Abstract
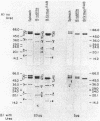
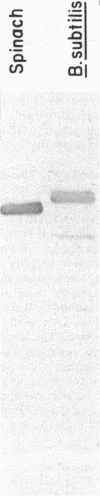
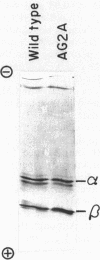
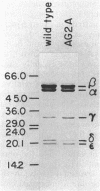
The F1 ATPase of Bacillus subtilis BD99 was extracted from everted membrane vesicles by low-ionic-strength treatment and purified by DEAE-cellulose chromatography, hydrophobic interaction chromatography, and anion-exchange high-performance liquid chromatography. The subunit structure of the enzyme was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in the absence and presence of urea. In the absence of urea, the alpha and beta subunits comigrated and the ATPase was resolved into four bands. The mobility of the beta subunit, identified by immunoblotting with anti-beta from Escherichia coli F1, was altered dramatically by the presence of urea, causing it to migrate more slowly than the alpha subunit. The catalytic activity of the ATPase was strongly metal dependent; in the absence of effectors, the Ca2+-ATPase activity was 15- to 20-fold higher than the Mg2+ -ATPase activity. On the other hand, sulfite anion, methanol, and optimally, octylglucoside stimulated the Mg2+ -ATPase activity up to twice the level of Ca2+ -ATPase activity (specific activity, about 80 mumol of Pi per min per mg of protein). The F1 ATPase was also isolated from mutants of B. subtilis that had been isolated and characterized in this laboratory by their ability to grow in the presence of protonophores. The specific activities of the ATPase preparations from the mutant and the wild type were very similar for both Mg2+- and Ca2+ -dependent activities. Kinetic parameters (Vmax and Km for Mg-ATP) for octylglucoside-stimulated Mg2+ -ATPase activity were similar in both preparations. Structural analysis by polyacrylamide gel electrophoresis and isoelectric focusing indicated that the five F1 subunits from ATPase preparations from the mutant and wild-type strains had identical apparent molecular weights and that no charge differences were detectable in the alpha and beta subunits in the two preparations. Thus, the increased ATPase activity that had been observed in the uncoupler-resistant mutants is probably not due to a mutation in the F1 moiety of the ATPase complex.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthon G. E., Jagendorf A. T. Evidence for multiple effects in the methanol activation of chloroplast coupling factor 1. Biochim Biophys Acta. 1986 Jan 28;848(1):92–98. doi: 10.1016/0005-2728(86)90164-7. [DOI] [PubMed] [Google Scholar]
- Cox G. B., Downie J. A., Fayle D. R., Gibson F., Radik J. Inhibition, by a protease inhibitor, of the solubilization of the F1-portion of the Mg2+-stimulated adenosine triphosphatase of Escherichia coli. J Bacteriol. 1978 Jan;133(1):287–292. doi: 10.1128/jb.133.1.287-292.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S. D., Tozer R. G., Antczak D. F., Heppel L. A. Monoclonal antibodies to Escherichia coli F1-ATPase. Correlation of binding site location with interspecies cross-reactivity and effects on enzyme activity. J Biol Chem. 1985 Sep 5;260(19):10418–10425. [PubMed] [Google Scholar]
- Feinstein D. L., Moudrianakis E. N. Hydrophobic and ionic effects upon the electrophoretic mobilities of the subunits of coupling factor 1 from mitochondria. Anal Biochem. 1984 Feb;136(2):362–371. doi: 10.1016/0003-2697(84)90231-8. [DOI] [PubMed] [Google Scholar]
- Futai M., Kanazawa H. Structure and function of proton-translocating adenosine triphosphatase (F0F1): biochemical and molecular biological approaches. Microbiol Rev. 1983 Sep;47(3):285–312. doi: 10.1128/mr.47.3.285-312.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti A. A., Clejan S., Falk L. H., Hicks D. B., Krulwich T. A. Isolation and characterization of uncoupler-resistant mutants of Bacillus subtilis. J Bacteriol. 1987 Oct;169(10):4469–4478. doi: 10.1128/jb.169.10.4469-4478.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks D. B., Krulwich T. A. The membrane ATPase of alkalophilic Bacillus firmus RAB is an F1-type ATPase. J Biol Chem. 1986 Sep 25;261(27):12896–12902. [PubMed] [Google Scholar]
- Hochman Y., Carmeli C. Correlation between the kinetics of activation and inhibition of adenosinetriphosphatase activity by divalent metal ions and the binding of manganese to chloroplast coupling factor 1. Biochemistry. 1981 Oct 27;20(22):6287–6292. doi: 10.1021/bi00525a001. [DOI] [PubMed] [Google Scholar]
- Krulwich T. A., Clejan S., Falk L. H., Guffanti A. A. Incorporation of specific exogenous fatty acids into membrane lipids modulates protonophore resistance in Bacillus subtilis. J Bacteriol. 1987 Oct;169(10):4479–4485. doi: 10.1128/jb.169.10.4479-4485.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- LeBel D., Poirier G. G., Beaudoin A. R. A convenient method for the ATPase assay. Anal Biochem. 1978 Mar;85(1):86–89. doi: 10.1016/0003-2697(78)90277-4. [DOI] [PubMed] [Google Scholar]
- Linnett P. E., Mitchell A. D., Partis M. D., Beechey R. B. Preparation of the soluble ATPase from mitochondria, chloroplasts, and bacteria by the chloroform technique. Methods Enzymol. 1979;55:337–343. doi: 10.1016/0076-6879(79)55042-3. [DOI] [PubMed] [Google Scholar]
- Penefsky H. S. Mechanism of inhibition of mitochondrial adenosine triphosphatase by dicyclohexylcarbodiimide and oligomycin: relationship to ATP synthesis. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1589–1593. doi: 10.1073/pnas.82.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick U., Bassilian S. Activation of magnesium ion specific adenosinetriphosphatase in chloroplast coupling factor 1 by octyl glucoside. Biochemistry. 1982 Nov 23;21(24):6144–6152. doi: 10.1021/bi00267a019. [DOI] [PubMed] [Google Scholar]
- Sedgwick E. G., Bragg P. D. Uncoupler-induced relocation of elongation factor Tu to the outer membrane in an uncoupler-resistant mutant of Escherichia coli. Biochim Biophys Acta. 1986 Mar 27;856(1):50–58. doi: 10.1016/0005-2736(86)90009-x. [DOI] [PubMed] [Google Scholar]
- Senior A. E., Wise J. G. The proton-ATPase of bacteria and mitochondria. J Membr Biol. 1983;73(2):105–124. doi: 10.1007/BF01870434. [DOI] [PubMed] [Google Scholar]
- Serrahima-Zieger M., Monteil H. Membrane ATPase of Bacillus subtilis. I. Purification and properties. Biochim Biophys Acta. 1978 Jun 8;502(3):445–457. doi: 10.1016/0005-2728(78)90077-4. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stan-Lotter H., Bragg P. D. Sulfhydryl groups of the F1 adenosine triphosphatase of Escherichia coli and the stoichiometry of the subunits. Arch Biochem Biophys. 1984 Feb 15;229(1):320–328. doi: 10.1016/0003-9861(84)90158-9. [DOI] [PubMed] [Google Scholar]
- Strotmann H., Hesse H., Edelmann K. Quantitative determination of coupling factor CF1 of chloroplasts. Biochim Biophys Acta. 1973 Aug 31;314(2):202–210. doi: 10.1016/0005-2728(73)90135-7. [DOI] [PubMed] [Google Scholar]
- Yu F., McCarty R. E. Detergent activation of the ATPase activity of chloroplast coupling factor 1. Arch Biochem Biophys. 1985 Apr;238(1):61–68. doi: 10.1016/0003-9861(85)90140-7. [DOI] [PubMed] [Google Scholar]