). They were originally discovered as autoimmune targets in patients suffering from primary biliary cirrhosis (PBC), a chronic progressive liver disease of systemic autoimmune character (Bernstein et al., 1984; Powell et al., 1984). Since NDs do not colocalize with other known subnuclear structures such as spliceosomes, coiled bodies, interchromatin granules, or DNA-replication sites, they represent novel nuclear domains, recently also designated as nuclear domain 10 (ND10), PML-containing oncogenic domains (PODs), or Kr-Bodies (Ascoli and Maul, 1991; Dyck et al., 1994; Weis et al., 1994).
The first protein component of NDs characterized biochemically as well as by cloning and sequencing of the cDNA was the Sp100 protein (Szostecki et al., 1987, 1990), an interferon (IFN)-inducible acidic protein with a highly aberrant electrophoretic mobility and transcription transactivating properties (Xie et al., 1993; Guldner, H.H., C. Szostecki, and H. Will, manuscript submitted for publication). Unlike the single copy human Sp100, the homologous gene in mice, mSp100, is highly amplified and in some populations visible as an inherited homogeneously staining region on chromosome 1 (Plass et al., 1995; Grötzinger et al., 1996a ). As recently demonstrated, several alternatively spliced mRNAs and proteins are expressed from the human Sp100 gene (Szostecki, 1991; Xie et al., 1993; Dent et al., 1996; Grötzinger et al., 1996c ; Guldner, H.H., C. Szostecki, and H. Will, manuscript submitted for publication) similar for the PML gene (Fagioli et al., 1992; Pandolfi et al., 1992).
NDs gained broader interest when the PML protein, known to be expressed as a fusion protein with the retinoic acid receptor α (RARα) in patients with acute promyelocytic leukemia (de Thé et al., 1990), was localized in NDs. The PML/RARα fusion protein is a neooncogene aberrantly expressed as a result of the t(15;17) chromosomal translocation that is the hallmark of this disease (Rowley et al., 1977). Expression of the PML/RARα protein disrupts the normal ND pattern and leads to a microspeckled and partially cytoplasmic distribution of the proteins (Koken et al., 1994; Weis et al., 1994). This is probably sufficient for blocking of promyelocytic differentiation and transformation of the cells (Altabef et al., 1996). PML is induced by type I and II IFNs with similar kinetics as Sp100 (Chelbi-Alix et al., 1995; Lavau et al., 1995; Grötzinger et al., 1996c ), probably due to a similar but not identical promoter configuration (Grötzinger et al., 1996b ; Stadler et al., 1995). Finally, both PML and Sp100 are autoantigens in PBC (Szostecki et al., 1992; Sternsdorf et al., 1995). Autoantibodies against both antigens frequently co-occur in the same patients, suggesting a direct or indirect association of the two proteins in the same complex (Sternsdorf et al., 1995). Recently, a leukocyte-specific ND-component with high sequence similarities to Sp100 was identified by two independent groups. This protein is also autoantigenic in PBC and has been designated Sp140 (Bloch et al., 1996) or LYSP100 (Dent et al., 1996).
Other less well-studied proteins were localized in NDs by immunostaining with monoclonal antibodies (Ascoli and Maul, 1991; Kamei, 1995) or autoimmune sera (Zuber et al., 1995). Besides Sp100, Sp140, and PML, three further putative ND proteins, Int-6, PIC1, and nuclear dot protein 52 (NDP52), have recently been characterized genetically and in part also functionally. Similarly to PML, the Int-6 protein is involved in tumorigenesis (Desbois et al., 1996). The PIC1 protein was discovered because of its direct interaction with PML (Boddy et al., 1996).
The NDP52 cDNA was isolated by immunoscreening of an expression library (Korioth et al., 1995) with a monoclonal antibody (mAb C8A2) obtained by immunization of mice with crude nuclei fractions (Martelli et al., 1992). NDP52 is a protein with a predicted molecular mass of 52 kD. The carboxy terminus of the protein shows homology with LIM domains, and the central portion exhibits an extended coiled-coil domain containing a leucine zipper motif (Korioth et al., 1995). Similarly to PML and Sp100 (Carvalho et al., 1995), NDP52 was reported to relocate after infection with herpes simplex virus type 1 (HSV 1), adenovirus (Korioth et al., 1995; Ishov and Maul, 1996), and cytomegalovirus (Korioth et al., 1996). Similarly to PML and Sp100, IFN γ was reported to enhance the steady-state levels of NDP52 mRNA as shown by Northern blotting, and IFN type I and II treatment increased the size and number of NDP52-specific dots when immunostained with monoclonal antibody C8A2 (Korioth et al., 1995).
To confirm the NDP52 location in NDs and to learn more about the architecture of NDs, we analyzed the NDP52 protein in more detail. We found that mAb C8A2 does not only react with the NDP52 protein but also with Sp100. Our polyclonal anti-NDP52 sera showed mainly cytoplasmic and diffuse nuclear staining but no ND staining. Furthermore, unlike the case for PML and Sp100, NDP52 mRNA and protein steady-state levels were not found to increase after treatment with IFN β and only weakly after IFN γ. Finally, strong homomeric NDP52 protein/protein interaction was demonstrated. Taken together, these data indicate that NDP52 is mainly a dimeric/multimeric cytoplasmic and nucleus-associated protein but not an ND-associated protein.
Materials and Methods
Isolation of RNA and Northern Blotting
Total RNA was isolated according to the acidic phenol/guanidiniumthiocyanate method using standard protocols (Chomczynski and Sacchi, 1987). Northern blotting and hybridization of RNAs with 32P-labeled probes were performed as described (Sambrook et al., 1989). Blots were exposed to imaging screens (Fuji Photo Film Co., Tokyo, Japan) and the signals were quantitated by a bio imaging analyzer (model Fujix BAS 2000; Fuji Photo Film Co.).
Reverse Transcription, PCR Amplification, Cloning, and DNA Sequencing
100 ng total RNA isolated from HeLa S3 cells were incubated with an anchored oligo (dT)-primer (5′-TTTTTTTTTTTAA-3′) for 10 min at 65°C and reverse transcribed using SuperScript reverse transcriptase (Stratagene, La Jolla, CA). PCR was performed in several independent 50-μl reaction mixtures containing 50 mM Tris-HCl, pH 9.1, 3.5 mM MgCl2, 16 mM (NH4)2SO4, 150 μg/ml BSA, 0.2 mM dNTPs, 20 pmol of each primer, and a mixture of 2.5 U of Taq DNA polymerase (Boehringer Mannheim, Penzberg, FRG) and 0.08 U Pfu DNA polymerase (Stratagene) (Barnes, 1994). As primers, oligonucleotides NDP-5′ (5′-GCCGCTGCTGGTTGCTGTCC-3′) and NDP-3′ (5′-TCAGGTTAGGCAGGATCTTGACTC-3′) were used. DNA fragments were amplified for 40 cycles in a Thermocycler (HyBaid Omnigene; MWG Biotech, Ebersberg, FRG). Reactions were performed with the profile 30 s 95°C, 1 min 56°C, and 3 min 72°C. PCR reaction products were pooled, gel purified, and cloned into the AT-cloning vector pXcm-Kan12, which resulted in plasmid pX-NDP. This vector is a pUC18-derivative containing the kanamycin (kan) resistance gene flanked by two synthetic XcmI restriction enzyme cleavage sites. Upon restriction cleavage of this plasmid with XcmI, the kan gene is released and single-T overhangs are created at the ends of the linearized vector. PCR fragments inserted into this vector were excised by BamHI digestion.
Three independently cloned NDP52 cDNAs were sequenced using the Taq-Cycle sequencing protocol with IRD-41–labeled primers (MWG Biotech) and a Li-COR automated sequencing device.
For expression of NDP52 as a fusion protein, the NDP52 cDNA obtained by BamHI digestion from plasmid pX-NDP was cloned in-frame into the prokaryotic expression vector pEx42 a, b, or c (Stemler et al., 1990) linearized by BamHI. The protein expressed contains at its NH2 terminus phage MS2-polymerase sequences (14 kD) followed by the full-length NDP52 sequence. Production of recombinant Sp100 protein fragments (Sp-AB, -CD, -DF, -GH, and -FL) used for immunoblotting with mAb C8A2 is described elsewhere (Szostecki et al., 1992). For in vitro transcription and expression in eukaryotic cells, the same BamHI fragment was inserted into the BamHI site of plasmid pSG5 (Green et al., 1988), which resulted in plasmid pSG5-NDP.
Cells, Cytokines, Transfections, and Indirect Immunofluorescence Microscopy
Human HEp-2, HeLa S3, MG63, and Rat R1H cells (Zywietz et al., 1995) were maintained in DME (GIBCO BRL, Gaithersburg, MD) supplemented with 10% FCS. Human IFN β and γ were used at a concentration of 1,000 U/ml and added to the medium for 16–20 h. Plasmid DNAs were introduced into cells by the calcium phosphate procedure (Sambrook et al., 1989). 1–5 μg of pSG5-NDP, pSG5-Sp100, or pSG5-PML plasmids adjusted to 10 μg DNA with pUC19 were precipitated per 6-cm dish. Calcium phosphate–DNA coprecipitates were left on the cells overnight and removed by changing the medium, and cells were then incubated for a further 24 h. For indirect immunofluorescence staining, cells were grown on coverslips and treated as indicated. Cells were fixed either in 1% freshly prepared paraformaldehyde in PBS (8 mM Na2HPO4, 1.5 mM KHPO4, 140 mM NaCl, 2.6 mM KCl, pH 7.3) for 5 min, followed by permeabilization in PBS containing 0.2% Triton X-100 and 20 mM glycine for 20 min (at room temperature), or at −20°C for 5 min in methanol and for 20 s in acetone. Rabbit or rat anti-NDP52 antisera were diluted from 1:200– 1:1,600 in PBS as indicated. The rat anti-Sp100 (Sp26) and rat anti-PML (anti–PML-N) antibodies were diluted 1:400. The dilutions of the monoclonal antibodies (in PBS) are indicated in the text. Cells were incubated with antibodies for 30 min at room temperature. For detection, DTAF- or LRSC-conjugated donkey anti–mouse, goat anti–rabbit, or donkey anti– rat IgG antibodies (Dianova GmbH, Hamburg, FRG) were diluted 1:200 in PBS and incubated as above.
Mammalian Two-Hybrid Interaction Assay
For in vivo interaction studies, the CLONTECH MATCHMAKER mammalian two-hybrid assay system (CLONTECH Labs, Palo Alto, CA) and HuH-7 hepatoma cells were used. Full-length NDP52, PML, and Sp100 cDNAs were each cloned in-frame into plasmids pM and pVP16 for expression of the corresponding fusion proteins with the Gal4 DNA binding domain and the VP16 transactivation domain, respectively. Full-length PML and NDP52 were cloned as EcoRI-BamHI fragments and Sp100 cDNA as EcoRI-SalI fragment. Reporter gene assays were performed using plasmid pG5CAT according to the manufacturer's protocol (CLONTECH Labs). Cells on a 60-mm Petri dish were transfected with 0.2 pmol pM and pVP16 constructs, 0.7 pmol of the reporter construct, and 1 μg of pCMV-βGal to standardize transfection efficiencies. CAT activity was determined with a commercial CAT-ELISA (Boehringer Mannheim).
Cell Extracts, Immunoblotting, and Antibodies
Preparation of total cellular protein extracts, SDS-PAGE (10 or 12.5% polyacrylamide) of proteins, and transfer onto nitrocellulose filters were carried out according to standard protocols (Sambrook et al., 1989). Blots were blocked in 5% (wt/vol) dry milk in water and incubated with primary antibodies in the same buffer. After washing four times in TBS (10 mM Tris-HCl, pH 7.6, 150 mM NaCl) or PBS, blots were incubated with horseradish peroxidase–conjugated secondary antibody (Dianova GmbH) as described above. For visualization of proteins, enhanced chemiluminescence was used (Amersham, Buckinghamshire, UK). Anti-NDP antibodies and human autoimmune sera were diluted 1:1,000 and 1:2,000, respectively. The mAb C8A2 was diluted 1:5 in 5% dry milk (in water) or 0.5% dry milk (in PBS). Secondary antibodies were diluted 1:10,000. Antibodies against gel-purified MS2–NDP52 fusion protein were produced by immunization of rats and rabbits with the antigen emulsified in Freund's adjuvants. Sera from patients with clinically and histologically proven PBC were provided by Dr. E.J. Heathcote (The Toronto Hospital, Toronto, Ontario, Canada); Drs. M.C. Jung, U. Spengler, and G.R. Pape (Institut für Immunologie, Universität München, FRG); R. Klein and P.A. Berg (Medizinische Klinik und Poliklinik, Universität Tübingen, FRG); H.-J. Lakomek (Klinik für Rheumatologie und physikalische Medizin, Minden, FRG); and M. Manns (Medizinische Hochschule, Hannover, FRG).
ELISA
Enzyme-linked immunosorbent assay (ELISA) was essentially carried out as described previously (Szostecki et al., 1992). Briefly, ELISA plates were coated with recombinant gel-purified NDP52 protein in 8 M urea for 2 h at room temperature. After washing the plates were blocked overnight at 4°C, washed, and incubated with a 1:100 dilution of PBC sera in PBS. Bound autoantibodies were detected with peroxidase-coupled antibodies against human IgG and orthophenylenediamine as substrate. OD492 values of 0.3 or below after 20 min of incubation obtained with control sera from healthy persons were defined as negative. In this test, an anti-Sp100 positive control PBC-serum resulted in an OD492 of 2.5.
Subcellular Fractionation
Cells grown as monolayer were harvested, washed once with PBS, and resuspended in hypotonic buffer A (10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM PMSF) (Dignam et al., 1983). After 15 min of swelling on ice, cells were homogenized carefully by 20–25 strokes in a Kontes all-glass Dounce homogenizer (Type B pestil). After centrifugation at 800 g for 10 min (4°C), the resulting supernatant was transferred into a new tube, cleared by a second centrifugation step at 10,000 g, and precipitated by addition of 10 vol ice-cold acetone. After drying, the precipitate was boiled for 10 min in 4 vol 2× SDS-loading buffer of the initial cell pellet. The pellet containing crude nuclei was washed in a large volume of buffer A and resuspended in 2 vol buffer A′ (buffer A supplemented with 0.5 mM DTT and 1% NP-40) of the initial cell pellet. After incubation on ice for 10 min, the supernatant was removed, cleared as described above, mixed with an equal volume of 2× SDS-loading buffer (Sambrook et al., 1989), and boiled for 10 min. The remaining nuclei were again washed in buffer A′, pelleted at 10,000 g, resuspended in 4 vol 2× SDS-loading buffer of the initial cell pellet, sonicated three times for 20 s, and boiled for 10 min. Cell extracts were stored at −20°C.
Synthesis of Radiolabeled PML, Sp100, NDP52, and Luciferase Proteins and Immunoprecipitation
In vitro transcription/translation and immunoprecipitation was carried out as described previously (Sternsdorf et al., 1995). The plasmids pSG5-PML (de Thé et al., 1991), pSp64-Sp100 (Sp100 cDNA inserted as HindIII-EcoRI fragment into plasmid pSp64), and pSG5-NDP were linearized by digestion with appropriate restriction enzymes. From these plasmids, run-off RNAs encoding full-length PML, Sp100, and NDP52 proteins, respectively, were produced using standard protocols for T7 (pSG5-PML, pSG5-NDP) and Sp6 (pSp64-Sp100) RNA polymerase. In vitro translation was carried out in rabbit reticulocyte lysate (Promega Corp., Madison, WI) with or without the presence of [35S]methionine according to the manufacturer's protocol. In addition, [35S]methionine-labeled luciferase translated in this system from the corresponding mRNA (Promega Corp.) was used as a control. An aliquot of each translation mixture was analyzed by SDS-PAGE to determine the amounts of radiolabeled protein. For interaction studies, the immunoprecipitates were washed in PBS. For immunoprecipitation with autoimmune sera, NETGel Buffer (Sambrook et al., 1989) was used. Gels were washed in 5% sodium acetate, dried, and exposed to x-ray films or imaging screens (Fuji Photo Film Co.).
Results
Cloning and Sequencing of the NDP52 cDNA
A full-length NDP52 cDNA was obtained by reverse transcriptase PCR using RNA from HeLa S3 cells. To exclude artificial mutations introduced during amplification, three independent reactions were performed. The resulting 1.4-kb NDP52 cDNA fragments were pooled and cloned, and finally three independent cDNA inserts were sequenced. One of the three clones was identical in sequence to that isolated previously from osteosarcoma cell line MG63 (Korioth et al., 1995), except for a single C to G nucleotide change at position 1219. The point mutation predicts an amino acid substitution of proline by alanine at amino acid position 389. Since this substitution was present in all three clones, it corresponds most likely to an allelic sequence variation of the NDP52 gene of HeLa S3 cells. This clone was used for further analysis, including the expression of recombinant NDP52 protein for the generation of polyclonal antisera in rabbit and rat. The other two clones contained one and four additional point mutations, respectively, presumably introduced during PCR reaction. The allelic exchange is located downstream of the predicted coiled-coil region (amino acids 134–350) and upstream of the LIM-homologous region (amino acids 395–446). Therefore, it is unlikely to affect these two putative functional domains. Furthermore, no obvious potential nuclear localization signal is present in the vicinity of the amino acid substitution. Taken together, it is unlikely that the NDP52 proteins expressed from the allelic genes in HeLa S3 and MG63 cells behave differently.
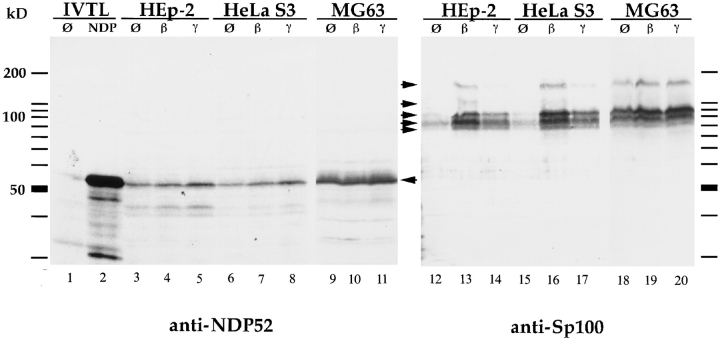
IFN Inducibility of NDP52
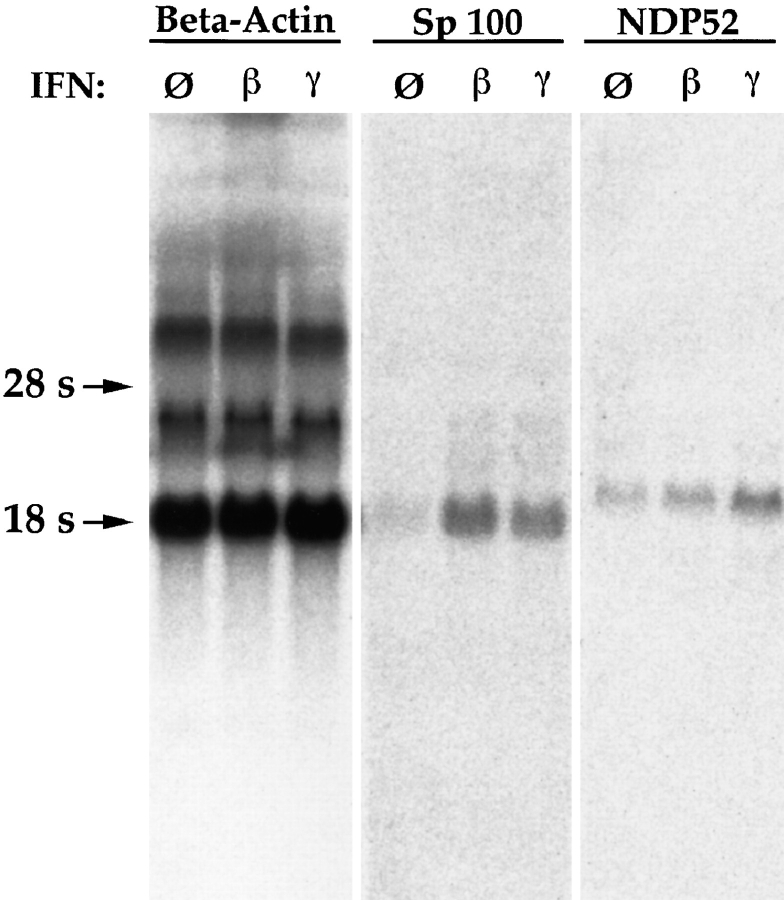
In the previous report (Korioth et al., 1995), enhanced NDP52 gene expression after IFN treatment of cells was demonstrated by immunofluorescence staining and Northern blotting. To demonstrate IFN-enhanced NDP52 protein expression also by immunoblotting and to test the specificity of the raised antisera, we used total cell extracts of untreated and IFN-stimulated cells and, as a control, in vitro–translated NDP52. Both sera generated by us recognized a protein of ∼55–58 kD in extracts of HEp-2, HeLa S3, and MG63 cells that comigrated with the in vitro– translated NDP52 protein (shown for the rabbit antiserum in Fig. 1, left; NDP52 indicated by arrow). This experiment shows that detection of NDP52 by immunoblotting can be achieved with total cell lysates. Taken together, our data demonstrate the specificity of the sera that allow highly sensitive detection of NDP52 and suggest that the NDP52 protein is not subject to major posttranslational modifications. Unexpectedly, these antibodies revealed no increase of NDP52 in three different IFN β–treated cell lines compared to nontreated cells and showed only a very minor increase in IFN γ–treated cells (Fig. 1, left, lanes β and γ). In contrast, immunoblots performed with the same extracts, and an Sp100-specific antibody showed the expected strong inducibility of Sp100 by IFN β and less strong inducibility by IFN γ in HEp-2 and HeLa S3 cells (Fig. 1, right; Sp100-specific bands indicated by arrows). In MG63 cells, IFN-increased Sp100 expression was hardly detectable, presumably because of a constitutive high level before treatment (Fig. 1, right). To test whether only the NDP52 mRNA (Korioth et al., 1995) but not the NDP52 protein levels are enhanced by IFN, we measured the NDP52 and, as a control, the Sp100 mRNA levels in IFN-treated and untreated HEp-2 cells. HEp-2 cells were chosen for this experiment since they were found to be highly responsive to IFN treatment on the protein level (see above). Northern blotting did not reveal an increase of NDP52 mRNA levels by IFN β (Fig. 2, NDP52) and revealed only a slight increase by IFN γ (twofold, as determined by quantitation of the image normalized by β-actin), whereas the Sp100 mRNA levels increased as expected (Fig. 2, Sp100). Taken together, these data demonstrate that IFN γ increases NDP52 gene expression only slightly and IFN β does not increase expression at all in any of the three cell lines tested.
Figure 1.
Immunoblot analysis of in vitro–translated protein and total cell extracts of HEp-2, HeLa S3, and MG63 cells, separated by 12.5% SDS-PAGE. In vitro–translated NDP52 detected by the polyclonal rabbit anti-NDP52 antiserum exhibits an electrophoretic mobility corresponding to a protein of 56–58 kD (lane 2). No specific reaction is seen using reticulocyte lysate alone (lane 1). In HEp-2, HeLa S3, and MG63 cells, endogenous NDP52 has the same electrophoretic mobility (lanes 3–11, marked by an arrow). Treatment of cells with IFN β (lanes 4, 7, and 10) and γ (lanes 5, 8, and 11) does not increase the amount of NDP52 (IFN β), or only increases it marginally (IFN γ). Immunoblot analysis of the same extracts using a polyclonal rat anti-Sp100 antiserum (lanes 12–20) in the HEp-2 and HeLa S3 cell lines shows a strong induction of Sp100 proteins (specific complexes indicated by arrows) by IFN β (lanes 13 and 16) and a weaker induction by IFN γ (lanes 14 and 17). MG63 cells, in contrast, showed only weak inducibility by both IFN β (lane 19) and IFN γ (lane 20).
Figure 2.
IFN inducibility of NDP52 and Sp100 mRNAs as determined by Northern blot analysis. Total RNA of untreated (lanes labeled ∅), IFN β– (lanes labeled β), and IFN γ– (lanes labeled γ) treated HEp-2 cells was analyzed using radiolabeled full-length NDP52 (lanes labeled NDP52), Sp100 (lanes labeled Sp100), and β-actin (lanes labeled Beta-Actin) cDNAs as probes. The amount of Sp100 mRNA is increased by IFNs β and γ (fifth and sixth lanes, respectively). The NDP52 mRNA level is not significantly increased by IFN β and only weakly by IFN γ (eighth and ninth lanes, respectively).
Intracellular Localization of NDP52 Protein as Studied by Indirect Immunofluorescence Staining
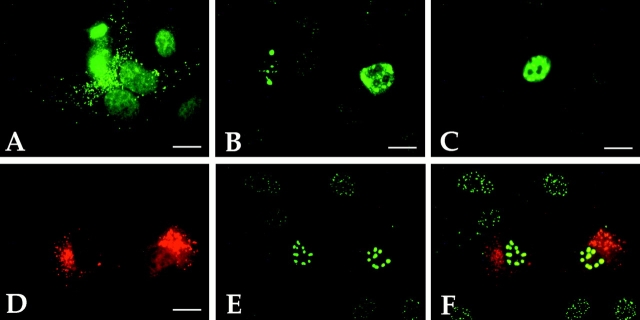
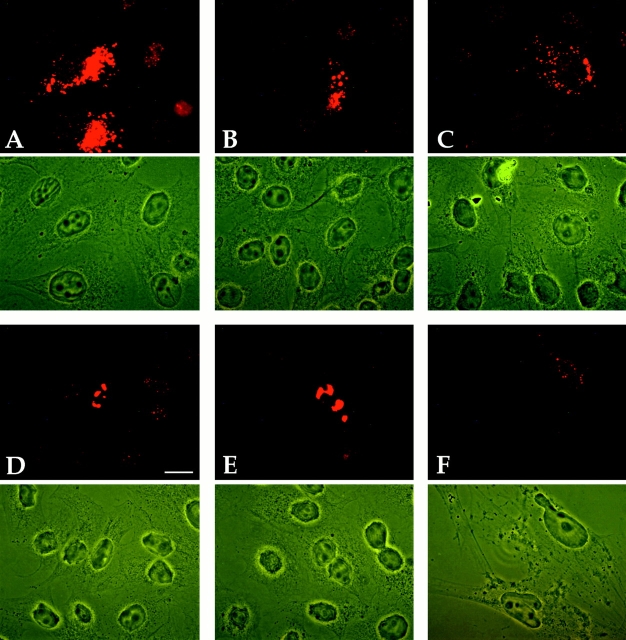
As a prerequisite for studies on the interaction of NDP52 with other ND proteins, we analyzed the suitability of the polyclonal anti-NDP52 antisera for detection of NDP52 protein expressed from the endogenous genes or from transfected plasmids in HeLa S3 and HEp-2 cells by immunofluorescence staining. In addition, the MG63 osteosarcoma cell line used for the initial localization studies (Korioth et al., 1995) was also tested here because it was reported to express high levels of endogenous NDP52. Untransfected MG63 cells treated with IFN γ for maximal expression of NDP52 showed weak staining only, which required long exposure times for photographic documentation (Fig. 3). Both sera used resulted in a coarse-grained cytoplasmic staining often concentrated adjacent to the nucleus and in addition, depending on cell type and fixation conditions, a diffuse nuclear staining (Fig. 3 B, rat anti-NDP52 and Fig. 3 C, rabbit anti-NDP52). No ND staining could be observed, irrespective of the fixation conditions and the cell type used. The diffuse nuclear staining was most prominent in HEp-2 cells after paraformaldehyde fixation (Fig. 3 B) and hardly visible in HeLa S3 cells after methanol-acetone fixation (Fig. 3 C, nontransfected cells). Control staining with preimmune sera also showed some nonspecific nuclear staining in paraformaldehyde-fixed cells when exposed for the same time. This indicates that the nuclear staining observed is in part or totally caused by binding of unspecific antibodies already present in the preimmune sera (Fig. 3 A). Both the intensity and the staining patterns were not influenced by IFN β or γ treatment in HeLa S3, HEp-2, and MG63 cells (data not shown). In no case could ND staining be detected with any of our NDP52-specific polyclonal antisera. Note that the same cells of Fig. 3 B stained in addition with a polyclonal rat anti-Sp100 serum showed the typical ND pattern in the nucleus (Fig. 3 B′).
Figure 3.
Immunofluorescence staining of IFN γ–treated MG63 cells fixed with paraformaldehyde (A and B), methanol/acetone-fixed HeLa S3 cells (C) transiently transfected with plasmid pSG5-NDP, and HEp-2 cells fixed with paraformaldehyde. Cells were incubated with rat preimmune serum (diluted 1:200) (A), polyclonal rat anti-NDP52 (diluted 1:200) (B and D), rabbit anti-NDP52 (diluted 1:800) (C), rabbit anti-Sp100 (diluted 1:400) (B′), and mAb C8A2 (diluted 1:3) (E). The anti-NDP52 polyclonal antiserum fails to detect NDs but shows diffuse nuclear and granular cytoplasmic staining (B and D), whereas the anti-Sp100 antiserum shows the typical ND staining in the same cells (B′). The corresponding rat-preimmune serum shows nonspecific diffuse nuclear staining (A) when photographic exposure was identical to B. Hardly any nuclear staining is detected in methanol/ acetone-fixed HeLa S3 cells (C). NDP52 overexpressed in transfected cells localizes predominantly in the cytoplasm (C). HEp-2 cells labeled with mAb C8A2 show the typical ND staining with additional diffuse nuclear and weaker coarse-grained cytoplasmic labeling (E, long exposure). Except for the lack of ND staining, a very similar pattern is observed when HEp-2 cells are labeled with the polyclonal rat anti-NDP52 antiserum (D). In A–C, phase contrast pictures are adjacent to the corresponding immunofluorescence staining photographs. Bars, 10 μm.
These data raised the question as to whether our sera recognize NDP52 in immunofluorescence analysis at all. We first compared the staining of HEp-2 cells by mAb C8A2 and our polyclonal rabbit anti-NDP52 serum. With both antibodies, coarse-grained cytoplasmic and diffuse nuclear staining was observed (Fig. 3, E and D), but mAb C8A2 in addition stained NDs. This finding implies that our serum either does not recognize NDP52 or that mAb C8A2 also cross-reacts with an ND-associated protein. To examine this directly, we first transiently transfected HeLa S3 and HEp-2 cells with the pSG5 expression vector containing the NDP52 cDNA under the control of the SV-40 promoter (pSG5-NDP) and analyzed the cells by indirect immunofluorescence. In the vast majority of these cells, overexpressed NDP52 was localized in the cytoplasm, and in very few cells, it was also found diffusely in the nucleus (Figs. 3 C and 4, A and D). In contrast, Sp100 or PML proteins transiently overexpressed in cells transfected with the corresponding plasmids localized, as expected, predominantly in the nucleus (Fig. 4, B and C, respectively). Serial dilutions of our NDP52 antisera unequivocally detected transiently overexpressed NDP52 protein in transfected cells up to the highest dilution tested (1:1,600; data not shown). These results show that the polyclonal sera react with NDP52 in eukaryotic cells even at high serum dilutions and strongly indicate that the staining pattern seen in nontransfected cells reflects the distribution of the endogenous NDP52 protein.
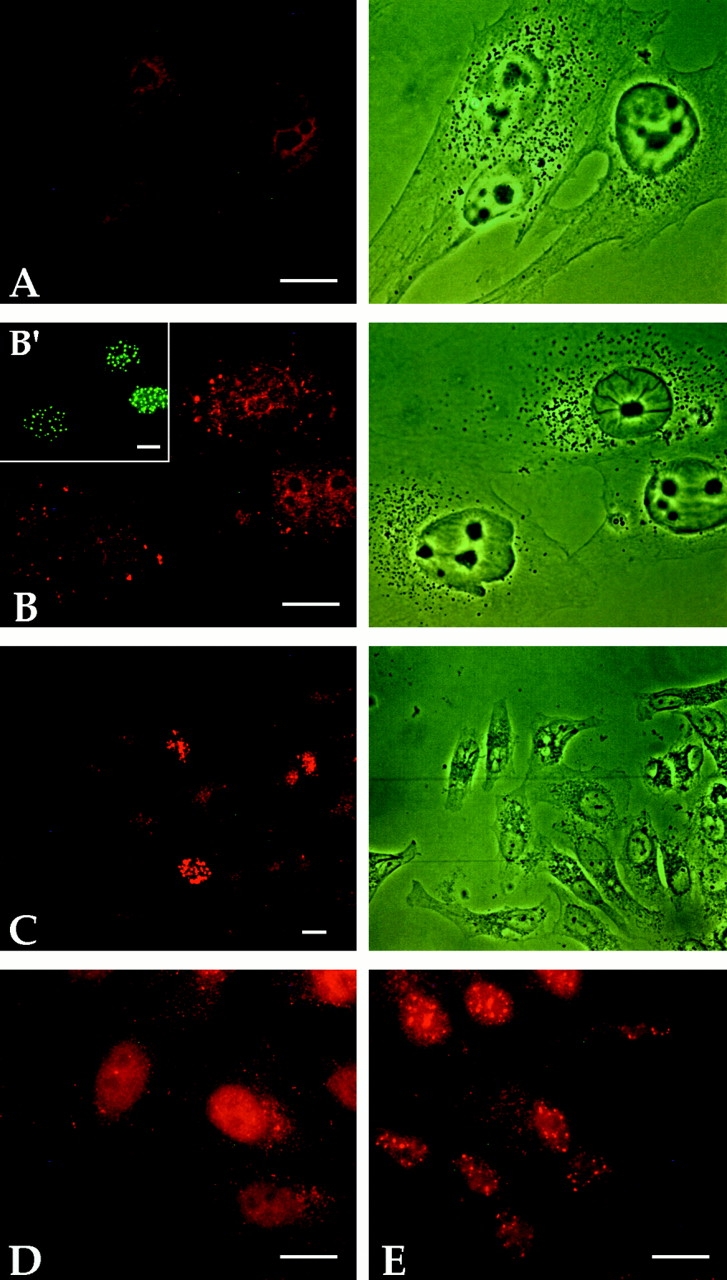
Figure 4.
Immunofluorescence staining of paraformaldehyde-fixed HEp-2 cells transfected with NDP52 (A), Sp100 (B), and PML (C) expression vectors and of methanol/acetone-fixed HEp-2 cells cotransfected with NDP52 and PML expression vectors (D–F). For detection of expressed proteins, the following polyclonal sera (diluted 1:400) have been used: rat anti-NDP52 (A), rat anti-Sp100 (B), rat anti-PML (C), and a mixture of rabbit anti-NDP52 and rat anti-PML (D–F). Sp100 and PML expressed in transfected cells show nuclear localization (B and C, respectively) whereas NDP52 localizes in the cytoplasm (A). Cells cotransfected with NDP52- and PML-expression vectors also exhibit nuclear localization of PML (E, green label) and predominantly cytoplasmic localization of NDP52 (D, red label) within the same cell. The transfected cell on the right in D shows, in addition, some NDP52 diffusely distributed in the nucleus. F is an overlay of the staining patterns shown in D and E.
To address the question of whether the cytoplasmic localization of NDP52, when expressed from a recombinant plasmid, might be due to an overload of the nuclear import mechanism of the cell, we cotransfected HEp-2 cells with NDP52 and PML expression vectors (Fig. 4). Cells expressing moderate levels of NDP52 showed mostly coarse-grained cytoplasmic staining (Fig. 4 D, left cell), whereas cells expressing high levels of the protein also exhibited diffuse nuclear staining (Fig. 4 D, right cell). At this exposure time, endogenous NDP52 is not visible. As expected, the same cells stained with a PML-specific antibody showed PML localization exclusively in nuclear dots in transfected and nontransfected cells (Fig. 4 E). Fig. 4 F shows both anti-NDP52 and anti-PML staining patterns superimposed. Cotransfection experiments using NDP52 and Sp100 expression vectors yielded similar results (data not shown). These data indicate that cytoplasmic localization of NDP52 expressed in transfected cells is not due to impaired nuclear import.
Subcellular Localization of NDP52 by Biochemical Fractionation
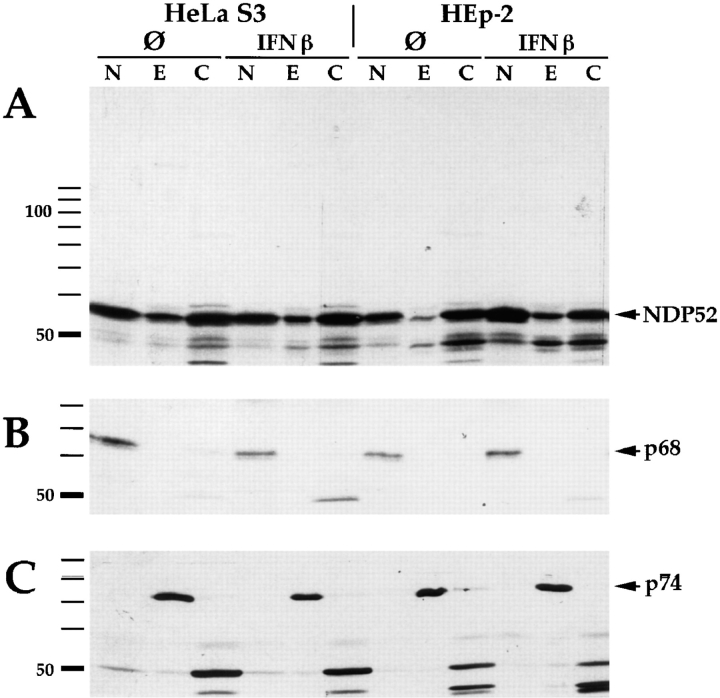
Because of the intriguing discrepancies between the intracellular localization of the NDP52 protein and the ND proteins PML as well as Sp100 in transfected cells and because of the lack of ND-specific staining with the polyclonal antisera also in nontransfected cells, we determined the intracellular localization of endogenous NDP52 by nucleo-cytoplasmic fractionation and immunoblotting. Since ND-specific staining after IFN treatment, as described for HeLa cells, could be a result not only of increased amounts of the protein but also of a different distribution within the cell as speculated previously (Korioth et al., 1995), both untreated and IFN-treated HEp-2 as well as HeLa S3 cells were biochemically subfractionated. Endogenous NDP52 was detected in the soluble cytoplasmic, the nuclear, and, to a minor extent, the detergent-extracted cytoskeleton-associated fraction (Fig. 5 A, lanes C, N, and E, respectively). No major NDP52 protein redistribution was observed after treatment with IFN β or γ (data not shown). The purity of subcellular fractions was checked by control incubations with human autoimmune sera recognizing either the snRNP protein p68 localizing in the nucleus (Theissen et al., 1986; Netter et al., 1990; Lamond and Carmo-Fonseca, 1993) or the dihydrolipoamide acetyltransferase (74 kD, p74), which is a mitochondrial protein (Van de Water et al., 1988). As expected, p68 was found exclusively in the nuclear fraction (Fig. 5 B, arrow), and p74 localized in the cytoskeleton-associated fraction (Fig. 5 C, arrow). These data indicate that NDP52 is localized in the cytoplasm but is also associated with the nucleus. Our data further indicate that no significant redistribution of NDP52 takes place between the cellular compartments after IFN treatment. In addition, preliminary nucleoplasma/ nuclear matrix fractionation experiments using an established procedure (Fey et al., 1986) also showed no redistribution after IFN treatment (tested for IFN β, data not shown).
Figure 5.
Nucleo-cytoplasmic fractionation of HEp-2 and HeLa S3 cells. Cell fractions were separated by SDS-PAGE and analyzed by immunoblotting: Lanes labeled N, nuclear proteins; lanes labeled E, nucleus/cytoskeleton-associated proteins (designated E for extraction); lanes labeled C, the soluble cytoplasmic fraction. A shows detection of NDP52 (indicated by arrow) using rabbit anti-NDP52 antiserum. NDP52 is detected in all three subfractions. No redistribution is observed after treatment of the cells with IFN β. As a control for purity of the subcellular fractions, immunoblotting of the same extracts was performed using human autoimmune sera recognizing the snRNP p68 protein, which localizes in the nucleus (B, arrow) or the mitochondrial p74 protein, which localizes in the cytoskeleton fraction (C, arrow).
NDP52 Interaction Studies Using Immunoprecipitation
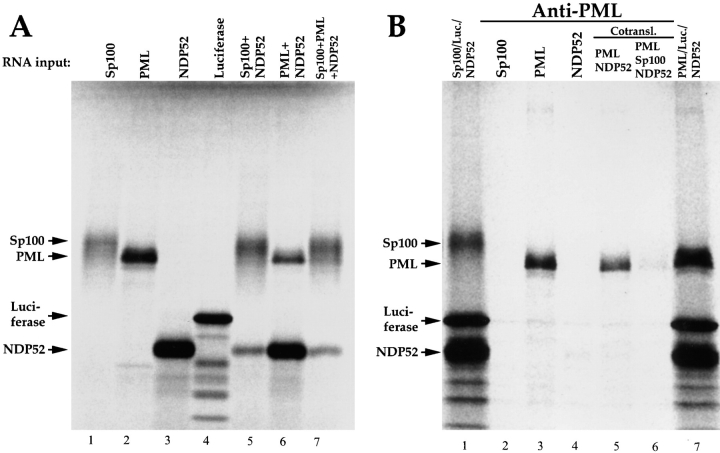
We next examined whether our failure to detect NDP52 in NDs is due to the fact that only a very limited number of epitopes is accessible to antibodies in immunofluorescence analysis because of direct binding of NDP52 to PML and/ or Sp100, which would make NDP52 detection with our polyclonal sera difficult. Sp100, PML, and NDP52 were cotranslated with [35S]methionine in vitro in various combinations to allow complex formation in statu nascendi. Proteins translated individually or together in one translation reaction were separated by SDS-PAGE (Fig. 6 A). When Sp100 was cotranslated with PML or NDP52, the amount of the latter two proteins was reduced, presumably because of the particularly strong translation initiation sequence context of the synthetic Sp100 mRNA used. As a specificity control in the immunoprecipitation experiments, in vitro–translated luciferase was added to the reaction after cotranslation. Immunoprecipitation was performed with rabbit anti-PML (Fig. 6 B) or rabbit anti-Sp100 (not shown) antibodies, and the precipitates were analyzed by SDS-PAGE and autoradiography. If a complex forms, one or both of the other ND proteins should coprecipitate. Using anti-PML antibodies, the PML protein was precipitated, but no coprecipitation of Sp100 or NDP52 occurred (Fig. 6 B). This argues against a direct interaction between any of the three in vitro–translated proteins.
Figure 6.
In vitro–translated NDP52 does not interact with PML or Sp100. 35S-labeled proteins were generated by in vitro transcription/ translation. A shows the translation products obtained with Sp100, PML, NDP52, or luciferase RNAs alone (lanes 1–4) and with combinations of some of these RNAs when translated in the same tube (lanes 5–7). All proteins were analyzed by SDS-PAGE and subsequent autoradiography. Immunoprecipitation with rabbit anti-PML serum results in precipitation of the PML protein only (B, lanes 3, 5, and 6). No coprecipitation of NDP52 or Sp100 is observed after cotranslation of the proteins (lanes 5 and 6). Luciferase was added to all precipitation reactions as a negative control. Lanes 1 and 7 represent mixtures of the translation reactions without precipitation as indicated.
NDP52 Interaction Studies Using the Two-Hybrid System in Eukaryotic Cells
To test potential interactions between NDP52 and PML and/or Sp100 in vivo, we used the two-hybrid assay in mammalian cells. All three proteins were expressed as fusion proteins with either the yeast Gal4 DNA binding domain (plasmids pM-PML, pM-Sp100, and pM-NDP) or the herpes simplex virus VP16 transactivating domain (plasmids pV-PML, pV-Sp100, and pV-NDP) containing an additional nuclear localization signal. As a reporter gene, a plasmid containing a Gal4-responsive promoter upstream of the CAT gene was used. Plasmids expressing p53 and SV-40 large T (TAg) fusion proteins served as positive controls (plasmids pM53 and pV-T). Results of the transfection experiments are summarized in Table I. Cotransfection of pM-NDP or pV-NDP with the corresponding PML or Sp100 construct did not result in CAT activity significantly above the negative controls (constructs with a viral core protein; data not shown). In contrast, cotransfection of pV-NDP/pM-NDP or pV-PML/ pM-PML showed a very strong and intermediate positive signal, respectively. These data suggest that none of the three proteins bind directly to each other and demonstrate that both NDP52 and PML homodimerize. Homodimerization of PML was shown previously by using different techniques (Perez et al., 1993).
Table I.
In Vivo Interaction Studies Using the Mammalian Two-Hybrid Assay
| pV-NDP | pV-Sp100 | pV-PML | pV-largeT | |||||
|---|---|---|---|---|---|---|---|---|
| pg | ||||||||
| pM-NDP | 290.3 | < | < | ND | ||||
| 296.5 | 1.1 | 1.7 | ||||||
| pM-Sp100 | < | ND | 1.6 | ND | ||||
| 0.4 | 1.8 | |||||||
| pM-PML | < | 2.4 | 28.0 | ND | ||||
| < | 2.5 | 20.2 | ||||||
| pM-53 | ND | ND | ND | 186.4 | ||||
| 251.1 | ||||||||
Values from two representative experiments are given. The Ga14 (pM-) and VP16 (pV-) constructs used in each respective cotransfection can be seen on the left side and on top of the scheme, respectively. Shown are the amounts of CAT protein in picograms in a 200-μl assay volume standardized to equal amounts of β-galactosidase expression. No NDP52 construct gives a signal above background when cotransfected with the PML or the Sp100 constructs. The pV-NDP/pM-NDP combination and the pV-PML/pM-PML combination, however, show a strong or intermediately strong positive signal, respectively, indicating that NDP52, like PML, homodimerizes. <, below detection limit.
Screening of PBC Sera for Anti-NDP52 Autoantibodies
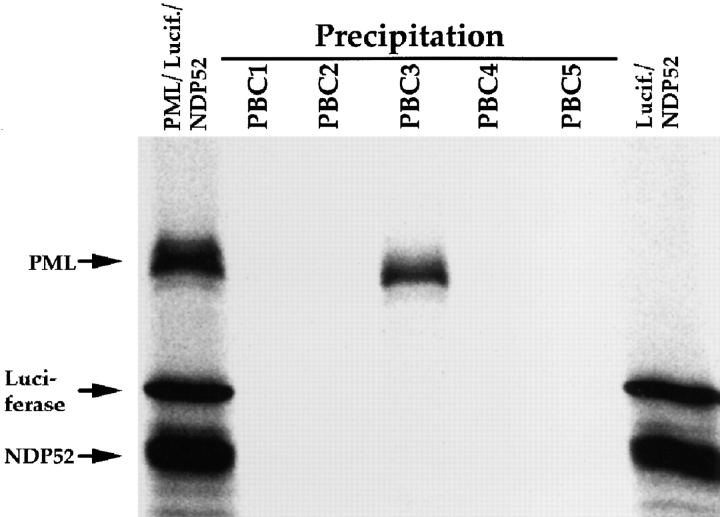
Two or several components are often coautoantigenic in autoimmune diseases when they are part of the same macromolecular complex (Tan, 1989, 1991). This is in fact also the case for the PML and Sp100 proteins, as shown previously (Sternsdorf et al., 1995; Züchner et al., 1997). Therefore, we tested whether PBC sera containing both PML and Sp100 autoantibodies also have anti-NDP52 autoantibodies. Radiolabeled NDP52 protein (Fig. 7, lane 1) mixed with radiolabeled luciferase as a specificity control (Fig. 7, lanes 2–6), were immunoprecipitated with each of five anti-Sp100/PML positive PBC sera (PBC1-PBC5). In one precipitation experiment, radiolabeled PML was added as a control (Fig. 7, lane PBC3), which was, as expected, efficiently precipitated by the anti-PML autoantibodies. In contrast, none of the five sera precipitated NDP52 protein. These data indicate that autoantibodies that recognize NDP52 synthesized by in vitro translation are not present in the PBC sera tested, whereas PML and Sp100 are recognized very well (Sternsdorf et al., 1995).
Figure 7.
Failure of detection of NDP52 autoantibodies in sera from PBC patients containing anti-PML and anti-Sp100 autoantibodies by immunoprecipitation. Radiolabeled NDP52 and, as a negative control, luciferase proteins were mixed and immunoprecipitated using five anti-Sp100/anti-PML double-positive PBC sera (lanes PBC1–PBC5). In lane PBC3, radiolabeled PML was included in the precipitation reaction as a positive control. The first and last lanes represent mixtures of the translation reactions as indicated.
To be able to screen a larger collection of PBC sera for anti-NDP52 autoantibodies and also for antibodies that recognize epitopes on denatured NDP52 proteins only, 48 PBC sera containing anti-Sp100 and/or anti-PML autoantibodies were tested by ELISA using recombinant NDP52 protein produced in Escherichia coli as template (see Materials and Methods). Moreover, 45 additional anti-PML/ anti-Sp100 negative PBC sera showing a dotlike pattern in immunofluorescence analysis were also screened. With the exception of two sera, none showed NDP52 reactivity above background (defined as <0.3 OD by using non-PBC sera as controls). The two sera with reactivity above background (OD = 1.3 and 0.8, respectively) were shown by immunoblotting to contain high levels of antibodies against E. coli proteins and showed no specific anti-NDP52 reactivity (data not shown). Taken together, these data strongly suggest that NDP52 is not coautoimmunogenic with PML and Sp100 in patients with primary biliary cirrhosis, which provides further indirect evidence against an association of NDP52 with a macromolecular ND protein complex.
mAb C8A2 Cross-reacts with the Sp100 Protein
To explain the discrepancy between the staining pattern of our anti-NDP52 polyclonal serum and mAb C8A2, we examined whether the mAb C8A2 cross-reacts with PML or Sp100. Therefore, we transfected HEp-2 and rat R1H cells with NDP52, PML, or Sp100 expression vectors and combinations thereof and analyzed the cells by immunofluorescence staining with the mAb C8A2 (Fig. 8, A–D). In these experiments, we intentionally did not double-label the cells with two different types of antibodies but rather labeled only with mAb C8A2 since double-labeling can result in artifacts due to insufficient wavelength cutoffs of the microscope filters or due to cross-species reactivity of the secondary antibodies.
Figure 8.
Cross-reaction of mAb C8A2 with Sp100 but not PML in paraformaldehyde-fixed cells as revealed by immunofluorescence staining. (A–C) Immunofluorescence analysis of HEp-2 cells transfected with NDP52 (A), Sp100 plus NDP52 (B), or PML plus NDP52 (C) expression vectors, respectively, using the mAb C8A2 at a dilution of 1:3. NDP52 localizes in the cytoplasm, and cells transfected with Sp100 plus NDP52 also show an increased nuclear dotlike staining (B). Overexpressed PML is not recognized (C). D and E show HEp-2 cells transfected with Sp100 expression vector alone (5 μg per 60-mm dish) and stained with the mAb C8A2 at dilutions of 1:3 and 1:50, respectively. Using a 1:50 dilution of mAb C8A2, overexpressed Sp100 is recognized, whereas the endogenous ND pattern is hardly visible (E). F shows rat R1H cells transfected with the Sp100 expression vector (1 μg per 60-mm dish) and stained with mAb C8A2 at a dilution of 1:3. For documentation, a cell with only weak expression was intentionally chosen. Sp100 is clearly recognized by the mAb. No endogenous ND pattern is detectable as evident from the untransfected cell visible in the same picture. The corresponding phase contrast pictures are given below the immunofluorescence pictures. Bar, 10 μm.
When the mAb C8A2 (20 μg/ml) was used for immunostaining of HEp-2 cells, NDP52 synthesized from the transfected vector was readily detected at a dilution of 1:200 and localized primarily in the cytoplasm (not shown). An ND pattern was observed in neither transfected nor untransfected cells when using this dilution of antibody. However, at higher mAb concentrations (dilutions 1:3 or 1:10) the typical ND staining pattern became apparent in paraformaldehyde-fixed cells (Fig. 8 A). Methanol/acetone-fixed cells did not show ND staining even when undiluted mAb C8A2 was used (not shown), suggesting a loss of epitope recognition due to the fixation process. When HEp-2 cells transfected with NDP52, with both NDP52 and Sp100, or both NDP52 and PML were immunostained with a 1:3 dilution of mAb C8A2, the following patterns were observed. NDP52 overexpressing cells showed bright cytoplasmic staining with exclusion of the nucleus and, in addition, a weak nuclear ND pattern both in the transfected and the nontransfected cells (Fig. 8 A). In cells overexpressing NDP52 and Sp100 simultaneously, enlarged nuclear dotlike staining in addition to the cytoplasmic NDP52 staining was observed (Fig. 8 B). This strongly suggests that mAb C8A2 not only recognizes NDP52 but also recognizes Sp100. In contrast, in cells cotransfected with NDP52 and PML expression vectors, no staining of overexpressed PML protein by mAb C8A2 in any of the NDP52 overexpressing cells (Fig. 8 C) was observed. Since NDP52 and PML proteins were coexpressed in ∼90% of the transfected cells (data not shown) when examining a second coverslip from the same transfection immunostained with polyclonal anti-NDP52 and anti-PML antibodies, we conclude that mAb C8A2 does not cross-react with PML. In cells transfected only with the Sp100 expression vector and immunostained with various concentrations of the mAb C8A2 (dilutions 1:3 and 1:50 are shown in Fig. 8, D and E), an enlarged, bright ND pattern typical for overexpressed Sp100 protein was observed. Note that the endogenous ND staining in the adjacent nontransfected cells was strong using the 1:3 dilution (Fig. 8 D) but drastically reduced in the 1:50 dilution (Fig. 8 E). Sp100 overexpressed from recombinant plasmids could be detected easily with this antibody diluted down to 1:100 (not shown). This shows that the mAb C8A2 recognizes overexpressed Sp100 even at dilutions where it fails to stain endogenous NDs. To exclude the possibility that the ND structure itself is enlarged by the overexpression of the Sp100 protein, which may mimic recognition of Sp100 by the NDP52-specific mAb, and to further exclude the possibility that the large amount of Sp100 expressed from the expression vector might lead to artificial staining, we also analyzed R1H rat cells. These cells showed neither endogenous ND staining with mAb C8A2 nor with any of our Sp100- or PML-specific sera. These cells were transfected with PML- (5 μg) and with different amounts of Sp100- expression vector (5 and 1 μg) to be able to stain Sp100 expressed at physiological levels. In R1H cells transfected with the PML expression vector, no staining of PML was observed when using mAb C8A2 (data not shown), whereas Sp100 was stained with the same antibody in many cells, even when transiently expressed at low levels (Fig. 8 F). However, the sensitivity of Sp100 detection by mAb C8A2 is considerably lower than that achieved with our polyclonal anti-Sp100 sera. In summary, these results clearly demonstrate that mAb C8A2 recognizes in addition to NDP52 the Sp100 protein but not the PML protein, implying that the ND staining with mAb C8A2 is due to cross-reaction with the Sp100 protein. This was further investigated by performing immunoblotting experiments with recombinant full-length Sp100 protein and a set of overlapping subfragments thereof and as a positive control with the recombinant NDP52 protein (see Materials and Methods). No significant reactivity of the mAb C8A2 with any of the Sp100 proteins was observed, whereas the recombinant NDP52 protein showed very strong reactivity (data not shown). These data suggest that recognition of the cross-reactive epitope on Sp100 is conformation dependent.
Discussion
In this study, we provided several independent lines of evidence that NDP52 is not associated with NDs but was erroneously localized in previous studies because mAb C8A2 cross-reacts with the Sp100 protein. NDP52 was localized here predominantly in the cytoplasm with a coarse-grained distribution preferentially close to the nucleus. Subcellular fractionation experiments are consistent with the cytoplasmic localization and in addition showed that a fraction of NDP52 is also associated with the nucleus. NDP52 was shown to form homodimers but no heterodimers with Sp100 and PML. Unlike what is known for Sp100 and PML, autoantibodies against NDP52 were not found in ND-staining PBC sera. These findings and our failure to demonstrate binding of NDP52 to Sp100 or PML argue that NDP52 neither mediates interaction of ND proteins nor is it part of the multiprotein ND complex. We also found no increase in NDP52 mRNA and protein levels after IFN β treatment and only little after IFN γ treatment of cells. Taken together, these data define (ND)P52 as a non–ND-associated dimeric or multimeric protein with a mainly coarse-grained cytoplasmic localization.
The cross-reactivity of mAb C8A2 with Sp100 shown in transfected cells by immunofluorescence staining offers a simple and convincing explanation for the previously reported erroneous localization of (ND)P52 to NDs (Korioth et al., 1995), and for some but not all discrepant results and observations. The increase in number and size of NDs in MG63, HEp-2, and HeLa S3 cells after IFN β and γ treatment, as observed previously by immunostaining with mAb C8A2 (Korioth et al., 1995) and confirmed in our study, can be fully explained by the cross-reactivity with the IFN-inducible Sp100 protein alone. Both the size and number of dots were shown to increase in IFN β– and γ–treated cells when stained with anti-Sp100 antibodies (Guldner et al., 1992). A significant increase in (ND)P52 protein expression in IFN γ– and IFN β–treated MG63, HEp-2, and HeLa S3 cells was seen by neither immunofluorescence staining (data not shown) nor immunoblotting. These findings are consistent with our failure to detect a strong increase in NDP52 mRNA levels by Northern analysis in HEp-2 cells. The increase in NDP52 mRNA levels reported for MG63 cells (Korioth et al., 1995) is not consistent with our findings. The reason for this discrepancy with previous data is not clear.
Because a weak nuclear staining was seen both with our preimmune sera from the rabbit and the rat immunized with recombinant (ND)P52 protein, we could not determine unequivocally whether (ND)P52 is an exclusively cytoplasmic protein or, in part, is also diffusely distributed in the nucleus. The diffuse staining of nuclei of various cells by mAb C8A2, which is found in addition to ND staining (Korioth et al., 1995, and our data, Fig. 8), may be taken as evidence against a purely cytoplasmic localization of (ND)P52. However, the diffuse nuclear form of (ND)P52 in COS cells, which was reported to reorganize into NDs after IFN treatment (Korioth et al., 1995), can also be explained by the cross-reactivity of mAb C8A2 with the Sp100 protein. In COS cells, IFNs increase the amount of Sp100 (our unpublished data), which presumably makes Sp100 detectable by mAb C8A2. This assumption is compatible with the observation that human HuH-7 hepatoma cells show detectable ND staining with Sp100-specific sera only after IFN treatment (unpublished data). Considering all the above-mentioned points and our failure to demonstrate a major intracellular redistribution of (ND)P52 in biochemical cell compartment fractionation experiments, we have strong reasons to believe that the previously described IFN-mediated change in cellular distribution of (ND)P52 actually represents IFN-enhanced Sp100 protein expression.
After infection with HSV-1 mutant FXE, and adenovirus Ad5, the redistribution of the ND antigen recognized by mAb C8A2 was reported to differ from that of Sp100 and PML (Korioth et al., 1995). Specifically, after infection with HSV-1 mutant FXE, C8A2 staining was shown to disappear in early stages of infection, whereas Sp100 staining was still detectable (Korioth et al., 1995). This finding could be taken as evidence against a possible cross-reaction of mAb C8A2 with Sp100 and/or PML. However, an alternative explanation for this finding could be that the cross-reactive Sp100 epitope is either not accessible, covered, or even destroyed after virus infection because of a change in Sp100 protein conformation. Such a conformational change could well abolish Sp100 staining by mAb C8A2 but not necessarily by polyclonal sera. Alternatively or in addition, the lower sensitivity in staining of Sp100 by mAb C8A2 compared to polyclonal anti-Sp100 (our data) could play a role. The removal of mAb C8A2-reactive protein together with Sp100 from PML-containing tracks induced by Ad5 virus and the colocalization of both antigens in the infected cells (Korioth et al., 1995; Ishov and Maul, 1996) corroborate our assumptions.
Two sets of data indicate that mAb C8A2 recognizes, in addition to NDP52, a conformation-dependent epitope on the Sp100 protein. First, fixation of cells with methanol/ acetone completely abolished ND staining by mAb C8A2, and Sp100 transiently expressed at high levels was recognized less well (data not shown). Second, recombinant subfragments of Sp100 expressed in E. coli showed no significant reactivity in immunoblotting. Note that our study does not exclude that the mAb C8A2 recognizes an Sp100-specific epitope that is dependent on posttranslational modification of Sp100, as this may not be present on Sp100 proteins expressed in bacteria and may be lost after the fixation conditions used here. Infection with HSV or other viruses might likewise lead to a specific loss of the modification and therefore to loss of staining by mAb C8A2. Taking these observations and considerations into account, loss of a single cross-reactive Sp100 epitope after redistribution/displacement of proteins from NDs as mediated by known viral proteins is very well conceivable.
Our conclusion that (ND)P52 is not a component of NDs is supported by strong direct evidence, but also by indirect pieces of evidence. Unlike that described for Sp100 and PML, against which autoantibodies co-occur very frequently (70%) in a subpopulation of PBC patients (Sternsdorf et al., 1995; Züchner et al., 1997), autoantibodies against (ND)P52 protein were not detected in any of the 93 PBC sera tested. Together with the fact that autoantibodies against several components of a macromolecular complex are induced in many autoimmune diseases, including PBC (Tan, 1989, 1991; Fatenejad et al., 1994), the absence of (ND)P52 autoantibodies in ND-staining PBC sera provides additional, though indirect, evidence against the localization of the (ND)P52 protein in NDs. The lack of (ND)P52 antibodies appears not to be due to low immunogenicity of (ND)P52, as indicated by the high titered (ND)P52 antibodies obtained by active immunization of rabbits and rats with recombinant (ND)P52 protein used by us. These data also argue against the presence of (ND)P52 in NDs. This conclusion is also in line with our failure to detect binding of (ND)P52 to PML or Sp100 protein by coprecipitation or by using the two-hybrid assay. However, the coprecipitation experiments provide no strong argument, as we have shown here and previously (Koken et al., 1994; Sternsdorf et al., 1995), that the PML and Sp100 proteins, when synthesized in vitro, do not bind directly to each other. Similar reasoning can be applied to our failure to show heterodimerization between any of the three proteins in the two-hybrid system. Note that a positive result in this system could theoretically also be obtained when heterodimerization would be dependent on additional cellular bridging proteins. Since a negative result in the two-hybrid system may always be caused by improper folding of the fusion proteins, our data do not firmly exclude heterodimerization of the three proteins when expressed without a fusion partner in the cell. However, the homodimerization of PML, as seen in the two-hybrid system and previously in coprecipitation experiments (Perez et al., 1993) and which is known to be mediated by the coiled-coil domain in the PML protein, argues against a strongly aberrant conformation of PML when expressed as a Gal4- or VP16-fusion protein. The same is probably true for the (ND)P52 protein, for which strong homodimerization (stronger than SV-40 T-antigen/p53 heterodimerization) was evident in the two-hybrid system. The putative central coiled-coil/leucine zipper and/or the LIM domain predicted from the (ND)P52 protein sequence most likely trigger its dimerization. Taken together, the homodimerization of (ND)P52 and PML and the lack of (ND)P52 heterodimerization with PML or Sp100 in this system strongly argue against a role for (ND)P52 in the interaction between different ND proteins.
One of the controls for the localization of (ND)P52 to NDs in the previous report was the staining of cells with a polyclonal antibody against recombinant (ND)P52 protein expressed in E. coli (Korioth et al., 1995). This polyclonal antibody decorated ND-like structures similar to mAb C8A2 (mentioned as data not shown in the paper). However, the authors have neither convincingly excluded cross-reactivity of the anti-(ND)P52 polyclonal antibody with Sp100 or PML nor have they demonstrated whether the same NDs reacted with both types of antibodies. For the following reasons we consider such controls as important. First, a weak ND-like staining in few cells by some polyclonal antibodies directed against proteins unrelated to ND proteins, as occasionally observed in our laboratory (unpublished data), suggests that ND proteins may become autoantigenic upon vigorous unspecific stimulation of the immune system. This may be due to the cell inflammation induced by the adjuvants used for immunization. Second, staining of cells with some polyclonal antibodies directed against proteins unrelated to ND proteins occasionally results in dot staining patterns that are different from Sp100/PML-specific NDs. Third, the reaction of mAb C8A2 both with (ND)P52 and Sp100 as well as the fact that mAb C8A2 was produced by conventional immunization with a nuclear protein fraction proves that there are similar structures on both proteins against which cross-reactive antibodies can be produced. Note, however, that the lack of cross-reactivity of our anti-(ND)P52 and anti-Sp100 polyclonal antisera with Sp100 and (ND)P52 proteins, respectively, indicates that the cross-reactive epitopes were not notably immunogenic in the animals we used.
mAbs recognizing different epitopes on more than one protein or selectively only one epitope on a single protein when one particular technique is used but two with another technique are rather common (Dighiero et al., 1983). Therefore, discrepancies between immunoblot and immunofluorescence staining data encountered with mAbs are not surprising. Interestingly, data published for two mAbs directed against putative ND proteins indicate that they react with more than one protein. First, mAb AP435 was reported to stain not only NDs but also to react with desmin, vimentin, and keratin 18 (Kamei, 1995). This mAb reacted with neither the PML nor the Sp100 proteins used in our studies (data not shown). Second, mAb 1150 (Epstein, 1984), which also stains NDs (Ascoli and Maul, 1991), reacts with the Sp100 protein (Doucas et al., 1996; our unpublished observations) when staining fixed cells in immunofluorescence analysis, but it reacts with a 55-kD protein (designated NDP55) on immunoblots (Ascoli and Maul, 1991). In summary, the data presented emphasize that extreme care has to be taken for the localization of proteins to specific cellular compartments and to NDs in particular.
Acknowledgments
This work was supported in part by a grant from the Thyssen Stiftung and the Deutsche Krebshilfe. The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
Abbreviations used in this paper
- HSV-1
herpes simplex virus type 1
- IFN
interferon
- ND(s)
nuclear dot(s) containing PML and Sp100 proteins
- NDP52
nuclear dot protein 52
- PBC
primary biliary cirrhosis
Footnotes
We are very grateful to Dr. F. Korioth for fruitful discussion and thank Dr. J. Frey for generously providing C8A2 hybridoma supernatant and MG63 osteosarcoma cells. Dr. H. Kamei kindly supplied mAb AP435, and Dr. H.H. Guldner provided p68- and p74-specific autoimmune sera. We thank Dr. F. Zywietz for providing the R1H cell line. We are also indebted to Dr. H. de Thé for providing plasmid pSG5-PML and rabbit antiserum and to Dr P. Chambon for cDNA of a variant of PML. We greatly appreciate the critical reading of the manuscript by T. Grötzinger, V. Radwitz-Will, and P. Forster. This article is based on a doctoral study by T. Sternsdorf in the Faculty of Biology, University of Hamburg.
Address all correspondence to Hans Will, Heinrich-Pette-Institut für experimentelle Virologie und Immunologie an der Universität Hamburg, Martinstraße 52, D-20251 Hamburg, FRG. Tel. and Fax: 49 40 48051221. E-mail: will@hpi.uni-hamburg.de
References
- Altabef M, Garcia M, Lavau C, Bae S-C, Dejean A, Samarut J. A retrovirus carrying the promyelocyte-retinoic acid receptor PML-RARa fusion gene transforms haematopoietic progenitors in vitroand induces acute leukaemias. EMBO (Eur Mol Biol Organ) J. 1996;15:2707–2716. [PMC free article] [PubMed] [Google Scholar]
- Ascoli CA, Maul GG. Identification of a novel nuclear domain. J Cell Biol. 1991;112:785–795. doi: 10.1083/jcb.112.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein RM, Neuberger JM, Bunn CC, Callender ME, Hughes GR, Williams R. Diversity of autoantibodies in primary biliary cirrhosis and chronic active hepatitis. Clin Exp Immunol. 1984;55:553–560. [PMC free article] [PubMed] [Google Scholar]
- Bloch DB, de la Monte SM, Guigaouri P, Filippov A, Bloch KD. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem. 1996;271:29198–29204. doi: 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Howe K, Etkin LD, Solomon E, Freemont PS. PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- Brasch K, Ochs RL. Nuclear bodies (NBs): a newly “rediscovered” organelle. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- Carvalho T, Seeler JS, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelbi-Alix MK, Pelicano L, Quignon F, Koken MHM, Venturini L, Stadler M, Pavlovic J, Degos L, de Thé H. Induction of the PML protein by interferons in normal and APL cells. Leukemia. 1995;9:2027–2033. [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- de Thé H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature (Lond) 1990;347:558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- de Thé H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- Dent AL, Yewdell J, Puviondutilleul F, Koken MHM, de Thé H, Staudt LM. LYSP100-associated nuclear domains (LANDs): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood. 1996;88:1423–1436. [PubMed] [Google Scholar]
- Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I tax oncoprotein. Science (Wash DC) 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- Dighiero G, Guilbert B, Fermand JP, Lymberti P, Danon F, Avrameas S. Thirty-six human monoclonal immunoglobulins with antibody activity against cytoskeleton proteins, thyroglobulin, and native DNA: immunologic studies and clinical correlations. Blood. 1983;62:264–270. [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- Dyck JA, Maul GG, Miller WJ, Chen JD, Kakizuka A, Evans RM. A novel macromolecular structure is a target of the promyelocyte-retinoic acid receptor oncoprotein. Cell. 1994;76:333–343. doi: 10.1016/0092-8674(94)90340-9. [DOI] [PubMed] [Google Scholar]
- Epstein AL. Immunobiochemical characterization with monoclonal antibodies of Epstein-Barr virus-associated early antigens in chemically induced cells. J Virol. 1984;50:372–379. doi: 10.1128/jvi.50.2.372-379.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagioli M, Alcalay M, Pandolfi PP, Venturini L, Mencarelli A, Simeone A, Acampora D, Grignani F, Pelicci PG. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene. 1992;7:1083–1091. [PubMed] [Google Scholar]
- Fatenejad S, Brooks W, Schwartz A, Craft J. Pattern of anti-small nuclear ribonucleoprotein antibodies in MRL/Mp-lpr/lpr mice suggests that the intact U1 snRNP particle is their autoimmunogenic target. J Immunol. 1994;152:5523–5531. [PubMed] [Google Scholar]
- Fey EG, Krochmalnic G, Penman S. The nonchromatin substructures of the nucleus: the ribonucleoprotein (RNP)–containing and RNP- depleted matrices analyzed by sequential fractionation and resinless section electron microscropy. J Cell Biol. 1986;102:1654–1665. doi: 10.1083/jcb.102.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green S, Issemann I, Sheer E. A versatile in vivo and in vitroeucaryotic expression vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grötzinger T, Jensen K, Guldner HH, Sternsdorf T, Szostecki C, Schwab M, Savelyeva L, Reich B, Will H. A highly amplified mouse gene is homologous to the human interferon-responsive Sp100 gene encoding an autoantigen associated with nuclear dots. Mol Cell Biol. 1996a;16:1150–1156. doi: 10.1128/mcb.16.3.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grötzinger T, Jensen K, Will H. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-γ activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility . J Biol Chem. 1996b;271:25253–25260. doi: 10.1074/jbc.271.41.25253. [DOI] [PubMed] [Google Scholar]
- Grötzinger T, Sternsdorf T, Jensen K, Will H. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML) Eur J Biochem. 1996c;238:554–560. doi: 10.1111/j.1432-1033.1996.0554z.x. [DOI] [PubMed] [Google Scholar]
- Guldner HH, Szostecki C, Grötzinger T, Will H. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol. 1992;149:4067–4073. [PubMed] [Google Scholar]
- Ishov AM, Maul GG. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J Cell Biol. 1996;134:815–826. doi: 10.1083/jcb.134.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei H. A nuclear dot-like structure that has a relationship with perinuclear intermediate filaments. Exp Cell Res. 1995;218:155–165. doi: 10.1006/excr.1995.1143. [DOI] [PubMed] [Google Scholar]
- Koken MHM, Puvion DF, Guillemin MC, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO (Eur Mol Biol Organ) J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korioth F, Gieffers C, Maul GG, Frey J. Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J Cell Biol. 1995;130:1–13. doi: 10.1083/jcb.130.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res. 1996;229:155–158. doi: 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. Localisation of splicing snRNPs in mammalian cells. Mol Biol Rep. 1993;18:127–133. doi: 10.1007/BF00986767. [DOI] [PubMed] [Google Scholar]
- Lavau C, Marchio A, Fagioli M, Jansen J, Falini B, Lebon P, Grosveld F, Pandolfi PP, Pelicci PG, Dejean A. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene. 1995;11:871–878. [PubMed] [Google Scholar]
- Martelli AM, Gilmour RS, Bertagnolo V, Neri LM, Manzoli L, Cocco L. Nuclear localization and signalling activity of phosphoinositidase C in Swiss 3T3 cells. Nature (Lond) 1992;358:242–245. doi: 10.1038/358242a0. [DOI] [PubMed] [Google Scholar]
- Netter HJ, Guldner HH, Szostecki C, Will H. Major autoantigenic sites of the (U1) small nuclear ribonucleoprotein-specific 68-kDa protein. Scand J Immunol. 1990;32:163–176. doi: 10.1111/j.1365-3083.1990.tb02906.x. [DOI] [PubMed] [Google Scholar]
- Pandolfi PP, Alcalay M, Fagioli M, Zangrilli D, Mencarelli A, Diverio D, Biondi A, Lo CF, Rambaldi A, Grignani F, et al. Genomic variability and alternative splicing generate multiple PML/RAR alpha transcripts that encode aberrant PML proteins and PML/RAR alpha isoforms in acute promyelocytic leukaemia. EMBO (Eur Mol Biol Organ) J. 1992;11:1397–1407. doi: 10.1002/j.1460-2075.1992.tb05185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez A, Kastner P, Sethi S, Lutz Y, Reibel C, Chambon P. PMLRAR homodimers: distinct DNA binding properties and heteromeric interactions with RXR. EMBO (Eur Mol Biol Organ) J. 1993;12:3171–3182. doi: 10.1002/j.1460-2075.1993.tb05986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass C, Weichenhan D, Kunze B, Hellwig T, Schneider C, Bautz FA, Grzeschik KH, Traut W, Winking H. A member of the mouse LRR transcript family with homology to the human Sp100 gene. Hereditas. 1995;122:245–256. doi: 10.1111/j.1601-5223.1995.00245.x. [DOI] [PubMed] [Google Scholar]
- Powell F, Schroeter AL, Dickson ER. Anti-nuclear antibodies in primary biliary cirrhosis. Lancet. 1984;1:288–289. doi: 10.1016/s0140-6736(84)90164-8. [DOI] [PubMed] [Google Scholar]
- Raska I, Dundr M, Koberna K. Structure-function subcompartments of the mammalian cell nucleus as revealed by the electron microscopic affinity cytochemistry. Cell Biol Int Rep. 1992;16:771–789. doi: 10.1016/s0309-1651(05)80021-9. [DOI] [PubMed] [Google Scholar]
- Rowley JD, Golomb HM, Dougherty C. 15/17 translocation, a consistent chromosomal change in acute promyelocytic leukemia. Lancet. 1977;1:549–550. doi: 10.1016/s0140-6736(77)91415-5. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Second Edition. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York. 545 pp.
- Stadler M, Chelbi-Alix MK, Koken MHM, Venturini L, Lee C, Saib A, Quignon F, Pelicano L, Guillemin MC, Schindler C, de Thé H. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene. 1995;11:2565–2573. [PubMed] [Google Scholar]
- Stemler M, Weimer T, Tu ZX, Wan DF, Levrero M, Jung C, Pape GR, Will H. Mapping of B-cell epitopes of the human hepatitis B virus X protein. J Virol. 1990;64:2802–2809. doi: 10.1128/jvi.64.6.2802-2809.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf T, Guldner HH, Szostecki C, Grötzinger T, Will H. Two nuclear-dot associated proteins, PML and Sp100, are often co-autoimmunogenic in patients with primary biliary cirrhosis. Scand J Immunol. 1995;42:257–268. doi: 10.1111/j.1365-3083.1995.tb03652.x. [DOI] [PubMed] [Google Scholar]
- Strouboulis J, Wolffe AP. Functional compartmentalization of the nucleus. J Cell Sci. 1996;109:1991–2000. doi: 10.1242/jcs.109.8.1991. [DOI] [PubMed] [Google Scholar]
- Szostecki, C. 1991. Studien zur Struktur, Expression und Funktion des nukleären Sp100-Autoantigenes und Analyse der humoralen Immunantwort. PhD thesis. Ludwig-Maximilians-Universität, München, Germany. 237 pp.
- Szostecki C, Krippner H, Penner E, Bautz FA. Autoimmune sera recognize a 100 kD nuclear protein antigen (Sp100) Clin Exp Immunol. 1987;68:108–116. [PMC free article] [PubMed] [Google Scholar]
- Szostecki C, Guldner HH, Netter HJ, Will H. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol. 1990;145:4338–4347. [PubMed] [Google Scholar]
- Szostecki C, Will H, Netter HJ, Guldner HH. Autoantibodies to the nuclear Sp100 protein in primary biliary cirrhosis and associated diseases: epitope specificity and immunoglobulin class distribution. Scand J Immunol. 1992;36:555–564. doi: 10.1111/j.1365-3083.1992.tb03224.x. [DOI] [PubMed] [Google Scholar]
- Tan EM. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- Tan EM. Autoantibodies in pathology and cell biology. Cell. 1991;67:841–842. doi: 10.1016/0092-8674(91)90356-4. [DOI] [PubMed] [Google Scholar]
- Theissen H, Etzerodt M, Reuter R, Schneider C, Lottspeich F, Argos P, Lührmann R, Philipson L. Cloning of the human cDNA for the U1 RNA-associated 70K protein . EMBO (Eur Mol Biol Organ) J. 1986;5:3209–3217. doi: 10.1002/j.1460-2075.1986.tb04631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Water J, Fregeau D, Davis P, Ansari A, Danner D, Leung P, Coppel R, Gershwin ME. Autoantibodies of primary biliary cirrhosis recognize dihydrolipoamide acetyltransferase and inhibit enzyme function. J Immunol. 1988;141:2321–2324. [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Xie K, Lambie EJ, Snyder M. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol. 1993;13:6170–6179. doi: 10.1128/mcb.13.10.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber M, Heyden TS, Lajouspetter AM. A human autoantibody recognizing nuclear matrix-associated nuclear protein localized in dot structures. Biol Cell. 1995;85:77–86. [PubMed] [Google Scholar]
- Züchner, D., T. Sternsdorf, C. Szostecki, E.J. Heathcote, K. Cauch-Dudek, and H. Will. 1997. Prevalence, kinetics, and therapeutic modulation of autoantibodies against Sp100 and PML in a large cohort of patients with primary biliary cirrhosis. Hepatology. In press. [DOI] [PubMed]
- Zywietz F, Reeker W, Kochs E. Tumor oxygenation in a transplanted rat rhabdomyosarcoma during fractionated irradiation. Int J Radiat Oncol Biol Phys. 1995;32:1391–1400. doi: 10.1016/0360-3016(94)00653-3. [DOI] [PubMed] [Google Scholar]