Abstract
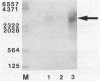
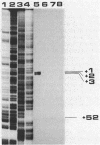
We characterized the in vivo transcription of the Cellulomonas fimi cenB gene, which encodes an extracellular endo-beta-1,4-glucanase (EC 3.2.1.4). By Northern blot (RNA blot) analysis, cenB mRNA was detected in C. fimi RNA preparations from glycerol-, glucose-, and carboxymethyl cellulose (CMC)-grown cells. The relative abundance of the specific mRNAs in these preparations appeared to depend on the carbon source provided, with the preparations from CMC-grown cells having the greatest amount of cenB transcripts, followed by glycerol- and glucose-grown cells. Therefore, the transcription of this gene could be regulated by the carbon source provided to C. fimi. High-resolution nuclease S1 protection studies were used to map cenB mRNA 5' termini with a unique 5'-labeled DNA probe and C. fimi RNA isolated in vivo. With this procedure, three 5' termini were found in abundance upstream of the translational initiation ATG codon in RNA preparations from C. fimi grown on CMC, while less-abundant 5' termini were found 52 bases closer to the ATG codon in RNA prepared from C. fimi grown on any one of the three substrates. These results are indicative of a tandem promoter arrangement, with the ATG-proximal promoter directing constitutive low-level cenB transcription and the more distal promoter directing higher levels of transcription under the inducing effects of the cellulosic substrate. The corresponding transcripts were not detected in S1 mapping experiments with RNA isolated in vivo from Escherichia coli clones harboring recombinant plasmids carrying C. fimi genomic inserts. Comparative analysis of the 5' -flanking DNA sequences of the cenB gene and the cenA and cex genes of C. fimi (N. M. Greenberg, R. A. J. Warren, D. G. Kilburn, and R. C. Miller, Jr., J. Bacteriol. 169:646-653, 1987) revealed a region of 50 bases in which these sequences displayed at least 64% homology.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Fornwald J. A., Schmidt F. J., Adams C. W., Rosenberg M., Brawner M. E. Two promoters, one inducible and one constitutive, control transcription of the Streptomyces lividans galactose operon. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2130–2134. doi: 10.1073/pnas.84.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg N. M., Warren R. A., Kilburn D. G., Miller R. C., Jr Regulation, initiation, and termination of the cenA and cex transcripts of Cellulomonas fimi. J Bacteriol. 1987 Feb;169(2):646–653. doi: 10.1128/jb.169.2.646-653.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOB F., MONOD J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961 Jun;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- O'Neill G., Goh S. H., Warren R. A., Kilburn D. G., Miller R. C., Jr Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene. 1986;44(2-3):325–330. doi: 10.1016/0378-1119(86)90197-6. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wong W. K., Gerhard B., Guo Z. M., Kilburn D. G., Warren A. J., Miller R. C., Jr Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene. 1986;44(2-3):315–324. doi: 10.1016/0378-1119(86)90196-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]